Abstract
DNA damage created by reactive oxygen species includes the prototypic oxidized pyrimidine, thymine glycol (Tg), which exists in oxidatively damaged DNA as two diastereoisomeric pairs. In Escherichia coli, Saccharomyces cerevesiae and mice, Tg is preferentially excised by endonuclease III (Endo III) and endonuclease VIII (Endo VIII), yNTG1 and yNTG2, and mNTH and mNEIL1, respectively. We have explored the ability of these DNA glycosylases to discriminate between Tg stereoisomers. Oligonucleotides containing a single, chromatographically pure (5S,6R) or (5R,6S) stereoisomer of Tg were prepared by oxidation with osmium tetroxide. Steady-state kinetic analyses of the excision process revealed that Endo III, Endo VIII, yNTG1, mNTH and mNEIL1, but not yNTG2, excise Tg isomers from DNA in a stereoselective manner, as reflected in the parameter of catalytic efficiency (kcat/Km). When DNA glycosylases occur as complementary pairs, failure of one or both enzymes to excise their cognate Tg stereoisomer from oxidatively damaged DNA could have deleterious consequences for the cell.
INTRODUCTION
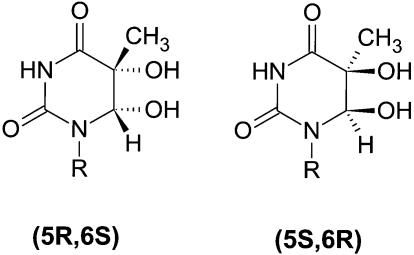
Genomic DNA is subjected to continuous damage by reactive oxygen species generated by cellular metabolism. Thymine glycol (Tg) resulting from chemical oxidation or ionizing radiation (1,2) contains two chiral carbon atoms, forming the (5S,6R:5S,6S) cis-trans and (5R,6S:5R,6R) cis-trans diastereoisomeric pairs (3,4). Following γ-irradiation of DNA, the (5R,6S) and (5S,6R) stereoisomers are present in approximately equal amounts (5); cis stereoisomers (Fig. 1) predominate over the trans forms, which are also detected in aqueous solution (6).
Figure 1.
Thymine glycol cis stereoisomers.
Structural analysis of (5R,6S) Tg embedded in duplex DNA reveals localized distortion of the helix (7). The presence of Tg blocks progression of high-fidelity DNA polymerases in vitro (8–11) and translesion synthesis may be error-free or error-prone (8,9,12–17). While DNA polymerase α shows no stereoselectivity (16) and stalls, following incorporation of adenine opposite both Tg isomers, Y-family DNA polymerases appear to differentiate between stereoisomers. DNA polymerase η bypasses the (5R,6S) isomer more efficiently than the (5S,6R) isomer (16), and DNA polymerase κ may have a higher fidelity when synthesizing across the (5S,6R) isomer (15). These findings suggest that stereoisomers may have differential toxicity for the cell. Site-specific mutagenesis studies in Escherichia coli reveal Tg to be weakly (0.3% T→C mutations) or non-mutagenic (18). Under certain experimental conditions this lesion is lethal to bacteria (2); in animal studies, Tg serves as a biomarker for oxidative DNA damage (19).
Oxidative DNA damage is repaired primarily by base excision repair (BER) (20,21). In E.coli, endonuclease III (Endo III; Nth) and endonuclease VIII (Endo VIII; Nei) initiate repair of oxidized pyrimidines, including Tg, by catalyzing excision of the damaged base and subsequent β-elimination, which leads to cleavage of the phosphodiester backbone. The E.coli nth mutant is a weak mutator and is not hypersensitive to the lethal effects of H2O2 or X-irradiation, suggesting redundancy in the pathways of oxidized pyrimidine repair. In contrast, the nth nei double mutant is a strong mutator, producing predominantly G:C→A:T transitions, and exhibits increased sensitivity to the effects of H2O2 and X-irradiation (22,23). These observations suggest that Endo VIII may serve as a back-up for Endo III during DNA repair (24); however, the activity of Endo VIII in E.coli is only 10% of the total endonuclease activity and the removal of Tg in E.coli appears to depend heavily on Endo III (25).
yNTG1 and yNTG2 are orthologs of Endo III, with overlapping substrate specificities that remove oxidized pyrimidines from Saccharomyces cerevisiae (26–30). Ntg1 is inducible by H2O2, while Ntg2 is constitutively expressed (27,31). Deletions in either gene increase the sensitivity of S.cerevisiae to H2O2-induced mutagenesis (26). The rate of spontaneous mutation remains unchanged, even when mutations in Ntg1 and Ntg2 are combined with deletion of Apn1, the major AP endonuclease in yeast, suggesting that other pathways are involved in the repair of oxidized pyrimidines in yeast (32). Characterization of yNTG1 and yNTG2 activity on DNA exposed to γ-radiation revealed that these enzymes have different kinetic parameters for release of Tg (29). Using site-specifically modified oligonucleotides, Alseth et al. found that while both purified enzymes could cleave a mixture of Tg isomers, only the interaction of yNTG2 and Tg-containing oligonucleotide duplex could be observed by gel shift analysis (26).
In the mouse, mNTH removes a variety of oxidized pyrimidines, including Tg, from damaged DNA. Deletion of the gene coding for this protein (NTH) revealed the presence of at least three other oxidized pyrimidine DNA glycosylase activities (33–35), one of which was subsequently identified as mNEIL1(36). This enzyme is one of three recently discovered homologs of Endo VIII in mammalian cells (37–40). mNEIL1 efficiently excises both stereoisomers of Tg from DNA (40), whereas reduction of mNEIL1 activity in mouse ES cells by siRNA knockdown technology increases the sensitivity of such cells to the cytotoxic effects of ionizing radiation (40). In contrast, mice deficient in mNTH have no discernable phenotype (33–35).
During the past 30 years, virtually all biological studies involving Tg were conducted with DNA containing a mixture of stereoisomers. In this paper, we demonstrate that the major DNA glycosylases that act on oxidatively damaged pyrimidines in bacteria, yeast and mammalian cells excise the (5S,6R) and (5R,6S) forms of Tg isomers from duplex DNA in a stereoselective manner, these enzymes appear to occur as complementary pairs in their respective organisms.
