Abstract
Developmental biology is challenged to reveal the function of numerous candidate genes implicated by recent genome-scale studies as regulators of organ development and diseases. Recapitulating organogenesis from purified progenitor cells that can be genetically manipulated would provide powerful opportunities to dissect such gene functions. Here we describe systems for reconstructing pancreas development, including islet β-cell and α-cell differentiation, from single fetal progenitor cells. A strict requirement for native genetic regulators of in vivo pancreas development, such as Ngn3, Arx, and Pax4, revealed the authenticity of differentiation programs in vitro. Efficient genetic screens permitted by this system revealed that Prdm16 is required for pancreatic islet development in vivo. Discovering the function of genes regulating pancreas development with our system should enrich strategies for regenerating islets for treating diabetes mellitus.
Keywords: stem cell, transcription factor, exocrine, endocrine, insulin
The pancreas is a solid organ with vital endocrine and exocrine functions and is the root of devastating human diseases such as diabetes mellitus and pancreatic adenocarcinomas (1, 2). Many genes have emerged as candidate regulators of pancreas development as a result of modern studies of pancreas development and diseases that use high-throughput and genome-scale methods (3). Similar to many fetal organs deep in the abdomen, the developing pancreas is relatively inaccessible to visualization and efficient conditional genetic manipulations, and experimental systems for recapitulating pancreas development to assess gene function have long been sought. Reconstituting islet differentiation using cultured embryonic stem cells or induced pluripotent stem (iPS) cells has been challenging, and the authenticity of developmental programs in putative pancreatic progenitors derived from these approaches has not been established (1). Likewise, discovery of gene function with classical fetal pancreas cultures has inherent challenges, including limited or ineffective ways to screen genetic loss of function in progenitor cells (4–6). Thus, in vitro reconstitution of pancreas development from native multipotent progenitor cells could substantially advance efforts to discover developmental gene function in this organ, but such a system has remained unachieved.
A subset of early pancreatic epithelial cells expresses the transcription factor Sox9, which prior studies showed can develop in vivo into exocrine duct or acinar cells, as well as into endocrine progenitor cells that generate all pancreatic islet cells, including β-cells (7–10). Thus, Sox9+ cells are multipotent progenitor cells. A key regulator of endocrine specification in these cells is Neurogenin3 (Ngn3), a basic helix-loop-helix (bHLH) transcription factor (11–15). Mice lacking the Ngn3 gene revealed its absolute requirement for islet cell development (12). Ngn3 initiates the genetic program for endocrine differentiation, culminating in differentiation of islet cell types producing distinct hormones, such as insulin-producing β-cells and glucagon-producing α-cells. Studies of humans have revealed Ngn3 and its targets as factors regulating diabetes susceptibility or pathogenesis (16–18). The allocation of Ngn3+ cells to each islet cell type is genetically regulated and important for maintaining metabolic homeostasis. For example, the homeodomain repressor Arx promotes differentiation of glucagon- and pancreatic polypeptide-producing islet cells, whereas Pax4 promotes development of insulin- and somatostatin-producing cells (reviewed in ref. 19). However, the mechanisms underlying islet cell allocation remain poorly understood, and it is unclear whether factors other than Ngn3, Arx, and Pax4 control this key step in pancreas development.
Here we describe a unique system for isolating, expanding, and differentiating native progenitor cells from fetal mouse pancreas. The accessibility of pancreatic progenitor cells to genetic manipulations and the authenticity of development in this system permitted an unprecedented short hairpin RNA (shRNA)-based genetic screen, as well as discovery of Prdm16 (20, 21) as a factor regulating in vivo islet cell differentiation and allocation.
Results and Discussion
Growth and Differentiation of Isolated Pancreatic Progenitor Cells in Pancreatic Spheres.
To reconstruct pancreatic differentiation from endogenous fetal progenitors cells, we focused on fetal pancreas cell populations expressing the transcription factor Sox9. We purified single Sox9+ pancreatic cells from mice on embryonic day (E) 11.5, a stage preceding the major periods of endocrine and exocrine differentiation (1, 2). Mice harboring a Sox9-eGFP reporter gene (Materials and Methods and ref. 22) permitted isolation of cells enriched for Sox9 mRNA and protein by flow cytometry (Fig. 1 A and B; see Fig. S2C). To remove endocrine cells derived from Sox9-eGFP+ cells, we developed the Ngn3-tdTomato transgenic line (Ngn3-tdT; Materials and Methods; Fig. S1). In this line, tdTomato expression marks fetal pancreatic endocrine progenitor cells expressing Ngn3; perdurance of the tdTomato signal also labels differentiated pancreatic islet cells (Figs. S1 and S2 A and B). Quantitative RT-PCR (qPCR) analysis of FACS-purified Sox9-eGFP+ Ngn3-tdTneg cells revealed that mRNAs encoding postulated markers of pancreatic progenitor cells (1, 2, 8), including Sox9, transcription factor 2 (Tcf2), Pdx1, hepatocyte nuclear factor 6 (Hnf6)/Onecut, and hairy and enhancer of split 1 (Hes1), were enriched (Fig.1B and Fig. S2 B and C). In contrast, mRNAs encoding differentiated endocrine cell markers, including NeuroD1, insulin2 (Ins2), glucagon (Gcg), or chromogranin A (ChgA), were reduced or undetectable (Fig. S2 B and C). Thus, Sox9-eGFP+ Ngn3-tdTneg cells resembled pancreatic progenitors.
