Abstract
Oxysterol binding protein (OSBP) is the only protein known to bind specifically to the group of oxysterols with potent effects on cholesterol homeostasis. Although the function of OSBP is currently unknown, an important role is implicated by the existence of multiple homologues in all eukaryotes so far examined. OSBP and a subset of homologues contain pleckstrin homology (PH) domains. Such domains are responsible for the targeting of a wide range of proteins to the plasma membrane. In contrast, OSBP is a peripheral protein of Golgi membranes, and its PH domain targets to the trans-Golgi network of mammalian cells. In this article, we have characterized Osh1p, Osh2p, and Osh3p, the three homologues of OSBP in Saccharomyces cerevisiae that contain PH domains. Examination of a green fluorescent protein (GFP) fusion to Osh1p revealed a striking dual localization with the protein present on both the late Golgi, and in the recently described nucleus-vacuole (NV) junction. Deletion mapping revealed that the PH domain of Osh1p specified targeting to the late Golgi, and an ankyrin repeat domain targeting to the NV junction, the first such targeting domain identified for this structure. GFP fusions to Osh2p and Osh3p showed intracellular distributions distinct from that of Osh1p, and their PH domains appear to contribute to their differing localizations.
INTRODUCTION
The mechanisms underlying the synthesis and uptake of sterols by eukaryotic cells are now relatively well characterized. However, much less is understood about how cells regulate their intracellular sterol levels, and how they maintain a nonhomogenous distribution of sterols between different internal membranes. Sterol homeostasis requires that there must be mechanisms to sense cellular sterol levels, and although there has been much recent progress in identifying some of the key regulators of cholesterol metabolism (Brown and Goldstein, 1999), less is understood about how sterol sensing occurs. The intracellular traffic of cholesterol appears to be important in this feedback (Lange and Steck, 1996). The majority of cholesterol is found in the plasma membrane, but it is in the endoplasmic reticulum (ER), which itself has low levels of cholesterol, where the changes in cellular cholesterol levels are responded to by the sterol regulatory element-binding protein (SREBP) system that controls the transcription of genes encoding cholesterol biosynthetic enzymes (Brown and Goldstein, 1999; Lange et al., 1999). Although it might be expected that the systems controlling cholesterol metabolism would recognize cholesterol itself, there has been a long-standing interest in the possibility that oxysterols, a group of oxidized derivatives of sterols, are important second messengers in sterol homeostasis (Brown and Goldstein, 1974; Kandutsch and Chen, 1974; Accad and Farese, 1998). Indeed, oxysterols such as 25-hydroxycholesterol are up to a 1000 times more potent than cholesterol itself as down-regulators of cholesterol synthesis (Kandutsch et al., 1978; Goldstein and Brown, 1990).
Because many oxysterols can be generated readily from the nonenzymatic oxidation of cholesterol, their physiological relevance has until recently been uncertain (Smith, 1996). However, there is now increasing evidence that oxysterols play important roles in vivo. First, intracellular hydroxylases have been discovered that can convert cholesterol into specific oxysterols, including 25-hydroxycholesterol (Lund et al., 1998, 1999; Russell, 2000). Second, it has recently been shown that the enzymes responsible for the hepatic conversion of excess cholesterol into bile acids are regulated by a nuclear hormone receptor (LXRa) that binds a specific subset of oxysterols, in particular 24-hydroxycholesterol, which are synthesized when cholesterol levels rise (Janowski et al., 1996). Third, although most mammalian cells export cholesterol to high-density lipoprotein particles in the plasma, at least two cell types, macrophages and neurons, export the bulk of sterol as 27- and 24-hydroxycholesterol, respectively (Bjorkhem et al., 1999).
If intracellular oxysterols serve as second messengers there must be particular proteins that recognize them. The only protein known to bind specifically to the group of oxysterols that are active in the down-regulation of cholesterol synthesis is oxysterol binding protein (OSBP) (Dawson et al., 1989a). OSBP was identified as being the most abundant cytosolic protein that bound to such regulatory oxysterols (Taylor et al., 1984; Dawson et al., 1989b). Characterization of mammalian OSBP showed that it is associated with the periphery of the Golgi and other intracellular membranes, and that this Golgi localization is stimulated by the presence of oxysterols (Ridgway et al., 1992). The function of OSBP is unclear, but overexpression in tissue culture cells has multiple effects on cholesterol homeostasis and sphingolipid synthesis (Lagace et al., 1997, 1999).
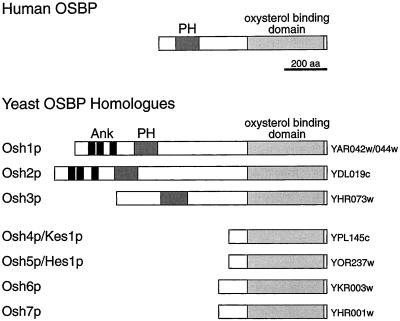
Although the precise function of OSBP has remained elusive, it at least seems certain that this function is required in all eukaryotes, because multiple OSBP homologues have been found in the genomes of all eukaryotes so far examined. These proteins all share a conserved 400 amino acid domain found at the C terminus of OSBP, which has been shown to bind oxysterols (Ridgway et al., 1992). For convenience we will refer to this shared, characteristic, domain as the “oxysterol binding domain,” although its binding specificity in other species has not been investigated. The existence of multiple OSBP homologues raises the question of whether the different proteins in a given organism have related but distinct functions. Some evidence that this is the case comes from the fact that OSBP homologues can be divided into two general classes, ones that comprise an oxysterol binding domain alone, and longer ones such as OSBP itself that have a pleckstrin homology (PH) domain at the N terminus. Most PH domains in other proteins direct localization to the plasma membrane, often by interaction with phosphatidylinositol phosphates (PIPs). We have found that, in contrast, the PH domain of OSBP specifies targeting to the trans-Golgi network (TGN) of mammalian cells, and this interaction requires the presence of Golgi PIPs (Levine and Munro, 1998).
To learn more about the functional relevance of the intracellular targeting of oxysterol binding proteins, we have studied the situation in the yeast Saccharomyces cerevisiae, which contains seven OSBP homologues (OSH genes) (Beh et al., 2001). Three of these genes (OSH1, OSH2, and OSH3) encode proteins that, like OSBP, have a large N-terminal region that includes a PH domain (Figure 1). Of these, Osh1p and Osh2p also have three ankyrin repeats, which are not found in the mammalian protein. We were interested in whether the presence of this N-terminal extension reflected a common site of action in the cell. We report here that green fluorescent protein (GFP) fusions to the three proteins are located to different parts of the cell. In particular Osh1p has a striking dual localization being found on both the Golgi and the nucleus-vacuole (NV) junction, a recently described specialization of these two organelles (Pan et al., 2000). Targeting of Osh1p to the Golgi depends upon the PH domain, whereas targeting to the NV junction is specified by the ankyrin repeat region, the first targeting domain identified for this unusual structure. This suggests that OSBP homologue function is required in multiple parts of the cell, and that as a consequence the different members of the family contain distinct targeting determinants.
Figure 1.
Structure of OSBP and its homologues in S. cerevisiae. The PH domain (PH), ankyrin repeats (Ank), and the portions homologous to the oxysterol binding domain of OSBP are indicated, along with the ORF names for the yeast proteins (Beh et al., 2001).
MATERIALS AND METHODS
Strains and Plasmids
The genotypes of yeast strains used are listed in Table 1. Deletions of OSH1, OSH2, and OSH3 were made by homologous recombination with the use of promoter and terminator fragments, between which were placed dual loxP sites flanking HIS3, LEU2, and URA3 for OSH1, 2, and 3, respectively (Sauer, 1994). In some cases markers were excised from strains containing OSH gene deletions by expression of Cre recombinase from the GAL1/10 promoter (CEN TRP1 plasmid, growth for 3 h with galactose), followed by plating on nonselective plates and then replica plating to identify colonies in which markers were lost. Loss of markers without unwanted chromosomal rearrangement around the loxP sites was confirmed by polymerase chain reaction (PCR).
Table 1.
