Abstract
The positive transcription elongation factor b (P-TEFb) is involved in physiological and pathological events including inflammation, cancer, AIDS, and cardiac hypertrophy. The balance between its active and inactive form is tightly controlled to ensure cellular integrity. We report that the transcriptional repressor CTIP2 is a major modulator of P-TEFb activity. CTIP2 copurifies and interacts with an inactive P-TEFb complex containing the 7SK snRNA and HEXIM1. CTIP2 associates directly with HEXIM1 and, via the loop 2 of the 7SK snRNA, with P-TEFb. In this nucleoprotein complex, CTIP2 significantly represses the Cdk9 kinase activity of P-TEFb. Accordingly, we show that CTIP2 inhibits large sets of P-TEFb- and 7SK snRNA-sensitive genes. In hearts of hypertrophic cardiomyopathic mice, CTIP2 controls P-TEFb-sensitive pathways involved in the establishment of this pathology. Overexpression of the β-myosin heavy chain protein contributes to the pathological cardiac wall thickening. The inactive P-TEFb complex associates with CTIP2 at the MYH7 gene promoter to repress its activity. Taken together, our results strongly suggest that CTIP2 controls P-TEFb function in physiological and pathological conditions.
Discovered in 1995 (1), P-TEFb (CyclinT1/Cdk9) is involved in physiological and pathological transcriptionally regulated events such as cell growth, differentiation, cancer, cardiac hypertrophy, and AIDS (for review, see refs. 2 and 3). It has been suggested to be required for transcription of most RNA polymerase II-dependent genes. However, a recent study suggests that a subset of cellular genes are distinctively sensitive to Cdk9 inhibition (4). P-TEFb is dynamically regulated by both positive and negative regulators. In contrast to Brd4, which is associated with the active form of P-TEFb (5, 6), the 7SK small nuclear RNA (7SK snRNA) and HEXIM1 inhibit Cdk9 activity in the inactive P-TEFb complex (7–10). P-TEFb elongation complexes are crucial for HIV-1 gene transactivation and viral replication. Recently, new P-TEFb complexes containing the HIV-1 Tat protein have been characterized (11, 12), providing evidence for the recruitment of an inactive Tat/P-TEFb complex to the HIV-1 promoter (13). However, defining the diverse nature and functions of the different P-TEFb complexes will require further investigations. The cellular protein CTIP2 (Bcl11b) has been highlighted as a key transcription factor for thymocyte (14, 15) and neuron development (16), odontogenesis (17), cancer evolution (18), and HIV-1 gene silencing (19). Besides AIDS, hypertrophic cardiomyopathy is a well-described P-TEFb–dependent pathology (for review, see refs. 20 and 21).
Here, we report that CTIP2 represses P-TEFb function as part of an inactive P-TEFb complex. In hearts of hypertrophic cardiomyopathic mice, CTIP2 controls P-TEFb-sensitive pathways involved in the establishment of this pathology. Together with the inactive P-TEFb complex, CTIP2 associates with the β-myosin heavy chain promoter to repress its activity. Thereby, CTIP2 might contribute to the regulation of the size of heart sarcomeres in physiological or pathological conditions.
Results
CTIP2 Is Associated with the Inactive P-TEFb Complex.
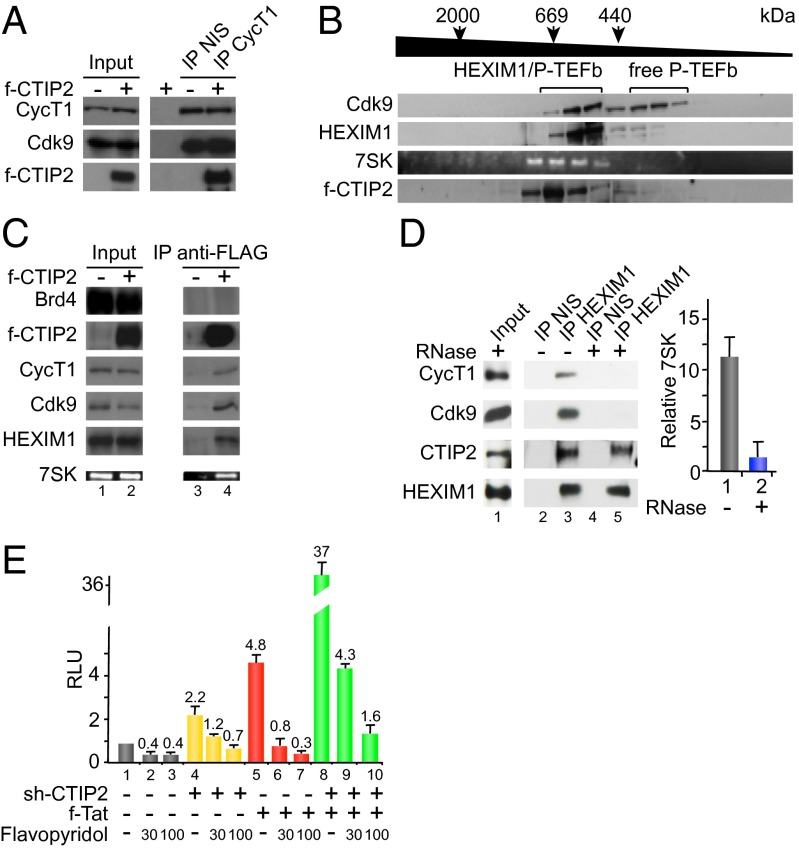
First, we investigated, whether or not CTIP2 associates with one of the previously described P-TEFb complexes. We performed immunoprecipitation experiments, revealing that CTIP2 coimmunoprecipitates with the CyclinT1 and Cdk9 components of the P-TEFb complex (Fig. 1A). To further define the CTIP2-containing complexes, we separated the previously described P-TEFb complexes by gel filtration chromatography (Fig. 1B). As shown in Fig. 1B, Cdk9 was detected in the low molecular weight (LMW) complex (”free” P-TEFb complex) and in the high molecular weight (HMW) complex that coeluted with HEXIM1 and the 7SK snRNA. Interestingly, we found CTIP2 in those latter fractions containing the HMW P-TEFb complex (Fig. 1B). These observations suggested that CTIP2 may be part of an inactive HEXIM1/7SK-including P-TEFb complex. To confirm this hypothesis, we performed additional coimmunoprecipitation experiments targeting the active and the inactive form of P-TEFb. We found that CTIP2 copurified with CyclinT1, Cdk9, HEXIM1, and 7SK but not with Brd4, making CTIP2 a previously undescribed component of the inactive P-TEFb complex. Confocal observations further confirmed the colocalization of CTIP2, P-TEFb, and HEXIM1 in the previously described CTIP2-induced nuclear structures (22) (Fig. S1). As suggested by the gel filtration elution profile, CTIP2 was not found in the active Brd4/P–TEFb complexes. The 7SK snRNA functions as a scaffold RNA facilitating the interaction between HEXIM1 and P-TEFb. To investigate how CTIP2 associates with the inactive P-TEFb complex, we performed coimmunoprecipitation experiments following RNase treatment to degrade the 7SK snRNA (Fig. 1D). As previously described, degradation of the 7SK snRNA (Fig. 1D, Right) induced the dissociation of P-TEFb and HEXIM1 (Fig. 1D, Left, column 5 vs. 3). However, CTIP2 remained associated with HEXIM1 upon RNase treatment, suggesting that CTIP2 directly interacts with HEXIM1 and that the 7SK RNA is required to associate with P-TEFb. These biochemical observations were supplemented by functional experiments. Because the HIV-1 LTR is a bonafide P-TEFb–sensitive promoter, we quantified its activity upon CTIP2 knockdown, Tat transactivation, and P-TEFb inhibition. We observed that knocking down CTIP2 increased the Tat-dependent transcriptional activity of the HIV-1 promoter, suggesting that endogenous CTIP2 might contribute to the repression of Tat by repressing P-TEFb function (Fig. 1E). In support of this hypothesis, treatments with Flavopyridol, a potent P-TEFb inhibitor, abrogated this synergistic transcriptional activation. Therefore, we conclude that CTIP2 may contribute to P-TEFb repression in the inactive form of the P-TEFb complex. Along this line, we further investigated CTIP2 function in this P-TEFb-, HEXIM1-, and 7SK snRNA-including nucleoprotein complex (Fig. 2).
