Abstract
The human microbiome plays a key role in human health and is associated with numerous diseases. Metagenomic-based studies are now generating valuable information about the composition of the microbiome in health and in disease, demonstrating nonneutral assembly processes and complex co-occurrence patterns. However, the underlying ecological forces that structure the microbiome are still unclear. Specifically, compositional studies alone with no information about mechanisms of interaction, potential competition, or syntrophy, cannot clearly distinguish habitat-filtering and species assortment assembly processes. To address this challenge, we introduce a computational framework, integrating metagenomic-based compositional data with genome-scale metabolic modeling of species interaction. We use in silico metabolic network models to predict levels of competition and complementarity among 154 microbiome species and compare predicted interaction measures to species co-occurrence. Applying this approach to two large-scale datasets describing the composition of the gut microbiome, we find that species tend to co-occur across individuals more frequently with species with which they strongly compete, suggesting that microbiome assembly is dominated by habitat filtering. Moreover, species’ partners and excluders exhibit distinct metabolic interaction levels. Importantly, we show that these trends cannot be explained by phylogeny alone and hold across multiple taxonomic levels. Interestingly, controlling for host health does not change the observed patterns, indicating that the axes along which species are filtered are not fully defined by macroecological host states. The approach presented here lays the foundation for a reverse-ecology framework for addressing key questions concerning the assembly of host-associated communities and for informing clinical efforts to manipulate the microbiome.
The human body is home to numerous microbial species and several complex microbial ecosystems. Advances in sequencing technologies and metagenomics now allow researchers to characterize the composition of species that inhabit the human body and the variation these communities exhibit in health and in disease (1–3). Specifically, recent studies of the microbiome have found tremendous variation among healthy individuals (1) and demonstrated clear associations between species composition and several host phenotypes including obesity (4, 5), inflammatory bowel disease (IBD) (2), and diabetes (6), as well as with external factors such as diet (7). These studies further demonstrated that, as in many other ecosystems, the composition of species in the microbiome exhibits distinct patterns that clearly deviate from a random distribution. For example, species composition in the human microbiome exhibits a significant checkerboard pattern, indicating pairs of taxa that exclude one another from shared environments (8, 9). These patterns are similar to those seen in macroecological communities, suggesting that similar pressures may act upon such microbial communities (10). Analysis of species composition in the gastrointestinal microbiomes of domesticated animals similarly revealed that deterministic interactions and niche processes, rather than stochastic neutral forces, dominate community assembly (11).
These studies provide valuable insights into potentially important regularities in the structure of host-associated communities. Just as important, however, is to reveal the underlying ecological forces that give rise to such regularities. Identifying these forces and the processes at play in structuring human-associated communities is crucial for developing a principled understanding of the mechanisms that maintain microbiome composition and drive disease-related compositional shifts, and will ultimately inform clinical efforts to manipulate the microbiome.
However, revealing the specific underlying forces that govern the structure of ecosystems and that give rise to specific patterns is a challenging task (10). Fundamentally different processes and distinct assembly rules can produce similar patterns (12). Specifically, two alternative processes can account for an observed checkerboard pattern. Cody and Diamond (13) suggested a species assortment model, in which competitive interactions between species lead to mutual exclusion. Alternatively, a checkerboard pattern can be attributed to a habitat-filtering model, in which species have affinities for nonoverlapping niches (14, 15). Compositional studies alone, therefore, cannot clearly distinguish between a species-interaction model and a habitat-filtering model and may not be able to pinpoint the driving forces that structure a community.
One way to elucidate community-structuring forces is to supplement compositional studies with prior knowledge or mechanistic models of the interaction between species (15). For example, if in a given set of communities species that exclude one another are known to compete for the same set of resources, one could argue that these communities are structured by species assortment. Conversely, knowing that species with similar nutritional requirements tend to co-occur suggests that these species are sorted by habitat filtering (14). Such information is often available in macroecological contexts from phenotypic traits or from feeding habits. In contrast, however, most species of the human microbiota have only recently been identified and lack a detailed biochemical description of their nutritional requirements and metabolic interactions.
Here, we use recent advances in systems biology and metabolic modeling to address this challenge, augmenting species composition and co-occurrence data with computational predictions of metabolic species interaction. Specifically, the reverse-ecology framework recently introduced (16–18) provides tools for obtaining insights into the ecology of microorganisms and their environments directly from genomic data and reconstructed metabolic models. Extending this reverse-ecology framework and integrating derived mechanistic models of species interactions with co-occurrence data allows us to determine forces driving species composition in the gut microbiome.
Results
Reverse-Ecology Framework for Predicting Species Interaction.