Yeast strains and plasmids used in this study
| Yeast strains | ||
| SEY6210 | MATα ura3-52 leu2-3,112 trp1-Δ901 his3-Δ200 lys2-801 suc2-Δ9 | Robinson et al. (1988) |
| TLY211 | SEY6210 Δosh1∷loxP | |
| TLY212 | SEY6210 Δosh2∷loxP | |
| TLY213 | SEY6210 Δosh3∷loxP | |
| RS453 | MATa/α ura3/ura3 leu2/leu2 trp1/trp1 his3/his3 ade2/ade2 | Siniossoglou et al. (2000) |
| RS453B | MATa ura3 leu2 trp1 his3 ade2 | |
| TLY221 | RS453B Δosh1∷HIS3 | |
| TLY227 | RS453B Δosh1∷HIS3 osh2∷PHO5 GFP OSH2 URA3KI | |
| TLY231 | RS453B Δnvj1∷HIS5Sp | |
| TLY232 | RS453B Δvac8∷HIS5Sp | |
| JRY6326 | SEY6210 TRP1∷PMET3-OSH2 osh1∷kan-MX4 osh2∷kan-MX4 osh3∷LYS2 osh4∷HIS3 osh5∷LEU2 osh6∷LEU2 osh7∷HIS3 | Beh et al. (2001) |
| Plasmids | ||
| pRS406 | Integration at URA3 | Sikorski and Hieter (1989) |
| pRS416 | CEN URA3 | Sikorski and Hieter (1989) |
| pRS426 | 2μ URA3 | Sikorski and Hieter (1989) |
| pTLUPG | URA3KI, PHO5 promoter (168 bp), GFP, myc | |
| pTL301 | pRS406 OSH1 promoter (978 bp), Osh1p (1–1190 aa), OSH1 terminator (299 bp) | |
| pTL302 | pRS416 OSH2 promoter (814 bp), Osh2p (1–1283 aa), OSH2 terminator (392 bp) | |
| pTL303 | pRS416 OSH3 promoter (699 bp), Osh3p (1–997 aa), OSH3 terminator (416 bp) | |
| pTL311 | pRS406 PHO5 promoter, GFP, Osh1p, OSH1 terminator | |
| pTL312 | pRS416 PHO5 promoter, GFP, Osh2p, OSH2 terminator | |
| pTL313 | pRS416 PHO5 promoter, GFP, Osh3p, OSH3 terminator | |
| pTL321 | pRS406 TPI promoter (412 bp), Osh1p (1–690 aa), GFP | |
| pTL322 | pRS406 TPI promoter, Osh1p (1–279 aa), PHspectrin, Osh1p (385–690 aa), GFP | |
| pTL323 | pRS406 TPI promoter, myc, PHspectrin, Osh1p (385–690 aa), GFP | |
| pTL324 | pRS406 OSH1 promoter, Osh1p (1–279 aa), GFP | |
| pTL325 | pRS406 TPI promoter, GFP, Osh1p (280–1190 aa), OSH1 terminator | |
| pTL327 | pRS406 TPI promoter, myc, Osh1p (280–690 aa), GFP | |
| pTL331 | pRS406 PHO5 promoter, GFP, PHOsh1p (280–384 aa) | |
| pTL342 | pRS416 PHO5 promoter, GFP, PHOsh2 (256–424 aa), GFP | |
| pTL343 | pRS416 PHO5 promoter, GFP, PHOsh3 (199–361 aa), GFP | |
| pTL352 | pRS426 OSH2 promoter (838 aa), Osh2p (1–722 aa), GFP | |
| pTL353 | pRS426 OSH3 promoter, Osh3p (1–563 aa), GFP | |
| pTL356 | as pTL331: K299→E R301→E | |
| pTL357 | as pTL352: K307→E R309→E | |
| pTL358 | as pTL353: K240→E K241→E R242→E | |
| pTL362 | pRS406 OSH1 promoter, Osh1p (1–690 aa), GFP | |
| pTL371 | pRS406 OSH1 promoter, GFP, Osh1p (689–1190 aa), OSH1 terminator | |
| pTL376 | pRS406 PHO5 promoter, GFP, Osh1p (689–1190 aa), OSH1 terminator |
NVJ1 and VAC8 were deleted with the use of the PCR method with the heterologous marker gene Schizosaccharomyces pombe HIS5 (Wach et al., 1997). GFP-Osh2p was constructed in a Δosh1 background (strains TLY227 and TLY228) by a similar integration of a PCR product comprising Kluyveromyces lactis URA3 (Langle-Rouault and Jacobs, 1995), PHO5 promoter, GFP and a Myc epitope tag upstream of the first codon of the OSH2 open reading frame (ORF), generated with the use of pTLUPG as a template.
Plasmids used are listed in Table 1. OSH1-3 open reading frames were cloned by allele rescue with the use of flanking promoters and terminators (Rothstein, 1991), and checked by restriction digest mapping and sequencing of junctions. PHspectrin sequence was as used previously (Levine and Munro, 1998). Linkers that resulted from the restriction sites between the individual components of the chimeric proteins described in Table 1 were as follows (in single-letter amino acid notation, with residues numbered according to their position in the relevant OSH ORF): GFP to Osh1p1 (pTL311) = LPG; GFP to Osh2p1 (pTL312) = LGAGA; GFP to Osh3p1 (pTL313) = SNSLG; Osh1p690 to GFP and Osh3563 to GFP (pTL321, pTL322, pTL323, pTL327, pTL353, pTL358, and pTL362) = TKLPMVTSPVEK; flanking PHspectrin and PHOsh1 (pTL322, pTL323, pTL325, pTL331, and pTL356) = KLGS at N terminus, KNS at C terminus; Osh1p279 to GFP (in pTL324) = HKLGSTKLPMVTSPVEK; GFP to Osh1p280 (pTL325) = LGS; GFP to Osh2p256 and Osh3p199 (in pTL342 and pTL343) = L; Osh2p424 to GFP (in pTL342, pTL352 and pTL357) = VEK; Osh3p361 to GFP (in pTL343) = GSQESTNTPVEK. Mutations were introduced by site-directed methods (Quick-Change; Stratagene, La Jolla, CA) into plasmids pTL356/pTL357/pTL358. In each case, the mutated area and flanking region were subcloned and checked by sequencing.
Assay for Δosh1 Growth Phenotype
Two microliters of 20-fold serial dilutions of cells was spotted on to minimal media containing tryptophan either at normal levels (40 μg/ml) or at suboptimal levels (15 μg/ml) and grown at 25°C for 72 or 96 h, respectively. Cells from multiple colonies were examined independently (typically 4–8), and results shown are from a single representative colony.
Microscopy
For imaging of GFP-chimeras in live cells by confocal microscopy (Bio-Rad MRC-600), log phase cultures (OD 600 nm = 1) were collected by brief centrifugation, and 0.25 μl of resuspended pellet was placed between a slide and a coverslip. To stain DNA in live cells, cultures were incubated with 4′-6-diamidino-2-phenylindole (DAPI) (2.5 μg/ml from a 5-mg/ml stock in dimethyl sulfoxide) for 3 h. To stain vacuoles, cultures were incubated with FM4-64 (40 μM from a 32 mM stock in dimethyl sulfoxide) for 15 min, followed by 45-min chase in fresh medium. Cells stained with DAPI and FM4-64 were mounted as described above, and photographed sequentially on an Axioscop microscope (Carl Zeiss Inc., Thornwood, NY) with the use of a CCD-1300 camera (Princeton Instruments, Trenton, NJ).
Immunofluorescence of formaldehyde fixed cells was carried out as described previously, except for the omission of extraction in methanol/acetone (Levine et al., 2000). Affinity purified rabbit antisera against Anp1p (Jungmann and Munro, 1998), Pep12p, and Tlg1p (Lewis et al., 2000) were detected with Alexa-568 secondary antibodies (Molecular Probes, Eugene, OR) and examined by confocal microscopy as described above.
RESULTS
The Three Full-Length OSBP Homologues in Yeast Are Functionally Distinct
Osh1p was previously identified as a full-length yeast homologue of mammalian OSBP (although it was initially categorized in error as two distinct ORFs, SWH1/YAR042w and OSH1/YAR044w) (Jiang et al., 1994; Schmalix and Bandlow, 1994). The similarities of Osh1p and OSBP include a similar overall size and domain structure, with a PH domain near the N terminus. The yeast genome contains six further OSBP homologues, four of which are much shorter than OSBP and lack PH domains, but two of which are full-length homologues that, like Osh1p, have a large N-terminal region containing a PH domain (Figure 1). Of these latter two, YDL019c is closely homologous to OSH1 (51% identity), whereas YHR073w is more divergent (25% identity). These two genes are referred to hereafter as OSH2 and OSH3, respectively (Beh et al., 2001).