MATERIALS AND METHODS
All chemicals and solvents were of the highest purity available commercially and were used without further purification. [γ-32P]ATP (3000 Ci/mmol) was obtained from Amersham Corp. and T4 polynucleotide kinase was from New England Biolabs. Escherichia coli Endo III was purified using published methods (41), while E.coli Endo VIII was cloned, overexpressed and purified as described previously (42). mNTH and mNEIL1 were overexpressed in E.coli and purified as described previously (40). yNTG1 and yNTG2 were kindly provided as a gift by E.Seeberg (26).
Synthesis and purification of oligonucleotides
The oligodeoxynucleotide 5′-GACAAGCGCAGTCAGCC GAACAC-3′ (I) containing a single thymine residue, and its complementary strand, were synthesized on an Applied Biosystems 394 DNA/RNA synthesizer (Foster City, CA), using standard phosphoramidite chemistry. Products of synthesis were deprotected and released from the controlled-pore glass support by treatment with 28% ammonium hydroxide for 16 h at 55°C. Subsequently, modified oligomers were purified on a Varian ProStar 6.0 high performance liquid chromatography (HPLC) system using a 5 micron, 250 × 10 mm, LUNA phenyl-hexyl column (Phenomenex, Torrence, CA). The solvent system consisted of 0.1 M triethylammonium acetate, pH 6.8, and acetonitrile; the flow rate was 4 ml/min. A gradient of 16–36% acetonitrile was used to elute the dimethoxytrityl-protected product over 30 min, the dimethoxytrityl group was removed with 80% acetic acid and a 5–15% gradient of acetonitrile was used for the deprotected oligomer. Oligonucleotide concentrations were calculated from A260 using molar extinction coefficients.
Preparation and characterization of oligonucleotides containing a single thymine glycol stereoisomer
Oligodeoxynucleotide I (0.2 mg) was incubated at room temperature for 24–48 h in a 2% aqueous solution of OsO4. Reaction products were extracted three times with chloroform to remove excess OsO4. The aqueous phase was evaporated to dryness and dissolved in 250 µl of water. Ten microliters of 0.3 mM NaHSO4 (20-fold molar excess) were added, and the mixture was stirred at room temperature for 16 h to reduce excess OsO4. The solution was again evaporated to dryness and reconstituted in 200 µl of water. A Waters Corp. HPLC system (Milford, MA), consisting of a 600 E Multisolvent Delivery system, a U6K injector and a 996 Photodiode Array Detector, was used in conjunction with an X-Terra, 10 × 50 mm, 5 micron column (Waters Corp.) run at 4.70 ml/min to isolate the cis stereoisomers. Column temperature was maintained at 60°C. After initially eluting isocratically with 5% acetonitrile in 0.1 M ammonium acetate, pH 6.8, for 5 min, a gradient of 5–9% acetonitrile was applied over 30 min. Fractions eluting at 15.8 and 29.2 min were collected, dried and re-chromatographed twice under the same conditions to obtain the pure stereoisomers. The fraction eluting at 15.8 min corresponds to the cis (5S,6R) stereoisomer and the later eluting fraction corresponds to the cis (5R,6S) stereoisomer. The mass of the purified products was confirmed by electrospray ionization mass spectrometry (ESI-MS). Assignment of stereoisomers was based on previous work, where shorter oligonucleotides were digested enzymatically, analyzed by ESI-MS-MS, and the HPLC elution pattern of the released thymine glycol cis-stereoisomers was compared to authentic standards provided by Dr J.Cadet (43).
To confirm that there were no other oxidized bases present in the Tg oligonucleotides, each [19 pmol (5S,6R) or 25 pmol (5R,6S)] was treated with 59 mM piperidine at 90°C for 35 min and purified before analysis on a Voyager-DE STR (Applied Biosystems, Framingham. MA) matrix assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer system operated in the linear mode. The MALDI mass spectrum from the sample (5S,6R) revealed two major peaks at m/z 3374.6 and 3454.4, along with minor peaks representing potassium and ammonium adducts. These fragments are consistent with cleavage and loss of the modified base, resulting in protonated fragments of 5′-PO3H2-CAGCCGAACAC-3′ (calc. 3373.5) and 5′-GACAAGCGAG-PO4H2-3′ (calc. 3453.6), respectively. No other fragment ions were observed in the spectrum, indicating that thymine glycol is the only oxidized base present in either oligonucleotide.
Assay of DNA glycosylase activities
A 23mer oligodeoxynucleotide substrate containing a single Tg residue was prepared by 5′-labeling with T4 polynucleotide kinase (PNK) and [γ-32P]ATP, followed by hybridizing to the complementary strand (1:1.3 ratio) in 1 mM Tris–HCl, pH 8.0, 0.1 mM disodium ethylenediaminetetraacetic acid (Na-EDTA). The reaction mixture for kinetic studies included this [γ-32P]-labeled duplex dissolved in a buffer designed for the glycosylase to be assayed (see below) in a total volume of 10 µl. Enzymes were diluted to working concentrations in 0.5× reaction buffer containing 0.5 mg/ml bovine serum albumin. Reactions used for kinetic studies of Endo III (0.5–2 nM) and Endo VIII (0.15–1.5 nM) contained 50 mM Tris–HCl, pH 7.5, 50 mM KCl, 2 mM Na-EDTA; for mNTH (1–5 nM) contained 50 mM Tris–HCl, pH 7.4, 50 mM KCl, 2 mM Na-EDTA; for mNEIL1 (0.6–2.5 nM) contained 25 mM Na phosphate, pH 7.5, 100 mM NaCl, 2 mM Na-EDTA, 1 mM 1,4-dithiothreitol (DTT); and for yNTG1 (21–105 nM) and yNTG2 (1–2 nM) contained 70 mM MOPS, pH 7.5, 1 mM DTT, 1 mM Na-EDTA, 5% glycerol. Reactions were initiated by addition of enzyme and, after 0–30 min at 37°C, terminated by the addition of 5 µl formamide loading buffer (44) and heating for 1 min at 95°C. An aliquot (5 µl) of each reaction mixture was subjected to polyacrylamide gel electrophoresis (PAGE) in 20% polyacrylamide/8 M urea.