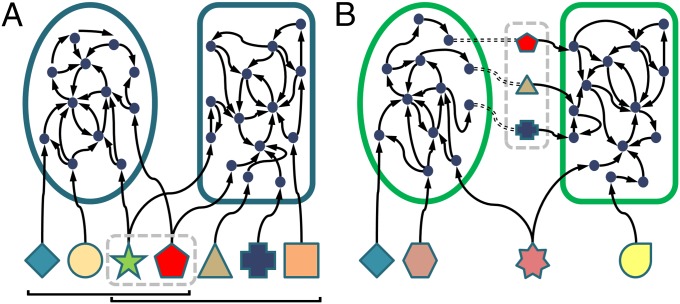
We use genome-scale metabolic network models to predict the interactions between pairs of microbial species. Networks are reconstructed based on available full genomes coupled with metabolic annotations (Methods). Such network-based models are clearly a simplified representation of the underlying metabolic pathways and dynamics, yet they have proved extremely powerful in elucidating various aspects of microbial metabolism (19, 20). Specifically, the reverse-ecology framework (18) has successfully used such models to predict important ecological attributes, including an organism’s biochemical environment (16), its interaction with its host or with other species (21–23), and ecological strategies for coping with cohabiting species (19) (see refs. 20 and 24 for additional applications). Following this approach, we use the seed set detection algorithm described in ref. 16 to analyze the metabolic network of each species. This graph theory-based algorithm identifies the set of compounds an organism exogenously acquires from its environment, representing the organism’s nutritional profile. Given the predicted nutritional profile of each species, we introduce two pairwise indices of metabolic interaction (Methods). We define the metabolic competition index as the fraction of compounds in a species’ nutritional profile that are also included in its partner’s nutritional profile (Fig. 1A). This provides a proxy for niche overlap and for the potential level of competition one species may experience in the presence of the other. We additionally define the metabolic complementarity index as the fraction of compounds in one species’ nutritional profile appearing in the metabolic network but not in the nutritional profile of its partner (Fig. 1B). Such compounds are used by both species, such that one acquires them exogenously whereas its partner synthesizes them from metabolic precursors, suggesting niche complementarity and potential syntrophy between the two species.
Fig. 1.
An illustration of model-based prediction of species interaction. The metabolic network of each species is reconstructed with nodes representing metabolites and edges connecting substrates to products. The shaped nodes represent exogenously acquired nutrients (seeds). (A) Evaluating metabolic competition. The brackets indicate the four and five seed nutrients exogenously acquired by the ellipse- and rectangle-shaped species, respectively. The two metabolites enclosed in a dashed contour denote shared nutrients for which the two species may compete. Accordingly, in this illustration, the competition index experienced by the first species in the presence of the second is 2/4, whereas the competition index of the second species in the presence of the first is 2/5. (B) Evaluating metabolic complementarity. The compounds enclosed in a dashed contour denote nutrients required by the second species that can be synthesized by the first species. In this example, the complementarity index of the second species in the presence of the first is 3/5.
Contrasting these predicted interaction indices with species co-occurrence patterns allows us to distinguish communities assembled by species assortment from communities assembled by habitat filtering. Specifically, as described above, a negative correlation between co-occurrence and metabolic competition (or a positive correlation between co-occurrence and metabolic complementarity) suggests that community assembly is strongly affected by species interactions: Species that compete for limited resources exclude one another from shared habitats, whereas species with complementary (and potentially cooperative) nutritional requirements tend to co-occur. In contrast, a positive correlation between co-occurrences and metabolic competition suggests community assembly by habitat filtering: A specific environment that offers some set of resources will be inhabited by species that require these resources (and that accordingly have similar nutritional requirements), whereas a different environment (e.g., a different sample) offering a different set of nutrients will select for a different set of species.
Predicted Interactions Recapitulate Species Interaction Between Oral Microorganisms.
To validate our framework, we first applied it to predict metabolic interactions among several human oral microbiota species whose interactions were carefully characterized. The human oral microbiota is relatively well described, and many oral species have already been cultured (25). These species interact via signaling as well as metabolic mechanisms, leading to a characteristic colonization pattern. Late-colonizing species are dependent on the presence of early colonizers that attach to the salivary pellicle for survival in the mouth. Pathogens typically arrive later in the cycle, once conditions favorable for their growth are established.
We focused on seven oral species known to influence one another’s growth in shared environments (Methods) (Table S1A). These species appear during different periods of dental plaque formation, ranging from initial colonizers to late-arriving pathogens (26). We reconstructed the metabolic networks of these species and determined their nutritional profiles, which were then used to calculate the metabolic competition index and metabolic complementarity index for each pair (Methods). We found that our predicted metabolic interaction indices (Table S1 B and C) capture species’ roles within the community and their behavior with interacting partners. Specifically, the pair Streptococcus oralis and Streptococcus gordonii have the lowest metabolic complementarity and the highest metabolic competition among all pairs. These two initial colonizers were shown to behave antagonistically (27, 28) and are expected to exploit similar niches. Furthermore, in relation to all other species, Porphyromonas gingivalis is the most complemented and poses the least competition to other species, which reflects its ability to grow mutualistically with a wide array of species from all phases of colony formation (28) (SI Text).
