SUMMARY
Methylxanthines, including caffeine and theophylline are among the most widely consumed stimulant drugs in the world. These effects are mediated primarily via blockade of adenosine receptors. Xanthine analogues with improved properties have been developed as potential treatments for diseases such as Parkinson’s disease. Here we report the structures of a thermostabilised adenosine A2A receptor in complex with the xanthines xanthine amine congener and caffeine, as well as the A2A selective inverse agonist ZM241385. The receptor is crystallised in the inactive state conformation as defined by the presence of a salt bridge known as the ionic lock. The complete third intracellular loop, responsible for G protein coupling, is visible consisting of extended helices 5 and 6. The structures provide new insight into the features which define the ligand binding pocket of the adenosine receptor for ligands of diverse chemotypes as well as the cytoplasmic regions which interact with signal transduction proteins.
INTRODUCTION
Plant-derived methylxanthines which include caffeine (from the coffee bean), theophylline (from the tea leaf) and theobromine (from the cocoa bean) are among the most widely consumed stimulant substances in the world with Americans consuming an average of 200mg of caffeine per day (Daly, 2007). In 1981, it was demonstrated that the behavioural stimulant effects of methylxanthines were mediated by blockade of adenosine receptors (Snyder et al., 1981), although at higher concentrations methylxanthines also have effects on several other target proteins such as phosphodiesterases (Daly, 2007). There are 4 receptors for adenosine (A1, A2A, A2B, A3) (Fredholm et al., 2011) which are all members of the G protein-coupled receptor (GPCR) family of membrane spanning proteins. The receptors are widely expressed in the brain, cardiovascular and immune system and there is growing evidence that drugs acting at adenosine receptors represent promising approaches in a wide range of diseases (Jacobson and Gao, 2006). Activation of adenosine receptors results in a conformational change propagated to the intracellular surface where the receptors interact either with heterotrimeric G proteins (Gilman, 1987) or through β-arrestin (DeWire et al., 2007) to regulate signalling to ion channels and enzyme pathways.
The naturally occurring methylxanthines such as caffeine are non-selective and have micromolar affinities at adenosine receptors (Muller and Jacobson, 2011). A large number of derivatives and analogues of these compounds have been made, with the aim of obtaining higher affinity and more selective ligands as research tools to characterise the function of adenosine receptors as well as for therapeutic purposes. Xanthine-based drugs have been evaluated clinically in diseases including asthma, Parkinson’s disease and pain (Daly, 2007). 8-Aryl derivatives of the xanthine core include the amine congener 8-[4-[[[[(2-aminoethyl)amino]carbonyl]methyl]oxy]phenyl]-l,3-dipropylxanthine (XAC), which has a greatly enhanced affinity for adenosine receptors and an increased solubility compared to caffeine. This compound has proved a very useful tool in the study of adenosine receptors since it has been radiolabelled for use as a tracer and has been immobilised for affinity purification of adenosine receptors (Muller and Jacobson, 2011).
The adenosine A2A receptor is of particular interest as a drug target for Parkinson’s disease, with the drug preladenant currently in clinical trials (Salamone, 2010). The design of drugs for GPCRs is hampered by the lack of structural information and hence obtaining the structure of this receptor in complex with a range of different ligand chemotypes was required to assist in the discovery of novel drugs targeted at this receptor. Obtaining high resolution structures of GPCRs is hampered by their intrinsic flexibility and their instability when removed from the plasma membrane (Tate, 2010). A number of approaches have recently been developed to overcome this problem. The first non-rhodopsin GPCR structure to be determined was the β2-adrenergic receptor (Rasmussen et al., 2007) in complex with an antibody fragment bound to the third intracellular loop (ICL) - a critical domain of the receptor that mediates G protein coupling, but also contributes to receptor flexibility. A higher resolution structure of the β2AR was obtained by fusing T4 lysozyme into ICL3 (Cherezov et al., 2007) and the same methodology was used to obtain the first structure of the adenosine A2A receptor (A2A-T4L) (Jaakola et al., 2008) in complex with ZM241385, as well as more recently the structures of the chemokine receptor CXCR4 (Wu et al., 2010) and the dopamine D3 receptor (Chien et al., 2010). The A2A-T4L receptor has also been crystallised in the presence of the high affinity agonist UK-432097 (Xu et al., 2011). The conformational state of these receptors can be difficult to determine since insertion of the T4 lysozyme can alter the pharmacology and prevents signalling (Rosenbaum et al., 2007). Nevertheless clear differences can be seen between the two A2A-T4L structures bound to the inverse agonist ZM241385 and the agonist UK-432097 which resemble some of the changes associated with receptor activation (Xu et al., 2011).
An alternative approach to obtaining structures is conformational thermostabilisation. This method involves the introduction of a small number of point mutations into the receptor, which increases the thermostability whilst altering the equilibrium between the agonist and antagonist conformation. The thermostabilisation of a particular conformation is directed by the pharmacology of the ligand used during the selection of residues for mutation (Tate and Schertler, 2009). The first structure obtained using this approach was the β1-adrenergic receptor (β1AR) (Warne et al., 2008). This was the first non-rhodopsin structure to clearly show features of the cytoplasmic regions of the receptor and revealed the presence of a short well defined helix in ICL2. However, in this structure ICL3 was truncated to assist in crystallisation. A thermostabilised neurotensin receptor has also been described (Shibata et al., 2009). Such receptors are known as StaRs for ‘stabilised receptors’ (Robertson et al., 2011).
The adenosine A2A receptor was previously thermostabilised in both agonist and inverse agonist conformations (Magnani et al., 2008), however these engineered proteins, were not considered stable enough for crystallization. A structure of the A2A receptor stabilised in an agonist conformation bound to adenosine and adenosine-5′-(N-ethylcarboxamide (NECA) has recently been obtained (Lebon et al, 2011a,b). Here we report the stabilisation of the A2A receptor in the inverse agonist conformation and subsequent X-ray structures of this receptor in complex with ZM241385 and the xanthines XAC and caffeine. The structures of the adenosine A2A receptor described here provide new insight into the binding mode of ligands of different chemical classes. Furthermore this is the first structure of the adenosine A2A receptor in the fully inactive state with the ionic lock present and the complete intracellular loop permitting for the first time a detailed view of the structural activation spectrum for this receptor.
RESULTS AND DISCUSSION
Thermostabilisation of the human adenosine A2A Receptor
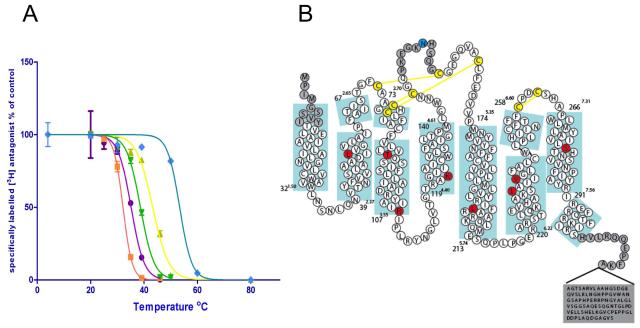
The adenosine A2A receptor was previously stabilised in both agonist and inverse agonist conformations (Magnani et al., 2008) however the stabilised inverse agonist receptor known as Rant21 or A2A-StaR1 (containing the stabilising mutations A54L2.52, T88A3.36, K122A4.43, V239A6.41; superscripts refer to Ballesteros-Weinstein numbering17) was not considered of sufficient stability for structural studies. Further mutagenesis in the presence of the inverse agonist ligand ZM241385 (Poucher et al., 1995) resulted in the identification of an additional 4 stabilising mutations (R107A3.55, L202A5.63, L235A6.37, S277A7.42) giving an apparent thermostability of 47°C in 0.1% decylmaltoside (Figure 1a) resulting in A2A-StaR2. For crystallisation, A2A-StaR2 was truncated at the C-terminus by 96 amino acids up to Ala316 and included a C-terminal decameric His-tag for purification. An N154A mutation was introduced to remove the glycosylation site (Figure 1b).
