Abstract
A chemical procedure was developed to functionalize poly(methyl methacrylate) (PMMA) substrates. PMMA is reacted with hexamethylene diamine to yield an aminated surface for immobilizing DNA in microarrays. The density of primary NH2 groups was 0.29 nmol/cm2. The availability of these primary amines was confirmed by the immobilization of DNA probes and hybridization with a complementary DNA strand. The hybridization signal and the hybridization efficiency of the chemically aminated PMMA slides were comparable to the hybridization signal and the hybridization efficiency obtained from differently chemically modified PMMA slides, silanized glass, commercial silylated glass and commercial plastic Euray™ slides. Immobilized and hybridized densities of 10 and 0.75 pmol/cm2, respectively, were observed for microarrays on chemically aminated PMMA. The immobilized probes were heat stable since the hybridization performance of microarrays subjected to 20 PCR heat cycles was only reduced by 4%. In conclusion, this new strategy to modify PMMA provides a robust procedure to immobilize DNA, which is a very useful substrate for fabricating single use diagnostics devices with integrated functions, like sample preparation, treatment and detection using microfabrication and microelectronic techniques.
INTRODUCTION
High-density oligonucleotide arrays have emerged as a powerful and promising tool for genetic analysis with a lower cost, higher throughput and reproducibility than the traditional gel-based methods. These so-called DNA chips, or microarrays, have nowadays numerous applications in expression studies or sequencing by hybridization (1–4). However, for these microarrays to replace conventional DNA diagnostic methods, substrates have to be well characterized in relation to thermal and chemical stability, absolute density of immobilized DNA, functional availability of DNA probes and reproducibility of the attachment chemistry. Direct synthesis on the chip of nucleic acids (5–7) and immobilization of pre-synthesized oligonucleotides on solid supports (8,9) are the two predominant methods of producing DNA microarrays. Technologies such as photolithographic DNA synthesis allow the manufacture of high-density oligonucleotide microarrays, but are costly and time consuming, which limits their widespread implementation. Therefore, the use of pre-synthesized oligonucleotides is more common for many diagnostics tests. Currently, there are several methods described in the literature for covalent attachment of modified oligonucleotides to pre-activated solid supports. The procedures to make these functionalized substrates are well established and are described on glass (10–15), oxidized silicon (16,17), silicon wafers (18), gold surfaces (19,20), filter membranes (21,22), optical fibers (23) and polyacrylamide gel pads (24). Plastic (organic) polymers are alternatives to these solid supports for making DNA microarrays. One particular advantage of using polymers is that microarrays can be produced inexpensively and in large numbers, making their use in commercial applications particularly attractive. Another advantage is that a wide range of microfabrication methods can be selected to fabricate miniaturized devices, around the microarray module. However, the development of a simple, well defined and reproducible surface modification for plastics is still in the early stages (25), but are essential for the development of biochips in polymer-based substrates, such as poly(methyl methacrylate) (PMMA). PMMA has successfully been used (in the form of micro spheres) for the immobilization of enzymes (26), for the design of microfluidic devices, for metal deposition (27) and for immobilization of proteins and DNA (28,29). Henry et al. (28) showed that aminated PMMA sheets can be used for the immobilization of enzymes effective in the restriction digestion of dsDNA. Waddell et al. (29) have developed a method for lowering the detection limit of oligonucleotides using aminated PMMA slides as a substrate for testing the immobilization and hybridization in a microarray type format.
In this report, we describe a simple and quick modification procedure for PMMA surfaces yielding primary amino groups, suitable for immobilizing different types of biomolecules, like DNA. The described procedure is compared with the PMMA modification described by Bulmus et al. (26).
MATERIALS AND METHODS
All chemicals and solvents were purchased from Sigma (Denmark) or Merck (Denmark), unless stated otherwise, and used without additional purification. All DNA oligonucleotides were ordered from TAQ Copenhagen (Denmark).