Fig. 1.
CTIP2 is associated with the inactive P-TEFb complex. (A) HEK293T cells expressing FLAG-CTIP2 were lysed and immunoprecipitated with anti-CyclinT1 antibody or with nonimmune serum IgG (NIS). Input and immunoprecipitated proteins were probed with anti-FLAG (f-CTIP2), anti-CyclinT1, and anti-Cdk9 antibodies. (B) Protein complexes from microglial cell nuclear extracts were separated in 29 fractions by HPLC gel filtration. Each collected fraction was subjected to Western blot analysis with the indicated antibodies and processed for 7SK detection by RT-PCR. Before all separations, the column was calibrated with protein standards (dextran blue, 2,000 kDa, fraction 9; thyroglobulin, 669 kDa, fraction 14; ferritin, 440 kDa, fraction 17). (C) HEK293T cells expressing FLAG-CTIP2 were lysed and immunoprecipitated with anti-FLAG antibody. Input (lines 1 and 2) and immunoprecipitated proteins (lines 3 and 4) were probed with anti-FLAG, anti-Brd4, anti-CyclinT1, anti-Cdk9, and anti-HEXIM1 antibodies. The presence of the 7SK snRNA was assessed by RT-PCR amplification. (D) Cellular extracts were treated (+) or not (−) with RNase before being subjected to immunoprecipitation with anti-Hexim1 antibody or nonimmune serum (NIS) as indicated. Inputs (line 1) and immunoprecipitated proteins (lines 2, 3, 4, and 5) were probed with anti-CTIP2, anti-CyclinT1, anti-Cdk9, and anti-HEXIM1 antibodies. 7SK snRNA degradation was controled by RT-PCR. (E) Cells were transfected with the LTR-LUC (HIV-1 promoter) epizomal reporter construct in the presence of Tat and/or sh-CTIP2 plasmids or the respective control constructs. Cells were treated with 30 or 100 nM flavopyridol 24 h posttransfection and subjected to luciferase assays 48 h posttransfection. Efficiencies of knockdown and overexpression were controlled by Western blot as in Fig. 2 F and G. Results are representative for at least three independent experiments performed in triplicates.
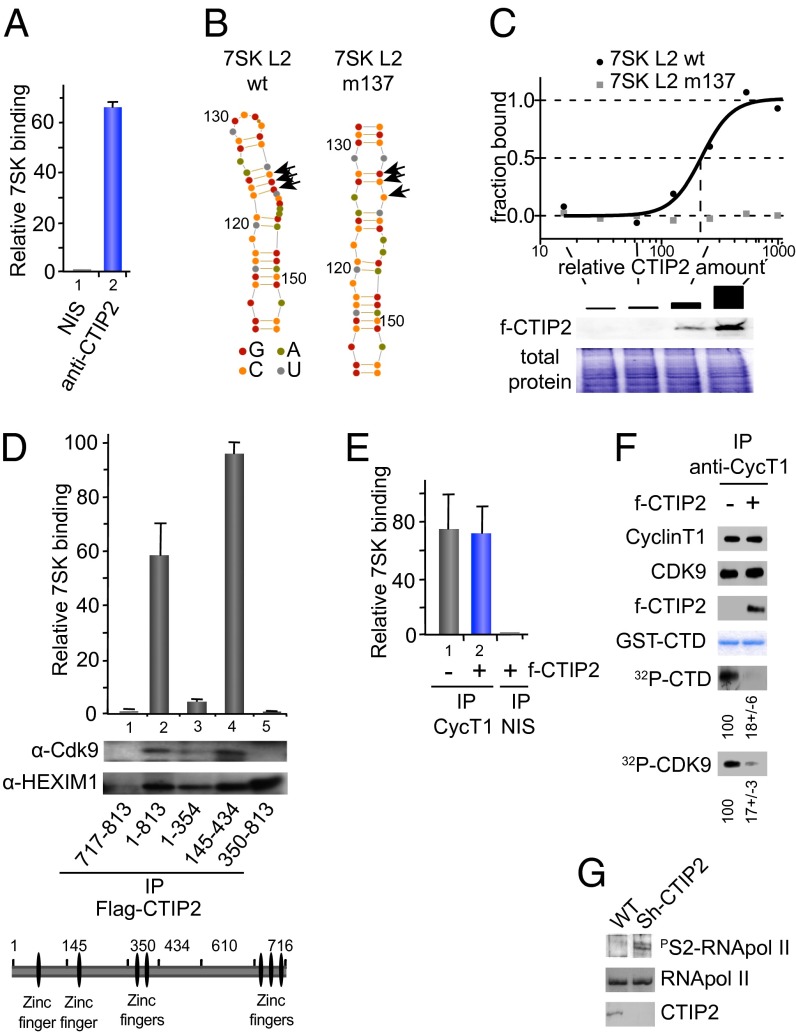
Fig. 2.