Following PAGE, radioactivity contained in each band on the polyacrylamide gel was measured using a PhosphorImager (Molecular Dynamics). The enzyme concentration and length of incubation was adjusted to cleave no more than 20% of the substrate, thereby maintaining initial velocity conditions. Substrate concentrations varied from 0.1 to 10 times the Km. Velocities were plotted against substrate concentration and the hyperbolic curve obtained fit to a rectangular hyperbola by least-squares non-linear regression. Apparent values were obtained for the Michaelis constant, Km, and the Vmax for cleavage; kcat was calculated by dividing the Vmax by the enzyme concentration. Standard errors derived from the curve-fitting analysis are reported. In determining the rate of cleavage of Tg by yNTG1, apparent values for the specificity constant, kcat/Km, were obtained by linear regression of the velocity versus substrate plot since, for this enzyme, we were unable to achieve saturation of substrate DNA. Thus, individual values for kcat and Km are not reported. At least three independent experiments were performed for all analyses.
Determination of KDapp by gel shift assay
Reaction mixtures contained 0.2–1 nM 32P-labeled oligonucleotide duplex, 25 mM sodium phosphate, pH 7.5, 100 mM NaCl, 2 mM Na-EDTA, 10% glycerol, and varying amounts of enzyme in a total volume of 10 µl. The enzyme was diluted with 0.5× reaction buffer containing 0.5 mg/ml bovine serum albumin. Reaction mixtures were preequilibrated at 4°C and all operations were performed at this temperature. Enzyme was added to the reaction and allowed to bind to the DNA substrate for 3 min. Aliquots (5 µl) were subjected to 8% non-denaturing PAGE (17-cm long), pre-run in 0.5× TBE at 300 V for at least 2 h. Samples were loaded at 300 V, with a tracer dye (bromophenol blue, 0.5× TBE, 10% glycerol) placed in a separate lane. After 10 min, the voltage was reduced to 190 V and electrophoresis was continued until the dye had migrated ∼10 cm from the origin. Radioactivity in the gels was measured with a Molecular Dynamics PhosphorImager. Binding constants were calculated from three independent experiments using a Jandel SigmaPlot version 5.00 non-linear fit routine.
RESULTS AND DISCUSSION
Although Tg is a prominent form of oxidative DNA damage and has been investigated in a number of laboratories for many years, virtually all studies of its mutagenic properties and repair have been performed on DNA or oligodeoxynucleotides containing a mixture of stereoisomers. Recent advances in HPLC made it possible to separate oligodeoxynucleotides containing each Tg stereoisomer (45) and we have used these methods to prepare Tg-containing substrates for the study reported here.
In DNA, and as a mononucleotide, Tg exists as a diastereomeric mixture of two cis-trans forms in which the equilibrium favors the cis form [80% cis for the (5S,6R) pair and 87% cis for the (5R,6S) pair] (3). The rate of cis-trans interconversion is relatively slow (3). Thus, in discussing the kinetic studies reported here, all enzymes are considered to act on the cis form.
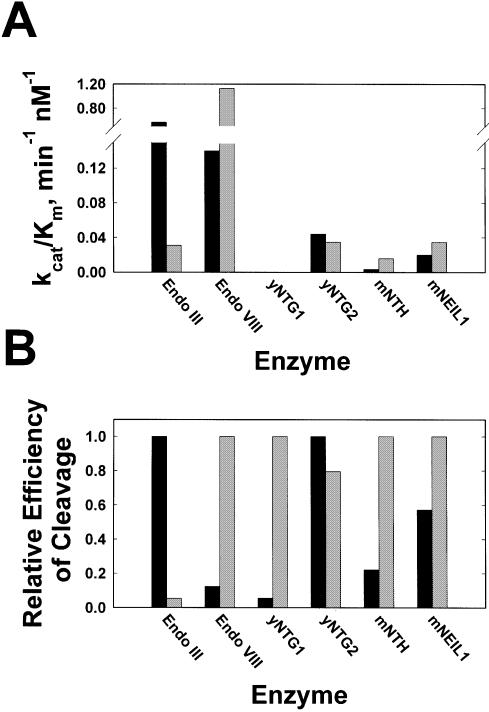
To explore the stereospecificity of DNA glycosylases in bacteria, yeast and mammals, we conducted binding and strand-nicking studies on purified duplex oligonucleotides containing the (5R,6S) and (5S,6R) stereoisomers of Tg. The results are summarized in Table 1, and Figure 2. Figure 2A compares graphically the values for specificity constants (kcat/Km) and Figure 2B contains normalized values for this parameter. The latter analysis facilitates the comparison of enzymes with markedly different catalytic efficiencies.
Table 1. Binding and kinetic parameters for cleavage of thymine glycol stereoisomers.
| Km (nM) | kcat (min–1) | kcat/Km (min–1 nM–1) | KDapp (nM) | |
|---|---|---|---|---|
| Endo III | ||||
| (5S,6R) | 10 ± 0.93 | 5.7 ± 0.11 | 0.57 ± 0.054 | 0.74 ± 0.27 |
| (5R,6S) | 146 ± 19 | 4.6 ± 0.24 | 0.031 ± 0.004 | 5.2 ± 0.81 |
| Endo VIII | ||||
| (5S,6R) | 9.6 ± 1.3 | 1.3 ± 0.06 | 0.14 ± 0.20 | 0.90 ± 0.18 |
| (5R,6S) | 1.8 ± 0.43 | 2.1 ± 0.19 | 1.13 ± 0.25 | 0.55 ± 0.13 |
| YNTG1 | ||||
| (5S,6R) | NM | NM | 1.67 × 10–5 | ND |
| (5R,6S) | 9.3 ± 1.4 | 0.0028 ± 0.0001 | 0.0003 ± 0.00005 | ND |
| YNTG2 | ||||
| (5S,6R) | 1.6 ± 0.6 | 0.70 ±0.009 | 0.044 ± 0.017 | ND |
| (5R,6S) | 7.1 ±1.5 | 0.25 ± 0.015 | 0.035 ±0.0075 | ND |
| mNTH | ||||
| (5S,6R) | 0.44 ± 0.09 | 0.0015 ± 0.000087 | 0.0035 ± 0.00021 | 10 ± 1.7 |
| (5R,6S) | 36 ± 6.8 | 0.57 ± 0.0004 | 0.016 ± 0.0016 | 5.8 ± 0.8 |
| mNEIL1 | ||||
| (5S,6R) | 3.7 ± 0.77 | 0.071 ± 0.0043 | 0.020 ± 0.002 | 5.8 ± 1.3 |
| (5R,6S)a | 0.62 ± 0.14 | 0.022 ± 0.013 | 0.035 ± 0.023 | 3.0 ± 0.9 |
aRosenquist et al. (40).