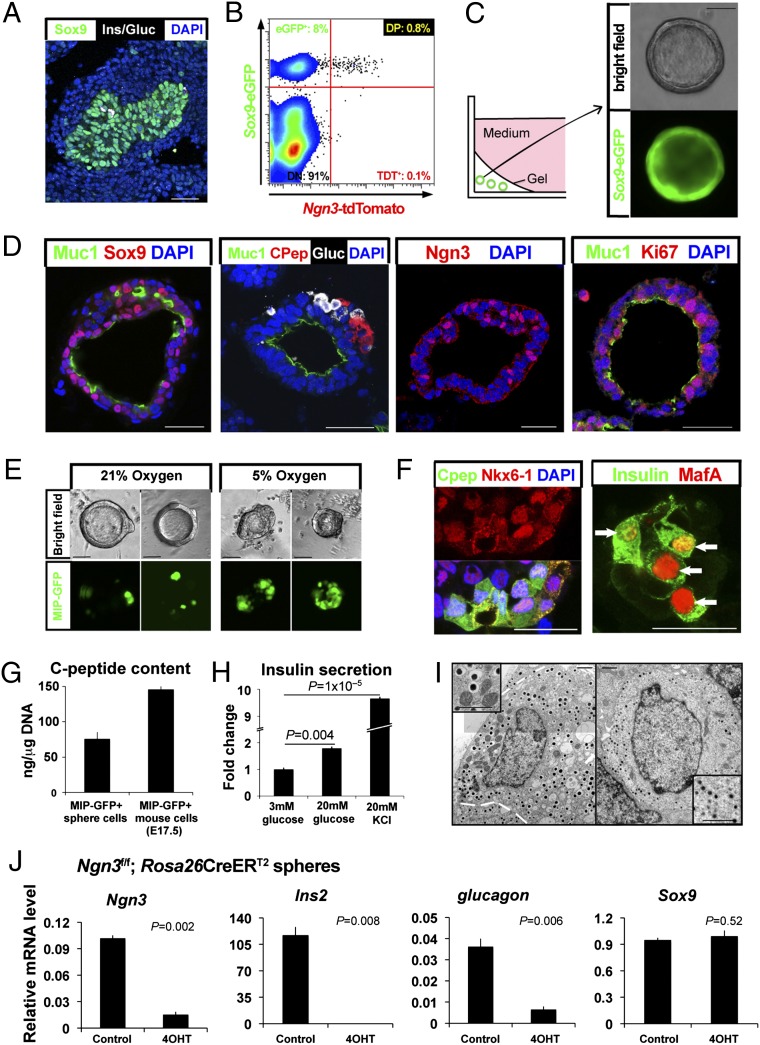
Fig. 1.
Growth and differentiation of isolated pancreatic progenitor cells in pancreatic spheres. (A) Immunohistologic detection of Sox9+ cells (green), the major population of epithelia in E11.5 fetal mouse pancreas. Sox9neg glucagon+ or Sox9neg Insulin+ endocrine cells (white) are observed. (Scale bar, 40 μm.) (B) FACS of enzymatically dissociated E11.5 pancreas from Sox9-eGFP; Ngn3-tdT dual reporter mice. Progenitor cells are the Sox9-eGFP+ Ngn3-tdTneg. (C) A schematic summarizing conditions for culturing purified pancreatic progenitors. (Scale bar, 50 μm.) (D) Immunohistology of pancreatic spheres. (Scale bar, 40 μm.) (E) Microscopic images of MIP-GFP expression in pancreatic spheres exposed to ambient (21%) or physiological (5%) oxygen. (Scale bar, 50 μm.) (F) Immunohistology of pancreatic spheres cultured in physiological oxygen. Arrows indicate MafA+ nuclei. (Scale bar, 25 μm.) (G) C-peptide content measured by ELISA, normalized to DNA from purified MIP-GFP+ cells (n = 3). (H) Insulin secretion after glucose or potassium challenge measured by ELISA (n = 6). (I) Transmission electron micrograph revealing cells with β-cell dense core vesicles (Left) and α cell vesicles (Right). Magnified images are shown in insets. Figure is a composite to create the full image. (Scale bar, 1 μm.) (J) Quantitative RT-PCR analysis of Ngn3 gene deletion by Cre recombinase-based inactivation of conditional Ngn3 alleles (n = 3). Error bars, SEM.
To test whether single Sox9-eGFP+ Ngn3-tdTneg cells are capable of expansion and multilineage differentiation, we established conditions (Materials and Methods and refs. 23 and 24) promoting clonal proliferation and differentiation of these cells. Isolated Sox9-eGFP+ Ngn3-tdTneg cells diluted to clonal density expanded into multicellular epithelial spheres containing a subset of cells that maintained Sox9-eGFP and Sox9 protein (Fig. 1 C and D; Datasets S1 and S2). During their formation over the course of 7 d, primary spheres (G1) derived from Sox9-eGFP+ Ngn3-tdTneg cells increased in size and number, consistent with detection of the proliferation antigen Ki67 in 21% ± 2% of sphere epithelial cells (Fig. 1D). Spheres were composed of epithelial cells surrounding a central lumen (Fig. 1C) and varied in size, with an average of 440 ± 48 cells (n = 43; Fig. 1D). We calculate that primary pancreatic progenitors are capable of at least eight to nine doublings. Video microscopy demonstrated sphere development from the growth of single cells (Fig. S3A). Likewise, cell mixing and limiting dilution analyses provided additional evidence for clonal sphere formation (Fig. S3 B and C) and ruled out sphere generation by mechanisms such as cell migration and aggregation. The achievement of sphere formation with more than 200 consecutive litters demonstrated the robustness of these methods. In contrast, we detected little to no sphere formation from Ngn3-tdT+ or Sox9-eGFPneg cells (Fig. S2D).