To further evaluate our predicted interactions on a large scale, we collected from the literature the growth rates of these species alone and in combinations using saliva as a sole nutrient source (25). To avoid comparison of absolute growth rate across potentially different conditions, we used this data in a comparative manner, generating a list of cases in which a given species was shown to grow better with one species than with another (Methods and Table S1D). Notably, the well-controlled environments in which these experiments were performed and our focus on growth rate comparative analysis allow us to control for all factors influencing growth of a species (such as habitat heterogeneity) except for the presence or absence of interacting partners. Accordingly, in these growth assays we expected that species would flourish when their interacting partners exploit nonoverlapping niches, reducing the potential effects of competition. As expected, we found that species that improve growth of the partner also tend to have higher metabolic complementarity and lower metabolic competition with those partners (P < 0.027 and P < 4 × 10−4, respectively; Methods). A more stringent analysis of this data yielded similar results (SI Text). Combined, the findings above demonstrate that our metabolic interactions indices successfully reflect the effect of species interaction on growth.
Predicted Metabolic Interactions and Co-Occurrences in the Gut Microbiome.
We next turned to investigate species interactions in the gut microbiome. In contrast to the controlled growth assays described above, here we considered the composition of naturally occurring communities as measured by metagenomic sequencing and aimed to elucidate the forces governing the assembly of these communities. Specifically, we focused on a set of 154 prevalent gut species, whose abundances across 124 individuals were obtained from shotgun metagenomic analysis (2) (Methods and Table S2A). To quantify the co-occurrence of the various species we calculated the abundance-based Jaccard similarity index between all pairs of species (Methods and Dataset S1B). Using alternative co-occurrence metrics did not qualitatively change the results reported below (SI Text and Fig. S1). Genome annotations for all species were collected from ref. 29 (Methods). Following the modeling and analysis procedure discussed above, the metabolic competition and metabolic complementarity indices were calculated for all pairs of species (Dataset S1A and Methods).
Comparing Predicted Interactions and Co-occurrence Patterns Suggests That Habitat-Filtering Shapes the Gut Microbiome.
We used these data to investigate the association between metabolic interaction and co-occurrence across all samples and all species. Specifically, we wished to determine whether species that compete with one another tend to co-occur or to exclude. We found that the metabolic competition index is positively correlated with co-occurrence, whereas the metabolic complementarity index is negatively correlated with co-occurrence (ρ = 0.211, P < 10−4 and ρ = −0.193, P < 10−4, respectively, Mantel correlation test; Methods; Table S3A). Notably, although the correlation is relatively mild, it is extremely significant, with none of the permuted null models (Methods) producing an equal or higher correlation value. This association between metabolic interaction and co-occurrence is even stronger when the analysis is limited to species pairs with coherent interaction indices (SI Text). As discussed above, these findings suggest that habitat filtering, rather than species assortment, is the dominant structuring force in the intestinal microbiome.
Metabolic Interactions of Species’ Partners and Excluders.
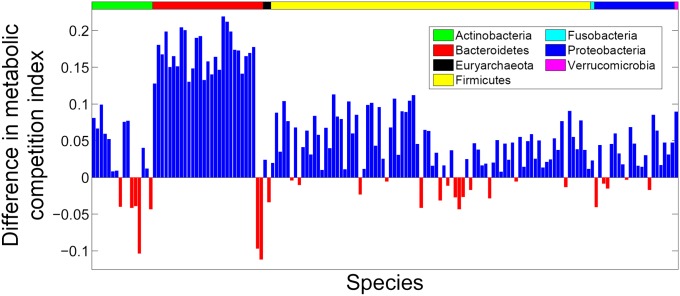
Given this observed correlation, we next sought to determine whether our framework could distinguish species that tend to significantly co-occur with a given species from those that tend to exclude it. For every species in our set, we defined as partners those 25% of species with which it has the highest co-occurrence index, and excluders as the 25% with which it has the lowest co-occurrence index. Using different threshold values for defining partners and excluders did not qualitatively change the findings reported below (SI Text). We compared the mean competition and complementarity indices of partners and excluders for each species. We found that in 82% of the species (127 out of 154; P < 2 × 10−4, permutation analysis; SI Text) the mean competition index with partners is higher than with excluders and that in 86% of the species (133 out of 154; P < 1 × 10−4, permutation analysis; SI Text) the mean complementarity index is lower with partners than with excluders (Fig. 2). Moreover, this partners and excluder separation is particularly strong when the analysis is limited to species pairs that exhibit consistent co-occurrence patterns across different host health states (SI Text and Fig. S2). Examining various ecological attributes, we additionally verified that this separation of partners and excluders is consistent across species and does not typify species with any specific ecological label (SI Text and Table S4). We further demonstrated that metabolic versatility does not explain the observed association between co-occurrence and metabolic competition (SI Text).
Fig. 2.
Partner species have higher metabolic competition than excluder species. Each bar represents a target species with the bar height representing the difference between the mean competition index with its partners and the mean competition index with its excluders. In total, 82% of species have higher metabolic competition index with partners (blue bars).
Habitat Filtering in the Gut Microbiome Cannot Be Explained by the Co-Occurrence of Phylogenetically Related Species.