Chemical modification of PMMA substrates
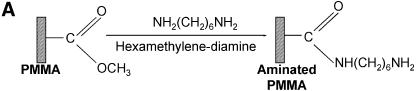
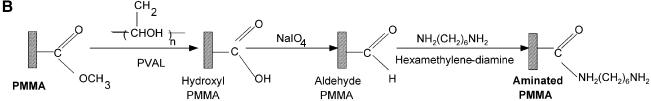
Commercial PMMA sheets were obtained from Röhm, Germany. The slides of 75 × 25 mm were obtained by cutting the PMMA with a CO2 laser. Two different protocols were used to modify and immobilize DNA probes on the plastic surface–PMMA (Fig. 1). The new method proposed here (Fig. 1A), is made by first washing the PMMA slides with isopropanol (99%), by soaking the slides on a slab at room temperature, and then rinsing thoroughly with MilliQ water. Subsequently, the plastic surface is incubated with a solution of 10% hexamethylene diamine in 100 mM borate buffer pH 11.5, for 2 h, followed by two washes with MilliQ water for 10 min each. Finally, PMMA slides are dried overnight at 30°C. The second strategy (Fig. 1B), described by Bulmus et al. (26), involved briefly: a first washing of the substrate with NaOH 10% (w/v) followed by ethanol 50% (v/v). Then, the PMMA was immersed for 20 min in a solution of 1 g/l polyvinyl alcohol, followed by an oxidation step with a solution of 1% NaIO4 for 1 h at room temperature. The amino functionality was then added by using a solution of hexamethylene diamine 10% (w/v) in 100 mM borate buffer pH 11.5, for 2 h. The final washing was then performed with borate buffer solutions with pH 11.5 and 8.2, for 15 min each.
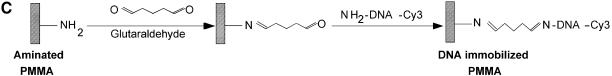
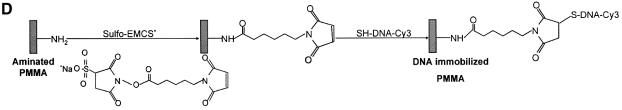
Figure 1.
Chemistries used to modify the PMMA substrates and to covalently immobilize 5′-end modified DNA oligonucleotides to aminated terminated surfaces. (A) The available methyl esters of the PMMA, under basic pH conditions, are reacted with an electron donor (N) present on the hexamethylene- diamine, yielding primary amines on the surface. (B) Protocol described by Bulmus et al. (26). (C) Attaching aminated DNA to aminated PMMA surfaces. The aminated PMMA was treated with the homobifunctional cross-linker glutaraldehyde, having a terminal aldehyde group reacting to the primary amino groups of the PMMA, through an imine bond. The NH2-DNA probes are subsequently reacted with the other aldehyde terminal of the cross-linker, establishing a covalent bond between the surface and the oligonucleotide probe. (D) Attaching thiolated DNA to aminated PMMA surfaces. The aminated PMMA is reacted with the NHS ester group of the heterobifunctional cross-linker sulfo-EMCS and subsequently the maleimide portion of the sulfo-EMCS is reacted with 5′-end thiolated DNA probe, to achieve a covalent immobilized DNA.
Detection of NH2 groups on the surface
To measure the amino groups on the PMMA substrates modified by the methods described above, a reaction with fluorescein-5-isothiocyanate (FITC) was performed. The FITC dye reacts specifically with NH2 groups to form covalent bonds. The fluorescence was detected by laser scanning of the surface (ScanArray Lite Microarray Analysis System; Packard BioScience, Billerica, MA). The density of NH2 groups was derived by comparing the fluorescent signal to a standard curve. This calibration curve was done by spotting, on PMMA, different concentrations of hexamethylene-diamine conjugated with 0.1 mg/ml FITC and reading the fluorescent output signal for each dilution of the amine compound (30). By plotting the fluorescence intensity as a function of the amino compound concentration, a calibration curve is defined.
Chemical modification (silanization) of glass slides
The glass slides (75 × 25 mm) were first cleaned and oxidized in 5% nitric acid, for 1 h at 90°C and then washed three times in MilliQ water for 5 min each. The glass slides were silanized by incubating the slides with 8% (v/v) 3-aminopropyltriethoxysilane pH 3.0, for 2 h at 80°C. After washing with MilliQ water (twice, 5 min each), the substrates were dried for 16 h at 115°C (31).