Immunohistology revealed that a subset of cells maintained Sox9 protein, whereas the others were Sox9neg. A subset of these Sox9neg cells expressed pancreatic endocrine markers such as insulin C-peptide, glucagon, and Ngn3 (Fig. 1D and Fig. S2E). Thus, we observed multilineage differentiation during primary sphere formation. Characteristically, duct-like cells expressing apical Mucin (Muc1) lined the sphere interior, separating hormone-positive cells from the central lumen, reminiscent of the tissue organization in the embryonic pancreas. Cells in sphere cultures did not express acinar markers such as amylase or carboxypeptidaseA (Cpa1). Although other methods demonstrated that Sox9-eGFP+ Ngn3-tdTneg cells retained the potential to form Amylase+ or Cpa1+ acinar cells (Fig. S4), these produced multicellular nonspherical organoids that we could not propagate; thus, we focused our investigations on sphere cultures. G1 spheres could be passaged in two subsequent rounds to produce G2 and G3 spheres, which maintained Sox9-eGFP expression and Sox9 protein in a subset of cells (Fig. 1C and D and Fig. S5). mRNAs encoding endocrine and ductal markers were maintained in G1 and G2 spheres; however, mRNA levels of endocrine markers declined in G3 spheres (Fig. S5C), indicating a reduced potential to generate multiple pancreatic cell types during progenitor expansion, consistent with prior in vivo mouse studies (25). We therefore focused on reconstructing pancreatic endocrine differentiation in G1 and G2 spheres.
The accessibility of this system allowed us to identify conditions that recapitulated the hallmark features of pancreatic endocrine differentiation in spheres (Materials and Methods; Dataset S3). We found that physiological oxygen levels (5% O2; ref. 26) significantly increased expression of Ngn3, Pdx1, Ins2, and glucagon mRNA compared with the levels found in spheres developing at ambient (21%) O2 levels (Fig. 1E and Fig. S6 A and B). Thus, the effect of oxygen on the development of isolated fetal pancreatic epithelium and cultured whole-organ rudiments (27, 28) may differ. FACS analysis demonstrated that ∼25% of epithelial cells producing the marker epithelial cell adhesion molecule (EpCAM) expressed the mouse insulin promoter-GFP reporter transgene (MIP-GFP; Materials and Methods) in spheres derived from MIP-GFP mice (Fig. S6 C and D). As in native pancreas, a subset of NK6 homeobox 1 (Nkx6-1+) cells coexpressed C-peptide and a subset of insulin+ cells coexpressed MafA (Fig. 1F). Insulin C-peptide levels in purified MIP-GFP+ sphere cells were comparable with those in purified β-cells from E17.5 MIP-GFP pancreas (Fig. 1G; SI Materials and Methods). Compared with spheres exposed to 3 mM glucose, we detected increased insulin release by spheres exposed to 20 mM glucose (Fig. 1H) or to the depolarizing agent potassium chloride (Fig. 1H). Transmission electron microscopy revealed that cell subsets harbored abundant granules with an electron-dense core, characteristic of β-cells (Fig. 1I, Left, and Fig. S6F), whereas others contained uniformly dense granules characteristic of α-cells (Fig. 1I, Right). We did not observe multihormonal cells either by immunodetection (Fig. 1D) or by transmission electron microscopy. Collectively, these findings demonstrate that cells developed cardinal features of native α-cells and β-cells, further verifying the authenticity of pancreatic developmental programs in our system.
Functional Genetic Screening for Essential Regulators of Islet Development, Using Cultured Progenitor Cells.
To assess whether genetic programs in our system reflected those controlling pancreas development in vivo, we used conditional genetics to inactivate Ngn3, which is required for pancreatic islet development (12). We generated G1 spheres from Ngn3f/f; Rosa26CreERT2 mice (Fig. 1J; Materials and Methods; ref. 29). 4-hydroxytamoxifen exposure activated CreERT2 recombinase, leading to deletion of a floxed Ngn3 allele in the genome and loss of Ngn3 mRNA in spheres (Fig. 1J), without affecting sphere growth or number (Fig. S7A). After Ngn3 loss in Ngn3f/f; Rosa26CreERT2 cells, levels of mRNA encoding chromogranin A, Ins2, NeuroD1 (a direct target of Ngn3; reviewed in ref. 2), and glucagon were reduced or undetectable compared with controls (Fig. 1J and Fig. S7 and S8). To confirm that Ngn3 loss prevented islet cell differentiation in this system, we infected 4-hydroxytamoxifen-exposed Ngn3f/f; Rosa26CreERT2 spheres (Ngn3-null spheres hereafter) with adenovirus-expressing mouse Ngn3 (Ad-Ngn3; Fig. S7 C and D). Compared with control Ad-Red Fluorescence Protein (Ad-RFP) virus or Ad-Ngn3-N89D virus expressing a Ngn3 null allele (17, 29), Ad-Ngn3 infection increased Ngn3 expression and restored endocrine development of progeny cells (Fig. S7C and D). Thus, genetic programs governing islet development in vivo are similarly required in our system.
We then constructed genetic screens with our system to identify regulators of islet development. We confirmed that shRNA delivered by lentiviral vector infection during G1 sphere development efficiently reduced expression of Ngn3, Pax4, and a transgene encoding eGFP (Fig. 2A–C). Deficiency for Ngn3 or Pax4 impairs pancreatic β-cell development and Ins2 expression in vivo (2), and induction of shNgn3 and shPax4 during sphere development reduced Ins2 expression (Fig. 2 B and C and Fig. S9A). In contrast, shRNAs targeting GFP or islet amyloid polypeptide (IAPP), which are not known to regulate Ins2 expression in vivo, did not modulate sphere Ins2 mRNA levels (Fig. 2A and Fig. S9B). Deficiency for Ngn3 or Arx impairs pancreatic α-cell development and glucagon expression in vivo (2), and induction of shNgn3 or shArx in spheres led to reduced glucagon mRNA levels (Fig. 2 B and D). Together, these results demonstrate that an shRNA-based screen in our system could identify regulators of pancreatic endocrine fate, such as Ngn3, or crucial regulators of islet cell differentiation and allocation, such as Pax4 and Arx.