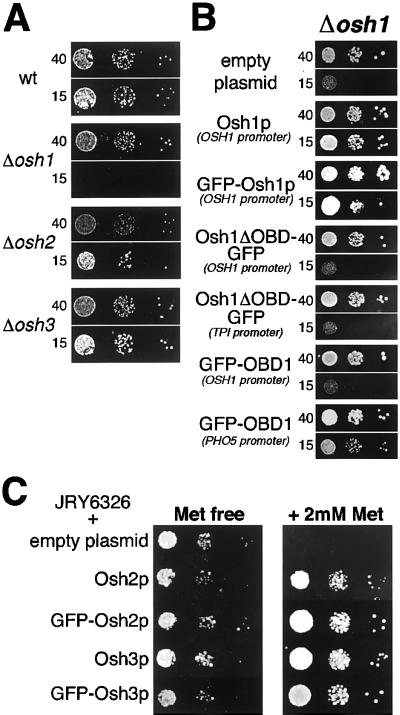
To initiate a characterization of these genes, we created strains carrying all possible combinations of Δosh1, Δosh2, and Δosh3 in a wild-type background (SEY6210). All were viable, showing equal growth on rich media at 25–37°C. The growth phenotype previously observed in Δosh1 is a cold-sensitive inhibition of growth in the presence of reduced levels of tryptophan, a phenotype also associated with erg mutations that have alterations in ergosterol structure or levels (Jiang et al., 1994). Because the auxotrophic markers used to construct the deletion strains interact with this phenotype (Skrzypek et al., 1998), the marker genes inserted during OSH gene deletion were excised by Cre recombinase to create marker-free strains that were genetically identical except for the presence or absence of the coding regions of OSH1-3. Δosh1 cells grew as wild-type cells on rich and minimal media at temperatures from 25 to 37°C (our unpublished results; Figure 2A). However, growth of Δosh1 cells was impaired at 25°C on minimal media with reduced tryptophan, as previously described (Figure 2A). In contrast, neither Δosh2 nor Δosh3 showed this phenotype, growing as wild-type under these conditions (Figure 2A). In addition, Δosh2 and Δosh3 showed no interaction with the growth phenotype of Δosh1, because this was not altered by the additional deletion of either, or both, of Δosh2 or Δosh3 (our unpublished results). In addition, one allele each of both OSH1 and OSH2 was deleted in a diploid strain. After sporulation, tetrad analysis showed 2:2 segregation of the growth phenotype, always cosegregating with the Δosh1 allele, and unaffected by cosegregation of the Δosh2 allele (our unpublished results).
Figure 2.
Assays of Osh protein activity in vivo. (A) Growth phenotype of Δosh strains. Δosh1 cells (TLY211) fail to grow at 25°C on suboptimal levels of tryptophan (15 μg/ml), compared with normal growth at normal levels of tryptophan (40 μg/ml). No growth phenotype is seen for strains that are wild type (SEY6210), Δosh2 (TLY212), and Δosh3 (TLY213). (B) Complementation of Δosh1 phenotype. Δosh1 cells (TLY221) were transformed either with an empty plasmid (pRS406); or with plasmids expressing either untagged Osh1p (pTL301), Osh1p tagged at the N terminus with GFP (pTL311); or GFP-tagged constructs containing portions of Osh1p expressed from the promoter stated in brackets. Osh1ΔOBD (1–690) was tagged at the C terminus with GFP and was expressed from either the OSH1 promoter (pTL362) or the TPI1 promoter (pTL321); OBD1 (689–1190) was tagged at the N terminus with GFP, and expressed from either the OSH1 promoter (pTL371) or the PHO5 promoter (pTL376). By quantitative protein blotting with anti-GFP the expression levels of the four constructs were, respectively, 3.0-, 20-, 0.7-, and 17-fold that of GFP-Osh1p. Growth conditions were as described in MATERIAL AND METHODS. Note that in the absence of OSH1, very slight growth was occasionally seen on low tryptophan. (C) In vivo activity of GFP fusions to Osh2p and Osh3p. JRY6326, a strain in which all seven OSH open reading frames are deleted but which is rescued from lethality by a copy of Osh2p expressed under the MET3 promoter (Beh et al., 2001), was transformed with an empty plasmid (pRS416), or with plasmids expressing Osh2p and Osh3p either untagged or GFP-tagged (pTL302, pTL303, pTL312, pTL313). Growth of JRY6326 was inhibited by repression of the MET3 promoter, but GFP tagged Osh2p and Osh3p were able to rescue the lethality as effectively as the untagged proteins.
The functional significance of the growth defect seen with Δosh1 cells was investigated by examining the ability of truncated versions of Osh1p to rescue the phenotype of Δosh1 cells. As expected, reintroduction of intact OSH1 rescued the growth defect (Figure 2B). In contrast, a version of the protein lacking the oxysterol binding domain was unable to restore growth when expressed from either the OSH1 promoter or overexpressed from the TPI1 promoter (Figure 2B). In contrast the Osh1p oxysterol binding domain alone was able to at least partially rescue growth, although this required overexpression with the use of a constitutive PHO5 promoter (Bajwa et al., 1987). These results suggest that growth at 25°C on low tryptophan requires the Osh1p oxysterol binding domain, i.e., this domain of Osh1p carries out the function(s) measured by this assay. In contrast, the N-terminal region of the protein appears to have no direct activity in this assay, but is required for the oxysterol binding domain to act at native levels of expression.
Osh Proteins Have Distinct Intracellular Distributions
We next investigated the intracellular localization of Osh1p, Osh2p, and Osh3p by tagging the proteins at their N termini with GFP. When the GFP-Osh1p construct was expressed from the OSH1 promoter in Δosh1 cells, it rescued the Δosh1 phenotype, indicating that it was functional (Figure 2B). Although this same phenotype cannot be used to assay the activity of Osh2p and Osh3p, it has recently been shown that either of proteins is sufficient to restore growth to cells from which all seven OSH genes have been deleted (Beh et al., 2001). When GFP-Osh2p and GFP-Osh3p were expressed in a strain in which the only other OSH gene was under a regulated promoter, the fusions were able to maintain growth when this promoter was repressed, indicating that these GFP fusions retain functional activity (Figure 2C).
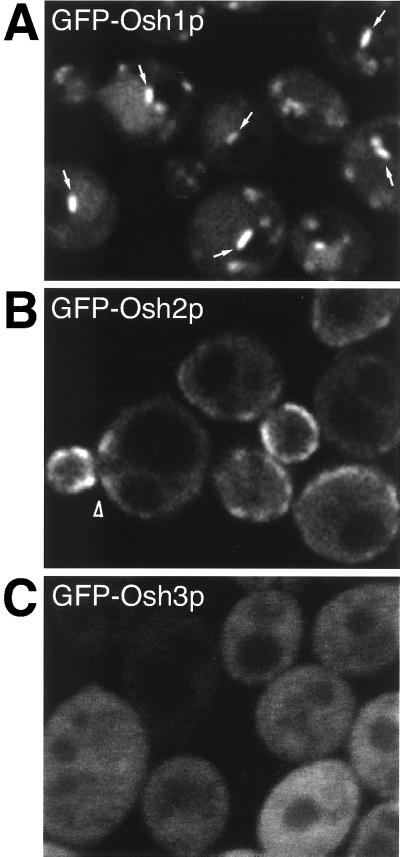
When live yeast expressing the GFP fusions were examined by fluorescence microscopy in midlog phase, GFP-Osh1p was localized in punctate structures that are discussed in detail below (Figure 3A). In contrast, GFP-Osh2p was apparently localized to the plasma membrane, concentrated in the budding area of G1 phase cells, and around the mother-daughter bud-neck of S phase cells, as well as in a diffuse cytoplasmic pool (Figure 3B). At higher levels of expression, GFP-Osh2p remained localized to the plasma membrane, although it then redistributed around the whole plasma membrane (our unpublished results). GFP-Osh3p was apparently diffusely distributed throughout the cytoplasm (Figure 3C), although it is possible that there may be a functional targeted population masked by this cytosolic pool. These results show that the three full-length homologues of OSBP in yeast are spatially distinct, which suggests that they may have differing roles in the cell.