NM, not measurable; ND, not determined.
Binding and kinetic parameters for each substrate were calculated from at least three independent experiments, as described in Materials and Methods.
Figure 2.
Stereoselectivity of bacterial, yeast and mammalian Endo III and Endo VIII homologs. (A) Cleavage efficiencies (kcat/Km) for the DNA glycosylases studied. Duplex DNA oligonucleotides containing thymine glycol paired with dA. (B) Relative cleavage efficiencies. The efficiencies are normalized with the most efficiently cleaved stereoisomer set to 1.0 to illustrate the selectivity of some of the enzymes studied. Therefore the relative efficiencies between enzymes are not comparable.
Discrimination between Tg stereoisomers by Endo III and Endo VIII
Endo III and Endo VIII are actively involved in the repair of oxidative DNA damage in prokaryotes; their relative efficiencies in excising the two Tg stereoisomers are complementary (Fig. 2). Discrimination between stereoisomers by Endo III is prominently reflected in the Km, which is nearly 15-fold higher for the (5R,6S) form. The kcat is similar for both stereoisomers. Interestingly, the difference in affinity, represented by the apparent Km, is only partially reflected in the experimentally determined dissociation constant (KDapp), suggesting that a step subsequent to binding contributes significantly to the observed stereoisomeric discrimination. For Endo VIII, both kcat and Km contribute to the difference in specificity constants, although the difference in kcat is <2-fold, while the difference in Km is 10-fold. KDapps for both substrates are in the subnanomolar range. These kinetic data suggest that Tg is most efficiently removed from the bacterial genome when Endo III and Endo VIII act in concert. Results from our kinetic analysis conflict with the conclusions of a recent report, which stated that Endo III failed to discriminate between the two Tg stereoisomers (46). In that study, oligonucleotides containing Tg were prepared by oxidation with KMnO4, and assignment of the (5S,6R) and (5R,6S)-Tg stereoisomers was based on the relative yield of each stereoisomer. The activity of Endo III in excising each stereoisomer was measured at a single substrate concentration under conditions that do not adhere to constraints for determining initial velocities (<20% cleavage). Furthermore, kcat and kcat/Km were reported only for the (5R,6S) stereoisomer. The values reported here for Km and kcat differ significantly from those determined by Asagoshi et al. (46); however, the kcat/Km ratio for Endo III calculated by these authors (0.041) is similar to our value of 0.031. The differences in the individual kinetic parameters may also be a result of the Tg residing in a different sequence context and/or the oligonucleotide length difference.
Recently, Alanazi et al. (47) reported that removal of Tg in E.coli is almost completely dependent on the presence of Endo III; they conclude that there is little redundancy in Tg repair. However, in their study, Tg was generated by H2O2 treatment of the bacteria and the level of the adduct in DNA was assayed using a monoclonal antibody coupled with ELISA. The nth mutant used in these experiments was markedly deficient in repairing Tg, but a low level of repair, consistent with the amount of Endo VIII present in E.coli (25), was observed. Importantly, the antibody used for this analysis (48) was not characterized with respect to its reactivity toward individual stereoisomers of Tg and questions have been raised about its authenticity (49,50).
Discrimination between Tg stereoisomers by yeast orthologs of Endo III, yNTG1 and yNTG2
yNTG1, one of several Endo III orthologs found in yeast (26,27), displays a high degree of stereoselectivity in excising Tg isomers from DNA (Fig. 2; Table 1). The apparent Km of this enzyme for the (5S,6R) stereoisomer is sufficiently high that we were unable to saturate the enzyme and accurately determine the kcat. However, a value for the specificity constant (kcat/Km) can be determined under the experimental conditions used here, as this parameter is represented by the slope of the line derived from a plot of velocity versus substrate concentration. We conclude that at least part of the stereospecificity of yNTG1 lies in the Km. This conclusion supports an earlier observation that, while the interaction of yNTG2 with Tg-containing DNA can be observed by gel shift, no shift was seen when yNTG1 was incubated with the same substrate (26). In contrast, yNTG2 exhibits almost no stereoselectivity toward the Tg stereoisomers (Fig. 2B) as minor differences in Km (4-fold) and kcat (3-fold) compensate for one other and result in nearly identical kcat/Km values for both isomers. Overall, yNTG2 displays higher activity than yNTG1 against Tg (Fig. 2A). Conflicting reports have appeared regarding the effects of deletion of Ntg1 and Ntg2 in cells. While all three reports show no increase in sensitivity to oxidizing agents (26,30,32), one report showed an increase in spontaneous and H2O2-induced mutagenesis (26), while another did not (27). The study that showed increased mutagenesis used exponentially growing cells, while in the other study the cells were in stationary phase. This difference in experimental conditions may explain the seemingly contradictory results. Our in vitro data suggest that deletion of Ntg2 should significantly decrease the level of repair of the (5S,6R) stereoisomer, since the complementary enzyme, yNTG1, is very inefficient at repairing this isomer.