Activation of the PMMA surface
The aminated surfaces were activated with two cross-linkers, glutaraldehyde and sulfo-EMCS (Fig. 1). The amino functionalized substrates were modified with a freshly prepared solution of 2.5% (v/v) glutaraldehyde in 0.1 M phosphate buffer pH 7.0, for 2 h at room temperature. Afterwards, the slides were washed twice, for 10 min each, with 0.1 M phosphate buffer pH 7.0 and finally dried at 30°C. Alternatively, the activation was done by immersion of the amino modified slides into a solution of 0.1 mM of sulfo-EMCS in borate buffer (100 mM, pH 9.0), for 2 h at room temperature. Finally, the slides were washed twice in borate buffer, 5 min each, rinsed thoroughly with MilliQ water and dried.
Spotting and DNA immobilization
Spotting was performed using a Cartesian Technologies PixSys spotting robot with a Stealth pin that deposited 0.6 nl/spot on the substrates. The size of the spots were ∼100 µm in diameter and 300 µm apart, using 100 mM phosphate buffer pH 7 as spotting solution. The spotted slides were incubated between 15 min and 24 h in a humidified chamber at 30°C and washed with 5× SSC + 0.1% Tween-20, for 15 min, rinsed with MilliQ H2O, and dried.
DNA hybridization
Prior to hybridization, glutaraldehyde-activated slides were blocked with Na2BH4, while maleimide-activated substrates were blocked with 2% BSA in 10 mM PBS. Cy5-labeled target-DNA was diluted between 0.2 and 1 µM in 6× SSC + 0.6% SDS + 0.1% salmon sperm DNA + 2% BSA. The microarrays were hybridized with 40 µl of the hybridization solution under a coverslip. The temperature of hybridization was between 30 and 40°C and the reaction was performed in a humidified chamber, for between 30 min and 24 h. After hybridization, the coverslips were gently removed and the slides were washed in 0.1× SSC + 0.5% SDS for 15 min, followed by a second wash in 0.1× SSC for 5min.
DNA quantification
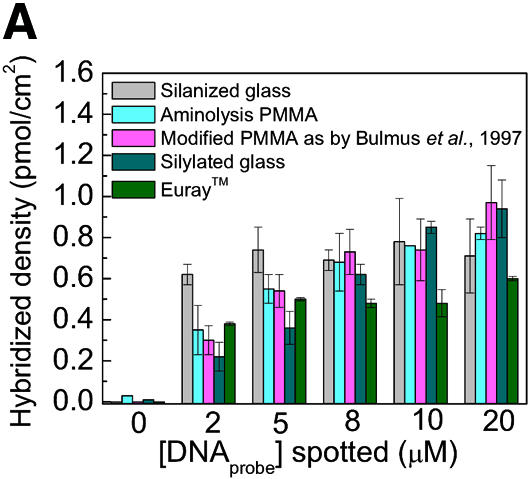
Thiol-labeled (-SH) or amino-labeled (-NH2) capture probes were diluted in 100 mM phosphate buffer (pH 7.0) to a dilution series ranging from 0.05 to 5 µM DNA. Ten replicas of each dilution were spotted on PMMA slides chemically modified by hexamethylene as described in this publication, as by Bulmus et al. (26), on silanized glass or on commercial plastic Euray™ (Exiqon, Vedbæk, Denmark) slides. The fluorescent intensity of the spots was quantified by laser scanning and quantified using Optiquant™ software to obtain a standard curve for the DNA capture probes. Similar calibration curves for the Cy5-labeled target DNA were made; however, dilutions were between 5 nM and 2 µM. After immobilization and hybridization, the fluorescence signal of the spots was quantified and the amount of immobilized and hybridized DNA was extracted from the respective calibration curves.
For each experimental condition tested on the microarrays, 10 replica spots of each solution were spotted on the same substrate and the experiment was repeated three to five times. The immobilization and hybridization data presented are the average of these repetitions and the error bars represent the standard deviation observed on this average.