Associated with the 7SK snRNA, CTIP2 inhibits Cdk9-mediated phosphorylation. (A) HEK293T cells were lysed and subjected to immunoprecipitations with indicated antibodies. The presence of 7SK snRNA was quantified by RT-Q-PCR after complex elution and RNA extraction. (B) Secondary structures of 7SK L2 and the mutant 7SK L2 m137 were calculated using Assemble software (version 2.1); nucleotides are indicated by color code and numbered with respect to their position in full-length 7SK RNA and arrows indicate mutations in m137. (C) MST analyses were performed with cellular extracts containing increasing amounts of FLAG-CTIP2 and Fam6-labeled 7SK L2 and 7SK L2 m137. The FLAG-CTIP2 amount was monitored by Western blot using anti-FLAG antibody, and the total protein amount was visualized by Coomassie staining. The highest CTIP2 concentration was abitrarily set to 1,000. (D) Mock, FLAG-CTIP2 wild-type, or FLAG-CTIP2 constructs transfected HEK293T cells were lysed and subjected to immunoprecipitation with anti-FLAG antibody. The presence of 7SK snRNA was quantified by RT-Q-PCR after complex elution and RNA extraction. Detection of immunoprecipitated proteins was determined by Western blot analysis with the indicated antibodies. (E) Mock or FLAG-CTIP2 transfected cells were lysed and subjected to immunoprecipitations with the indicated antibodies. The presence of 7SK snRNA was quantified by RT-Q-PCR after complex elution and RNA extraction. (F) FLAG-CTIP2 or mock transfected cells were lysed and subjected to immunoprecipitation with anti-CyclinT1 antibody. Immunoprecipitated proteins were incubated for 1 h at 30 °C with GST-CTD and 1 μCi of 32P-labeled ATP. After SDS/PAGE, labeled GST-CTD and endogenous Cdk9 were detected by autoradiography. Quantification of the signals was performed using ImageJ software. The presence of the indicated proteins was determined by Western blot analysis with the indicated antibodies. The amount of GST-CTD was assessed by Coomassie staining. Specificity for the GST-CTD substrate was controlled by using mutated GST-CTD (Fig. S5). (G) Extracts from WT or CTIP2 knockdown cell lines were subjected to Western blot analysis with the indicated antibodies.
CTIP2 Interacts with the 7SK snRNA.
First, gel shift experiments were performed to define whether CTIP2 interacts with the 7SK snRNA in vitro. As shown in Fig. S2, increasing amounts of purified GST-CTIP2 protein shifted increasing amounts of 32P-labeled 7SK snRNA, indicating that CTIP2 interacts with the 7SK in vitro. As a control, GST-CTIP2 did not bind to TAR RNA in vitro. To define whether this interaction also exists in vivo, we performed immunoprecipitations of CTIP2-associated complexes and probed for the presence of 7SK RNA by RT-PCR. We found that the 7SK snRNA copurified with CTIP2, suggesting that this association exists in vivo (Fig. 2A). Micro scale thermophoresis experiments confirmed this result and allowed us to precise the domain of 7SK involved in this interaction. CTIP2 associates with the Loop 2 domain of the 7SK snRNA, but not with a point-mutated Loop 2 (Fig. 2 B and C). We next investigated which domain of CTIP2 is involved in the interaction with 7SK and P-TEFb. To do so, we performed immunoprecipitation experiments with truncated forms of CTIP2 (Fig. 2D). This mapping confirmed the need of the 7SK snRNA for the interaction with P-TEFb but not with HEXIM1 (Fig. 2D and Fig. S3). Indeed, a CTIP2 mutant lacking amino acids 355–813 was unable to associate with 7SK snRNA and P-TEFb, but still associated with HEXIM1 (Fig. 2D, column 3). Moreover, we identified the domain from 349 to 475 aa of CTIP2 as the potential interface for P-TEFb binding and thereby for the 7SK snRNA interaction (Fig. S4). Surprisingly, the deletion of the 717 first amino acids (out of 813) of CTIP2 was needed to release the interaction with HEXIM1 (Fig. 2D, column 1). Next, we tested whether or not CTIP2 regulates P-TEFb activity.
CTIP2 Inhibits Cdk9-Mediated Phosphorylation.
To explain CTIP2-mediated repression of the P-TEFb function, we first hypothesized that CTIP2 may favor P-TEFb recruitment into the inactive complex. CyclinT1 immunoprecipitation experiments were performed to confirm this hypothesis. Surprisingly, CTIP2 overexpression did not favor recruitment of P-TEFb into the inactive 7SK snRNA-containing complex (Fig. 2E). Because the amount of P-TEFb in the inactive complex was not impacted by CTIP2, we next tested the inhibition of the P-TEFb kinase activity. We first quantified the Cdk9 kinase activity of the CyclinT1-immunoprecipitated complex in the presence or absence of CTIP2 overexpression. As shown in Fig. 2F, CTIP2 overexpression significantly inhibits Cdk9 activity (Fig. 2F and Fig. S5). To confirm that this repression also occurs in physiological conditions, we analyzed the global level of RNA Pol II serine 2 phosphorylation in CTIP2 knockdown cells. Accordingly, higher levels of RNA Pol II serine 2 phosphorylation were observed in CTIP2-depleted cells (Fig. 2G). These observations suggest that CTIP2 represses P-TEFb function by inhibiting Cdk9 activity.
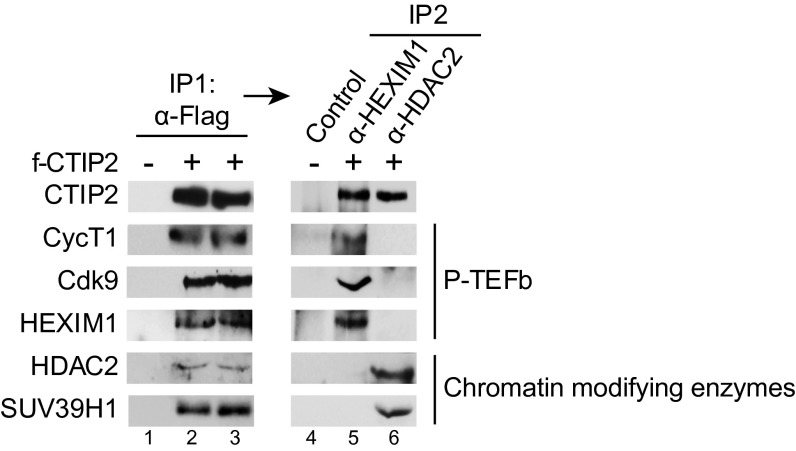
CTIP2-Including Complexes Containing P-TEFb– and Chromatin-Modifying Enzymes Are Mutually Exclusive.
We have reported that CTIP2 silences HIV-1 gene expression by the recruitment of HDAC1, HDAC2, and SUV39H1 to the viral promoter (19). To examine the existence of multiple CTIP2 complexes, we performed sequential immunoprecipiation experiments. For this purpose, CTIP2 associated complexes (IP1) were further immunoprecipitated with anti-HEXIM1 or anti-HDAC2 antibodies to discriminate between potential P-TEFb– and chromatin-modifying enzyme-containing complexes, respectively (IP2). As shown in Fig. 3, HDAC2 and Suv39H1 were excluded from the P-TEFb complex whereas neither Cdk9, CyclinT1, nor HEXIM1 was found in the complex containing the chromatin-modifying enzymes. These data demonstrate that CTIP2 associates at least with two distinct nuclear complexes.
Fig. 3.
CTIP2-including complexes containing P-TEFb and chromatin modifying enzymes are mutually exclusive. Mock or FLAG-CTIP2 transfected HEK293T cells were lysed and subjected to immunoprecipitation with anti-FLAG antibody. After washing, antibody-bound complexes were eluted with FLAG peptide and immunoprecipitated with the indicated antibodies. The presence of the indicated proteins was determined by Western blot analysis.