Discrimination between Tg isomers by mammalian orthologs of Endo III (mNTH) and Endo VIII (mNEIL1)
To explore the stereospecific repair of Tg in mammalian cells, we performed steady-state kinetic analyses of strand-nicking catalyzed by mNTH, the mouse ortholog of Endo III, and by mNEIL1, one of three mouse orthologs of Endo VIII. It has been reported that the human ortholog of NTH does not follow Michaelis–Menten kinetics (51); however, at the concentrations of enzyme and substrate used in our experiments, the increase in velocity was proportional to the increase in enzyme concentration. Therefore, Km and kcat are useful parameters for comparing the activity of mNTH on stereoisomers of Tg.
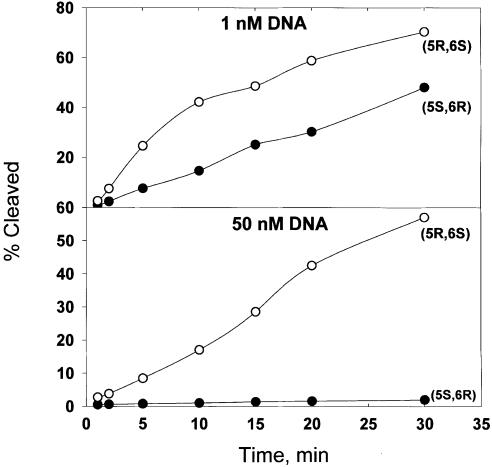
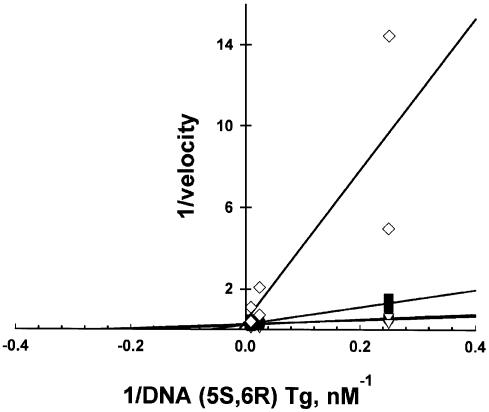
Overall, the (5R,6S) stereoisomer promotes nicking nearly 5-fold more efficiently by mNTH than does the (5S,6R) isomer (Fig. 2; Table 1). The higher specificity constant is reflected in the 380-fold higher kcat for the (5R,6S) isomer (Table 1). In contrast, the Km for the (5S,6R) stereoisomer is 82-fold lower than the Km for its (5R,6S) counterpart. This dichotomy suggests that, at very low substrate concentrations, where saturation of the enzyme active site by either stereoisomer is similar, differences between these two substrates with respect to rates of Tg isomer excision will be less pronounced, as shown in Figure 3. Experiments in which one isomer was end-labeled with 32P while the other isomer was used as an unlabeled competitor reveal that competitive kinetics prevail (Fig. 4). Moreover, the value for the inhibition constant, Ki, for each stereoisomer is comparable to that of the Km determined in steady-state reactions containing a single stereoisomer. The (5S,6R) stereoisomer exhibits a lower Km and Ki and, therefore, is expected to compete with the (5R,6S) stereoisomer at low substrate concentrations. However, in genomic DNA, where the search is by one-dimensional diffusion, the rate of encounter will not depend on the nature of the lesion, and the chemical step will likely be ‘rate-limiting’, provided that—as with oligonucleotide substrates—the base-flipping and other conformational rearrangements occur faster than chemical steps. The lower the kcat, the longer the enzyme will remain at the lesion site. Thus, if these conditions prevail, the (5S,6R) stereoisomer will compete with (5R,6S) stereoisomer by virtue of its lower kcat.
Figure 3.
Excision of Tg stereoisomers by mNTH as a function of time at 37°C. Oligonucleotide duplexes containing (5R,6S) or (5S,6R) stereoisomers of Tg (concentrations indicated) were incubated with 5 nM mNTH for 0–30 min under standard reaction conditions (Materials and Methods).
Figure 4.
The (5S,6R) Tg stereoisomer is a competitive inhibitor of the (5R,6S) isomer. mNTH activity on 32P-labeled (5R,6S) was determined in the presence of increasing concentrations of competing unlabeled (5S,6R) stereoisomer. The activities represent initial rates of dephosphorylation (<20% substrate cleavage). In the Lineweaver–Burk plot, the reciprocal of the velocity is plotted versus the reciprocal of the 32P-labeled substrate concentration in the presence of (filled circle), 0 nM (5S,6R) Tg; (open triangle), 0.1 nm (5S,6R) Tg; (filled square), 0.4 nM (5S,6R) Tg; and (open diamond), 4 nM (5S,6R) Tg.
Like Endo VIII, mNEIL1 possesses both DNA glycosylase and β,δ-lyase activities, displaying robust activity against DNA containing oxidized pyrimidines (36,40,52). The specificity constant for cleavage of the (5R,6S) stereoisomer by mNEIL1 is nearly 2-fold higher than for its (5S,6R) counterpart (Fig. 2; Table 1). In this case, the specificity constant is a product of opposing Kms and kcats. Although both mammalian enzymes are more efficient in removing the (5R,6S) than the (5S,6R) stereoisomer, the specificity constants (0.035/nM/min and 0.02/nM/min, respectively) for mNEIL1 cleavage of both are higher than for incision of the preferred (5R,6S) stereoisomer (0.016/nM/min) by mNTH (Fig. 2A; Table 1). Mouse cells in which levels of mNEIL1 have been reduced by 70% using RNA interference techniques show increased sensitivity to the effects of ionizing radiation (40). Both Tg stereoisomers are generated by ionizing radiation (5) and mNEIL1 is significantly more efficient than mNTH in removing the (5S,6R) stereoisomer. Thus, accumulation of this stereoisomer alone, or together with other oxidative lesions, may contribute to the observed increased radiosensitivity of mNEIL1-deficient ES cells (40).