Thermal cycling stability
DNA probes immobilized through the glutaraldehyde cross-linker, were thermocycled under PCR-like conditions and submitted to 0, 2, 5, 10 and 20 cycles, using a Gene Frame (Abgene, Epsom, UK) on the top of the surface filled with PCR buffer and MgCl2 solution (Roche). After the thermocycling the slides were washed and hybridized as described above. The thermocycling was performed using a PTC-200 PCR Cycler (MJ Research, Kebolab, Roskilde, Denmark). Each thermal cycle consisted of a ‘denaturing step’ at 94°C for 30 s, an ‘annealing step’ at 54°C for 30 s and an ‘extension step’ at 72°C for 30 s.
RESULTS AND DISCUSSION
Surface modification
The most widely used substrate for DNA microarrays is glass whose chemical modification is well established (12). However, well developed chemistries on polymeric surfaces are still in their infancy. So far, PMMA has been modified by Bulmus et al. (26) and by Henry et al. (28) to yield aminated PMMA sheets. In the present work, we have developed a surface chemistry that yields primary amines on a PMMA surface, and compared with the previously described chemical methods is both simpler and faster (Fig. 1A and B).
A fluorescent-based technique was used to quantify the number of NH2 groups on PMMA slides after chemical modification. FITC is a fluorescent dye that is specifically reactive towards primary amines (30). The NH2 surface density was estimated from a standard curve (see Materials and Methods). The procedure presented here on PMMA (both for 1 and 2 h of reaction) resulted in a density of primary amine groups that was twice as high as for the reaction described by Bulmus et al. (26) (Table 1). The uniformity of primary amines on chemically treated slides was determined by immersion of a modified and non-modified PMMA surface into a 0.1 mg/ml FITC solution and by subsequently scanning the substrates. The fluorescence was homogeneous on activated PMMA sheets while the fluorescent signal was almost absent on non-modified PMMA surfaces (data not shown). The density of NH2 groups on the PMMA surface was four to five times lower than the values reported for silanization of glass or silica surfaces (32) and 20-fold lower than the values reported by others for the modification of PMMA (6.85 nmol/cm2) (29). However, the values of primary amines reported here are still in excess when compared with the density of closely packed ssDNA strands (150 pmol/cm2) oriented normal to the surface, considering a cross-sectional diameter of 20 Å for the ssDNA chain (20).
Table 1. Quantification of amino groups on chemically modified PMMA.
| Chemistry incubation conditions | Density (nmol NH2/cm2) | |
|---|---|---|
| A | 1 h | 0.28 ± 0.03 |
| 2 h | 0.29 ± 0.02 | |
| B | Aqueous medium | 0.16 ± 0.01 |
A, aminolysis reaction described in this paper; B, chemistry by Bulmus et al. (26).
DNA immobilization
The effect of immobilization kinetics, probe concentration, cross-linker types and chemical modifications procedures, on the density of DNA capture probes achieved on aminated PMMA sheets was investigated.
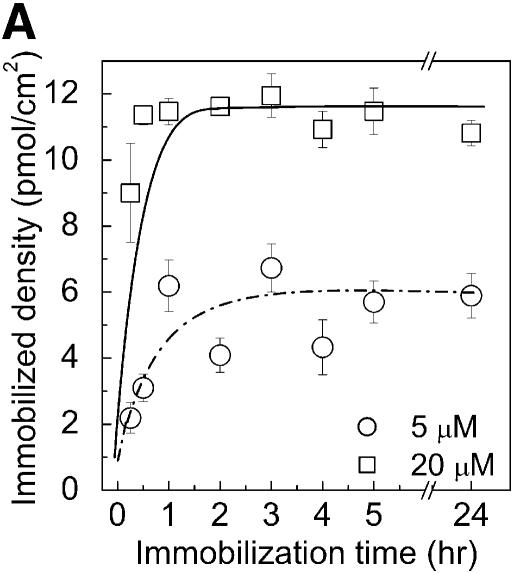
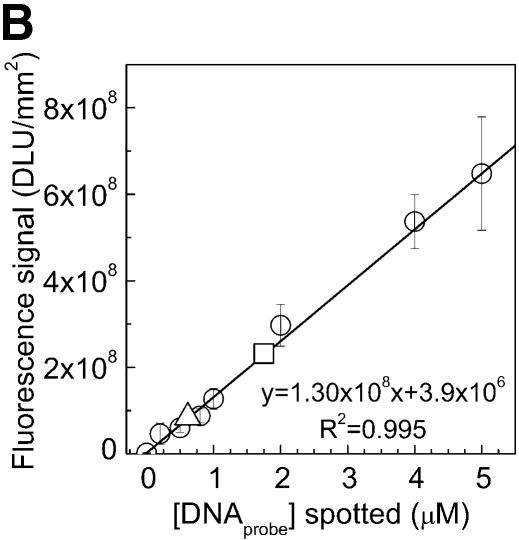
To find the conditions that resulted in the highest probe density on the surface, the kinetics of probe immobilization, for various probe concentrations, was investigated on aminated PMMA cross-linked with glutaraldehyde (Fig. 1C). The results showed that the attachment density reaches a maximum between 1 and 2 h of immobilization (Fig. 2A). The highest DNA capture probe density (12 ± 0.8 pmol/cm2) was obtained using a 20 µM DNA solution for spotting and 2 h of immobilizing time. One of the calibration curves derived from a dilution series of the aminated capture probes, on chemically aminated PMMA (according to Fig. 1A), is shown in Figure 2B. There is a strong linear correlation between spotted DNA concentration and obtained fluorescence in the standard curve.