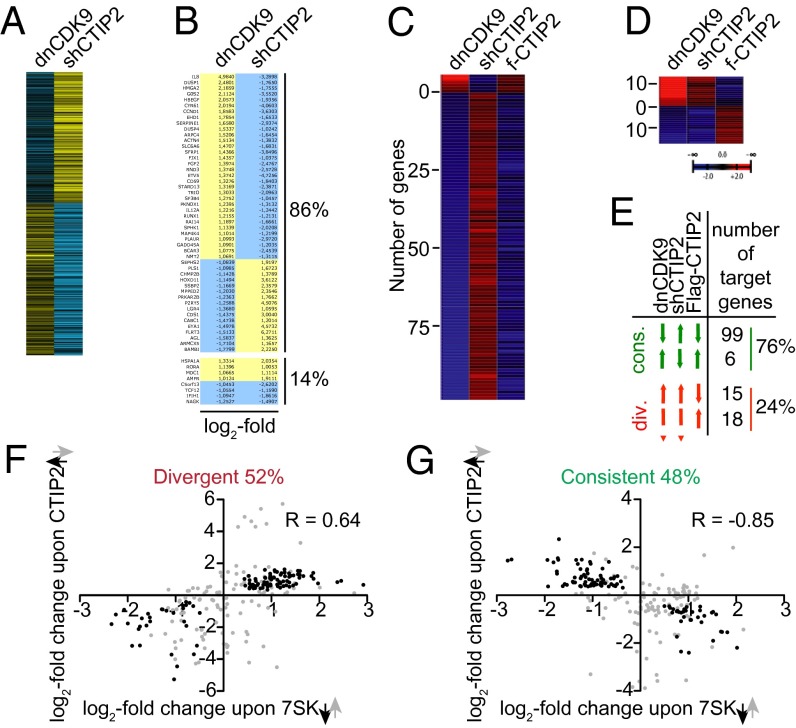
CTIP2 Regulates P-TEFb–Sensitive Genes.
To validate the model emerging from our observations, establishing CTIP2 as a negative regulatory component of P-TEFb, we examined the genome-wide transcriptional consequences of a CTIP2 knockdown in microglial and HEK293 cells. Using DNA microarray analysis, genes whose expression was modulated by siRNA-mediated deletion of CTIP2 (Dataset S1) were compared with those regulated by the expression of a dominant negative mutant of Cdk9 (dnCdk9) (4). Hierarchical clustering of these genes revealed a significant anticorrelation between the two gene expression profiles (Fig. 4A). Clustering of highly coregulated genes (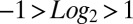 in both gene profiles) revealed that 86% of the Cdk9 sensitive genes are inversely regulated by the CTIP2 knockdown (Fig. 4B and Fig. S6). The comparison of the genes significantly
in both gene profiles) revealed that 86% of the Cdk9 sensitive genes are inversely regulated by the CTIP2 knockdown (Fig. 4B and Fig. S6). The comparison of the genes significantly 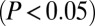 regulated by CTIP2 overexpression, knockdown, and dnCdk9 expression in HEK293 cells confirmed the observations made in microglial cells (Figs. 4 C–E). The 25% of the genes whose expression was significantly
regulated by CTIP2 overexpression, knockdown, and dnCdk9 expression in HEK293 cells confirmed the observations made in microglial cells (Figs. 4 C–E). The 25% of the genes whose expression was significantly 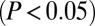 altered following CTIP2 knockdown or overexpression were also sensitive to dnCdk9, and, among them, 76% were regulated consistently with a Cdk9-inhibitory activity of CTIP2. Note that the vast majority of these consistently regulated genes are repressed upon dnCdk9 expression, in concordance with the elongation-activating role of P-TEFb. Besides that, we identified 14% (microglial cells) and 24% (HEK293 cells) of the CTIP2 target genes to be divergently regulated upon dnCdk9 expression, pointing at a different mechanism, where CTIP2 can potentially also contribute to Cdk9 activation (Fig. 4D). Whether this mode of action is direct or indirect needs to be further investigated.
altered following CTIP2 knockdown or overexpression were also sensitive to dnCdk9, and, among them, 76% were regulated consistently with a Cdk9-inhibitory activity of CTIP2. Note that the vast majority of these consistently regulated genes are repressed upon dnCdk9 expression, in concordance with the elongation-activating role of P-TEFb. Besides that, we identified 14% (microglial cells) and 24% (HEK293 cells) of the CTIP2 target genes to be divergently regulated upon dnCdk9 expression, pointing at a different mechanism, where CTIP2 can potentially also contribute to Cdk9 activation (Fig. 4D). Whether this mode of action is direct or indirect needs to be further investigated.
Fig. 4.
CTIP2 regulates P-TEFb–sensitive genes. (A) Genes modulated by the knockdown of CTIP2 in microglial cells were compared with the genes modulated by an inhibition of Cdk9 activity (expression of the dnCdk9 construct) (4). Shades of yellow and blue represent relative activation and repression, respectively. (B) Genes highly sensitive to both dnCdk9 expression and CTIP2 knockdown (modulation average: 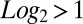 ) were clustered and identified. (C) Transcriptome heatmap of genes significantly
) were clustered and identified. (C) Transcriptome heatmap of genes significantly 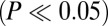 oppositely regulated by Cdk9 and CTIP2: comparison of the expression of dnCdk9 with CTIP2 overexpression and knockdown in HEK293 cells. (D) As in C, but for the statistically significantly coregulated genes. (E) Table showing the number of target genes differentially expressed in each of the comparisons from C and D. (F) Scatter plot of LogQ values for the 112 genes significantly
oppositely regulated by Cdk9 and CTIP2: comparison of the expression of dnCdk9 with CTIP2 overexpression and knockdown in HEK293 cells. (D) As in C, but for the statistically significantly coregulated genes. (E) Table showing the number of target genes differentially expressed in each of the comparisons from C and D. (F) Scatter plot of LogQ values for the 112 genes significantly 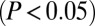 coregulated by CTIP2 overexpression (black arrow up) and 7SK knockdown (black arrow down). The LogQ values of the same set of genes upon CTIP2 knockdown (gray arrow down) and 7SK overexpression (gray arrow up) are illustrated in gray. (G) Same as in F, but for the 105 genes antiregulated in these conditions.
coregulated by CTIP2 overexpression (black arrow up) and 7SK knockdown (black arrow down). The LogQ values of the same set of genes upon CTIP2 knockdown (gray arrow down) and 7SK overexpression (gray arrow up) are illustrated in gray. (G) Same as in F, but for the 105 genes antiregulated in these conditions.
CTIP2 Regulates 7SK snRNA-Sensitive Genes.