Previous studies have found that phylogenetically related species tend to co-occur in the gut (2, 30). Because functional capacity and nutritional preferences are strongly linked to phylogeny (16, 31), we wished to confirm that the above association between co-occurrence and nutritional profile overlap is not a simple derivative of phylogenetic relatedness. To this end, we used 16s rRNA sequence similarity to estimate the phylogenetic distance between the various species in our analysis. We found that metabolic interaction and co-occurrence are still significantly correlated even when controlling for phylogenetic distance (Table S3A). Thus, although phylogenetically related species do co-occur in the gut (2), this alone cannot account for the observed habitat-filtering signature. To further control for phylogeny, we additionally examined the correlation between metabolic interaction and co-occurrence within each phylum separately. We observed a similar trend, wherein co-occurrence correlates positively with metabolic competition and negatively with metabolic complementarity (Table S3B). Notably, the magnitude of the correlation between metabolic interaction and co-occurrence within phyla is markedly higher compared with the correlation observed across all species, suggesting that the impact of various structuring forces varies at different phylogenetic scales (Discussion).
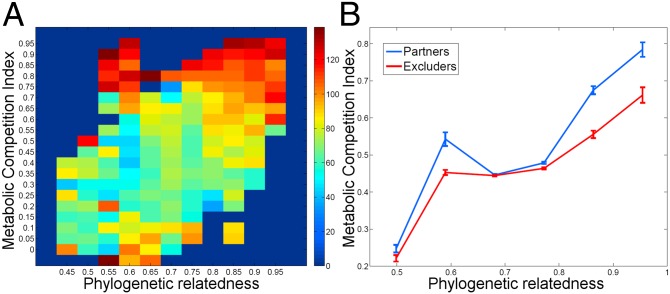
To further examine the link between metabolic interaction, phylogenetic relatedness, and co-occurrence in detail, we binned all species pairs by both metabolic competition index and phylogenetic distance and calculated the average co-occurrence in each such bin. As demonstrated in Fig. 3A, phylogenetic relatedness is correlated with metabolic competition index (ρ = 0.457, P < 10−4, Mantel correlation test). However, for a given phylogenetic distance, we still observed an increase in co-occurrence as the level of competition increases. To more rigorously validate this finding, we additionally examined whether the competition index with partners (as defined above) differs from the competition index with excluders across different phylogenetic distances. We again found that partners are associated with significantly higher metabolic competition than excluders across all phylogenetic distances (Fig. 3B). Additional analysis comparing competition, complementarity, and phylogeny in distinguishing partners vs. excluders can be found in SI Text and Fig. S3.
Fig. 3.
Habitat filtering in the gut microbiome across varying phylogenetic distances. (A) Heat map of ranked co-occurrence score, binned by phylogenetic distance (x axis) and metabolic competition index (y axis). The color of each bin represents the mean co-occurrence of all pairwise associations within it. Evidently, even among species pairs with a given phylogenetic relatedness, mean co-occurrence tends to increase with metabolic competition (red regions at the top of the heat map). (B) Average metabolic competition index and SE of partners and excluders vs. phylogenetic relatedness. At any level of phylogenetic relatedness, species have more similar nutritional profiles (significantly higher metabolic competition index) with partners than with excluders (P < 0.05 in all bins, one-tailed Mann–Whitney U test; see also Table S3E).
Compositional Shifts Associated with Host Health and Body Mass Index Do Not Fully Account for Observed Habitat-Filtering Patterns.
The above findings suggest a habitat-filtering model, wherein some properties of the gut environment govern variation in species composition. Notably, previous studies of the gut microbiome identified a strong association between species composition and both obesity (4, 5, 24) and IBD (2, 24), suggesting these may be major environmental filters influencing community composition. Here, we examined whether these host states can solely account for the observed habitat-filtering patterns. To this end, we portioned the 124 samples into four groups: healthy/lean, healthy/obese, IBD/lean, and IBD/obese. If host state is indeed the sole environmental determinant affecting species filtering, the correlation reported above between co-occurrence and metabolic interaction should disappear when considering samples from each of these controlled groups separately. We determined the co-occurrence of all species pairs within each group (Dataset S1 C–F) and calculated again the correlation between metabolic interaction indices and co-occurrence. We found that in all groups, co-occurrence still correlates positively with metabolic competition and negatively with metabolic complementarity (Table S3C). We similarly found that controlling for additional host attributes, including nationality and enterotype, does not change this pattern (SI Text). Taken together, these findings imply that the host factors examined do not fully explain the impact of the host gut environment on the composition of the microbiota and that other (and potentially yet unknown) factors contribute to habitat filtering in the gut environment and to observed species co-occurrence patterns.
Analysis of Data from the Human Microbiome Project Validates a Habitat-Filtering Model.
Finally, to validate and extend our results, we set out to examine whether the various patterns reported above can be observed in an additional and independent dataset describing the composition of the human microbiome. To this end, we used recently obtained data from the Human Microbiome Project (HMP), a large-scale effort to characterize human-associated microbial communities across five major body areas and ∼300 healthy individuals (1). We collected the relative abundances of 335 species (Dataset S2A) across 690 HMP shotgun metagenomic samples (Methods). From these data, the co-occurrence of all species pairs was determined (Dataset S2C). The metabolic competition and complementarity indices of all species pairs were determined as described above (Dataset S2B).