Based on these in vitro experiments it is of interest to consider the complementary roles these two enzymes may play in the cell. Although mNTH exhibits lower activity than mNEIL1 against both substrates (Fig. 2A), the kinetics of Tg cleavage using extracts of mNTH+/+ and mNTH–/– cells and a mixture of Tg stereoisomers as substrate suggest that mNTH is responsible for the majority of Tg cleavage activity (34). Using nuclear extracts of mouse cells, McTigue et al. (43) observed stereoselectivity between the Tg stereoisomers similar to that reported for purified mNTH. In fact, in one study of mNTH–/– mitochondrial liver extracts, incision of Tg activity was not detected (53). However, two other groups detected Tg incision activity in mNTH–/– cells and extracts (33,34,36). None of these studies used purified Tg stereoismers, however, so the stereoselectivity of these reactions is not known. Additionally, it is possible that these enzymes are involved in repair in different tissues or at different times during the cell cycle. mNTH1 is expressed ubiquitously in mice and humans, but mRNA levels are higher in some tissues (54,55) and expression in humans is cell cycle dependent (56). In most tissues, mRNA levels of NEIL1 are lower than those of NTH1, with the possible exception of spleen, heart and skeletal muscle (36).
Furthermore, other proteins may alter the selectivity of mNTH. For example, human NTH1 has been shown to interact with APE1 (57), Y-box binding protein 1 (51) and with itself to form dimers (58), all of which increase its overall turnover rate. Other glycosylases have also been shown to be covalently modified by the ubiquitin-like SUMO proteins which, in the case of thymine DNA glycosylase, decrease its substrate binding affinity and increase the rate of turnover (59). While we did not examine the activity of mNTH or mNEIL1 in the presence of modifiers, they may play some role in increasing or altering stereoselectivity of Tg repair in cells.
CONCLUSIONS
Two cis stereoisomers of Tg are present, in approximately equal concentrations, in DNA oxidized by osmium tetroxide or exposed to ionizing radiation. DNA glycosylases involved in the repair of these lesions in cells are stereoselective in vitro, with the striking preference shown by E.coli Endo III for the (5S,6R) stereoisomer being complemented by a preference of Endo VIII for its (5R,6S) counterpart. One of the NTH orthologs involved in Tg repair in yeast, yNTG1, is also stereoselective. In mammals, both of the NTH orthologs have an overall preference for the (5R,6S) stereoisomer, with mNTH showing the stronger stereoselectivity. Depending upon their relative concentration, both enzymes may be required for efficient excision of Tg. The failure of one or both glycosylases to excise their cognate stereoisomer from oxidatively damaged DNA could have deleterious consequences for the cell.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Robert Rieger and Avalyn Lewis for mass spectrometry analysis. We are indebted to Dmitry Zharkov and Carlos de los Santos for helpful suggestions and critical reading of the manuscript. We are grateful to Richard Cunningham and Erling Seeberg for stimulating discussions and for generously providing the endonuclease III (R.C.), yNTG1 (E.S.) and yNTG2 (E.S.) used in this research. This research was supported by grants CA47995 and CA17395 from the National Cancer Institute.
REFERENCES
- 1.Frenkel K., Goldstein,M.S. and Teebor,G.W. (1981) Identification of the cis-thymine glycol moiety in chemically oxidized and γ-irradiated deoxyribonucleic acid by high-pressure liquid chromatography analysis. Biochemistry, 20, 7566–7571. [DOI] [PubMed] [Google Scholar]
- 2.Wallace S.S. (2002) Biological consequences of free radical-damaged DNA bases. Free Radic. Biol. Med., 33, 1–14. [DOI] [PubMed] [Google Scholar]
- 3.Lustig M.J., Cadet,J., Boorstein,R.J. and Teebor,G.W. (1992) Synthesis of the diastereomers of thymidine glycol, determination of concentrations and rates of interconversion of their cis–trans epimers at equilibrium and demonstration of differential alkali lability within DNA. Nucleic Acids Res., 20, 4839–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miaskiewicz K., Miller,J. and Osman,R. (1993) Ab initio theoretical study of the structures of thymine glycol and dihydrothymine. Int. J. Radiat. Biol., 63, 677–686. [DOI] [PubMed] [Google Scholar]
- 5.Teebor G., Cummings,A., Frenkel,K., Shaw,A., Voituriez,L. and Cadet,J. (1987) Quantitative measurement of the diastereoisomers of cis thymidine glycol in γ-irradiated DNA. Free Rad. Res. Commun., 2, 303–309. [DOI] [PubMed] [Google Scholar]
- 6.Vaishnav Y.N., Kan,L.S. and Swenberg,C.E. (1992) Application of high-performance liquid chromatography assay for monitoring kinetics of interconversions of stereoisomers of thymidine glycol. J. Liq. Chromatogr., 15, 2385–2396. [Google Scholar]
- 7.Kao J.Y., Goljer,I., Phan,T.A. and Bolton,P.H. (1993) Characterization of the effects of a thymine glycol residue on the structure, dynamics and stability of duplex DNA by NMR. J. Biol. Chem., 268, 17787–17793. [PubMed] [Google Scholar]
- 8.Ide H., Kow,Y.W. and Wallace,S.S. (1985) Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic Acids Res., 13, 8035–8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark J.M. and Beardsley,G.P. (1986) Thymine glycol lesions terminate chain elongation by DNA polymerase I in vitro. Nucleic Acids Res., 14, 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark J.M. and Beardsley,G.P. (1987) Functional effects of cis-thymine glycol lesions on DNA synthesis in vitro. Biochemistry, 26, 5398–5403. [DOI] [PubMed] [Google Scholar]