Figure 3.
Osh proteins are spatially distinct. GFP-tagged full-length Osh proteins expressed from the PHO5 promoter in log phase cells. (A) GFP-Osh1p: in many cells a single bright linear structure can be seen adjacent to the vacuole (arrows). Similar localization was also seen with expression at lower levels from the OSH1 promoter (our unpublished results). (B) GFP-Osh2p: in cells with small buds, Osh2p is concentrated in the bud and the neck region of mother cell (arrowhead). It has proved impossible to fix GFP-Osh2p to compare its distribution to plasma membrane and ER markers, but in live cells the fluorescence was coincident with the lipophilic dye FM4-64 when added to cells at 4°C to label the plasma membrane (our unpublished results). However, we cannot formally exclude the possibility that the protein is located to that portion of the endoplasmic reticulum that is adjacent to the plasma membrane. (C) GFP-Osh3p: diffusely cytoplasmic with no discernible subcellular concentration. GFP-tagged Osh1p and Osh3p were expressed from pTL311- and pTL313. GFP-tagged Osh2p was expressed by tagging the ORF in the genome of Δosh1 cells (strain TLY227).
Osh1p Localizes to Two Separate Organelles
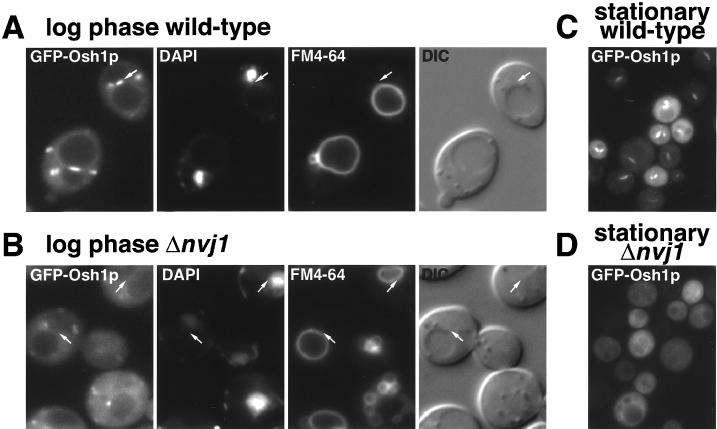
Most of the punctate structures labeled by GFP-Osh1p were small and randomly distributed throughout the cytoplasm, a pattern typical of yeast Golgi membranes (as confirmed below). However, an unusual single linear structure could also be seen at one side of the vacuole (arrows in Figure 3A). The linear structures appeared in multiple adjacent confocal sections, hence they are disk-shaped, and were occasionally seen as discs lying above or below the vacuole in the horizontal plane (our unpublished results). When live cells expressing GFP-Osh1p were labeled with DAPI to detect DNA, and with FM4-64 to visualize the vacuole, these linear structures were found to be located directly between the vacuole and the nucleus (Figure 4A). Although most cells had a single linear structure, some cells with multilobed vacuoles contained two linear structures between the nucleus and two separate vacuoles, but not between the vacuoles themselves. This distribution is the same as the recently described NV junction, a region of close contact between the nuclear envelope and the vacuole whose formation requires the ER membrane protein Nvj1p and the vacuole protein Vac8p (Pan et al., 2000).
Figure 4.
Osh1p localizes to the NV junction. (A) In log phase, GFP-Osh1p is localized to punctate structures and a single linear structure, lying between the DAPI-stained nucleus and the FM4-64-stained vacuole (arrow). (B) In Δnvj1 cells, GFP-Osh1p is no longer in linear structures between the nucleus and vacuole. (C) In stationary phase, punctate localization is lost but GFP-Osh1p stays localized to the NV junction. (D) In stationary phase Δnvj1 cells have lost both localizations, with cytoplasmic staining only. GFP-Osh1p was expressed from plasmid pTL311 in either Δosh1 cells (“wild type”, TLY221), or in Δnvj1 cells (TLY231). Living cells were labeled with DAPI (nucleus and mitochondria, seen as dots and curvilinear structures) and FM4-64 (vacuoles). The vacuole in the Δnvj1 cells was often more fragmented than in wild-type cells, an aspect of this deletion not investigated in the original study. It is perhaps not surprising that release from the nucleus should affect the structure of the vacuole, an organelle whose morphology is highly variable depending on growth conditions and strain background (Roberts et al., 1991; Pan et al., 2000).
To confirm that the linear structures containing GFP-Osh1p are NV junctions, the gene NVJ1 was deleted, because loss of this gene has been shown to lead to the loss of the NV junction (Pan et al., 2000). In Δnvj1 cells, which still contain both nucleus and vacuole, GFP-Osh1p was no longer found between these two organelles, although it was still located in punctate structures (Figure 4B). These observations were strengthened by our finding of conditions under which the only localization of Osh1p is to the NV junction. When cells were examined at stationary phase we observed a complete loss of localization to the multiple punctate structures, but a clear preservation of staining of the NV junction (Figure 4C). The basis of this altered distribution is unknown, but it does not appear to reflect dispersal of the Golgi because a GFP fusion to the late-Golgi t-SNARE Tlg1p was unaffected in these circumstances (our unpublished results). Δnvj1 cells in stationary phase showed delocalization of GFP-Osh1p from both of its target organelles (Figure 4D), as expected. These results show that under normal growth conditions Osh1p has a bipartite localization to two separate organelles, one of which is the NV junction, and that by manipulation of growth conditions or genetic background it is possible to target Osh1p solely to one or other of these two locations.
The Ankyrin Repeat Region of Osh1p Localizes to the NV Junction
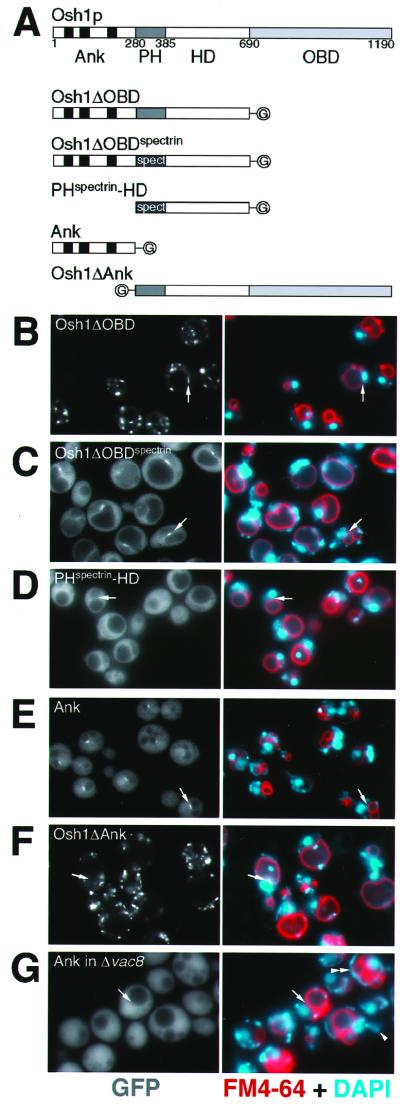
The dual localization of Osh1p might result either from a single targeting domain binding the same ligand found in two sites, or from two targeting domains each with a distinct ligand specific to a single site. A molecular dissection of Osh1p was undertaken to investigate the basis for its dual localization. Osh1p can be divided into four sections along its primary sequence (Figure 5A). These are a region containing three ankyrin repeats, the PH domain (PHOsh1), and the oxysterol binding domain (OBD). In addition, there is a less well conserved region between the PH domain and the oxysterol binding domain, which in both Osh1p and OSBP is predicted to contain short stretches of amphipathic helix (Ridgway et al., 1992), and so we will refer to it as the helical domain (HD).
Figure 5.