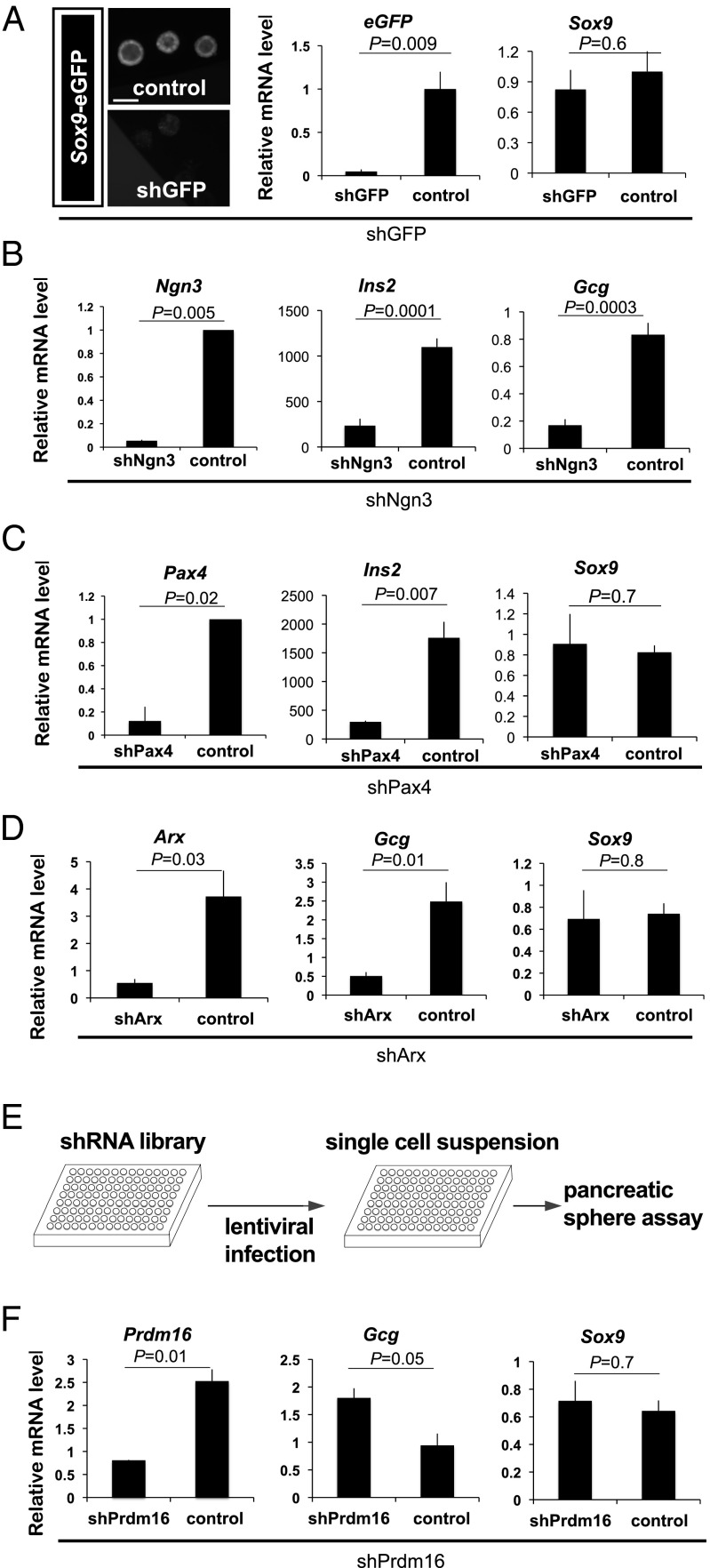
Fig. 2.
Functional genetic screening for essential regulators of islet development, using cultured pancreatic progenitor cells. (A) shGFP-mediated knock-down of Sox9-eGFP was confirmed by eGFP fluorescence intensity under a microscope (Left) and eGFP mRNA levels by qRT-PCR (Right), whereas Sox9 mRNA levels were maintained. (Scale bar, 50 μm.) (B–D) shRNA-mediated knock-down of Ngn3, Pax4, or Arx, assessed by qRT-PCR quantification of indicated mRNAs, including Ins2, glucagon, and Sox9. (E) Schematic showing the lenti-viral delivery of a shRNA library to pancreatic spheres cultured in a 96-well format. (F) Quantification of Prdm16, glucagon, and Sox9 mRNA levels in spheres after Prdm16 shRNA–mediated knockdown. (A–F) n = 3. Error bars, SEM.
To identify additional islet developmental regulators, we used a library of lentiviral shRNAs to knock down expression of 20 genes encoding transcriptional regulators that are expressed in the developing pancreas (Materials and Methods) but have no known function in this developmental program, and for which there are extant mutant mouse strains (Dataset S4). We evaluated shRNA effects on sphere development by measuring key islet cell regulators or hormones (Materials and Methods). Induction of shPrdm16 during sphere development efficiently reduced Prdm16 mRNA levels and increased glucagon mRNA without affecting the expression of Sox9 (Fig. 2F). We did not observe consistent changes in the expression of islet regulators or hormones after knockdown of the other 19 genes (Materials and Methods). Thus, our screen predicted that loss of Prdm16 function would disrupt islet cell differentiation in pancreas development.
Prdm16 Is Required for Pancreatic Islet Development.