Because 7SK snRNA has been described as a key inhibitor of P-TEFb, we next compared the 7SK- and the CTIP2-dependent transcriptome (Fig. 4 F and G and Datasets S2–S5). About 48% of the genes were inversely affected by CTIP2 overexpression or 7SK knockdown. This observation is consistent with a P-TEFb–repressive role of CTIP2 and coincides with our model, in which both 7SK snRNA and CTIP2 contribute to the inactivation of Cdk9. Surprisingly, 52% of the genes were found to be similarly regulated following CTIP2 overexpression or 7SK knockdown, suggesting that CTIP2 regulates a subset of 7SK-sensitive genes by a still unknown, P-TEFb–independent mechanism (Fig. 4F).
CTIP2 Regulates Cdk9-Sensitive Genes Modulated During Cardiac Hypertrophy in Mouse.
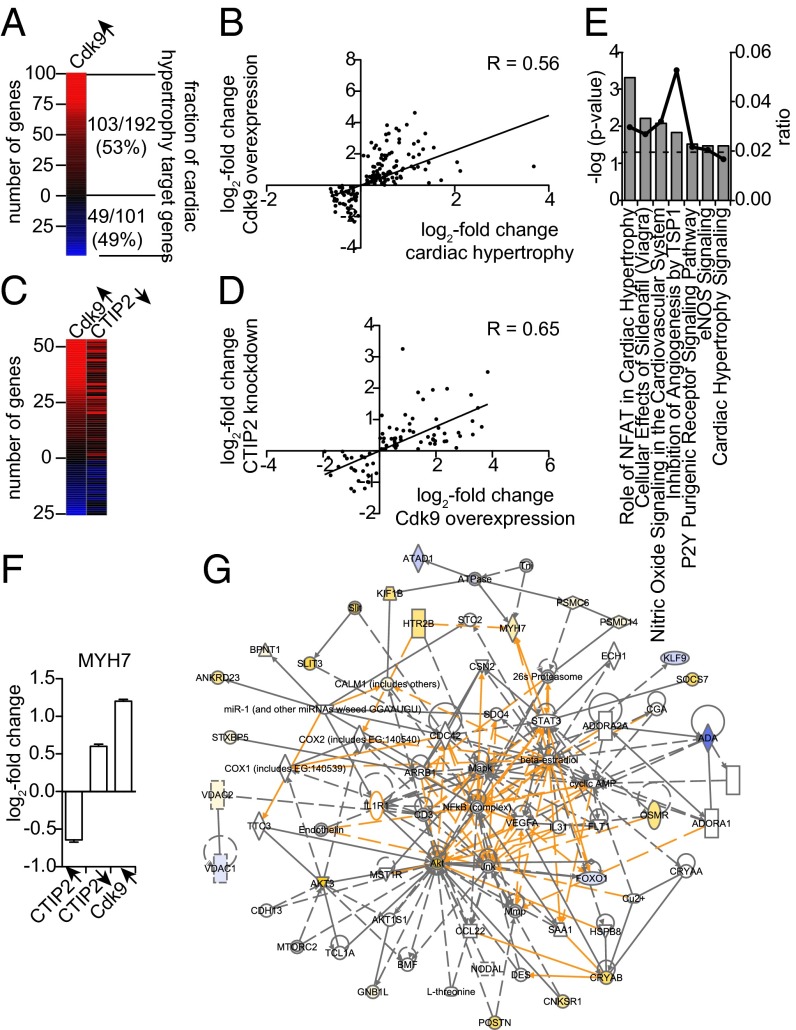
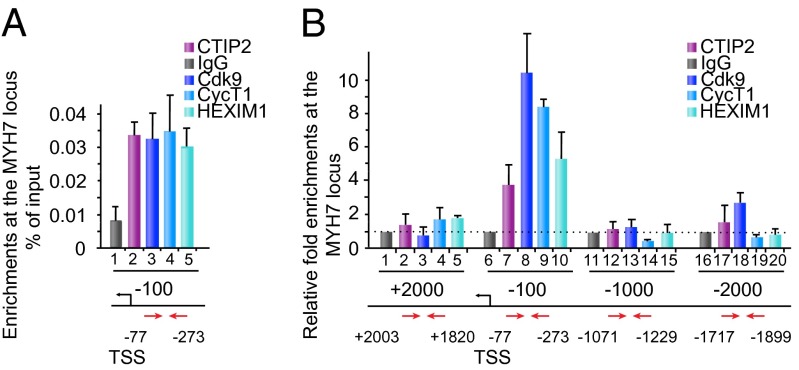
As a compound of the inactive P-TEFb complex, CTIP2 should contribute to the regulation of P-TEFb–related physiopathological events. To investigate the physiological relevance of our observations, we focused on one of the best-characterized P-TEFb–associated pathologies: hypertrophic cardiomyopathy (HCM). HCM was triggered by phenylephrin (PE) administration to adult mice for 15 d. PE is an alpha-adrenergic receptor agonist known to trigger cardiac hypertrophy in mice. Gene expression profiles from HCM mice were compared with those obtained upon Cdk9 overexpression and CTIP2 knockdown (Fig. 5). Cdk9 overexpression regulated half of the HCM-modulated genes, confirming the influence of P-TEFb on this specific pathological gene expression pattern (Fig. 5A and Dataset S6). We observed a significant correlation between the gene expression levels from both conditions (Fig. 5B). By comparing this cluster of genes with CTIP2-sensitive genes, we observed that 50% of the Cdk9-sensitive HCM genes were consistently regulated by CTIP2 knockdown (Fig. 5 C and D and Dataset S7). Interestingly, key HCM pathways are enriched in the CTIP2/Cdk9 cluster of modulated genes (Fig. 5E). We found that the gene of the sarcomeric β-myosin heavy chain (MYH7) was regulated by CTIP2 consistently with a repression of Cdk9 function (Fig. 5F). Moreover, a network analysis of the HCM Cdk9- and CTIP2-sensitive genes highlights the major impact of CTIP2 on the regulation of cardiac hypertrophy (Fig. 5G). Indeed, key HCM pathways, such as the MAPK, the Ca2+/Calmodulin, the NF-κB/NFAT, and the PI3K/AkT pathways, are regulated by CTIP2 (Fig. 5G). We found endogenous CTIP2, CyclinT1, Cdk9, and HEXIM1 bound to the MYH7 promoter (Fig. 6A). Moreover, overexpressed CTIP2 bound to the MYH7 proximal promoter but not to −1,000 bp and −2,000 bp upstream or +2,000 bp downstream of the transcription start site (Fig. 6B). Accordingly, with our model, the recruitment of Cdk9 (Fig. 6, column 3), CyclinT1 (Fig. 6, column 4), and HEXIM1 (Fig. 6, column 5) is favored by the presence of CTIP2. These observations argue for a CTIP2-mediated anchorage of an inactive P-TEFb complex at the MYH7 promoter and suggest a major influence of CTIP2 in the control of the size of heart sarcomeres. The proposed mechanistic model is presented in Fig. 7.
Fig. 5.