Ankyrin (Ank) repeats of Osh1p specify targeting to the NV junction. (A) Structure of Osh1p and of the constructs examined (plasmids pTL321-325). Osh1p can be divided into four regions: Ank, containing three ankyrin repeats; PH, PHOsh1 (in some constructs replaced by PHspectrin); HD, a domain predicted to contain helical regions; and OBD, oxysterol binding domain. The constructs are tagged with GFP (“G”). (B–F) Triple label fluorescent micrographs of live log phase Δosh1 cells (TLY221) expressing the indicated GFP-tagged constructs and stained with FM4-64 (red) and DAPI (blue) as in Figure 4. Arrows indicate typical NV junctions. The same localizations were seen in wild-type cells (our un published results). The NV junctions are relatively small, and so are not always visible in any given focal plane. When cells expressing GFP-Osh1p were counted the percentage of cells showing clear staining in a linear structure between opposed nuclei and vacuoles was typically 85–95%. The exceptions were almost always small daughter cells where the vacuole is still highly fragmented and presumably the NV junction has yet to properly form. A comparable frequency of NV junction structures was seen with all constructs containing the N-terminal ankyrin repeat domain of Osh1p (G) Δvac8 cells (TLY232) expressing pTL324 treated as in B–F except FM4-64 staining was for 45 min, followed by a 3-h chase to examine transfer of vacuolar material to daughter cells, confirming that loss of Vac8p leads to reduced vacuolar segregation into daughter cells (arrowhead), and highly fragmented vacuoles in some cells (double arrowhead) (Fleckenstein et al., 1998; Pan and Goldfarb, 1998; Wang et al., 1998).
Deletion of the oxysterol binding domain did not affect the bipartite localization seen with full-length Osh1p (Figure 5B), indicating that both targeting signals are found somewhere within the N-terminal three domains. To examine the role of the PH domain in the context of the whole protein, PHOsh1 was replaced with the PH domain of human spectrin, a PH domain that does not show any specific localization when expressed on its own in yeast (our unpublished results). This hybrid construct was no longer localized to punctate structures, but remained localized to the NV junction (Figure 5C), indicating that PHOsh1 is not required for targeting to the NV junction. Subdividing the remaining Osh1p sequences further showed that, while removing the ankyrin repeat region resulted in diffuse fluorescence (PHspectrin-helical domain, Figure 5D), the ankyrin repeats alone localized to the NV junction (Figure 5E). Moreover, a version of Osh1p missing just the ankyrin repeats was found only in small punctate structures, and not the NV junction (Figure 5F). These results demonstrate that the ankyrin repeat domain of Osh1p is necessary and sufficient for targeting of Osh1p to the NV junction.
Formation of the NV junction requires the ER membrane protein Nvj1p, and so we investigated whether the Osh1p ankyrin repeats were binding directly to Nvj1p itself. The NV junction is apparently held together by Nvj1p binding to Vac8p on the vacuolar membrane, so that in Δvac8 cells the NV junction is lost, and Nvj1p redistributes around the whole nuclear envelope (Pan et al., 2000). However, in Δvac8 cells the ankyrin repeat region of Osh1p produced entirely diffuse staining (Figure 5G) with no staining of the nuclear envelope where Nvj1p would be expected to be localized (Pan et al., 2000). Vac8p is found in both the NV junction, and on the rest of the vacuolar membrane, but in both wild-type and Δnvj1 neither intact Osh1p nor the ankyrin repeats were seen on the vacuolar membrane outside of the NV junction in the wild-type cells. Taken together, these results indicate that the feature recognized by the ankyrin repeats is neither Vac8p nor Nvj1p alone, but rather a ligand that appears at the NV junction as a result of the interaction of the two proteins.
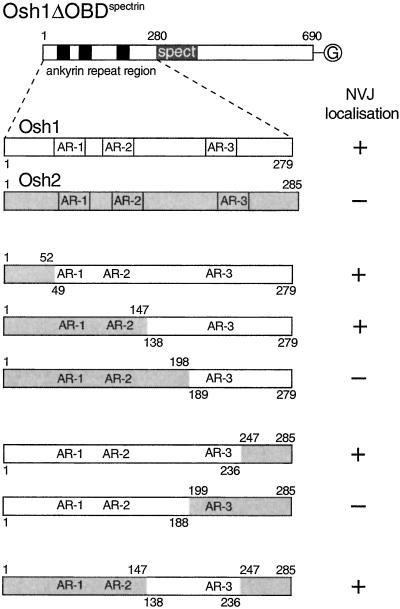
As shown above, Osh2p does not target to the NV junction, despite also having three ankyrin repeats near its N terminus (Figures 1 and 3). When the N-terminal 285 amino acids of Osh2p, which contain these repeats, was used to replace the equivalent region from Osh1p in the NV-junction–specific construct Osh1DOBDspectrin, it was now diffusely localized, indicating that the ankyrin repeat domains from the two proteins contain different targeting information (Figure 6). Examination of chimeras between the ankyrin repeat domains of the two proteins showed that a region of 100 amino acids of Osh1p, including the third repeat, was sufficient to confer NV-junction targeting activity to the Osh2p chimera (Figure 6). Dividing this region further resulted in loss of targeting, suggesting that the interactions most critical for targeting to the NV junction location are mediated over an extended region within the ankyrin repeat domain that includes repeat 3, and the linker between repeats 2 and 3.
Figure 6.
A portion of the ankyrin repeat domain is required for targeting to the NV junction. Constructs based on Osh1DOBDspectrin, a GFP-tagged version of Osh1p without the OBD, and with the PH domain replaced with that of spectrin (plasmid pTPL322, Figure 5C). The ankyrin repeat containing N-terminal region in plasmid pTPL322 was replaced with either the equivalent region of Osh2p, or with the indicated chimeras between Osh1p and Osh2p. The intracellular location of the GFP fusions was examined by fluorescence microscopy of live transfected cells, and the proteins were either entirely diffuse, or localized to the NV junction as indicated. The three ankyrin repeats in Osh1p are 51–80, 96–125, and 196–225 (SWISSPROT P39555).
TGN Targeting Is Specified by the PH Domain of Osh1p
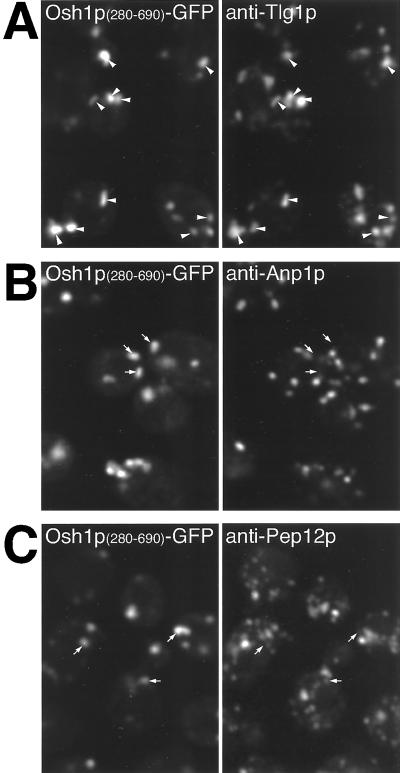
The above-mentioned results suggest that the PH domain is not required for targeting to the NV junction, but is necessary for targeting of Osh1p to the other punctate structures observed with the full-length protein. These punctate structures have an appearance characteristic of the yeast Golgi, and this was confirmed by with the use of double label immunofluorescence to compare the distribution of the PH-domain containing chimeras with specific compartmental markers. The unstacked nature of Golgi cisternae in S. cerevisiae allows different Golgi subcompartments to be distinguished by fluorescence microscopy in fixed cells. Although live cells expressing GFP-tagged PHOsh1 showed punctate localization (Figure 8), this targeting was lost on fixation. In contrast, a C-terminally extended construct consisting of PHOsh1+HD was still localized after fixation, although the HD does not add any extra targeting information (Figure 5D), and so this construct was used for colocalization studies. Figure 7A shows that structures containing GFP extensively overlap with Tlg1p, a resident of the TGN and early endosomes (Holthuis et al., 1998). Colocalization between proteins in the Golgi compartments of yeast is usually not absolute, presumably reflecting the dynamic nature of the organelle. In contrast, the distribution of PHOsh1+HD was completely distinct both from the cis-Golgi marker Anp1p and the late endosomal marker Pep12p (Figure 7, B and C) (Becherer et al., 1996; Jungmann and Munro, 1998). Thus, the PH domain of Osh1p primarily localizes to TGN membranes or closely related early endosomes. Given that the same construct targeted to the TGN when expressed in mammalian cells (Levine and Munro, 1998), it appears that the nature and localization of the membrane receptor for this family of PH domains has been conserved through evolution.