Figure 1.
(a) Thermostability plots of the wild type adenosine A2A receptor (purple circles), A2A-T4L (orange squares), A2A-StaR1 (previously Rant21, yellow triangles), A2A-T4L engineered to include the A2A-StaR1 mutations in combination with the T4L fusion (A54L2.52/T88A3.36/K122A4.43/V239A6.41) (green triangles) and vs A2A–StaR2 (blue diamonds). Error bars are derived from standard deviation and calculated from duplicated temperature points (n=2) within a single experiment (b) Sequence of the wild-type A2A receptor in relation to the secondary structure determined from the structure of A2A-StaR2. Thermostabilising residues are shown in red circles and the glycosylation site mutation N154A in blue. Disulphide bonds are in yellow. Numbers refer to the first and last residue in each TM (blue boxes) with the Ballesteros-Weinstein numbering in superscript (same for all subsequent figures).
Pharmacology of the inverse agonist state
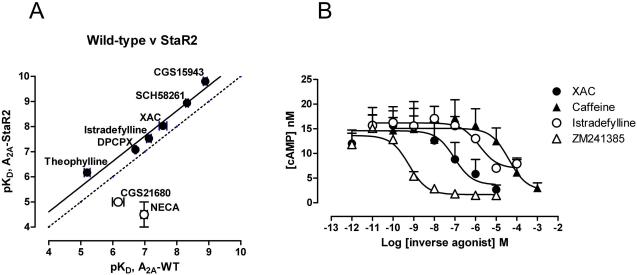
The engineered receptor A2A-StaR2 bound ZM241385 (KD=1.9 nM) and also a range of structurally diverse antagonists with a similar affinity to the wild type receptor (Figure 2a and supplementary Table S1). In contrast, the affinities of agonists including NECA and CGS21860 were reduced by greater than 100-fold and the receptor no longer activated G proteins. This pharmacology is consistent with trapping of the inverse agonist conformation and is similar to the change in pharmacology observed for the stabilised β1AR-m23 (Serrano-Vega et al., 2008).
Figure 2.
(a) Comparison of the pharmacology of A2A-StaR2 with the wild type adenosine A2A receptor. Radioligand competition binding assays for a range of antagonists (closed circles) and agonists (open circles) were carried out with [3H]-ZM-241385 on membranes from cells transiently transfected with receptors. (b) Effects of the ligands ZM241385 (filled squares), XAC (filled circles), caffeine (open circles) and istradefylline (open squares) on the levels of cAMP measured following over expression of the wild type A2A receptor which results in high levels of constitutive activity and elevated levels of cAMP. All ligands are inverse agonists although istradefylline is a weak partial inverse agonist compared to ZM241385, XAC and caffeine.
The thermostabilising residues Thr883.36 and Ser2777.42 lie at the bottom of the agonist binding pocket and have been shown to play a role in binding of the ribose ring of the agonist and in agonist activation (Ivanov et al., 2009; Jiang et al., 1996; Xu et al., 2011, Lebon et al, 2011b). Mutation of these residues is highly stabilising towards the inverse agonist bound but not the agonist bound conformation suggesting that they play a key role in the conformational selection of the receptor. In order to determine whether the loss of agonist affinity was directly related to mutation of these residues both T88A3.36 and S277A7.42 in A2A-StaR2 were mutated back to their wild type amino acids, both individually and together. The stabilised receptor containing the 6 mutations A54L2.52, K122A4.43, V239A6.41, R107A3.55, L202A5.63, L235A6.37 in the absence of T88A3.36 and/or S277A7.42 still had identical pharmacology with respect to loss of agonist binding (Supplementary Table S2). This suggests that the loss of agonist affinity is not just a direct result of these mutations but is also due to conformational selection as previously observed for the stabilised β1AR-m23 (Serrano-Vega et al., 2008). Although T88A3.36 and S277A7.42 have little effect on inverse agonist/antagonist pharmacology we cannot rule out the possibility that the presence of these mutations has an effect on the binding of the ligands presented here, however the consistency of the binding interactions described here with previous extensive mutagenesis data (Dal Ben et al., 2010) and the structures of A2A-T4L in complex with ZM241385 (Jaakola et al., 2008), UK-432097 (Xu et al., 2011) and adenosine and NECA (Lebon et al., 2011b) suggest that this is not the case. Additionally, all of the inverse agonist ligand structures presented here are found positioned over 5Å from either of these residues.
Prior to the structure determination of A2A-StaR2 bound to the ligands ZM241385, XAC and caffeine, their pharmacological profile was characterised. Stable inducible cell lines expressing the native adenosine A2A receptor were constructed using the Flp-In TREx system (Invitrogen). Induction of receptor expression resulted in gradually increasing basal levels of signalling, in the presence of adenosine deaminase which removes endogenous adenosine, indicative of constitutive activity. All the compounds used for subsequent structural studies ZM241385, XAC and caffeine were found to equally inhibit constitutive activity indicative of them being inverse agonists whereas istradefylline (Jenner, 2005) was found to be a weak partial inverse agonist (Figure 2b).
Structure of the thermostabilised adenosine A2A receptor in complex with ZM241385
The purified A2A-StaR2 in complex with ZM241385 was crystallised by vapour diffusion in sitting drops and data collected on the microfocus beamline I24 at the Diamond Light Source (Oxfordshire, UK) corresponding to a 99.9% complete dataset to 3.29Å. The structure was solved by molecular replacement with 3EML with 1 copy of the A2A-StaR2 per crystallographic asymmetric unit. Statistics for data collection and refinement are given in Table 1.
Table 1. Crystallographic Table of Statistics.
| DATA COLLECTION | A2A StaR2 – ZM241385 |
A2A StaR2 – XAC | A2A StaR2 – Caffeine |
|---|---|---|---|
|
| |||
| Space Group | I222 | I222 | I222 |
|
| |||
| Cell Dimensions a, b, c, ( Å ) |
111.93, 112.55, 125.68 | 111.86, 113.06, 126.80 |
112.33, 113.33, 129.30 |
|
| |||
| Cell Angles α, β, γ ( ° ) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
|
| |||
| Resolution ( Å ) | 83.95 – 3.29 | 48.69 – 3.30 | 50.0 – 3.60 |
|
| |||
| Rmerge | 0.11 ( 0.88) | 0.09 ( 0.69) | 0.13 ( 0.83) |
|
| |||
| I / σ I * | 10.3 ( 2.2 ) | 7.7 ( 1.5 ) | 6.8 ( 1.5 ) |
|
| |||
| Completeness ( % ) | 99.9 ( 100.0 ) | 89.8 ( 90.2 ) | 91.9 ( 93.5 ) |
|
| |||
| Redundancy | 10.7 ( 10.8 ) | 5.3 ( 5.2 ) | 6.7 ( 6.8 ) |
|
| |||
| REFINEMENT | |||
|
| |||
| Resolution ( Å ) | 20.00 – 3.29 | 19.93 – 3.30 | 20.00 – 3.60 |
|
| |||
| No. Reflections | 12,010 | 10,627 | 8,939 |
|
| |||
| Rwork / Rfree (%) | 27.6 / 31.5 | 29.8 / 31.9 | 29.7 / 31.1 |
|
| |||
| No. atoms | |||
| Protein | 2,250 | 2,250 | 2,250 |
| Ligand | 24 | 20 | 14 |
|
| |||
| B-factors Å2 | |||
| Protein | 138.2 | 157.1 | 148.9 |
| Ligand | 148.3 | 136.8 | 146.4 |
|
| |||
| R.m.s deviations | |||
| Bond lengths ( Å ) | 0.001 | 0.001 | 0.002 |
| Bond Angles ( ° ) | 0.384 | 0.394 | 0.389 |
|
| |||
| Ramachandran Plot: | |||
| Preferred (%) | 90.94 | 93.73 | 93.73 |
| Allowed (%) | 8.36 | 5.57 | 6.27 |
| Outlier (%) | 0.70 | 0.70 | 0.00 |
Statistics in parentheses throughout refer to outer resolution shell.