CTIP2 regulates P-TEFb–sensitive genes in hearts of hypertrophic cardiomyopathic mice. (A) Heatmap of genes significantly 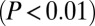 differentially expressed in hearts of hypertrophic cardiomyopathic compared to healthy mice, whose expression is affected in the same manner upon overexpression of Cdk9 in HEK293 cells. The fraction of up- and down-regulated cardiac hypertrophy target genes in the Cdk9 condition is indicated. (B) Scatter plot of the genes shown in A. The Pearson correlation coefficient (R) is indicated. (C) Heatmap of the fraction of cardiac hypertrophy target genes affected by Cdk9 overexpression and shRNA-mediated knockdown of CTIP2. (D) Scatter plot of the genes shown in C. (E) Canonical cardiovascular pathway enrichment among the genes shown in C. The P value threshold
differentially expressed in hearts of hypertrophic cardiomyopathic compared to healthy mice, whose expression is affected in the same manner upon overexpression of Cdk9 in HEK293 cells. The fraction of up- and down-regulated cardiac hypertrophy target genes in the Cdk9 condition is indicated. (B) Scatter plot of the genes shown in A. The Pearson correlation coefficient (R) is indicated. (C) Heatmap of the fraction of cardiac hypertrophy target genes affected by Cdk9 overexpression and shRNA-mediated knockdown of CTIP2. (D) Scatter plot of the genes shown in C. (E) Canonical cardiovascular pathway enrichment among the genes shown in C. The P value threshold 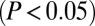 is indicated as a dotted line. (F) Expression changes of the sarcomeric protein MYH7 upon CTIP2 overexpression, knockdown, and overexpression of Cdk9. (G) Gene network of the genes shown in C. Up-regulated genes are shown in yellow, and down-regulated genes are shown in blue.
is indicated as a dotted line. (F) Expression changes of the sarcomeric protein MYH7 upon CTIP2 overexpression, knockdown, and overexpression of Cdk9. (G) Gene network of the genes shown in C. Up-regulated genes are shown in yellow, and down-regulated genes are shown in blue.
Fig. 6.
CTIP2 binds to the MYH7 gene promoter region. (A) Cells were subjected to chromatin immumoprecipitation experiments with the indicated antibodies. The associated promoter region of the MYH7 gene was quantified by Q-PCR. Enrichments are presented as % of input. (B) Cells transfected with the control FLAG-pcDNA3 or the FLAG-CTIP2 plasmids were subjected to chromatin immumoprecipitation experiments with the indicated antibodies 48 h posttransfection. The associated promoter, the −1,000 bp, the −2,000 bp, and the +2,000 bp regions of the MYH7 gene were quantified by Q-PCR. Fold enrichments are presented relative to the enrichments observed in the control condition. Results are representative of at least three independent experiments.
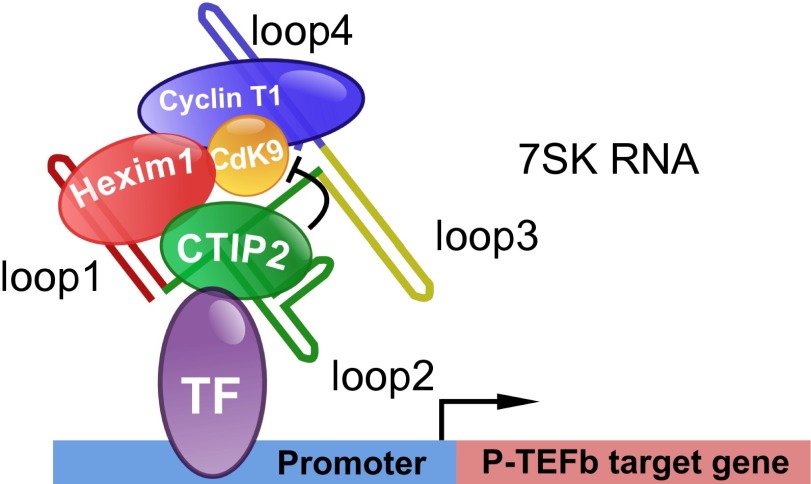
Fig. 7.
Model for CTIP2-mediated repression of P-TEFb target genes. Bound to target promoters, CTIP2 interacts with HEXIM1 and the 7SK snRNA to repress P-TEFb kinase activity and thus the expression of P-TEFb–sensitive genes.
Discussion
The cellular factor CTIP2 is a key transcriptional repressor involved in development (14–17, 23), T lineage commitment (15, 24), cancer (18, 25), and HIV-1 gene silencing (19). In association with chromatin modifying enzymes, CTIP2 promotes the establishment of a local heterochromatin environment at target promoters such as the cellular p21 gene promoter (26) and the viral HIV-1 gene promoter (19, 22, 27). Interestingly, both genes have been reported to be highly sensitive to the P-TEFb elongation complex (28–30). To secure the cell integrity, P-TEFb is dynamically regulated at physiological conditions. 7SK snRNA and HEXIM1 inhibit Cdk9 activity in an inactive HEXIM1/7SK/P-TEFb complex (7–10). We show here that CTIP2 associates with the inactive HEXIM1/7SK/P-TEFb complex, which is distinct from the previously described CTIP2-associated complex containing chromatin-modifying enzymes (19). Our genome-wide comparison of Cdk9- and CTIP2-sensitive cellular genes is consistent with CTIP2 being a P-TEFb–repressive component. Mechanistically, we observed that RNA Pol II Serine 2 phosphorylation levels increase in CTIP2-knockdown cells. In addition, in vitro kinase assays identified CTIP2 as a direct inhibitor of Cdk9 enzymatic activity. Based on these observations, we propose that, when associated with HEXIM1/7SK/P-TEFb, CTIP2 significantly represses Cdk9 kinase activity and thus contributes to the inhibition of P-TEFb function. Interestingly, the presence of the 7SK snRNA is crucial for recruiting CTIP2 to P-TEFb. CTIP2 is able to bind the 7SK snRNA in vitro and in vivo, suggesting a direct interaction between both components. The 7SK snRNA has been shown to have at least two major functions in gene expression regulation: it negatively regulates P-TEFb activity and it acts as a negative regulator of the architectural transcription factor HMGA1 (31–34). However, whether CTIP2 impacts other 7SK-including complexes such as the 7SK/hnRNP (K, A1/A2, or Q1/R) complexes (35) remains under question. Recently, multifunctional HIV-1 Tat-associated P-TEFb complexes were identified. Tatcom2 includes the 7SK snRNA, LARP7 but not HEXIM1, and Tatcom1 includes MLL-fusion partners and the PAF1 complex (12). Preliminary results suggest that CTIP2 may associate with PAF1 and LARP7 (Fig. S7). However, deciphering the influence of Tat expression on the CTIP2/7SK/HEXIM1 complex and defining the impact of CTIP2 on the Tat-associated complexes will need more investigations. Interestingly, CTIP2 and HMGA1 both associate with the loop 2 region of 7SK RNA. Moreover, preliminary data suggest that CTIP2 collaborates with HMGA1 to inhibit HIV-1 gene transcription and the viral replication. Because we have also observed that endogenous CTIP2 represses the Tat/P-TEFb–mediated transactivation of the HIV-1 promoter (Fig. 1E), the repressive activity of CTIP2 and HMGA1 on this P-TEFb–sensitive promoter is now attested. In addition, this control of P-TEFb might contribute to the persistence of the latently