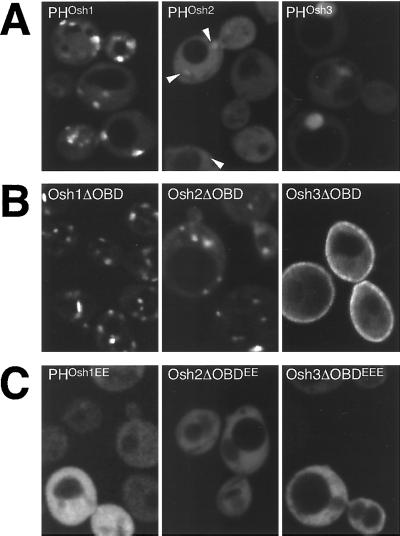
Figure 8.
Targeting by the three Osh PH domains. GFP-tagged portions of Osh1-3p visualized in log phase Δosh1 cells (TLY221). (A) PH domains of Osh proteins have different targeting strengths. PHOsh1 localizes readily to punctate structures, PHOsh2 is largely diffuse, except for a number of barely detectable punctate structures seen against the strong cytoplasmic background (arrowheads) and PHOsh3 is diffuse with some nuclear localization. Plasmids pTL331, pTL342, pTL343. (B) Constructs containing all but the oxysterol binding domains target to distinct intracellular localizations (plasmids pTL321, pTL352, pTL353). Compared with the PH domain alone, Osh1ΔOBD shows additional NV junction localization and reduced cytoplasmic background, Osh2ΔOBD shows localization to punctate structures, and Osh3ΔOBD shows some localization to the plasma membrane. Osh3ΔOBD shows no nuclear localization, probably because of its increased size. (C) Constructs as in A or B but with conserved basic residues of the variable loop 1 of the PH domain replaced with glutamate residues (plasmids pTL356, pTL357, pTL358). The levels of wild-type and mutant proteins were indistinguishable by protein blotting with anti-GFP antibodies (our unpublished results).
Figure 7.
Immunofluorescence localization of PHOsh1 domain to the TGN. Δosh1 yeast (TLY221) expressing GFP-tagged PHOsh1 and the helical domain (residues 280–690, pTL327) were fixed and stained with rabbit antibodies to Tlg1p (A), Anp1p (B), and Pep12p (C), markers for TGN/early endosomes, cis-Golgi, and prevacuolar compartment, respectively. Colocalization of GFP is only seen with Tlg1p (arrowheads), but not with the other two markers (arrows).
PH Domains of Osh Proteins Show Distinct Intracellular Targeting
Full-length Osh1p and Osh2p are localized to different parts of the cell, yet their PH domains are highly homologous (70% identical, 91% similar). To determine whether differences between these PH domains are responsible for the different intracellular distributions of the full-length proteins, targeting of GFP-tagged PH domains was examined. PHOsh1 was clearly visible in punctate structures, while PHOsh2 showed only barely discernible punctate staining, and PHOsh3 was entirely diffuse (Figure 8A). To ask whether this weak membrane localization by PHOsh2 could be enhanced by the presence of surrounding sequences, we also examined constructs comprising the entire N-terminal regions, i.e., the whole protein without the oxysterol binding domains. As mentioned above, Osh1ΔOBD showed a bipartite localization with less in the cytoplasm than seen for the PH domain alone (Figure 8B). However, Osh2ΔOBD surprisingly showed a strong punctate localization similar to PHOsh1, and clearly distinct from the plasma membrane localization of the full-length protein (Figure 8B). This punctate staining by Osh2ΔOBD did not directly result from either of the regions flanking the PH domain, which when expressed on their own were completely cytoplasmic (our unpublished results). In contrast to the punctate localizations of Osh1ΔOBD and Osh2ΔOBD, Osh3ΔOBD showed partial localization to the plasma membrane. To determine whether these interactions were mediated by the PH domains, conserved basic residues in variable loop 1 of the domain were altered to glutamate residues, mutations which, by analogy with other PH domains, would be expected to abrogate PIP binding (Hyvonen and Saraste, 1997; Levine and Munro, 1998). These mutations caused complete delocalization of PHOsh1, Osh2ΔOBD, and Osh3ΔOBD (Figure 8C), implying that the PH domains of all three Osh proteins can contribute to intracellular targeting. This result indicates that PHOsh2 can target to internal membranes, but the affinity appears weaker than for PHOsh1, and this targeting is apparently masked, or altered, in the context of full-length Osh2p, a situation similar to that seen for mammalian OSBP (Ridgway et al., 1992).
Precise localization of Osh2ΔOBD was not possible, because the construct delocalized under a variety of fixation conditions. However, it would appear to localize to a site closely related to the TGN, because in live cells PHOsh1, Osh1ΔOBD, and Osh2ΔOBD colocalize with FM4-64 at intermediate times after uptake (10–40 min; our unpublished results). It has been shown previously that before accumulating in the vacuole, this endocytic tracer enters the TGN, where it colocalizes with the late Golgi marker Sec7p (Lewis et al., 2000).
DISCUSSION
OSBP is the founder member of a protein family with multiple homologues in all eukaryotes so far examined. One possible explanation for such a multiplicity of related genes is that a degree of specialization has arisen on top of a basic function that is common to all. It is possible that such a family of related proteins is located at the same place in the cell, but carries out different functions, or alternatively the proteins could perform similar functions but be located in different places. We have previously found that the PH domain of mammalian OSBP specifies its location to the Golgi, and in this article we investigate the three yeast Osh proteins with PH domains, and find that they have different intracellular distributions. The localizations were determined in live cells expressing GFP-tagged proteins, but because the actual functions of the Osh proteins are still unclear it was not possible to directly examine these functions for each of the GFP fusions. However, for all three proteins it was possible to show that the GFP-fusion was at least able to rescue phenotypes of deletion strains. Thus, it seems likely that the localizations obtained for these fusions are an accurate representation of the distribution of the native proteins.
We find that Osh1p is located to both the late Golgi and the NV junction. Investigation of the domains required for this dual localization of Osh1p revealed that each intracellular site is specified by a distinct domain in the protein, with the PH domain being necessary and sufficient for Golgi targeting, and the ankyrin repeat domain specifying targeting to the NV junction. This is the first example of a peripheral membrane protein targeting to this specialization of the nucleus and vacuole. A related ankyrin repeat region is also found at the N terminus of some of the OSBP homologues from the yeasts Candida albicans, Kluyveromyces thermotolerans, and S. pombe, but is not present in OSBP from any other organisms, consistent with the notion that the NV junction is a yeast-specific structure. The NV junction is formed by a direct interaction between Nvj1p in the nuclear envelope, and a subpopulation of the Vac8p on the vacuolar membrane (Pan et al., 2000). However, the NV junction-targeting domain from Osh1p does not appear to bind to either of these proteins directly, as revealed by its diffuse distribution in strains lacking either Nvj1p or Vac8p. The targeting thus requires the correct formation of the junctional structure, and so it is possible that the domain binds only to the Nvj1p/Vac8p complex, or to some other factor recruited to the junction. Osh2p has similar ankyrin repeats, but the protein does not localize to the NV junction, and the role of its ankyrin repeats is unclear because this region shows only diffuse localization when fused to GFP (our unpublished results).
In contrast, the PH domain-dependent targeting to the Golgi seems to reflect a more general feature of the OSBP family. We have previously found that the PH domain of mammalian OSBP specifies targeting to the mammalian Golgi (Levine and Munro, 1998), and the same is the case for the Osh1p PH domain in yeast. Indeed this yeast PH domain also specifies Golgi targeting in mammalian cells (Levine and Munro, 1998). The PH domain of mammalian OSBP was found to require PIPs for binding to the Golgi (Levine and Munro, 1998), and such a labile ligand would be consistent with our observation that Osh1p rapidly relocates to the Golgi as cells move out of stationary phase (our unpublished results). A role for PIPs in Golgi targeting is also consistent with phosphatidylinositol 4-kinase having been found on the Golgi in mammalian cells (Godi et al., 1999), and being required for Golgi function in yeast (Hama et al., 1999; Walch-Solimena and Novick, 1999; Audhya et al., 2000), and we have initiated an investigation of the precise lipid kinase requirements for Osh1p targeting.