Note the XAC and caffeine bound diffraction data exhibited significant anisotropy
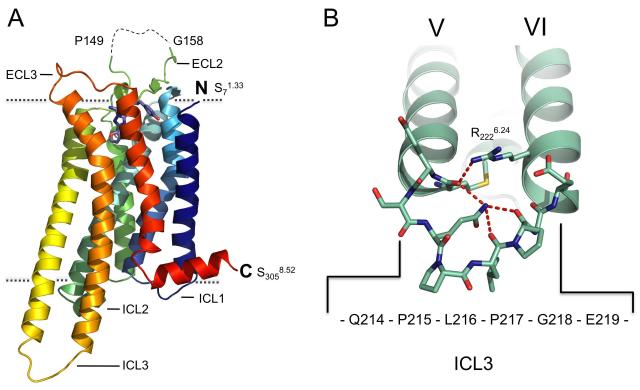
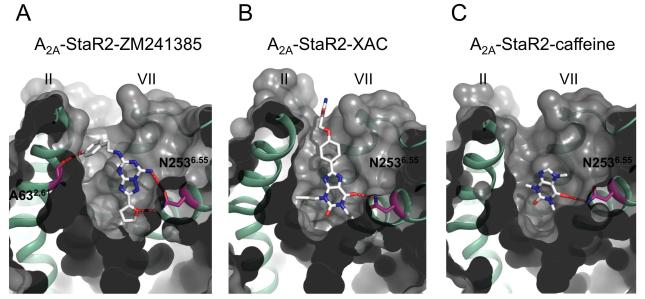
The overall structure (Figure 3a) includes residues 7-149 and 158-305 and the ligand ZM241385. A striking feature of the A2A-StaR2 structure is the extended nature of transmembrane helix (TM)5 and TM6 which project ~15Å into the cytoplasm. TM5 extends through to Ser2135.74 and is connected to TM6 by 6 residues to the helix of TM6 commencing at Arg2206.22 (Figure 3b and Supplementary Figure S1). The ordered and extended nature of the ICL3, which is also seen in squid rhodopsin (Murakami and Kouyama, 2008) is contributed to by Pro215 and Pro217 and a network of potential hydrogen bonds involving Arg2226.24, the main chain carbonyl of Met2115.72, and between Gln2145.75 and the main chain carbonyl of Pro217 and Leu216.
Figure 3. Overall Structure of the Adenosine A2A Receptor.
(a) Structure of A2A-StaR2-ZM241385 in rainbow colouration (blue to red) from the N-terminus to the C-terminus, ZM241385 is represented as a stick model (blue), oxygen atoms are red and nitrogen atoms deep blue. The visible extracellular and intracellular loops are labelled. The disordered portion of ECL2 is shown as a dashed line. Dotted lines denote approximate boundaries of the plasma membrane (b) TM5 and TM6 of the A2A-StaR2 (pale green) with ICL3 represented as a stick model. The potential H-bonding network connecting the two TM helices is shown (dashed red lines) involving Arg2226.24/Met2115.72/Gln214/Pro217. See also Figure S1. Individual TMs are numbered using Roman numerals and in all figures forthwith.
This is the first time structures of the same GPCR have been obtained using the different crystallisation strategies of thermostabilisation and T4L fusions and so provides a useful comparison of the techniques. The similarity of the structures provides confidence that engineering receptors through either fusion proteins or mutagenesis provides an effective approach to GPCR crystallization and does not result in major abnormalities of the structures. A comparison of the position of the thermostabilising mutations in the A2A-StaR2 with the position of these residues in the A2A-T4L structure shows no obvious perturbation of the structure around the mutations (Supplementary Figure S2), however we cannot rule out the possibility that individual mutations may alter the structure in some way. In the case of all GPCR structures care needs to be taken with more detailed interpretation of the structures, preferably in the context of pharmacological analysis of the crystallization constructs.
The closest structural agreement between the A2A-StaR2-ZM241385 and the A2A-T4L-ZM241385 structure occurs between TMs 1, 2, 3, 4 and 7 (Cα RMSD = 0.51Å) whereas superposition of TM5 and TM6 (residues 174-203 and 222-258) reveals significant differences (Cα RMSD = 1.62Å). In A2A-T4L the C-terminal portion of TM5 is displaced out of the helical bundle and moves laterally towards TM6 compared to A2A-StaR2 (Figure 4a). The intracellular end of TM6 in A2A-T4L is rotated towards TM5 by pivoting ~42° at Val2296.31 away from the helical bundle, whilst TM6 of A2A-StaR2 continues to pack with TM5. The global position of TM5 and TM6 of the A2A-StaR2 are in closer agreement to the ground state of rhodopsin (PDB code: 1F88) (Figure 4b) than A2A-T4L. Although both structures of the A2A receptor bind the same inverse agonist ligands ZM241385 there are significant differences in the pharmacology. The A2A-StaR2 has a pharmacology consistent with the inverse agonist state whilst A2A-T4L has a more agonist-like pharmacology with respect to agonist binding (Jaakola et al., 2008). Differences are also seen in the receptor structures that are consistent with the two structures representing different conformational states of the receptor.
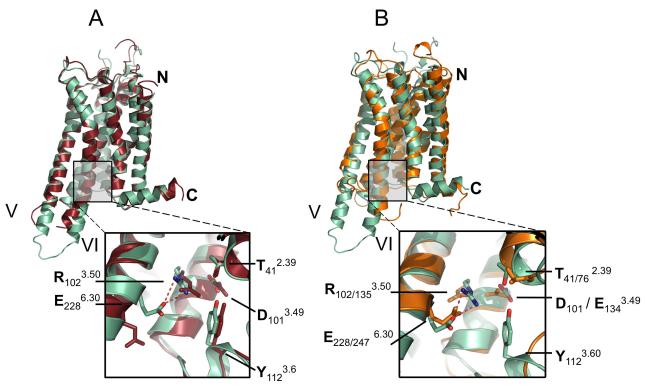
Figure 4. Comparison of A2A-StaR2-ZM241385 with A2A-T4L-ZM241385 and Rhodopsin.
(a) Superposition of A2A-StaR2-ZM241385 (pale green) with A2A-T4L-ZM241385 (dark red) using consensus Cα atoms of TM1-7, note the difference in TM5/6 helical trajectory. (Inset) Close-up of interactions across the DRY motif of A2A-StaR2 and A2A-T4L structures demonstrating ionic lock formation in the A2A-StaR2 (potential hydrogen bonds represented as dashed red lines). See also Supplementary Figure S3a. (b) Superposition of A2A-StaR2-ZM241385 (pale green) with bovine rhodopsin (orange – 1F88) using consensus Cα atoms of TM1-7. (Inset) close up of interactions across the A2A-StaR2 DRY and Rhodopsin ERY motif illustrating the similarity of the ionic lock (second subscript residue numbers refer to rhodopsin 1F88).