infected HIV-1 reservoirs by counteracting the major viral reactivation events. Although ∼50% of the cellular genes were similarly modified upon CTIP2 overexpression or 7SK knockdown, the remaining half showed an inverse correlation. This subset of genes was identified as activated by CTIP2, but repressed by 7SK RNA. These genes are likely regulated by CTIP2 in a Cdk9-independent manner. D’Orso and Frankel showed the recruitment of the inactive P-TEFb complex to the Sp1 binding sites of the HIV-1 promoter (13). However, no mechanistic evidence was provided on the way of this recruitment. Because CTIP2 is anchored to the Sp1 region via an association with Sp1 and LSD1 proteins (27, 36), CTIP2 may constitute a platform for the recruitment of inactive P-TEFb to the HIV-1 promoter. Together with AIDS, hypertrophic cardiomyopathy is a well-described P-TEFb–dependent pathology (for review, see refs. 20 and 21). Upon HCM, the overexpression of the β-myosin heavy chain induces an increase of the size of the sarcomeres and a pathological thickening of the heart muscle. This final target gene expression is controlled by multiple pathways, all contributing to HCM (20). Our results suggest that CTIP2 contributes to the control of the MAPK, the Ca2+/Calmodulin, the NF-κB/NFAT, and the PI3K/AkT pathways. In addition, we show that CTIP2 inhibits the expression of the β-myosin heavy chain (MYH7) consistently with the repression of Cdk9 activity. Moreover, the CTIP2-mediated anchorage of an inactive P-TEFb complex at the MYH7 promoter further argues for the presence of the inactive P-TEFb complex at promoters of P-TEFb sensitive genes. In fact, a recent study by Ji et al. has demonstrated the assembly of 7SK RNA-containing complexes at gene promoters (37). Taken together, our results suggest that CTIP2 is a potent inhibitor of P-TEFb function in two of the best charaterized P-TEFb sensitive pathologies: HCM and AIDS. As a part of the 7SK/HEXIM1/P-TEFb complex, CTIP2 controls P-TEFb–sensitive gene expression and thereby may constitute a pharmaceutical target for fighting P-TEFb–dependent pathologies.
Materials and Methods
Cell Culture.
The human microglial cell line (provided by M. Tardieu, Paris, France) (38) and HEK293T cell lines were maintained in Dulbecco’s modified Eagle medium (DMEM) containing 10% (vol/vol) FCS and 100 U/mL penicillin/streptomycin.
Coimmunoprecipitation Assays and Antibodies.
Two days posttransfection, immunoprecipitations were performed using the standard technique with M2 anti-FLAG (Sigma), anti-Hexim1 (Abcam), or anti-CyclinT1 (Santa Cruz) antibodies overnight at 4 °C with or without RNase or HMBA treatments. Finally, the immunoprecipitated complexes were processed for SDS/PAGE and Western blot analysis, real time quantitative PCR (qRT-PCR), or RT-PCR assay. Proteins were detected in Western blot analyses using antibodies directed against the FLAG epitope (M2 mouse monoclonal from Sigma), CyclinT1 and Cdk9 (Santa Cruz), and CTIP2, Brd4, and HEXIM1 proteins (Abcam).
Gel Filtration Experiments.
The 3 mg of microglial cellular nuclear extracts were concentrated by using a Microcon YM-10 Centrifugal Filter Unit (3,000 Nominal Molecular Weight Limit) and separated by gel filtration on a Superose 6 PC 3.2/30 column (Amersham Biosciences) as previously described (39). The 29 fractions collected were accessed by SDS/PAGE and Western blot analysis. Upon RT-PCR amplification, the presence of the 7SK was visualized by agarose gel electrophoresis.
Micro Scale Thermophoresis.
Micro Scale Thermophoresis (MST) experiments were conducted with Fam6-labeled 7SK L2 RNA or 7SK L2 m137 RNA and increasing concentrations of FLAG-CTIP2 using a Monolith NT.115 (Nanotemper Technologies) as described previously (40) (Laser-power 20%, Laser-on time 60s, LED-power 30%).
Transcriptome Analyses.
Transcriptome analyses were performed using either Agilent Human whole-genome array (G2534-60011), Applied Biosystems human arrays (Product nos. 4339628 and 4336875), or an Affymetrix GeneChip MOE 430 2.0 array according to the manufacturer’s instructions. Data quality was determined using a QC procedure (41). Data were normalized using NeONORM with k = 0.02 (42–44). Subtraction profiling was performed as in refs. 31 and 32 using the CDS test (45).
Generation of Cardiac Hypertrophic Mice.
Micropumps (Alzet 2002 Osmotic Pumps) were set up to deliver (R)-(-) Phenylephrine hydrochloride (Sigma P6126-5G) at 80 mg⋅kg−1⋅day−1 or vehicle buffer only (PBS, 0.002% ascorbic acid) for 15 d. The micropumps were implanted under the back skin of five C57BL/6N mice for each group under 2% Isofurane (Abbott), 2% in oxygen. Cardiac hypertrophy was validated by the increase of heart weight to body weight ratio in treated animals. Protocols are in accordance with the ethic recommendations of the Université Pierre et Marie Curie, Paris 6 University.
Supplementary Material
Acknowledgments
We thank Dr. Olivier Bensaude for scientific discussions and improvement of the manuscript. S.E. and R.R. are recipients of a postdoctoral fellowship from the Agence Nationale de Recherches sur le SIDA et les Hépatites Virales (ANRS). This work was supported by grants from Centre National de la Recherche Scientifique (CNRS, France), the Agence Nationale de Recherches sur le SIDA et les hépatites virale (ANRS, France), Sidaction, the Institut Universitaire de France (IUF), the Belgian Fund for Scientific Reseach (FRS-FNRS, Belgium), the Télévie Program of the FRS-FNRS, the Walloon Region (the Excellence Program ‘Cibles’ and WALEO 021/5110 Program), the European AIDS treatment network (NEAT) integration grant, the Internationale Brachet Stiftung, the Fondation Roi Baudouin, the University of Franche-Comté, the National Institute of Mental Health (R21 MH083585) and the Pennsylvania Department of Health. We thank the Institut Universitaire de Technologie (IUT) Louis Pasteur de Schiltigheim (O.R., C.S., and V.L.D.). B.V.D. and C.V.L. are Chargé de Recherches and Directeur de Recherches du FRS-FNRS, respectively.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220136110/-/DCSupplemental.