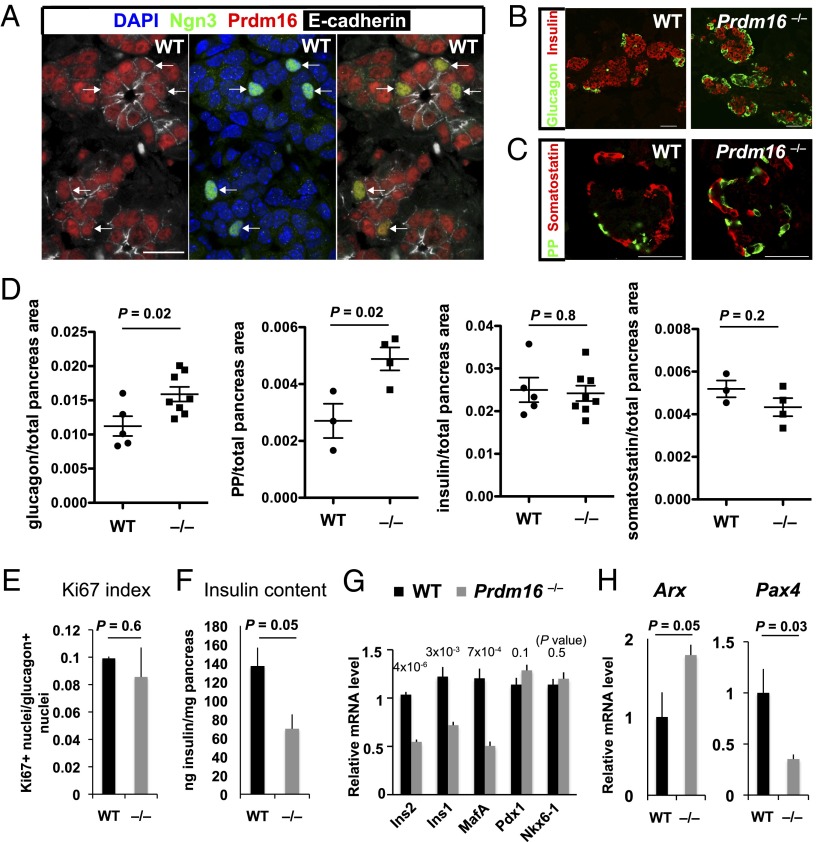
Prdm16 encodes a transcription factor and histone H3 methyltransferase (21) and is expressed in fetal mouse pancreatic epithelial cells including Ngn3+ cells (Fig. 3A and Fig. S9C). Prdm16 is required for development of adipose, brain, and other tissues, and Prdm16 mutants die soon after birth (20, 30, 31). To investigate whether Prdm16 is required for islet development in vivo, we analyzed pancreas development in Prdm16 mutants (31). Immunohistology and morphometry studies revealed a 50% increase of α-cells in Prdm16−/− mutant mice at E18.5 compared with their wild-type littermates (Fig. 3 B and D and Fig. S9D). In contrast, pancreas weight [control, 6.8 ± 2.9 mg (n = 4); Prdm16−/−, 8.6 ± 2.8 mg (n = 3); P = 0.4] and proliferation of Glucagon+ islet cells (Fig. 3E) were not significantly altered in mutants compared with wild-type controls. Prdm16−/− mutants also showed a 100% increase of cells expressing pancreatic polypeptide (PP); in contrast, we observed no detectable change in numbers of insulin+ and somatostatin+ islet cells (Fig. 3 B–D). However, levels of mRNA encoding the hallmark β-cell products Ins1, Ins2, and MafA were reduced and accompanied by a 50% reduction of total insulin content in Prdm16−/− mutant pancreas (Fig. 3 F and G). These findings support the possibility that Prdm16 might regulate expression of factors controlling islet fate and function.
Fig. 3.
Prdm16 is required for pancreatic islet development. (A) Prdm16 expression in pancreatic epithelial cells including Ngn3+ cells. Cofocal images of E15.5 wild-type FVB/NJ pancreas. A cryosection was stained with antibodies against Ngn3 (green), Prdm16 (red), and E-cadherin (white), counterstained with DAPI (blue). Most E-cadherin+ epithelial cells expressed Prdm16 in nuclei. Prdm16 staining with this antibody was undetectable in mutant tissues (Fig. S9C). Virtually all Ngn3+ cells were Prdm16+. Arrows, Ngn3+ nuclei. (Scale bar, 20 mm.) (B and C) Representative pancreatic sections from E18.5 Prdm16 mutants and control littermates were immunostained with indicated antibodies. (Scale bars, 50 μm.) (D) Quantification of total glucagon, PP, insulin and somatostatin area/total pancreatic area in E18.5 control and Prdm16 mutant pancreas (n = 3–8 per genotype). (E) Ki67 index in glucagon+ cells, measured by immunohistology (n = 3–4). (F) Insulin content in total pancreas at E18.5, measured by ELISA and normalized to total pancreas weight (n = 3–4). (G) Relative mRNA levels of the indicated β cell markers in total pancreas at E18.5, measured by qRT-PCR (n = 4). (H) mRNA levels of Arx and Pax4 in wild-type and Prdm16 mutant mice at E15.5 were measured by qRT-PCR (n = 4). (D–H) Error bars, SEM.
In a prior study (32), transgenic misexpression of the Arx homeodomain repressor from a Pdx1 regulatory sequence led to increased numbers of fetal-stage Glucagon+ and PP+ cells accompanied by reduced expression of the Arx target Pax4. Consistent with that study, Prdm16−/− cells had significantly increased expression of Arx and reduced expression of Pax4 (Fig. 3H; SI Materials and Methods). Together with our finding of α-cell and PP-cell hyperplasia, these data reveal an unsuspected requirement for Prdm16 in controlling expression of Arx and Pax4, two crucial regulators of islet cell fate and allocation (19) in pancreas development. Additional molecular studies are warranted to investigate the basis for Arx and Pax4 regulation by Prdm16 in islet development.