One of the most highly conserved sequence motifs in GPCRs is the E/DRY motif in TM3. In bovine rhodopsin the side chain of Arg1353.50 within the E/DRY motif hydrogen bonds to the side chain of Glu2476.30 at the N-terminus of TM6 to form the so-called ‘ionic lock’ (Vogel et al., 2008), part of a network of interactions bridging TM3 and TM6 and stabilising the inactive-state conformation. Inverse agonists are considered to preferentially bind to and stabilise this inactive conformation thus reducing any basal activity (Kenakin, 2004). During activation the ionic lock is broken allowing the outward movement of TM6. A2A-StaR2 has the potential ionic lock in place with the side chains of Glu2286.30 and Arg1023.50 in a similar conformation to that found in dark-state rhodopsin (Figure 4a & b and Figure S3a). The presence of the ionic lock in the A2A-StaR2 is similar to that in dark-state rhodopsin and is consistent with this structure representing the ground state of the receptor. The absence of the ionic lock in the A2A-T4L structure (see Figure 4a) appears to be the result of an outward movement and rotation in TM6 resulting from the T4 lysozyme fusion in ICL3. This effect of the T4L fusion is receptor specific since the ionic lock is present in in the dopamine D3 receptor-T4L structure (Chien et al., 2010). The impact of the T4L on the structure may also depend it’s the position within ICL3 and the length of the truncated ICL3 loop.
The extracellular surface of the A2A receptor consists primarily of the second and third extracellular loops (ECL2 and ECL3) with ECL2 ordered through disulphide linkages to ECL1 (Jaakola et al., 2008). Interestingly, changes at the extracellular surface of the ligand binding site involving a different rotamer conformation in His2646.66 and a displacement of the disulphide bonded CysProAspCys motif (still maintaining the disulphide link between Cys2596.61 and Cys2626.64) away from the entrance of the ligand binding cavity which may facilitate a more “open” entrance in comparison to A2A-T4L, facilitating access of the ligand to the binding pocket of the inactive receptor. Data from agonist bound structures of the adrenergic receptors suggests that upon ligand binding there is an inward movement around the binding pocket which results in the higher affinity binding of agonists (Rasmussen et al., 2011; Warne et al., 2011). Another feature of the extracellular surface of the A2A-StaR2 is a slightly extended anti-parallel β-sheet formed by Gly692.67 – Ala722.70 of ECL1 and Gln1635.25-Cys1665.27 of ECL2 (see Figure 4 a & b) which is similar to that observed in rhodopsin (PDB code: 1F88).
Ligand binding mode of ZM241385
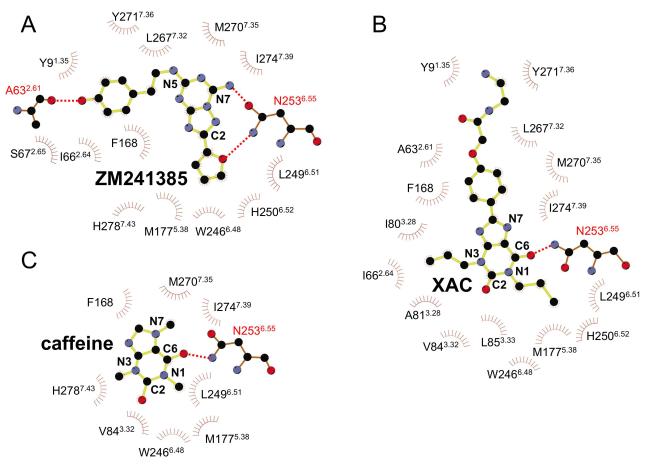
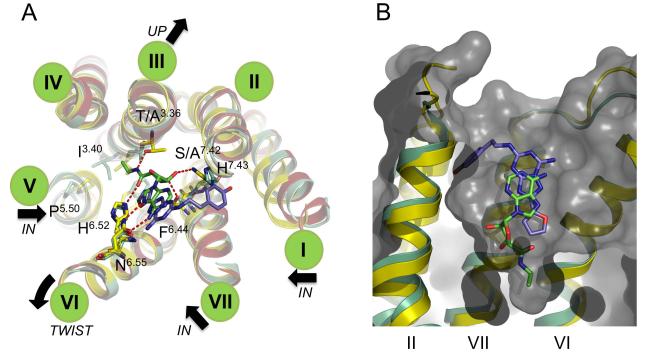
The structure of the A2A-StaR2 presents a highly open extracellular configuration exposing the entrance to the ligand binding cavity. Equivalent Cα atoms of ECL1 of A2A-StaR2 are a difference of ~3Å away from the entrance to the ligand binding pocket compared to ECL1 of A2A-T4L. This difference propagates from a kink at the top of TM3 at Phe833.31. Due to the disulphide bond between Cys712.69 and Cys1595.20, this perturbation in ECL1 also results in a ~3 Å difference of ECL2 (residues 157-164) laterally and towards the plasma membrane. The helical portion of ECL2 remains positioned to supply the π stacking between Phe168 and the triazolotriazine core of ZM241385. This bicyclic component is located between the aromatic Phe168 and a hydrophobic surface supplied by Ile2747.39 including, to a lesser extent, Leu2496.51 and Met2707.35. The carboxamide carbonyl of Asn2536.55 hydrogen bonds to the NH2 group of ZM241385 (Figure 5 a). Asn2536.55 is critical and Ile2747.39 important for ligand binding, as demonstrated in site directed mutagenesis studies (Dal Ben et al., 2010).
Figure 5. Residues involved in ligand binding of the xanthines and ZM241385.
2D schematics of the A2A-StaR2 ligand binding interactions, a) ZM241385, b) XAC and c) caffeine. Diagrams were generated using the LigPlot software (Wallace et al., 1995).
The position of the furan ring and the triazolotriazine are similar to that in the A2A-T4L structure and are supported by extensive previous modelling and mutagenesis data (Dal Ben et al., 2010; Ivanov et al., 2009; Kim et al., 2003). The furan ring of ZM241385 is positioned towards TM5 and TM7, making a hydrophobic interaction with Trp2466.48, and a π-π stack with His2506.52, and sits between two hydrophobic residues, Leu2496.51 and Met1775.38. Trp2466.48 and His2506.52 have been implicated in both antagonist and agonist binding from mutagenesis experiments (Dal Ben et al., 2010; Ivanov et al., 2009). The oxygen acceptor atom of the furan additionally forms a hydrogen bond to the amide NH2 group of Asn2536.55 (Figures 5a, 6a and Figure S3b). This hydrogen bond is likely to be important in the structure-activity relationship in this and related molecules, in where the furan moiety is known to be very important for high affinity binding (Shah and Hodgson, 2010).
Figure 6. Comparison of ligand positions in the A2A-StaR2 co-crystal structure binding site cavity.
The structure of A2A-StaR2 is depicted as a grey surface, cut away to reveal the ligand binding pocket with TM2 and TM5 depicted as green helices. The ligands are represented as sticks (grey), oxygen atoms are red and nitrogen atoms blue and amino acid side chains are similarly depicted with carbons in purple, oxygen in red and nitrogen in blue. Specific hydrogen bonds are marked as dashed red lines with Asn2536.55 and Ala632.61 shown. Structures are A2A-StaR2 in complex with a) ZM241385, (b) XAC and c) caffeine. See also Supplementary Figure S4.