References
- 1.Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270(21):12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 2.Brasier AR. Expanding role of cyclin dependent kinases in cytokine inducible gene expression. Cell Cycle. 2008;7(17):2661–2666. doi: 10.4161/cc.7.17.6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Q, Yik JH. The Yin and Yang of P-TEFb regulation: Implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev. 2006;70(3):646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garriga J, Xie H, Obradovic Z, Graña X. Selective control of gene expression by CDK9 in human cells. J Cell Physiol. 2010;222(1):200–208. doi: 10.1002/jcp.21938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19(4):523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19(4):535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 7.Michels AA, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23(13):2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414(6861):322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 9.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414(6861):317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 10.Yik JH, et al. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12(4):971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 11.He N, et al. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 2010;38(3):428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sobhian B, et al. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38(3):439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol. 2010;17(7):815–821. doi: 10.1038/nsmb.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikawa T, et al. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329(5987):93–96. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010a;329(5987):89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45(2):207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 17.Golonzhka O, et al. Ctip2/Bcl11b controls ameloblast formation during mammalian odontogenesis. Proc Natl Acad Sci USA. 2009;106(11):4278–4283. doi: 10.1073/pnas.0900568106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganguli-Indra G, et al. CTIP2 expression in human head and neck squamous cell carcinoma is linked to poorly differentiated tumor status. PLoS ONE. 2009;4(4):e5367. doi: 10.1371/journal.pone.0005367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marban C, et al. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 2007;26(2):412–423. doi: 10.1038/sj.emboj.7601516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harvey PA, Leinwand LA. The cell biology of disease: Cellular mechanisms of cardiomyopathy. J Cell Biol. 2011;194(3):355–365. doi: 10.1083/jcb.201101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinsey TA, Kass DA. Small-molecule therapies for cardiac hypertrophy: Moving beneath the cell surface. Nat Rev Drug Discov. 2007;6(8):617–635. doi: 10.1038/nrd2193. [DOI] [PubMed] [Google Scholar]
- 22.Rohr O, et al. Recruitment of Tat to heterochromatin protein HP1 via interaction with CTIP2 inhibits human immunodeficiency virus type 1 replication in microglial cells. J Virol. 2003;77(9):5415–5427. doi: 10.1128/JVI.77.9.5415-5427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albu DI, et al. Transcription factor Bcl11b controls selection of invariant natural killer T-cells by regulating glycolipid presentation in double-positive thymocytes. Proc Natl Acad Sci USA. 2011;108(15):6211–6216. doi: 10.1073/pnas.1014304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li P, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329(5987):85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Keersmaecker K, et al. The TLX1 oncogene drives aneuploidy in T cell transformation. Nat Med. 2010;16(11):1321–1327. doi: 10.1038/nm.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherrier T, et al. p21(WAF1) gene promoter is epigenetically silenced by CTIP2 and SUV39H1. Oncogene. 2009;28(38):3380–3389. doi: 10.1038/onc.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marban C, et al. COUP-TF interacting protein 2 represses the initial phase of HIV-1 gene transcription in human microglial cells. Nucleic Acids Res. 2005;33(7):2318–2331. doi: 10.1093/nar/gki529. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Giraud S, Hurlstone A, Avril S, Coqueret O. Implication of BRG1 and cdk9 in the STAT3-mediated activation of the p21waf1 gene. Oncogene. 2004;23(44):7391–7398. doi: 10.1038/sj.onc.1207972. [DOI] [PubMed] [Google Scholar]
- 29.Gomes NP, et al. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20(5):601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92(4):451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 31.Eilebrecht S, et al. 7SK small nuclear RNA directly affects HMGA1 function in transcription regulation. Nucleic Acids Res. 2011;39(6):2057–2072. doi: 10.1093/nar/gkq1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eilebrecht S, Bécavin C, Léger H, Benecke BJ, Benecke A. HMGA1-dependent and independent 7SK RNA gene regulatory activity. RNA Biol. 2011;8(1):143–157. doi: 10.4161/rna.8.1.14261. [DOI] [PubMed] [Google Scholar]
- 33.Eilebrecht S, Benecke BJ, Benecke A. 7SK snRNA-mediated, gene-specific cooperativity of HMGA1 and P-TEFb. RNA Biol. 2011;8(6):1084–1093. doi: 10.4161/rna.8.6.17015. [DOI] [PubMed] [Google Scholar]
- 34.Eilebrecht S, Schwartz C, Rohr O. 2013. Non-coding RNAs: Novel players in chromatin-regulation during viral latency. Curr Opin Virol, 10.1016/j.coviro.2013.04.001. [DOI] [PubMed]
- 35.Diribarne G, Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 2009;6(2):122–128. doi: 10.4161/rna.6.2.8115. [DOI] [PubMed] [Google Scholar]
- 36.Le Douce V, et al. LSD1 cooperates with CTIP2 to promote HIV-1 transcriptional silencing. Nucleic Acids Res. 2012;40(5):1904–1915. doi: 10.1093/nar/gkr857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji X, et al. SR Proteins Collaborate with 7SK and Promoter-Associated Nascent RNA to Release Paused Polymerase. Cell. 2013;153(4):855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janabi N, Peudenier S, Héron B, Ng KH, Tardieu M. Establishment of human microglial cell lines after transfection of primary cultures of embryonic microglial cells with the SV40 large T antigen. Neurosci Lett. 1995;195(2):105–108. doi: 10.1016/0304-3940(94)11792-h. [DOI] [PubMed] [Google Scholar]
- 39.Topark-Ngarm A, et al. CTIP2 associates with the NuRD complex on the promoter of p57KIP2, a newly identified CTIP2 target gene. J Biol Chem. 2006;281(43):32272–32283. doi: 10.1074/jbc.M602776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eilebrecht S, Wilhelm E, Benecke BJ, Bell B, Benecke AG. HMGA1 directly interacts with TAR to modulate basal and Tat-dependent HIV transcription. RNA Biol. 2013;10(3):436–434. doi: 10.4161/rna.23686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brysbaert G, Pellay FX, Noth S, Benecke A. Quality assessment of transcriptome data using intrinsic statistical properties. Genomics Proteomics Bioinformatics. 2010;8(1):57–71. doi: 10.1016/S1672-0229(10)60006-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noth S, Brysbaert G, Benecke A. Normalization using weighted negative second order exponential error functions (NeONORM) provides robustness against asymmetries in comparative transcriptome profiles and avoids false calls. Genomics Proteomics Bioinformatics. 2006;4(2):90–109. doi: 10.1016/S1672-0229(06)60021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noth S, Benecke A. Systems Epigenomics Group Avoiding inconsistencies over time and tracking difficulties in Applied Biosystems AB1700/Panther probe-to-gene annotations. BMC Bioinformatics. 2005;6:307. doi: 10.1186/1471-2105-6-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noth S, Brysbaert G, Pellay FX, Benecke A. High-sensitivity transcriptome data structure and implications for analysis and biologic interpretation. Genomics Proteomics Bioinformatics. 2006;4(4):212–229. doi: 10.1016/S1672-0229(07)60002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tchitchek N, et al. CDS: A fold-change based statistical test for concomitant identification of distinctness and similarity in gene expression analysis. Genomics Proteomics Bioinformatics. 2012;10(3):127–135. doi: 10.1016/j.gpb.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.