- 11.Hatahet Z., Kow,Y.W., Purmal,A.A., Cunningham,R.P. and Wallace,S.S. (1994) New substrates for old enzymes. 5-hydroxy-2′-deoxycytidine and 5-hydroxy-2′-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA N-glycosylase, while 5-hydroxy-2′-deoxyuridine is a substrate for uracil DNA N-glycosylase. J. Biol. Chem., 269, 18814–18820. [PubMed] [Google Scholar]
- 12.Rouet P. and Essigmann,J.M. (1985) Possible role for thymine glycol in the selective inhibition of DNA synthesis on oxidized DNA templates. Cancer Res., 45, 6113–6118. [PubMed] [Google Scholar]
- 13.Hayes R.C. and LeClerc,J.E. (1986) Sequence dependence for bypass of thymine glycols in DNA by DNA polymerase I. Nucleic Acids Res., 14, 1045–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson R.E., Yu,S.L., Prakash,S. and Prakash,L. (2003) Yeast DNA polymerase zeta (zeta) is essential for error-free replication past thymine glycol. Genes Dev., 17, 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischhaber P.L., Gerlach,V.L., Feaver,W.J., Hatahet,Z., Wallace,S.S. and Friedberg,E.C. (2002) Human DNA Polymerase κ bypasses and extends beyond thymine glycols during translesion synthesis in vitro, preferentially incorporating correct nucleotides. J. Biol. Chem., 277, 37604–37611. [DOI] [PubMed] [Google Scholar]
- 16.Kusumoto R., Masutani,C., Iwai,S. and Hanaoka,F. (2002) Translesion synthesis by human DNA polymerase η across thymine glycol lesions. Biochemistry, 41, 6090–6099. [DOI] [PubMed] [Google Scholar]
- 17.McNulty J.M., Jerkovic,B., Bolton,P.H. and Basu,A.K. (1998) Replication inhibition and miscoding properties of DNA templates containing a site-specific cis-thymine glycol or urea residue. Chem. Res. Toxicol., 11, 666–673. [DOI] [PubMed] [Google Scholar]
- 18.Basu A.K., Loechler,E.L., Leadon,S.A. and Essigmann,J.M. (1989) Genetic effects of thymine glycol: site-specific mutagenesis and molecular modeling studies. Proc. Natl Acad. Sci. USA, 86, 7677–7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cathcart R., Schwiers,E., Saul,R.L. and Ames,B.N. (1984) Thymine glycol and thymidine glycol in human and rat urine: a possible assay for oxidative DNA damage. Proc. Natl Acad. Sci. USA, 81, 5633–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallace S.S. (1998) Enzymatic processing of radiation-induced free radical damage in DNA. Radiat. Res., 150, S60–S79. [PubMed] [Google Scholar]
- 21.Memisoglu A. and Samson,L. (2000) Base excision repair in yeast and mammals. Mutat. Res., 451, 39–51. [DOI] [PubMed] [Google Scholar]
- 22.Jiang D., Hatahet,Z., Blaisdell,J.O., Melamede,R.J. and Wallace,S.S. (1997) Escherichia coli endonuclease VIII: cloning, sequencing and overexpression of the nei structural gene and characterization of nei and nei nth mutants. J. Bacteriol., 179, 3773–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito Y., Uraki,F., Nakajima,S., Asaeda,A., Ono,K., Kubo,K. and Yamamoto,K. (1997) Characterization of endonuclease III (nth) and endonuclease VIII (nei) mutants of Escherichia coli K-12. J. Bacteriol., 179, 3783–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallace S.S., Bandaru,V., Kathe,S.D. and Bond,J.P. (2003) The enigma of endonuclease VIII. DNA Repair, 2, 441–453. [DOI] [PubMed] [Google Scholar]
- 25.Melamede R.J., Hatahet,Z., Kow,Y.W., Ide,H. and Wallace,S.S. (1994) Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry, 33, 1255–1264. [DOI] [PubMed] [Google Scholar]
- 26.Alseth I., Eide,L., Pirovano,M., Rognes,T., Seeberg,E. and Bjoras,M. (1999) The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell. Biol., 19, 3779–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eide L., Bjoras,M., Pirovano,M., Alseth,I., Berdal,K.G. and Seeberg,E. (1996) Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. Proc. Natl Acad. Sci. USA, 93, 10735–10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.You H.J., Swanson,R.L., Harrington,C., Corbett,A.H., Jinks-Robertson,S., Sentürker,S., Wallace,S.S., Boiteux,S., Dizdaroglu,M. and Doetsch,P.W. (1999) Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry, 38, 11298–11306. [DOI] [PubMed] [Google Scholar]
- 29.Senturker S., Auffret van der Kemp,P., You,H.J., Doetsch,P.W., Dizdaroglu,M. and Boiteux,S. (1998) Substrate specificities of the ntg1 and ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Res., 26, 5270–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gellon L., Barbey,R., Auffret van der Kemp,P., Thomas,D. and Boiteux,S. (2001) Synergism between base excision repair, mediated by the DNA glycosylases Ntg1 and Ntg2 and nucleotide excision repair in the removal of oxidatively damaged DNA bases in Saccharomyces cerevisiae. Mol. Genet. Genomics, 265, 1087–1096. [DOI] [PubMed] [Google Scholar]
- 31.You H.J., Swanson,R.L. and Doetsch,P.W. (1998) Saccharomyces cerevisiae possesses two functional homologues of Escherichia coli endonuclease III. Biochemistry, 37, 6033–6040. [DOI] [PubMed] [Google Scholar]
- 32.Swanson R.L., Morey,N.J., Doetsch,P.W. and Jinks-Robertson,S. (1999) Overlapping specificities of base excision repair, nucleotide excision repair, recombination and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 2929–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takao M., Kanno,S.-i., Shiromoto,T., Hasegawa,R., Ide,H., Ikeda,S., Sarker,A.H., Seki,S., Xing,J.Z., Le,X.C. et al. (2002) Novel nuclear and mitochondrial glycosylases revealed by disruption of the mouse Nth1 gene encoding an endonuclease III homolog for repair of thymine glycols. EMBO J., 21, 3486–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ocampo M.T., Chaung,W., Marenstein,D.R., Chan,M.K., Altamirano,A., Basu,A.K., Boorstein,R.J., Cunningham,R.P. and Teebor,G.W. (2002) Targeted deletion of mNth1 reveals a novel DNA repair enzyme activity. Mol. Cell. Biol., 22, 6111–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elder R.H. and Dianov,G.L. (2002) Repair of dihydrouracil supported by base excision repair in mNTH1 knock-out cell extracts. J. Biol. Chem., 277, 50487–50490. [DOI] [PubMed] [Google Scholar]