The phenol group of the ZM241385 is found in a cleft formed by Glu131.39, Ala632.61, Ile662.64, Ser672.65, Leu2677.32, Met2707.35, Ile2747.39, His2787.43 and Tyr2717.36 at the extracellular ends of TM1, 2 and 7 with Tyr2717.36 displaying a rotation towards TM1 to incorporate this conformation of the phenolic moiety (Figure 6a and Figure 7a). The phenolic hydroxyl is also poised to make an additional hydrogen bond with the backbone carbonyl of Ala632.61 which itself chelates a water molecule in the previously reported A2A structure (Jaakola et al., 2008) (Figure 6a and 7a). Mutagenesis studies have shown that Glu131.39, Ile692.64 (in the A1 receptor), Ile2747.39 and His2787.43 are all involved in binding of ligands, suggesting the importance of this cleft for a range of ligands (Dal Ben et al., 2010; Ivanov et al., 2009). The position of the phenol group in this structure differs from the previous A2A-T4L structure where it was modelled to point vertically into the solvent-exposed part of the open binding cavity, however in the A2A-T4L structure this part of the ligand had higher temperature factors than other parts of the ligand reflecting its flexibility (Jaakola and Ijzerman, 2010; Katritch et al., 2010; Michino et al., 2009). Differences in the position of this portion of the ligand between the two A2A receptor structures reflect the difficulty of accurately placing small molecule ligands in current GPCR structures as well as the inherent flexibility of this portion of the molecule. It should also be noted that the two structures were obtained under different crystallization conditions, in particular the A2A-StaR2 structure was obtained at a pH of 8-8.75 whereas the A2A-T4L structure was obtained at pH 5.5-6.5. These differences may affect the protonation of ligand as well as amino acid side chains. The position of the phenol ring of the ZM241385 ligand observed in the A2A-StaR2 structure and its interaction with TM2 was predicted by several of the top scoring groups in GPCR Dock 2008 modelling competition (Katritch et al., 2010).
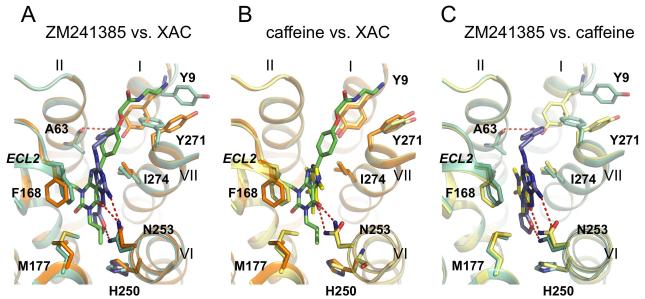
Figure 7. Comparison of specific interactions in the ligand binding site between the A2A-StaR2-ZM241385/XAC/caffeine co-structures.
a) Superposition of A2A-StaR2 (pale green) bound to ZM241385 (blue) with A2A-StaR2 (orange) bound to XAC (green) b) Superposition of A2A-StaR2 (orange) bound to XAC (green) with A2A-StaR2 (yellow) bound to caffeine (bright yellow) c) Superposition of A2A-StaR2 (pale green) bound to ZM241385 (blue) with A2A-StaR2 (yellow) bound to caffeine (bright yellow). A2A-StaR2 is shown in helical representation with specific side chains making important ligand receptor interactions marked. Specific hydrogen bonds are marked as dashed red lines. Ligands are represented as stick models (coloured according to the ligand) with oxygen atoms red and nitrogen atoms blue.
Ligand binding mode of the xanthines
The purified A2A-StaR2 in complex with either XAC and caffeine was crystallised by vapour diffusion in sitting drops and data collected on I24 at the Diamond Light Source (Oxfordshire, UK) corresponding to a 89.8% complete dataset to 3.3Å for XAC and 91.9% complete dataset to 3.6Å for the caffeine co-structures. Both structures were solved by molecular replacement using 3PWH with 1 copy of the A2A-StaR2 per crystallographic asymmetric unit followed by manual rebuilding between multiple rounds of refinement. Statistics for data collection and refinement are given in Table 1. Comparison of the two A2A-StaR2 xanthine co-structures with ZM241385 bound to A2A-StaR2 show they are in close agreement with RMSD values across the helical transmembrane regions of <0.5Å. Furthermore, both xanthine co-structures maintain the cardinal features of the ground state receptor, as discussed earlier for that of the A2A-StaR2-ZM241385, which is consistent with the conformational trapping of the StaR and also the similar pharmacological profile of the ligands. However, subtle differences across the binding sites of the three inverse agonist bound A2A-StaR2 structures indicate some degree of induced fit to accommodate ligands of different chemical classes.
XAC binds in the same site as ZM241385 (Figure 5b & 6b), forming the same two key interactions, the π stacking interaction between its heterocyclic core and Phe168 from ECL2 and a hydrogen bond with Asn2536.55 (Figure 5b). Superposition of XAC and ZM241385 shown after alignment of the receptor structures (Figure 7a) reveals that the xanthine intersects the plane of the ZM241385 triazolotriazine core at a relative angle of ~42°. Thus XAC binding to the receptor induces a shift of Phe168 towards Val172 resulting in a displacement of the helical portion of ECL2. The hydrogen bonding distance between the xanthine carbonyl and Asn2536.55 is 2.9Å and is consistent with a rotation of the amino acid side chain (relative to the ZM241385 structure) to allow the donor NH2 group to engage with the ligand, although a weaker hydrogen bond could still form if the amino acid side chain were in the reverse rotamer conformation. The two propyl substituents of the ligand extend towards the bottom of the binding site (Figure 6b). The propyl group on N1 forms hydrophobic contacts with Leu853.33, Leu2496.51 and induces rotameric changes of Met1775.38 and His2506.52 (Figure 7a). The propyl on N3 is in contact with Ala813.29, Ile662.64 and Val843.32, with the edge of the pocket flanked by Phe168 from ECL2 and to a lesser extent Ala632.61 and Ile803.28 (Figure 6b & 7a). At the top of the binding site the phenyl group of the ligand is coplanar with the xanthine core and sits against Ile2747.39, Leu2677.32, Met2707.35 and Tyr2717.36. The polar tail of the XAC ligand is towards the top of TM1 and 7 in a groove formed between Tyr91.35 and Tyr2717.36 (Figure 6b & 7a). This region of the receptor appears quite flexible, and as such these tyrosine residues adopt two different rotameric states dependent upon the ligand in the complex (Figure 7). The terminal polar tail of XAC, which serves as a water solubilising group, sits in the groove created by rotation of Tyr91.35 and Tyr2717.36 as indicated in Figure 7a, however electron density in this flexible region of the ligand is not complete suggesting several alternate conformations and the lowest energy conformation of the ligand is shown.