Interestingly, although full-length Osh2p is located at the plasma membrane, when just its PH domain was expressed as a fusion to GFP it was apparently targeted to the Golgi, albeit weakly. Longer fragments of Osh2p that contain all but the oxysterol binding domain were targeted more efficiently to internal membranes in a manner dependent on the PH domain. This situation is similar to that seen with mammalian OSBP, which is reported to be located on vesicular structures throughout the cytoplasm and to translocate to the Golgi only when oxysterols are added, or cholesterol traffic is perturbed (Ridgway et al., 1992, 1998). However, when the oxysterol binding domain of OSBP is removed, the remainder of the protein is constitutively targeted to the Golgi, suggesting that Golgi binding of the PH domain can be masked by other domains in the protein. Thus, it may be that although Osh2p is normally targeted to the plasma membrane, it has the capacity to translocate to the Golgi in appropriate circumstances.
Intracellular Localization and Function of the OSBP Family
At present the precise function of the OSBP family remains elusive. The necessity and, when overexpressed, the sufficiency of the Osh1p oxysterol binding domain to rescue the phenotype of Δosh1 suggests that the key effects of the protein are mediated by this domain. Although oxysterol binding has so far only been reported for mammalian OSBP, there are several suggestions that yeast OSBP homologues have roles in lipid metabolism and/or trafficking. First, deletion of OSH4 (KES1) is a by-pass suppressor of sec14ts, suggesting a role in regulating phospholipid metabolism in the Golgi (Fang et al., 1996). Second, the growth phenotype of Δosh1 is characteristic of many of the erg mutants of ergosterol synthesis, and has also been reported in disturbances of PIP metabolism (Gaber et al., 1989; Jiang et al., 1994; Stolz et al., 1998). Third, Δosh2, but not Δosh1, is associated with reduced ergosterol levels, although with no growth phenotype (Daum et al., 1999). However the clearest evidence for a role in ergosterol biology comes from a recent comprehensive analysis of the phenotypes of all 127 combinations of OSH gene deletions (Beh et al., 2001). Deletion of all seven OSH genes is lethal, with cells accumulating threefold elevated levels of ergosterol. Any one OSH gene is sufficient to rescue this lethality (with only OSH1 requiring overexpression), indicating that the seven proteins share a common essential function. However, when each of the OSH genes is deleted individually a specific range of phenotypes is observed, many consistent with mild perturbation of ergosterol synthesis or trafficking, indicating that the encoded proteins perform distinct functions, in addition to their common essential function.
Our localization data provide strong independent support for the idea that the different Osh proteins have specific functions, because it appears that individual proteins are targeted to distinct parts of the cell. A set of proteins acting in a range of different intracellular locations seems to us more compatible with a role in either the sensing of lipid levels, or the transport of lipids or precursors between organelles, rather than with a direct role in lipid synthesis. For instance one possible function is the intracellular traffic of oxysterols, which have been found in a variety of species, including yeast (Nes et al., 1989; Bocking et al., 2000; Gardner et al., 2001). Because oxidation of sterols makes them more water soluble, oxysterols will have a higher propensity to diffuse across the cytoplasm, making them more suitable as second messengers. The presence of high-affinity receptors on the periphery of specific organelles might increase the local concentration of oxysterols present at low levels, and so present a sterol-dependent signal to proteins restricted to the particular bilayer. It has been proposed from work in mammalian cells that OSBP regulates sphingolipid synthesis, which might be expected to be coregulated with the levels of sterols (Lagace et al., 1999). We have recently found that Aur1p, the inositol phosphophorylceramide synthase that initiates yeast sphingolipid synthesis, is localized to the medial Golgi (Levine et al., 2000). It is, therefore, unlikely that Aur1p is the direct target of the Osh proteins we have studied here, and indeed cells deleted for all three genes showed no change in the activity of Aur1p as measured with the use of a fluorescent substrate in whole cells (our unpublished results). It may be that the effect of OSBP on sphingomyelin synthase is indirect, but it is also possible that it is regulated differently from inositol phosphophorylceramide synthase.
The targeting to the NV junction is intriguing, and because the ER and the vacuole have critical roles in the synthesis and degradation of lipids, respectively, it is conceivable that a structure that connects the two could have a role in the transport of lipid metabolic precursors, or the regulation of lipid metabolism. Interestingly, it has recently been reported that an ER protein Tsc13p, which is required for fatty acid elongation is enriched in the NV junction (Kohlwein et al., 2001). Although the significance of this is somewhat unclear because Elo2p and Elo3p, which are involved in the same process, are not enriched in the junction, it is at least suggestive of a role for the NV junction in lipid metabolism or transport. The NV junction itself is not required for growth on low tryptophan because Δnvj1 cells grew as wild type (and Δosh1/Δnvj1 cells grew as Δosh1), and removal of the ankyrin repeats did not prevent Osh1p rescuing Δosh1 (our unpublished results).
In summary, we have found that 3 OSBP homologues in yeast are spatially distinct, despite all having PH domains, and that Osh1p has a novel targeting determinant specific for the NV junction. This diverse intracellular targeting suggests that the families of OSBP-related proteins present in all eukaryotes are likely to include homologues that perform similar tasks at multiple intracellular sites.
ACKNOWLEDGMENTS
We thank Rob Arkowitz, Mike Lewis, Ben Nichols, Robert Sauer, and Symeon Siniossoglou for antibodies, plasmids and strains; and Alison Gillingham and Hugh Pelham for comments on the manuscript. We thank Christopher Beh and Jasper Rine for communication of results before publication, and provision of strains. T.P.L. was supported by a fellowship from the British Heart Foundation.
REFERENCES
- Accad M, Farese RV. Cholesterol homeostasis: a role for oxysterols. Curr Biol. 1998;8:R601–R604. doi: 10.1016/s0960-9822(98)70386-0. [DOI] [PubMed] [Google Scholar]
- Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajwa W, Rudolph H, Hinnen A. PHO5 upstream sequences confer phosphate control on the constitutive PHO3 gene. Yeast. 1987;3:33–42. doi: 10.1002/yea.320030106. [DOI] [PubMed] [Google Scholar]
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh CT, Cool L, Phillips J, Rine J. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics. 2001;157:1117–1140. doi: 10.1093/genetics/157.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkhem I, Diczfalusy U, Lutjohann D. Removal of cholesterol from extrahepatic sources by oxidative mechanisms. Curr Opin Lipidol. 1999;10:161–165. doi: 10.1097/00041433-199904000-00010. [DOI] [PubMed] [Google Scholar]
- Bocking T, Barrow KD, Netting AG, Chilcott TC, Coster HG, Hofer M. Effects of singlet oxygen on membrane sterols in the yeast Saccharomyces cerevisiae. Eur J Biochem. 2000;267:1607–1618. doi: 10.1046/j.1432-1327.2000.01179.x. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. Suppression of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity and inhibition of growth of human fibroblasts by 7-ketocholesterol. J Biol Chem. 1974;249:7306–7314. [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Tuller G, Nemec T, Hrastnik C, Balliano G, Cattel L, Milla P, Rocco F, Conzelmann A, Vionnet C, Kelly DE, Kelly S, Schweizer E, Schuller HJ, Hojad U, Greiner E, Finger K. Systematic analysis of yeast strains with possible defects in lipid metabolism. Yeast. 1999;15:601–614. doi: 10.1002/(SICI)1097-0061(199905)15:7<601::AID-YEA390>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Dawson PA, Ridgway ND, Slaughter CA, Brown MS, Goldstein JL. cDNA cloning and expression of oxysterol-binding protein, an oligomer with a potential leucine zipper. J Biol Chem. 1989a;264:16798–16803. [PubMed] [Google Scholar]