We first examined the association between metabolic interaction indices and co-occurrence across all samples and all species. As observed above for the intestinal microbiome, co-occurrence correlates positively with the metabolic competition index and negatively with the metabolic complementarity index (Dataset S2D), suggesting that the human microbiome is globally structured by habitat filtering. This observation is somewhat expected, given the gross differences between the five major body sites sampled, the distinct characteristic organisms in each (1), and the tendency of species to co-occur across related specific subsites (30). The obtained correlations are relatively weak but are highly significant (Mantel correlation test; Methods) and further increase when controlling for phylogeny (Dataset S2D).
Considering data from intestinal samples alone, we again observed a similar correlation pattern, validating a habitat-filtering model as the dominant assembly mechanism in the gut in this second independent dataset (Dataset S2D). We further examined whether this model represents a general plan for structuring host-associated microbial communities or whether communities in other anatomical sites are potentially subject to different structuring forces. Partitioning samples according to body site and repeating our analysis we found that in communities inhabiting the airways, skin, and the urogenital tract, co-occurrence similarly correlates positively with metabolic competition and negatively with metabolic complementarity (Dataset S2D). These correlations remain significant when controlling for phylogeny. In the oral community, the observed correlation is generally weaker, probably owing to relatively low number of genomes available and the pooling of several subsites (SI Text).
Discussion
Much effort has recently been placed on using co-occurrence to predict interactions of microbial species, either globally (32) or within the human microbiome (1, 2, 30). These studies provide valuable insights into nonrandom regularities in community composition but may not be sufficient to pinpoint the underlying forces giving rise to these regularities. The framework presented in this study, combining species abundance information with mechanistic modeling of species interactions, renders feasible a more principled analysis of these structuring forces. Specifically, we showed that predicted metabolic interactions correlate with co-occurrence patterns and that species with similar nutritional profiles tend to co-occur, suggesting that habitat filtering is the dominant structuring force of the human microbiome. Groups of species that feed on the same compounds are directly influenced by the availability of those compounds in the environment and accordingly covary in abundance across hosts.
Clearly, community assembly in the gut is a complex process. Habitat filtering and species assortment are not mutually exclusive in structuring communities (14). For example, primary consumers of polysaccharides may compete over fiber such as cellulose (33), yet they also release oligosaccharides, which are consumed by other species (34). Our analysis identifies habitat filtering as a principal force but clearly does not imply that direct species interactions do not play a role. The detrimental effects of competition over nutrients may, for example, be mitigated by the sheer abundance of resources, coupled to the naturally high turnover rate in the intestine. Species may, however, still compete over other resources, resulting in lower overall growth (35).
Previous studies of the composition of the microbiome have highlighted phylogeny as a key determinant of co-occurrence patterns (2, 30). However, our analysis demonstrates that although phylogenetic relatedness is correlated with both co-occurrence and metabolic interaction, phylogeny cannot fully account for the observed habitat-filtering pattern. In fact, the intensity of the habitat-filtering signature increases within phyla, indicating that it may be stronger at finer phylogenetic resolutions. These findings potentially contrast with recent observations of bacterial diversity in the oral cavity, where significant community structure was demonstrated at the level of genera but not of species (8). Our results may further suggest a strong tendency toward convergent genomic evolution in the gut and potential pressure acting on the evolution of intestinal microbes away from functional diversification (31).
Clearly, however, care must be taken in interpreting these results. Scale, for example, is an important factor and must be taken into account. Considering the variation in pH, nutrient content, oxygen content, and other environmental attributes among the various body sites studied by the HMP, a signature of habitat filtering is probably expected when studying whole-body species co-occurrence patterns: Different body sites will clearly select for very different sets of organisms. Our findings are in line with previous studies demonstrating that body site has the greatest influence in determining species composition, with less variation observed across individuals (1). However, our analysis of each body site and specifically of the gut microbiome indicates that even when most variation in these factors is controlled, organisms are further filtered on a local scale by as yet undetermined environmental factors.
Specifically, focusing on the gut microbiome, we demonstrated that several host phenotypes that were suggested to affect composition such as obesity, IBD, or host nationality are not the sole determining axes along which species are filtered, suggesting subtler environmental and ecological determinants. A likely candidate is the biochemical content, to which host diet is the key contributor. Diet has been demonstrated to be a strong predictor of intestinal microbiota composition (7, 36) and may accordingly be the primary link between host macroecological state and community composition. Specifically, diets that provide a surplus of nutrients preferred by a subset of the community will increase the abundance of those species, in accordance with a habitat-filtering process (37).
Clearly, the models used in this study are a simplification of the underlying biology and have several limitations. First, connectivity-based models and topological analysis cannot fully quantify the strength of metabolic interactions. For example, our method weighs each overlapping compound equally in determining metabolic competition, ignoring the potential contribution of each compound to growth or constrains on reaction fluxes. Similarly, our method aims to quantify the set of compounds both species potentially require, but without prior knowledge about nutrient availability it is hard to determine which compound these species will actually compete for. Notably, constraints-based approaches can potentially overcome some of these limitations by explicitly modeling the environment and by incorporating constraints on fluxes and nutrient uptake (38). However, in contrast to the homology-based networks used in this study, such models require detailed biochemical data and a manually curated reconstruction process and are accordingly not yet available for the vast majority of gut species studied here.