Caffeine binds to the A2A-StaR2 at a similar position to the xanthine portion of XAC and the furan moiety/triazolotriazine core of ZM241385 (Figure 5c & 6c). The small (fragment-sized) caffeine molecule sits in the hydrophobic pocket formed by Phe168, Ile2747.39, Leu2496.51, Met2707.35, Trp2466.48, and Val843.32 with an additional polar contact to His2787.43. The position of the π stacking side chain of Phe168 from ECL2 and Met1775.38 sit in a similar to the ZM241385 structure (Figure 7b & 7c). The xanthine core of caffeine sits coplanar to the core of ZM241385, in contrast to the ~42° offset of the XAC xanthine core to that of ZM241385 (Figure 7c). The hydrogen bond from caffeine to the carboxamide of Asn2536.55 is via the exocyclic C6 carbonyl, forming a 3.4Å hydrogen bond. Caffeine and XAC differ in that caffeine has a methyl substituent on N7, whilst in XAC there is a hydrogen atom at this position which appears to alter the vector of the C6 carbonyl with Asn2536.55. The C6 carbonyl interaction with Asn2536.55 from caffeine is consistent with the rotamer of the Asn2536.55 being the same as in the ZM241385 structure. It should be noted that the lower resolution of the A2A-StaR2 – caffeine co-structure introduces a degree of uncertainty in the exact rotation of caffeine in the plane of the ligand. However, the position of the ligand in the binding site and the rotation around an axis through the C6 carbonyl to N1 (the ‘tilt’) is clearly defined by Fo-Fc omit map calculation (see Supplementary Figure S4).
Insights into the adenosine A2A receptor structural activation spectrum
Structures of two receptors, rhodopsin and the β2-AR, have previously been obtained in both active (Rasmussen et al., 2011; Scheerer et al., 2008; Standfuss et al.,2011) and inactive conformations (Cherezov et al., 2007; Palczewski et al., 2000). More recently, structures of the A2A receptor thermostabilised in the agonist conformation (A2AR-GL31) bound to adenosine or the synthetic agonist NECA have been solved (Lebon et al., 2011a), as has that of the A2A-T4L bound to the agonist UK-432097 (Xu et al., 2011). The adenosine A2A receptor now represents the third GPCR for which the ground and active states are structurally characterised. For the purposes of the discussion below we have focussed the comparison of our structure with the A2AR-GL31 receptor bound to NECA (which binds in a similar way to adenosine) (Lebon et al., 2011). We note that the helical movements in this receptor are very similar to those observed in the T4L structure (Xu et al., 2011) suggesting that these two agonist-bound structures represent the same conformational state.
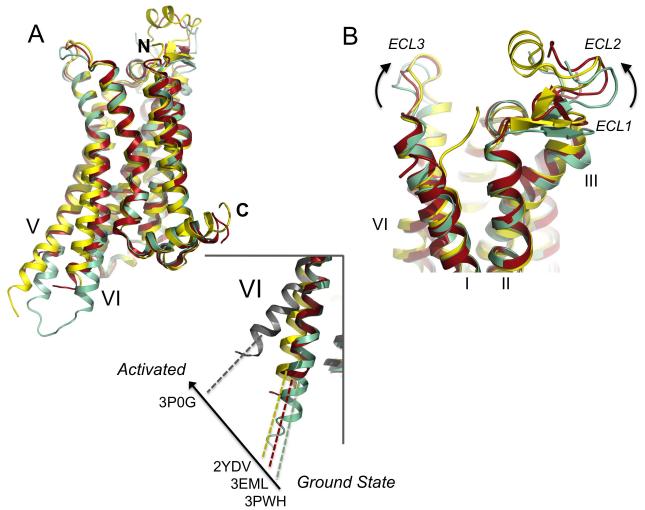
In the cases of rhodopsin and the β2-AR, receptor activation results in a contraction of the ligand binding site and an outward movement of the cytoplasmic ends of TM5 and 6, exposing the hydrophobic G protein binding platform comprised of the cytoplasmic ends of TM5 and 6 and the central receptor bundle (Rasmussen et al., Scheerer et al, 2008; Standfuss et al., 2011). These receptors were crystallized in the presence of a peptide from the G protein transducin (Scheerer et al, 2008; Standfuss et al., 2011) or a nanobody (Rasmussen et al., 2011) which mimics some features of the G protein. Superposition of the A2A-StaR2-ZM241385 structure (PDB code: 3PWH) with the A2A-T4L-ZM241385 structure (PDB code: 3EML) and A2AR-GL31-NECA (PDB code: 2YDV) (Figure 8a) reveals a similar mechanism for activation in the A2A receptor, and provides a structural basis for the agonist like pharmacology of the A2A-T4L-ZM241385 receptor.
Figure 8. Comparison of the conformations of different A2A receptor structures.
(a) Superposition of the A2A-StaR2-ZM241385 (pale green) (PDB code: 3PWH), A2A-T4L-ZM241385 (dark red) (PDB code: 3EML), A2AR-GL31 (yellow) (PDB code: 2YDV) and agonist-β2AR Nb80 (grey) (PDB code: 3P0G) structures in cartoon representation, ligands have been omitted. The N and C-termini are marked, and TM5 and 6 are labelled. (inset) close up view of TM6 illustrating the degree of helix rotation from the ground state (3PWH) to R* (3P0G). 3EML and 2YDV sit at intermediate points on the activation pathway. (b) Superposition of the A2AR structures, coloured and represented as in (a), close-up view of the extracellular side of the receptor, demonstrating the conformational changes occurring in ECL1 and 2 upon activation.
Comparing the structures of the agonist and inverse agonist thermostabilised receptors (A2A-StaR2 bound to ZM241385 and A2AR-GL31 bound to NECA) suggests that agonist binding to the A2A receptor results in a significant contraction of the binding site. This contraction results in the agonist conformation having an increased affinity for agonists and a decreased affinity for antagonists (Lebon et al, 2011a) relative to the inverse agonist conformation. This high affinity state for agonists is well documented in pharmacological studies on a wide range of GPCRs (Kenakin, 2004). Comparison of the two structures in the region of the binding site indicates that in the agonist structure there is a 3.3Å shift of ECL3 (measuring equivalent Cα’s of Cys262) and a 2.5 Å shift of the non-helical component (measuring equivalent Cα’s of Gly162) upwards and laterally towards the core of the receptor (Figure 8b), whilst Phe168 still remains suitably positioned to π stack with the adenine ring of NECA. A similar but smaller shift in the loops (ECL3 and ECL2 of 2.6Å and 2.1Å respectively) is also seen when comparing the ZM241385 bound structures of A2A-T4L with that of A2A-StaR2. The basis for this binding site contraction lies in structural rearrangements occurring at the extracellular end of TM2 and across TM3, 5, 6 and 7. In comparison the superposed agonist-β2AR Nb80 receptor displays the highest perturbation in these equivalent transmembrane regions. The A2A-T4L structure bound to ZM241385 appears to have begun these structural rearrangements and may represent an early step on the activation spectrum.
Upon activation of the A2A receptor there is an inward shift of TM7 towards the core of the receptor, an upward (towards the extracellular side of the receptor) movement of TM3 by ~2Å, a bulge in TM5 moving the helix towards the core of the receptor and a rigid body rotation of TM4 around Phe2426.44 (Figure 9a). At a molecular level recognition of NECA and changes in the binding site from the ground state A2A-StaR2 structure appear driven by hydrogen bond formation from the adenine core to Asn2536.55, and from the ribose group to Ser2777.42 and His2787.43. His2787.43 undergoes a rotameric change and concomitant 1.3Å movement of equivalent Cα’s across this region of TM7 contributing to the contraction of the ligand binding site.
Figure 9. Activation mechanism of the adenosine A2A receptor.