- Dawson PA, Vanderwesthuyzen DR, Goldstein JL, Brown MS. Purification of oxysterol binding-protein from hamster liver cytosol. J Biol Chem. 1989b;264:9046–9052. [PubMed] [Google Scholar]
- Fang M, Kearns BG, Gedvilaite A, Kagiwada S, Kearns M, Fung MK, Bankaitis VA. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 1996;15:6447–6459. [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein D, Rohde M, Klionsky DJ, Rudiger M. Yel013p (Vac8p), an armadillo repeat protein related to plakoglobin and importin alpha is associated with the yeast vacuole membrane. J Cell Sci. 1998;111:3109–3118. doi: 10.1242/jcs.111.20.3109. [DOI] [PubMed] [Google Scholar]
- Gaber RF, Copple DM, Kennedy BK, Vidal M, Bard M. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol Cell Biol. 1989;9:3447–3456. doi: 10.1128/mcb.9.8.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Shan H, Matsuda SP, Hampton RY. A positive oxysterol-derived signal for 3-hydroxy-3-methylglutaryl CoA reductase degradation in yeast. J Biol Chem. 2001;276:8681–8694. doi: 10.1074/jbc.M007888200. [DOI] [PubMed] [Google Scholar]
- Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA. ARF mediates recruitment of PtdIns-4-OH kinaseb and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Hama H, Schnieders EA, Thorner J, Takemoto JY, DeWald DB. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J Biol Chem. 1999;274:34294–34300. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- Holthuis JC, Nichols BJ, Dhruvakumar S, Pelham HR. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvonen M, Saraste M. Structure of the PH domain and Btk motif from Bruton's tyrosine kinase: molecular explanations for X-linked agammaglobulinaemia. EMBO J. 1997;16:3396–3404. doi: 10.1093/emboj/16.12.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signaling pathway mediated by the nuclear receptor LXRa. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- Jiang B, Brown JL, Sheraton J, Fortin N, Bussey H. A new family of yeast genes implicated in ergosterol synthesis is related to the human oxysterol binding-protein. Yeast. 1994;10:341–353. doi: 10.1002/yea.320100307. [DOI] [PubMed] [Google Scholar]
- Jungmann J, Munro S. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with α-1,6-mannosyltransferase activity. EMBO J. 1998;17:423–434. doi: 10.1093/emboj/17.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandutsch AA, Chen HW. Inhibition of sterol synthesis in cultured mouse cells by cholesterol derivatives oxygenated in the side chain. J Biol Chem. 1974;249:6057–6061. [PubMed] [Google Scholar]
- Kandutsch AA, Chen HW, Heiniger H-J. Biological activity of some oxygenated sterols. Science. 1978;201:498–501. doi: 10.1126/science.663671. [DOI] [PubMed] [Google Scholar]
- Kohlwein SD, Eder S, Oh CS, Martin CE, Gable K, Bacikova D, Dunn T. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:109–125. doi: 10.1128/MCB.21.1.109-125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace TA, Byers DM, Cook HW, Ridgway ND. Altered regulation of cholesterol and cholesteryl ester synthesis in Chinese-hamster ovary cells overexpressing the oxysterol-binding protein is dependent on the pleckstrin homology domain. Biochem J. 1997;326:205–213. doi: 10.1042/bj3260205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace TA, Byers DM, Cook HW, Ridgway ND. Chinese hamster ovary cells overexpressing the oxysterol binding protein (OSBP) display enhanced synthesis of sphingomyelin in response to 25-hydroxycholesterol. J Lipid Res. 1999;40:109–116. [PubMed] [Google Scholar]
- Lange Y, Steck TL. The role of intracellular cholesterol transport in cholesterol homeostasis. Trends Cell Biol. 1996;6:205–208. doi: 10.1016/0962-8924(96)20016-9. [DOI] [PubMed] [Google Scholar]
- Lange Y, Ye J, Rigney M, Steck TL. Regulation of endoplasmic reticulum cholesterol by plasma membrane cholesterol. J Lipid Res. 1999;40:2264–2270. [PubMed] [Google Scholar]
- Langle-Rouault F, Jacobs E. A method for performing precise alterations in the yeast genome using a recyclable marker. Nucleic Acids Res. 1995;23:3079–3081. doi: 10.1093/nar/23.15.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Munro S. The pleckstrin homology domain of oxysterol-binding protein recognizes a determinant specific to Golgi membranes. Curr Biol. 1998;8:729–739. doi: 10.1016/s0960-9822(98)70296-9. [DOI] [PubMed] [Google Scholar]
- Levine TP, Wiggins CA, Munro S. Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2267–2281. doi: 10.1091/mbc.11.7.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HR. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund EG, Guileyardo JM, Russell DW. cDNA cloning of cholesterol 24-hydroxylase, a mediator of cholesterol homeostasis in the brain. Proc Natl Acad Sci USA. 1999;96:7238–7243. doi: 10.1073/pnas.96.13.7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund EG, Kerr TA, Sakai J, Li WP, Russell DW. cDNA cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism. J Biol Chem. 1998;273:34316–34327. doi: 10.1074/jbc.273.51.34316. [DOI] [PubMed] [Google Scholar]
- Nes WD, Sihua X, Haddon WF. Evidence for similarities and differences in the biosynthesis of fungal sterols. Steroids. 1989;53:533–558. doi: 10.1016/0039-128x(89)90030-5. [DOI] [PubMed] [Google Scholar]
- Pan X, Goldfarb DS. YEB3/VAC8 encodes a myristylated armadillo protein of the Saccharomyces cerevisiae vacuolar membrane that functions in vacuole fusion and inheritance. J Cell Sci. 1998;111:2137–2147. doi: 10.1242/jcs.111.15.2137. [DOI] [PubMed] [Google Scholar]
- Pan X, Roberts P, Chen Y, Kvam E, Shulga N, Huang K, Lemmon S, Goldfarb DS. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol Biol Cell. 2000;11:2445–2457. doi: 10.1091/mbc.11.7.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway N, Dawson P, Ho Y, Brown M, Goldstein J. Translocation of oxysterol binding-protein to Golgi-apparatus triggered by ligand-binding. J Cell Biol. 1992;116:307–319. doi: 10.1083/jcb.116.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway ND, Lagace TA, Cook HW, Byers DM. Differential effects of sphingomyelin hydrolysis and cholesterol transport on oxysterol-binding protein phosphorylation and Golgi localization. J Biol Chem. 1998;273:31621–31628. doi: 10.1074/jbc.273.47.31621. [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. In: Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. In Methods in Enzymology, Volume 194: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink GR, editors. San Diego: Academic Press; 1991. pp. 281–301. [DOI] [PubMed] [Google Scholar]
- Russell DW. Oxysterol biosynthetic enzymes. Biochim Biophys Acta. 2000;1529:126–135. doi: 10.1016/s1388-1981(00)00142-6. [DOI] [PubMed] [Google Scholar]
- Sauer B. Recycling selectable markers in yeast. Biotechniques. 1994;16:1086–1088. [PubMed] [Google Scholar]
- Schmalix WA, Bandlow W. SWH1 from yeast encodes a candidate nuclear factor-containing ankyrin repeats and showing homology to mammalian oxysterol-binding protein. Biochim Biophys Acta. 1994;1219:205–210. doi: 10.1016/0167-4781(94)90273-9. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Hurt EC, Pelham HR. Psr1p/Psr2p, two plasma membrane phosphatases with an essential DXDX(T/V) motif required for sodium stress response in yeast. J Biol Chem. 2000;275:19352–19360. doi: 10.1074/jbc.M001314200. [DOI] [PubMed] [Google Scholar]
- Skrzypek MS, Nagiec MM, Lester RL, Dickson RC. Inhibition of amino acid transport by sphingoid long chain bases in Saccharomyces cerevisiae. J Biol Chem. 1998;273:2829–2834. doi: 10.1074/jbc.273.5.2829. [DOI] [PubMed] [Google Scholar]
- Smith LL. Review of progress in sterol oxidations - 1987–1995. Lipids. 1996;31:453–487. doi: 10.1007/BF02522641. [DOI] [PubMed] [Google Scholar]
- Stolz LE, Kuo WJ, Longchamps J, Sekhon MK, York JD. INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J Biol Chem. 1998;273:11852–11861. doi: 10.1074/jbc.273.19.11852. [DOI] [PubMed] [Google Scholar]
- Taylor FR, Saucier SE, Shown EP, Parish EJ, Kandutsch AA. Correlation between oxysterol binding to a cytosolic binding-protein and potency in the repression of hydroxymethylglutaryl coenzyme-a reductase. J Biol Chem. 1984;259:12382–12387. [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase PIK1 regulates secretion at the Golgi. Nat Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- Wang YX, Catlett NL, Weisman LS. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J Cell Biol. 1998;140:1063–1074. doi: 10.1083/jcb.140.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]