Moreover, it is important to note that although nutrient availability is an important factor, metabolic interactions are not the only determinants of partner preference among microbes. Adhesion, coaggregation, signaling, and antibiotic tolerance are critical to community assembly. For example, it has recently been shown that microbes form discrete ecological units that cooperate in the production of antibiotics (39). As molecular methods improve, multiple “meta-omic” data types (such as metaproteomic and metametabolomic data) are becoming available, providing insights into such complex interspecies processes. Developing advanced analytic and modeling frameworks that integrate these data types is one of the major challenges microbial ecology currently faces (40, 41). Specifically, modeling and predicting the full range of species interactions and validating predicted interactions via model systems (42) can dramatically improve our understanding of the microbiome in health and in disease.
Notably, elucidating the assembly rules of the microbiome goes beyond gaining a better understanding of basic ecological processes and has profound clinical implications. Specifically, one of the key challenges of human microbiome research is the development of intervention strategies for driving the intestinal microbiota to favorable states and for microbiome-based therapy (20, 43). In this context, our observation that habitat filtering dominates the assembly of the intestinal community suggests that certain species can be targeted with relatively little concern about their interaction with other members of the community. Similarly, high levels of niche overlap among community members may indicate that dietary supplements may not be precise enough to target species individually. An extended framework for analyzing species interactions within clinical settings could play a key role in the development of microbiome-based treatments. For example, identifying the set of compounds for which species compete could inform dietary-based intervention efforts, safe drug development, species isolation, and colonization studies (17, 44). This study and the framework introduced here are an important first step in this direction, highlighting the opportunities and challenges ahead.
Methods
Species and Community Data.
We obtained a list of seven oral microbial species from ref. 28. This list comprises species that have been isolated and had their growth on saliva assayed. A list of prevalent gut microbial species was obtained from ref. 2. This list comprises 155 bacterial species for which whole genome sequence is available and that had sequence coverage >1% in a metagenomic sample from at least 1 of 124 individuals analyzed (Table S2A). A set of ecological attributes for each species was obtained from the Prokaryotic Genome Project tables at the National Center for Biotechnology Information (Table S2B).
Abundance data for these metagenomic samples were obtained from ref. 2. Species abundance was calculated as the sum of sequence length from reads unambiguously mapped to a unique region of a species’ genome, normalized by the total length of the unique portion of the species’ genome sequence. To account for different sequencing depth across samples, genome coverage was normalized to 1 Gb of sequence. Using this shotgun sequencing-based method to estimate the abundance of each genome in the community provides a natural approach to coupling species abundance data with the genomic data used to reconstruct the species’ metabolic networks (discussed below). For each metagenomic sample, nationality, body mass index, and health state (IBD/healthy) of each contributing individual was recorded. For Danish individuals, the enterotype was also recorded. Species abundances were normalized to reflect relative abundances. Species co-occurrence was defined as the similarity in abundance profiles as measured by the continuous Jaccard similarity index (SI Text). We further demonstrated that our co-occurrence measures are robust to the number of individuals sampled (SI Text and Fig. S1).
Metabolic Network Reconstruction.
We obtained genomic data for all organisms from the Department of Energy Joint Genome Initiative's Integrated Microbial Genomes project (IMG, http://img.jgi.doe.gov) (29). For each species, the list of genes mapped to the Kyoto Encyclopedia of Genes and Genomes (45) orthologous groups (KOs) was downloaded (Table S2A). We used these data to reconstruct the genome-scale metabolic network of each species. Networks were represented as directed graphs with nodes representing compounds and edges representing reactions linking substrates to products. A detailed description of the reconstruction procedure can be found in ref. 16.
Analysis of Growth Data of Oral Species.
Growth rate of species was obtained from several previous studies (25) (SI Text) that describe growth assays of multiple oral species in various combinations. We generated a list of all species trios for which we can comparatively determine partners’ influence on growth (Table S1C). Specifically, each trio is defined as a target species (e.g., P. gingivalis) and two partner species: a favored partner (e.g., Aggregatibacter actinomycetemcomitans) and a disfavored partner (e.g., Fusobacterium nucleatum), such that the target species grows better with the favored partner than with the disfavored partner. We used the paired Student's t test to confirm that the metabolic interaction indices associated with favored partners are significantly different from those associated with the disfavored partners. To validate these results with increased stringency, we additionally used a manually curated dataset, obtaining qualitatively similar results (SI Text).
Predicting Metabolic Competition and Complementarity.