Conclusions
In summary, the accessibility and reliability of organogenesis reconstituted from purified pancreas progenitor cells described here allows for scalable studies to analyze gene function in pancreatic islet development. Current methods using embryonic stem cells or other multipotent cell lines generate polyhormonal cells that require xenotransplantation for months before the emergence of insulin-secreting cells resembling islet β-cells (33). In contrast, our system produced monohormonal cells within 1 wk of culture, including glucose-responsive insulin-secreting cells. Although some aspects of development, such as production of fully mature β-cells and complete morphogenesis of islets, await further optimization, our system permitted unbiased genetic loss-of-function analysis and predicted an unexpected in vivo role for Prdm16 in islet development. Taken together, this unique system represents a practical and conceptual advance for efficiently identifying genes, molecules, or conditions that control development, maturation, function, and regeneration of pancreatic cell subsets, including islet cells, and should accelerate progress toward treating diabetes (34, 35).
Materials and Methods
Animals.
All animal studies were performed in accordance with Stanford University Animal Care and Use guidelines. Mice harboring the Sox9-eGFP BAC transgene (from Mutant Mouse Regional Resource Center, University of California, Davis), a floxed allele of Ngn3, or the Rosa26CreERT2 transgene were previously described (22, 29, 36, 37). Because of eGFP perdurance, Sox9-eGFP+ cells in Sox9-eGFP mice contain a mixture of Sox9+ and Sox9neg progeny (22). MIP-GFP mice (38) were a gift from M. Hara (University of Chicago, Chicago, IL) and maintained in a CD-1 background. Actin-DsRed and FVB/NJ mice were purchased from Jackson Laboratory (Bar Harbor, ME). Prdm16 mutant mice (csp1; ref. 31) were purchased from Jackson Laboratory (stock number 013100). Ngn3-tdTomato transgenic mice were generated by pronuclear injection of a DNA construct comprised of tdTomato cDNA (without the ATG start codon) preceded by 6.5 kb DNA encompassing the mouse Ngn3 promoter, the first exon, the intron, and the Ngn3 ATG start codon, and were recovered by recombination from BAC RP23-121F10 (Children’s Hospital Oakland Research Institute, Oakland, CA). The tdTomato cDNA sequence was followed by bovine growth hormone poly(A) sequence in pCDNA (Invitrogen). The genetic background of the one-cell embryo used for injection was C57BL/6, and the offspring were backcrossed to CD-1 females for more than three generations. CD-1 mice were purchased from Charles River Laboratories.
Pancreatic Cell Dispersion.
We dissected the dorsal pancreas from E11.5 mouse embryos obtained by timed mating. Midday of the day on which the vaginal plug was detected was considered as stage E0.5 in the timing of embryo collection. Tissue was washed in Dulbecco’s PBS and then dissociated with 1× TrypLE Express (Invitrogen) at 37 °C for 5 min and triturated. TrypLE was neutralized with 10% (vol/vol) FBS (HyClone) in PBS. Cell viability at this stage exceeded 90%, as assessed by Trypan Blue exclusion. Dissociation into single cells was confirmed by light microscopy.
FACS.
Cells were treated for 15 min in a blocking solution composed of FACS buffer [2% (vol/vol) FBS in PBS] containing 300 ng/mL rat IgG (Jackson ImmunoResearch; modified from ref. 39; Fig. S9E). Either 10 μg/mL propidium iodide (PI; Sigma) or 10 μg/mL AQUA (Molecular Probes) was used to separate dead cells. In some experiments, we used biotin anti-EpCAM (G8.8, 1:100; BioLegend) to purify epithelial cells. We isolated Sox9-eGFPneg and EpCAMneg cells to purify pancreatic mesenchyme, excluding the possibility of mesenchymal cell contamination by epithelial cells using FACS and molecular analysis (Fig. S2). Streptavidin-APC (1:200; eBioscience) was used to visualize biotinylated antibodies. To sort cells, we used the FACS Aria II fitted with a 100-μM nozzle and the operational DIVA software (BD Biosciences). Drops containing more than one cell were removed by using forward scatter height. FACS data were analyzed by using FlowJo software (Tree Star). Cells were collected in 10% (vol/vol) FBS in PBS. Cells were then washed and resuspended in cell culture media.
Cell Culture.
We cultured spheres and organoids in 24-well tissue culture plates (BD Biosciences) except in the experiments for shRNA screening, in which we used 96-well tissue culture plates (BD Biosciences). For 24-well plates, cells isolated from two E11.5 pancreata (600 progenitor cells and 3 × 104 mesenchymal cells) were mixed on ice with a mixture of 30 μL culture media (see following text for the media composition) and 40 μL chilled growth factor-reduced Matrigel (BD) and were cultured as a cell suspension at the bottom rim of each well. Matrigel was solidified at 37 °C for 60 min and overlayed with 500 μL culture media. The culture plates were placed in a 21% or 5% oxygen tissue culture incubator (40). The media was changed every other day, and 0.33 μM retinoic acid was added daily. After 7 d, spheres were harvested or passaged. Organoid analysis was done in G1.
Cell Culture Media.
We used three different culture conditions in which basal media components were shared and slight optimizations were made for each purpose. The basal media contained PrEBM (Lonza) supplemented with 0.1% (vol/vol) insulin (Lonza), 0.1% (vol/vol) transferrin (Lonza), 20 ng/mL mouse IGF1 (R&D), and 0.4% (vol/vol) bovine pituitary extract (Lonza).
To expand and grow spheres, they were cultured at ambient oxygen (21% O2), and the basal media was supplemented with 150 ng/mL human FGF10 (R&D), 2% (vol/vol) B27 (Invitrogen), and 0.33 μM all-trans retinoic acid (Sigma) (S21 media). To induce differentiation of spheres, we cultured spheres in physiological oxygen (5% O2) and supplemented the basal media with 50 ng/mL FGF10, 10 mM nicotinamide (Sigma) (S5 media). Organoids were cultured at ambient oxygen (21% O2). The basal media was supplemented with 150 ng/mL FGF10, 10mM nicotinamide, and 0.5 mg/mL mouse R-spondin1-Fc (gift from A. Ootani and C. Kuo, Stanford University) or 200 ng/mL purified Wnt3a (T.A.B. and R.N.) (Fig. S9E).