- 36.Takao M., Kanno,S.-i., Kobayashi,K., Zhang,Q.-M., Yonei,S., van der Horst,G.T.J. and Yasui,A. (2002) A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J. Biol. Chem., 277, 42205–42213. [DOI] [PubMed] [Google Scholar]
- 37.Bandaru V., Sunkara,S., Wallace,S.S. and Bond,J.P. (2002) A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair, 1, 517–529. [DOI] [PubMed] [Google Scholar]
- 38.Hazra T.K., Izumi,T., Boldogh,I., Imhoff,B., Kow,Y.W., Jaruga,P., Dizdaroglu,M. and Mitra,S. (2002) Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl Acad. Sci. USA, 99, 3523–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morland I., Rolseth,V., Luna,L., Rognes,T., Bjoras,M. and Seeberg,E. (2002) Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res., 30, 4926–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenquist T.A., Zaika,E., Fernandes,A.S., Zharkov,D.O., Miller,H. and Grollman,A.P. (2003) The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair, 2, 581–591. [DOI] [PubMed] [Google Scholar]
- 41.Asahara H., Wistort,P.M., Bank,J.F., Bakerian,R.H. and Cunningham,R.P. (1989) Purification and characterization of Escherichia coli endonuclease III from the cloned nth gene. Biochemistry, 28, 4444–4449. [DOI] [PubMed] [Google Scholar]
- 42.Rieger R.A., McTigue,M.M., Kycia,J.H., Gerchman,S.E., Grollman,A.P. and Iden,C.R. (2000) Characterization of a cross-linked DNA-endonuclease VIII repair complex by electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom., 11, 505–515. [DOI] [PubMed] [Google Scholar]
- 43.McTigue M.M., Rieger,R.A., Rosenquist,T.A., Iden,C.R. and de la Santos,C.R. Stereoselective excision of thymine glycol lesions by mammalian cell extracts. DNA Repair, in press. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 45.Wang Y. (2002) HPLC isolation and mass spectrometric characterization of two isomers of thymine glycols in oligodeoxynucleotides. Chem. Res. Toxicol., 15, 671–676. [DOI] [PubMed] [Google Scholar]
- 46.Asagoshi K., Odawara,H., Nakano,H., Miyano,T., Terato,H., Ohyama,Y., Seki,S. and Ide,H. (2000) Comparison of substrate specificities of Escherichia coli endonuclease III and its mouse homologue (mNTH1) using defined oligonucleotide substrates. Biochemistry, 39, 11389–11398. [DOI] [PubMed] [Google Scholar]
- 47.Alanazi M., Leadon,S.A. and Mellon,I. (2002) Global genome removal of thymine glycol in Escherichia coli requires endonuclease III but the persistence of processed repair intermediates rather than thymine glycol correlates with cellular sensitivity to high doses of hydrogen peroxide. Nucleic Acids Res., 30, 4583–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leadon S.A. (1987) Production of thymine glycols in DNA by radiation and chemical carcinogens as detected by a monoclonal antibody. Br. J. Cancer Suppl., 8, 113–117. [PMC free article] [PubMed] [Google Scholar]
- 49.Leadon S.A. (2003) Retraction. DNA Repair, 2, 361. [DOI] [PubMed] [Google Scholar]
- 50.Gowen L.C., Avrutskaya,A.V., Latour,A.M., Koller,B.H. and Leadon,S.A. (2003) Retraction. Science, 300, 1657. [DOI] [PubMed] [Google Scholar]
- 51.Marenstein D.R., Ocampo,M.T., Chan,M.K., Altamirano,A., Basu,A.K., Boorstein,R.J., Cunningham,R.P. and Teebor,G.W. (2001) Stimulation of human endonuclease III by Y box-binding protein 1 (DNA-binding protein B). Interaction between a base excision repair enzyme and a transcription factor. J. Biol. Chem., 276, 21242–21249. [DOI] [PubMed] [Google Scholar]
- 52.Hazra T.K., Kow,Y.W., Hatahet,Z., Imhoff,B., Boldogh,I., Mokkapati,S.K., Mitra,S. and Izumi,T. (2002) Identification and characterization of a novel human DNA glycosylase for repair of cytosine derived lesions. J. Biol. Chem., 277, 30417–30420. [DOI] [PubMed] [Google Scholar]
- 53.Karahalil B., de Souza-Pinto,N.C., Parsons,J.L., Elder,R.H. and Bohr,V.A. (2003) Compromised incision of oxidized pyrimidines in liver mitochondria of mice deficient in NTH1 and OGG1 glycosylases. J. Biol. Chem., 278, 33701–33707. [DOI] [PubMed] [Google Scholar]
- 54.Sarker A.H., Ikeda,S., Nakano,H., Terato,H., Ide,H., Imai,K., Akiyama,K., Tsutsui,K., Bo,Z., Kubo,K. et al. (1998) Cloning and characterization of a mouse homologue (mNthl1) of Escherichia coli endonuclease III. J. Mol. Biol., 282, 761–774. [DOI] [PubMed] [Google Scholar]
- 55.Aspinwall R., Rothwell,D.G., Roldan-Arjona,T., Anselmino,C., Ward,C.J., Cheadle,J.P., Sampson,J.R., Lindahl,T., Harris,P.C. and Hickson,I.D. (1997) Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc. Natl Acad. Sci. USA, 94, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luna L., Bjørås,M., Hoff,E., Rognes,T. and Seeberg,E. (2000) Cell-cycle regulation, intracellular sorting and induced overexpression of the human NTH1 DNA glycosylase involved in removal of formamidopyrimidine residues from DNA. Mutat. Res., 460, 95–104. [DOI] [PubMed] [Google Scholar]
- 57.Marenstein D.R., Chan,M.K., Altamirano,A., Basu,A.K., Boorstein,R.J., Cunningham,R.P. and Teebor,G.W. (2003) Substrate specificity of human endonuclease III (hNTH1). Effect of human APE1 on hNTH1 activity. J. Biol. Chem., 278, 9005–9012. [DOI] [PubMed] [Google Scholar]
- 58.Liu X., Choudhury,S. and Roy,R. (2003) In vitro and in vivo dimerization of human endonuclease III stimulates its activity. J. Biol. Chem., 278, 50061–50069. [DOI] [PubMed] [Google Scholar]
- 59.Hardeland U., Steinacher,R., Jiricny,J. and Schar,P. (2002) Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J., 21, 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]