We use the seed set of each species as a proxy for its nutritional profile. The seed set represents the minimal set of compounds an organism exogenously acquires to synthesize all other compounds and can be inferred from the topology of its metabolic network using a previously published method (16). Given these nutritional profiles, two interaction indices were calculated for each pair of species: the metabolic competition index and the metabolic complementarity index. The metabolic competition index represents the similarity in two species’ nutritional profiles. It is calculated as the fraction of compounds of query species X’s seed set that are also present in the seed set of a target Y. Because seed compounds are associated with a confidence score (see ref. 16), this fraction is calculated as a normalized weighted sum. This index provides an upper bound for the amount of competition one species can encounter from another. Using an additional and previously described seed set-based metric for competition produced qualitatively similar results (SI Text). The metabolic complementarity index represents the complementarity in two species’ nutritional profiles and provides an upper limit for potential syntrophy. To this end, we modified the host–parasite biosynthetic support score (21) to reflect potential complementarity between pairs of microbial species. Specifically, the score is calculated as the fraction of seed compounds of a query species X that are producible by the metabolic network of a target Y but are not a part of Y’s seed set. These may also represent compounds essential to one organism that its partner may provide. Notably, neither of these indices is necessarily symmetric.
Estimation of Phylogenetic Relatedness.
We used the level of similarity between the 16s rRNA gene as a proxy for the evolutionary distance between species. The 16s rRNA gene sequences for 143 species were collected from IMG (29) or from the GreenGenes database (46). For 16s analysis, we followed the procedure described in ref. 31.
Evaluating the Correlation Between Co-Occurrence Scores and Metabolic Interaction Indices.
To calculate the correlation between co-occurrence and metabolic interaction, we generated two matrices, the first listing the co-occurrence scores between all species pairs and the second listing the predicted interaction index (either competition or complementarity). Because co-occurrence scores are generally symmetric whereas interaction indices are not (discussed above), we also generated a symmetric version by replacing each element in the interaction matrix with the mean of each value and that opposite the diagonal. The Spearman correlation between the upper triangles of the co-occurrence matrix and the interaction matrix was calculated. To determine the significance of this association, we used a permutation-based Mantel test. The rows and columns of the co-occurrence matrix were randomly permuted, preserving species identities (i.e., row and column orders are permuted similarly). For each of 10,000 permuted matrices, we again calculated the Spearman correlation, and the P value is the fraction of permutated matrices with correlations as high as or higher than the original. To control for phylogenetic relatedness, an additional matrix that describes the phylogenetic relatedness between all species was generated (discussed above), and the Spearman partial correlation of the interaction and co-occurrence matrices, controlling for phylogenetic relatedness, was calculated. Significance was determined using the same permutation approach described above.
Analysis of HMP Community Data.
We obtained shotgun metagenomic community profiling data from the Human Microbiome Project Data Analysis and Coordination Center Web site (http://hmpdacc.org/HMSMCP/). These data represent relative abundance of bacteria and archaea at different taxonomic levels, as determined by the MetaPhlAn pipeline (47). MetaPhlAn enables estimation of species abundances and comparison across metagenomic samples of different sequencing depths. In total, 397 species level taxa were classified among 690 samples. Each sample represents one of five major body sites. Because MetaPhlAn does not identify taxa at the strain level, representative genomes were selected from IMG. Where possible, genomes marked “Human Microbiome Project (HMP) Reference Genomes” were selected. In cases in which multiple genomes were available, the genome with the greatest number of KO annotations, and then the greatest number of genes, was selected. A list of the 335 species from the MetaPhlAn profile and representative genomes is available in Dataset S2A. Abundance of these species in each sample was renormalized in the same method as with the MetaHIT data. Percent similarity of the 16S rRNA gene was used to estimate phylogenetic relatedness as before, analyzing 314 species with representative 16S sequences.
Supplementary Material
Acknowledgments
We thank Dusko Ehrlich and Paul Kolenbrander for providing data on the gut and oral microbiomes and for their advice. R.L. is supported by a National Science Foundation Graduate Research Fellowship under Grant DGE-0718124. E.B. is an Alfred P. Sloan Research Fellow. This work was supported in part by New Innovator Award DP2 AT007802-01 (to E.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300926110/-/DCSupplemental.