Lenti-Viral-Based shRNA Screening.
Transcription factors expressed in the developing pancreas were identified by analysis of gene expression of isolated pancreatic cells (refs. 41–49; Dataset S4). Lentiviruses carrying shRNAs were purchased from the RNAi Consortium at the Broad Institute (Cambridge, MA) in a 96-well format. The virus titer was typically between 3 × 107 and 3 × 108 U/mL. Pancreata from E11.5 MIP-GFP; Ngn3-tdTomato transgenic mice were dissociated with TrypLE and resuspended in 100 μL S21 medium, and 0.5–1 pancreata were plated in one well of a 96-well plate (BD Biosciences). One hundred microliters of viruses were overlayed, and “spin-infection” was then performed by centrifuging the plates at 1020 × g for 90 min at 34 °C. Cells were washed three times with S21 medium. Diluted with 8 μL culture medium, 12 μL Matrigel was overlayed, and cells were cultured with 0.33 μM RA for 4 d to expand the progenitors as pancreatic spheres. RA was added every day, and fresh medium was added every other day. After 4 d, the plates were transferred to physiological oxygen levels to stimulate endocrine differentiation, and the medium was replaced with S5 culture medium. Infection efficiency was about 50%, and infected cells were selected by puromycin (1 μg/mL; Invitrogen). After 5 d culture at 5% O2, we visually scored the MIP-GFP and Ngn3-tdT fluorescence and counted spheres. After harvesting, we isolated RNA and performed qRT-PCR to measure mRNA levels of In2, Gcg, Sox9, and beta actin. Experiments were performed in triplicate, with results presented as mean ± SEM. The gene expression levels and a visual score of reporter expression level were compared with wells treated with shGFP or empty vector. We tested 4–5 different shRNA clones for each gene in independent wells (Dataset S4). When consistent results were obtained, genes were analyzed further.
Statistical Analyses.
Each variable was analyzed using the two-tailed Student t test. For all analyses, a P value of less than 0.05 was considered significant. Results are given as means ± SEM.
Supplementary Material
Acknowledgments
We thank A. Ootani and C. Kuo for R-spondin; J. Perrino for electron microscopy, T. Jacks [Massachusetts Institute of Technology (MIT)], M. Hara (University of of Chicago), and the MMRRC for mouse strains; D. Root and S. Silver (Broad Institute, MIT) for the shRNA library; Stanford Shared FACS facility, J. Mulholland and K. Lee (Stanford Cell Sciences Imaging Facility), Len and Lee Herzenberg, K. Atkuri, H. Chen, X. Gu, Y. Liu, R. Rodriguez, P. Wang, S. Wu, Y. Zeng, T. Leung, and J. Wang for assistance and advice; and D. Lawson and members of the S.K.K. laboratory for helpful discussions. Support for this project was given by the Berry Fellowship Program (to T.S.). This work was supported by awards from the Juvenile Diabetes Research Foundation, the Larry L. Hillblom Foundation, and the Howard Hughes Medical Institute (to S.K.K.). S.K.K. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304507110/-/DCSupplemental.
References
- 1.Puri S, Hebrok M. Cellular plasticity within the pancreas—lessons learned from development. Dev Cell. 2010;18(3):342–356. doi: 10.1016/j.devcel.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gittes GK. Developmental biology of the pancreas: A comprehensive review. Dev Biol. 2009;326(1):4–35. doi: 10.1016/j.ydbio.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 3.Bonnefond A, Froguel P, Vaxillaire M. The emerging genetics of type 2 diabetes. Trends Mol Med. 2010;16(9):407–416. doi: 10.1016/j.molmed.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Lenoir O, et al. Specific control of pancreatic endocrine β- and δ-cell mass by class IIa histone deacetylases HDAC4, HDAC5, and HDAC9. Diabetes. 2011;60(11):2861–2871. doi: 10.2337/db11-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mellitzer G, et al. IA1 is NGN3-dependent and essential for differentiation of the endocrine pancreas. EMBO J. 2006;25(6):1344–1352. doi: 10.1038/sj.emboj.7601011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prasadan K, et al. Endocrine-committed progenitor cells retain their differentiation potential in the absence of neurogenin-3 expression. Biochem Biophys Res Commun. 2010;396(4):1036–1041. doi: 10.1016/j.bbrc.2010.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seymour PA, et al. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci USA. 2007;104(6):1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynn FC, et al. Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc Natl Acad Sci USA. 2007;104(25):10500–10505. doi: 10.1073/pnas.0704054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuyama K, et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43(1):34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- 10.Kopp JL, et al. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development. 2011;138(4):653–665. doi: 10.1242/dev.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apelqvist A, et al. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400(6747):877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 12.Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci USA. 2000;97(4):1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwitzgebel VM, et al. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 2000;127(16):3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- 14.Jensen J, et al. Independent development of pancreatic alpha- and beta-cells from neurogenin3-expressing precursors: A role for the notch pathway in repression of premature differentiation. Diabetes. 2000;49(2):163–176. doi: 10.2337/diabetes.49.2.163. [DOI] [PubMed] [Google Scholar]
- 15.Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129(10):2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, et al. Mutant neurogenin-3 in congenital malabsorptive diarrhea. N Engl J Med. 2006;355(3):270–280. doi: 10.1056/NEJMoa054288. [DOI] [PubMed] [Google Scholar]
- 17. Jensen JN, Rosenberg LC, Hecksher-Sorensen J, Serup P (2007) Mutant neurogenin-3 in congenital malabsorptive diarrhea. N Engl J Med 356(17):1781–1782. [DOI] [PubMed]