(a) Close-up of the ligand binding sites of the A2AR structures superposed and coloured as in Figure 8 (ECL regions have been omitted for clarity). Only the ZM241385 and NECA ligands are shown in blue and green stick representation respectively. Specific hydrogen bonds made between A2AR-GL31 and NECA are marked as dashed red lines. Residues implicated in ligand binding and transmission of conformational changes through the receptor are labelled and only shown for 3PWH and 2YDV. Transmembrane helices are denoted by green discs. The black arrows show transmembrane helical movements occurring upon activation. In summary, agonist binding leads to inward shifts of TM1, 5 and 7, an upward movement (towards the extracellular side of the receptor) of TM3 and a rigid body rotation of TM4. (b) Superposition of A2AR-GL31-NECA (yellow cartoon and ligand in green stick representation) and A2A-StaR2-ZM241385 (green cartoon and ligand in blue stick representation) TM’s 2, 6 and 7 labelled. Only the A2AR-GL31 is depicted as a grey surface, cut away to reveal the NECA agonist binding deeper in the receptor ligand binding pocket compared to the ZM241385 inverse agonist.
Additionally, the amide from NECA (which extends 5.6Å deeper into the A2AR binding cavity than the furan moiety of ZM241385 in the A2A-StaR2 structure (Figure 9b) hydrogen bonds to Thr883.36 which moves 2.3Å as TM3 translates up towards the extracellular side of the receptor. The result of this is to pull Ile923.40 up and in towards Phe2426.44 creating a hydrophobic cavity that is “filled” by the movement of TM5 and the bulge stemming from Pro1895.50 effecting the inward movement of residues at the N-terminus of TM5. The rigid-body movement of TM6, pivoting about Phe2426.44 through the movement of Ile923.40 and also permitted by breaking of the ionic-lock then results in His2506.52 moving to sterically occlude the binding of ZM241385 while making a hydrogen bond with the amide carbonyl of NECA (Figure 9a). The hydrogen bonding interactions from Ser2777.42 and Thr883.36 to the agonists may explain the effect of the S277A and T88A mutants in the A2A-StaR2 conformational ground-state thermostabilisation.
One of the most significant differences occurring as a result of agonist binding is seen in the cytoplasmic conformation of the ends of TM5 and TM6 as the G protein binding site becomes exposed, allowing coupling and signal cascade activation. As is evident from the structural superposition, (Figure 8a) the position of TM5 and TM6 in the A2A-StaR2-ZM241385 represents the starting point or ground-state in this transition where these helices sit most closely associated to the core of the receptor, as held by the ionic-lock to TM3. Superposition of the agonist-β2AR Nb80 bound structure (PDB code: 3P0G) demonstrates equivalent residues at the end of TM6 have moved over 11Å in the stabilised and “induced-fit” of the Nb80 G protein mimic. Between these two ends of the spectrum lies the thermostabilised adenosine agonist structure (and the T4L structures), representing points on the activation pathway from both an intra and extracellular conformational perspective where A2A-T4L-ZM241385 appears as an early intermediate between A2A-StaR2-ZM241385 and A2AR-GL31-NECA. We are still missing the fully active R* conformation of the A2A receptor, which will likely require crystallization with a G protein or G protein mimetic. What is not yet known is how close the current agonist bound structures are to the fully active A2A receptor conformation. A2AR-GL31 is able to couple to G proteins and activate signalling and taken together with the high binding affinity, this suggests that this indeed represents an active state of the receptor. This is also supported by the fact that the agonist UK-432097 bound A2A-T4L structure obtained with a different ligand and using completely different methodology is so similar to the NECA bound A2AR-GL31 structure. One possibility is that the very large 11Å movement of TM6 observed for the β2AR Nb80 bound structure is specific to the β2AR or that helical movements have been exaggerated as a result of the nanobody binding. In contrast, in the opsin structure TM6 moves outward by only 6Å compared with dark-state rhodopsin (Scheerer et al., 2008).
The range of A2A structures now available in complex with multiple ligands and in multiple conformational states represents an invaluable resource to design improved agonist and antagonist drugs for this important therapeutic target. In addition these structures contribute to our general understanding of ligand binding to members of the GPCR protein family and their mechanism of activation.
EXPERIMENTAL PROCEDURES
Thermostability assays
HEK293T cells, transfected with the A2A receptor constructs were resuspended in ice cold buffer [50mM Tris pH7.4; 400mM NaCl; 1% n-dodecyl-ß-maltoside (DDM) and protease inhibitors (Complete, Roche)]. After incubation for 1h at 4°C, samples were centrifuged (16,000g, 20 min, 4°C) and the supernatant was detergent exchanged into 0.1% decylmaltoside (DM) from Ni-NTA resin. Thermostability was assessed by incubating [3H]-ZM241385 (100nM)-bound A2A receptor at increasing temperatures for 30 min followed by a 5 minutes incubation on ice. Receptor bound and free radioligand were separated by gel filtration as previously described (Robertson et al., 2011).
Membrane radioligand binding
Membranes from transfected HEK293 cells were incubated with [3H]-ZM241385 as previously described (Robertson et al., 2011) in the presence or absence of competing compounds. After 90 min incubation at room temperature assays were terminated by rapid filtration and bound ligand measured by scintillation spectroscopy.
G protein-coupling activity of the adenosine A2A receptor measured in whole cells
Stable HEK293 inducible cell lines of the A2AR-His6 (amino acid residues 1-316 of human A2AR) were constructed using the Flp-in T-Rex (Invitrogen). Colonies were combined and tested for doxycycline-induced receptor expression. Cells were seeded at a density of 25,000 per well and induced with doxycyline (3 ng/mL); after 16 h media was removed and replaced with fresh media containing 100μM Ro-201724 and 2 U/mL adenosine deaminase. Cells were incubated at 37°C for 30 min prior to addition of varying concentrations of test drug (25°C, 30 min). Cells were then lysed and cAMP produced detected using the HTRF technology (CisBio, France) according to manufacturer’s instructions before plates were read on a PolarStar fluorescence plate reader (BMG LabTech, Germany).
Purification and crystallization
A2A-StaR2 was expressed using the baculovirus system, purified in 0.1% decylmaltoside, with detergent exchange to n-nonyl-β-d-glucopyranoside via IMAC. Crystals for all three structures were grown by vapour diffusion in sitting drops after addition of an equal volume of reservoir solution (0.1M Tris-HCl (pH 8.0-8.75), 32-42% PEG 1000, 0.25M MgCl2, 0.3% w/v n-nonyl-β-D-glucopyranoside, 0.1% w/v 1-Butanol, and 0.05% CYMAL-6) following purification in the presence of the appropriate ligand. Crystals appeared after 24 h and took on average three weeks to reach maximum dimensions of 100 × 500 × 50μm. Diffraction data from cryo-cooled crystals were collected at I24 of the Diamond Light Source, Oxford, UK at a wavelength of 0.9777 Å using a Dectris Pilatus 6M detector. Images were integrated and scaled using the programs XDS (Kabsch, 2010) and SCALA in the CCP4 suite of programs (COLLABORATIVE COMPUTATIONAL PROJECT, 1994; Evans, 2006). Molecular replacement was performed using the PHASER program (McCoy et al., 2007). Crystallographic modelling and manual rebuilding was performed using the program COOT (Emsley et al., 2010), and refinement performed using simulated annealing, restrained refinement and TLS in the PHENIX package (Adams et al., 2010).
Supplementary Material
HIGHLIGHTS.
Structure of adenosine A2A receptor in ground state conformation with ionic lock.
Complete third intracellular loop present consists of extended helices 5 and 6
Complexes with xanthines and ZM241385 show overlapping binding interactions
Thermostabilisation enables a structure bound with the low affinity ligand caffeine.
ACKNOWLEDGEMENTS
The authors thank Gwyndaf Evans, Robin Owen and Danny Axford at I24, Diamond Light Source, Oxford, UK for their help and assistance. We thank Andrew Leslie for expert advice on the crystallographic data processing and refinement, Joao Dias for helping with data collection and Jon Hadden for helping to establish the crystallisation facility at Heptares. We also thank the following for their assistance with the manuscript: Christopher Langmead, Jonathan Mason and other colleagues at Heptares.