References
- 1.Huttenhower C, et al. Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26(1):5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- 6.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 7.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333(6038):101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bik EM, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4(8):962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horner-Devine MC, et al. A comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology. 2007;88(6):1345–1353. doi: 10.1890/06-0286. [DOI] [PubMed] [Google Scholar]
- 11.Jeraldo P, et al. Quantification of the relative roles of niche and neutral processes in structuring gastrointestinal microbiomes. Proc Natl Acad Sci USA. 2012;109(25):9692–9698. doi: 10.1073/pnas.1206721109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emerson BC, Gillespie RG. Phylogenetic analysis of community assembly and structure over space and time. Trends Ecol Evol. 2008;23(11):619–630. doi: 10.1016/j.tree.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Cody M, Diamond J. Ecology and Evolution of Communities. Cambridge, MA: Belknap Press Harvard Univ Press; 1975. [Google Scholar]
- 14.Weiher E, Clarke PGD, Keddy PA. Community assembly rules, morphological dispersion, and the coexistence of plant species. Oikos. 1998;81:309–322. [Google Scholar]
- 15.Cornwell WK, Schwilk LDW, Ackerly DD. A trait-based test for habitat filtering: Convex hull volume. Ecology. 2006;87(6):1465–1471. doi: 10.1890/0012-9658(2006)87[1465:attfhf]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 16.Borenstein E, Kupiec M, Feldman MW, Ruppin E. Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc Natl Acad Sci USA. 2008;105(38):14482–14487. doi: 10.1073/pnas.0806162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janga SC, Babu MM. Network-based approaches for linking metabolism with environment. Genome Biol. 2008;9(11):239. doi: 10.1186/gb-2008-9-11-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy R, Borenstein E. Reverse ecology: From systems to environments and back. Adv Exp Med Biol. 2012;751:329–345. doi: 10.1007/978-1-4614-3567-9_15. [DOI] [PubMed] [Google Scholar]
- 19.Freilich S, et al. Metabolic-network-driven analysis of bacterial ecological strategies. Genome Biol. 2009;10(6):R61. doi: 10.1186/gb-2009-10-6-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borenstein E. Computational systems biology and in silico modeling of the human microbiome. Brief Bioinform. 2012;13(6):769–780. doi: 10.1093/bib/bbs022. [DOI] [PubMed] [Google Scholar]
- 21.Borenstein E, Feldman MW. Topological signatures of species interactions in metabolic networks. J Comput Biol. 2009;16(2):191–200. doi: 10.1089/cmb.2008.06TT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freilich S, et al. The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res. 2010;38(12):3857–3868. doi: 10.1093/nar/gkq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cottret L, et al. Graph-based analysis of the metabolic exchanges between two co-resident intracellular symbionts, Baumannia cicadellinicola and Sulcia muelleri, with their insect host, Homalodisca coagulata. PLOS Comput Biol. 2010;6(9):e1000904. doi: 10.1371/journal.pcbi.1000904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci USA. 2012;109(2):594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolenbrander PE. Multispecies communities: Interspecies interactions influence growth on saliva as sole nutritional source. Int J Oral Sci. 2011;3(2):49–54. doi: 10.4248/IJOS11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8(7):471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 27.Palmer RJ, Jr, Kazmerzak K, Hansen MC, Kolenbrander PE. Mutualism versus independence: Strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect Immun. 2001;69(9):5794–5804. doi: 10.1128/IAI.69.9.5794-5804.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Periasamy S, Kolenbrander PE. Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. J Bacteriol. 2009;191(22):6804–6811. doi: 10.1128/JB.01006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markowitz VM, et al. IMG: The Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 2012;40(Database issue):D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faust K, et al. Microbial co-occurrence relationships in the human microbiome. PLOS Comput Biol. 2012;8(7):e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaneveld JR, Lozupone C, Gordon JI, Knight R. Ribosomal RNA diversity predicts genome diversity in gut bacteria and their relatives. Nucleic Acids Res. 2010;38(12):3869–3879. doi: 10.1093/nar/gkq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaffron S, Rehrauer H, Pernthaler J, von Mering C. A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 2010;20(7):947–959. doi: 10.1101/gr.104521.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louis P, Scott KP, Duncan SH, Flint HJ. Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol. 2007;102(5):1197–1208. doi: 10.1111/j.1365-2672.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- 34.Dehority BA. Effects of microbial synergism on fibre digestion in the rumen. Proc Nutr Soc. 1991;50(2):149–159. doi: 10.1079/pns19910026. [DOI] [PubMed] [Google Scholar]
- 35.Foster KR, Bell T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol. 2012;22(19):1845–1850. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolida S, Meyer D, Gibson GR. A double-blind placebo-controlled study to establish the bifidogenic dose of inulin in healthy humans. Eur J Clin Nutr. 2007;61(10):1189–1195. doi: 10.1038/sj.ejcn.1602636. [DOI] [PubMed] [Google Scholar]
- 38.Klitgord N, Segrè D. Environments that induce synthetic microbial ecosystems. PLOS Comput Biol. 2010;6(11):e1001002. doi: 10.1371/journal.pcbi.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cordero OX, et al. Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science. 2012;337(6099):1228–1231. doi: 10.1126/science.1219385. [DOI] [PubMed] [Google Scholar]
- 40.Turnbaugh PJ, Gordon JI. An invitation to the marriage of metagenomics and metabolomics. Cell. 2008;134(5):708–713. doi: 10.1016/j.cell.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 41.Greenblum S, Chiu HC, Levy R, Carr R, Borenstein E. Towards a predictive systems-level model of the human microbiome: Progress, challenges, and opportunities. Curr Opin Biotechnol. 2013 doi: 10.1016/j.copbio.2013.04.001. 10.1016/j.copbio.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taormina MJ, et al. Investigating bacterial-animal symbioses with light sheet microscopy. Biol Bull. 2012;223(1):7–20. doi: 10.1086/BBLv223n1p7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lemon KP, Armitage GC, Relman DA, Fischbach MA. (2012) Microbiota-targeted therapies: An ecological perspective. Sci Transl Med 4(137): 4:137rv5. [DOI] [PMC free article] [PubMed]
- 44.Röling WFM, Ferrer M, Golyshin PN. Systems approaches to microbial communities and their functioning. Curr Opin Biotechnol. 2010;21(4):532–538. doi: 10.1016/j.copbio.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 45.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Segata N, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9(8):811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.