- 18.Ashcroft FM, Rorsman P. Diabetes mellitus and the β cell: The last ten years. Cell. 2012;148(6):1160–1171. doi: 10.1016/j.cell.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kordowich S, Mansouri A, Collombat P. Reprogramming into pancreatic endocrine cells based on developmental cues. Mol Cell Endocrinol. 2010;323(1):62–69. doi: 10.1016/j.mce.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 20.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454(7207):961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinheiro I, et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell. 2012;150(5):948–960. doi: 10.1016/j.cell.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 22.Seymour PA, et al. A dosage-dependent requirement for Sox9 in pancreatic endocrine cell formation. Dev Biol. 2008;323(1):19–30. doi: 10.1016/j.ydbio.2008.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci USA. 2007;104(1):181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhushan A, et al. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development. 2001;128(24):5109–5117. doi: 10.1242/dev.128.24.5109. [DOI] [PubMed] [Google Scholar]
- 25.Stanger BZ, Tanaka AJ, Melton DA. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature. 2007;445(7130):886–891. doi: 10.1038/nature05537. [DOI] [PubMed] [Google Scholar]
- 26.Simon MC, Keith B. The role of oxygen availability in embryonic development and stem cell function. Nat Rev Mol Cell Biol. 2008;9(4):285–296. doi: 10.1038/nrm2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinis M, et al. Oxygen tension regulates pancreatic beta-cell differentiation through hypoxia-inducible factor 1alpha. Diabetes. 2010;59(3):662–669. doi: 10.2337/db09-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah SR, et al. Embryonic mouse blood flow and oxygen correlate with early pancreatic differentiation. Dev Biol. 2011;349(2):342–349. doi: 10.1016/j.ydbio.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S, et al. Myt1 and Ngn3 form a feed-forward expression loop to promote endocrine islet cell differentiation. Dev Biol. 2008;317(2):531–540. doi: 10.1016/j.ydbio.2008.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chuikov S, Levi BP, Smith ML, Morrison SJ. Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nat Cell Biol. 2010;12(10):999–1006. doi: 10.1038/ncb2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bjork BC, Turbe-Doan A, Prysak M, Herron BJ, Beier DR. Prdm16 is required for normal palatogenesis in mice. Hum Mol Genet. 2010;19(5):774–789. doi: 10.1093/hmg/ddp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collombat P, et al. Embryonic endocrine pancreas and mature beta cells acquire alpha and PP cell phenotypes upon Arx misexpression. J Clin Invest. 2007;117(4):961–970. doi: 10.1172/JCI29115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naujok O, Burns C, Jones PM, Lenzen S. Insulin-producing surrogate β-cells from embryonic stem cells: Are we there yet? Mol Ther. 2011;19(10):1759–1768. doi: 10.1038/mt.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322(5907):1490–1494. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliver-Krasinski JM, Stoffers DA. On the origin of the beta cell. Genes Dev. 2008;22(15):1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425(6961):917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 37.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445(7128):661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 38.Hara M, et al. Transgenic mice with green fluorescent protein-labeled pancreatic beta -cells. Am J Physiol Endocrinol Metab. 2003;284(1):E177–E183. doi: 10.1152/ajpendo.00321.2002. [DOI] [PubMed] [Google Scholar]
- 39.Sugiyama T, Rodriguez RT, McLean GW, Kim SK. Conserved markers of fetal pancreatic epithelium permit prospective isolation of islet progenitor cells by FACS. Proc Natl Acad Sci USA. 2007;104(1):175–180. doi: 10.1073/pnas.0609490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Atkuri KR, Herzenberg LA, Niemi AK, Cowan T, Herzenberg LA. Importance of culturing primary lymphocytes at physiological oxygen levels. Proc Natl Acad Sci USA. 2007;104(11):4547–4552. doi: 10.1073/pnas.0611732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu G, et al. Global expression analysis of gene regulatory pathways during endocrine pancreatic development. Development. 2004;131(1):165–179. doi: 10.1242/dev.00921. [DOI] [PubMed] [Google Scholar]
- 42.Soyer J, et al. Rfx6 is an Ngn3-dependent winged helix transcription factor required for pancreatic islet cell development. Development. 2010;137(2):203–212. doi: 10.1242/dev.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White P, May CL, Lamounier RN, Brestelli JE, Kaestner KH. Defining pancreatic endocrine precursors and their descendants. Diabetes. 2008;57(3):654–668. doi: 10.2337/db07-1362. [DOI] [PubMed] [Google Scholar]
- 44.Juhl K, Sarkar SA, Wong R, Jensen J, Hutton JC. Mouse pancreatic endocrine cell transcriptome defined in the embryonic Ngn3-null mouse. Diabetes. 2008;57(10):2755–2761. doi: 10.2337/db07-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffman BG, et al. Identification of transcripts with enriched expression in the developing and adult pancreas. Genome Biol. 2008;9(6):R99. doi: 10.1186/gb-2008-9-6-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarkar SA, et al. Synergizing genomic analysis with biological knowledge to identify and validate novel genes in pancreatic development. Pancreas. 2012;41(6):962–969. doi: 10.1097/MPA.0b013e31823d0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dorrell C, et al. Transcriptomes of the major human pancreatic cell types. Diabetologia. 2011;54(11):2832–2844. doi: 10.1007/s00125-011-2283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffman BG, Zavaglia B, Beach M, Helgason CD. Expression of Groucho/TLE proteins during pancreas development. BMC Dev Biol. 2008;8:81. doi: 10.1186/1471-213X-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pinho AV, Rooman I, Real FX. p53-dependent regulation of growth, epithelial-mesenchymal transition and stemness in normal pancreatic epithelial cells. Cell Cycle. 2011;10(8):1312–1321. doi: 10.4161/cc.10.8.15363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.