Footnotes
SUPPLEMENTAL DATA includes 4 figures and 2 tables and is available online with this paper.
ACCESSION NUMBERS
Co-ordinates and structure factors have been submitted to the rcsb Protein Data Bank. The PDB codes are 3PWH (ZM241385), 3REY (XAC) and 3RFM (caffeine).
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien EY, Liu W, Zhao Q, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COLLABORATIVE COMPUTATIONAL PROJECT, N The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr D Biol Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Dal Ben D, Lambertucci C, Marucci G, Volpini R, Cristalli G. Adenosine Receptor Modeling: What Does the A2A Crystal Structure Tell Us? Current Topics in Medicinal Chemistry. 2010;10:993–1018. doi: 10.2174/156802610791293145. [DOI] [PubMed] [Google Scholar]
- Daly JW. Caffeine analogs: biomedical impact. Cellular and molecular life sciences: CMLS. 2007;64:2153–2169. doi: 10.1007/s00018-007-7051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Beta-arrestins and cell signaling. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott W, Cowtan K. Features and Development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Fredholm BB, Ijzerman AP, Jacobson KA, Linden J, Müller CE. International Union of Basic and Clinical Pharmacology. LXXXI. Nomenclature and classification of adenosine receptors--an update. Pharmacol Rev. 2011;63:1–34. doi: 10.1124/pr.110.003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman AG. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Ivanov AA, Barak D, Jacobson KA. Evaluation of homology modeling of G-protein-coupled receptors in light of the A(2A) adenosine receptor crystallographic structure. J Med Chem. 2009;52:3284–3292. doi: 10.1021/jm801533x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola VP, Ijzerman AP. The crystallographic structure of the human adenosine A2A receptor in a high-affinity antagonist-bound state: implications for GPCR drug screening and design. Current opinion in structural biology. 2010;20:401–414. doi: 10.1016/j.sbi.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Jacobson KA, Gao ZG. Adenosine receptors as therapeutic targets. Nat Rev Drug Discov. 2006;5:247–264. doi: 10.1038/nrd1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner P. Istradefylline, a novel adenosine A2A receptor antagonist, for the treatment of Parkinson’s disease. Expert Opin Investig Drugs. 2005;14:729–738. doi: 10.1517/13543784.14.6.729. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Van Rhee AM, Kim J, Yehle S, Wess J, Jacobson KA. Hydrophilic side chains in the third and seventh transmembrane helical domains of human A2A adenosine receptors are required for ligand recognition. Mol Pharmacol. 1996;50:512–521. [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritch V, Rueda M, Lam PC, Yeager M, Abagyan R. GPCR 3D homology models for ligand screening: lessons learned from blind predictions of adenosine A2a receptor complex. Proteins. 2010;78:197–211. doi: 10.1002/prot.22507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T. Principles: receptor theory in pharmacology. Trends Pharmacol Sci. 2004;25:186–192. doi: 10.1016/j.tips.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Kim SK, Gao ZG, Van Rompaey P, Gross AS, Chen A, Van Calenbergh S, Jacobson KA. Modeling the adenosine receptors: comparison of the binding domains of A2A agonists and antagonists. J Med Chem. 2003;46:4847–4859. doi: 10.1021/jm0300431. [DOI] [PubMed] [Google Scholar]
- Lebon G, Bennett K, Jazayeri A, Tate CG. Thermostabilisation of an Agonist-Bound Conformation of the Human Adenosine A(2A) Receptor. J Mol Biol. 2011a 2011 Apr 9; doi: 10.1016/j.jmb.2011.03.075. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebon G, Warne T, Edwards PC, Bennett K, Langmead CJ, Leslie AG, Tate C. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011b doi: 10.1038/nature10136. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani F, Shibata Y, Serrano-Vega MJ, Tate CG. Co-evolving stability and conformational homogeneity of the human adenosine A2a receptor. Proc Natl Acad Sci U S A. 2008;105:10744–10749. doi: 10.1073/pnas.0804396105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michino M, Abola E, Brooks CL, 3rd, Dixon JS, Moult J, Stevens RC. Community-wide assessment of GPCR structure modelling and ligand docking: GPCR Dock 2008. Nat Rev Drug Discov. 2009;8:455–463. doi: 10.1038/nrd2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller CE, Jacobson KA. Xanthines as adenosine receptor antagonists. Handb Exp Pharmacol. 2011;200:151–199. doi: 10.1007/978-3-642-13443-2_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Poucher SM, Keddie JR, Singh P, Stoggall SM, Caulkett PW, Jones G, Coll MG. The in vitro pharmacology of ZM 241385, a potent, non-xanthine A2a selective adenosine receptor antagonist. Br J Pharmacol. 1995;115:1096–1102. doi: 10.1111/j.1476-5381.1995.tb15923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, et al. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Robertson N, Jazayeri A, Errey J, Baig A, Hurrell E, Zhukov A, Langmead CJ, Weir M, Marshall FH. The properties of thermostabilised G protein-coupled receptors (StaRs) and their use in drug discovery. Neuropharmacology. 2011;60:36–44. doi: 10.1016/j.neuropharm.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- Salamone JD. Preladenant, a novel adenosine A(2A) receptor antagonist for the potential treatment of parkinsonism and other disorders. IDrugs. 2010;13:723–731. [PubMed] [Google Scholar]
- Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Conformational thermostabilization of the β1-adrenergic receptor in a detergent-resistant form. Proc Natl Acad Sci U S A. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah U, Hodgson R. Recent progress in the discovery of adenosine A(2A) receptor antagonists for the treatment of Parkinson’s disease. Curr Opin Drug Discov Devel. 2010;13:466–480. [PubMed] [Google Scholar]
- Shibata Y, White JF, Serrano-Vega MJ, Magnani F, Aloia AL, Grisshammer R, Tate CG. Thermostabilization of the neurotensin receptor NTS1. J Mol Biol. 2009;390:262–277. doi: 10.1016/j.jmb.2009.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder SH, Katims JJ, Annau Z, Bruns RF, Daly JW. Adenosine receptors and behavioral actions of methylxanthines. Proc Natl Acad Sci U S A. 1981;78:3260–3264. doi: 10.1073/pnas.78.5.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standfuss J, Edwards PC, D’Antona A, Fransen M, Xie G, Oprian DD, Schertler GF. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–60. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate CG. Practical considerations of membrane protein instability for purification and crystallisation. Methods Mol Biol. 2010;2010:601–187. doi: 10.1007/978-1-60761-344-2_12. [DOI] [PubMed] [Google Scholar]
- Tate CG, Schertler GF. Engineering G protein-coupled receptors to facilitate their structure determination. Curr Opin Struct Biol. 2009;19:386–395. doi: 10.1016/j.sbi.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Vogel R, Mahalingam M, Ludeke S, Huber T, Siebert F, Sakmar TP. Functional role of the “ionic lock”--an interhelical hydrogen-bond network in family A heptahelical receptors. J Mol Biol. 2008;380:648–655. doi: 10.1016/j.jmb.2008.05.022. [DOI] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Warne T, Moukhametzianov R, Baker JG, Nehme R, Edwards PC, Leslie AG, Schertler GF, Tate CG. The structural basis for agonist and partial agonist action on a beta(1)-adrenergic receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao ZG, Cherezov V, Stevens RC. Structure of an Agonist-Bound Human A2A Adenosine Receptor. Science. 2011 doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.