Figure 2.
Immobilization kinetics of oligonucleotides onto chemically modified PMMA substrate (according to the protocol in Fig. 1A) using different probe concentrations and immobilization times (A). NH2-DNA oligo-probes were immobilized at concentrations of 5 and 20 µM. The density of immobilized DNA probes was obtained by scanning the surface after immobilization and reading the output fluorescent signal from each spot, and converted into a molecular density using a standard curve (circles) (B). Fluorescent NH2-DNA oligo probes were spotted in 0, 0.2, 0.5, 0.8, 1, 2, 4 and 5 µM concentrations and scanned directly without washing. The fluorescent signals obtained for NH2-DNA oligo probes immobilized for 2 h at 5 and 20 µM concentrations [from (A)] are represented in (B) by triangles (5 µM) and squares (20 µM), respectively. Both the calibration curve and the immobilization quantifications were done using the same settings for the laser power and photo multiplier tube gain (see Materials and Methods).
The chemical modification proposed by Bulmus et al. (26) and the one described here were evaluated by immobilization of aminated probes. The probes were attached for 2 h, on glutaraldehyde-activated surfaces with an initial spotted solution of 20 µM. Table 2 shows that the presented protocol resulted in approximately the same density of immobilized probes as the protocol described by Bulmus et al. (26).
Table 2. Surface density of immobilized aminated DNA probes and utilization of amine groups for immobilization on chemically modified PMMA and cross-linked with glutaraldehyde.
| Chemistry | Immobilized DNA (pmol/cm2) | 5′ Specifica (pmol/cm2) | Immobilizationb yield (%) | %NH2 usedc |
|---|---|---|---|---|
| A | 11.62 ± 0.75 | 9.75 | 8.7 | 3.4 |
| B | 10.23 ± 0.37 | 8.22 | 7.3 | 5.1 |
The probes were spotted in 20 µM concentration and immobilized for 2 h. A, aminolysis reaction described in this paper; B, chemistry by Bulmus et al. (26).
aThe 5′-end specificity is obtained by subtracting the density of immobilized probes without the NH2 group on the 5′ end to the value of immobilization density obtained with the NH2-DNA probe (Table 3).
bThe immobilization yield is the ratio between the covalently attached DNA probes and the initial DNA spotted.
cThe %NH2 used is defined as the fraction of primary amines on PMMA that were used for DNA immobilization.
To determine the contribution of the 5′-end reactive group of the DNA probes (NH2 or SH) to the immobilization density, PMMA substrates chemically modified by aminolysis were spotted with 20 µM solutions of DNA probes with or without NH2 or SH groups in the 5′ terminus (probe-Cy3, Table 3). The resulting densities for non-functionalized DNA probes were very similar: 2.0 pmol/cm2 for the PMMA aminated by the protocol described by Bulmus et al. (26) and 1.9 pmol/cm2 for the PMMA modified with the chemistry presented here. The 5′-end covalently immobilized DNA was calculated by subtracting these values to the attachment densities obtained after immobilization of DNA probes with the reactive amino and thiol groups on the 5′-end terminus resulting in ∼10 pmol/cm2 of specifically immobilized probes (Table 2).
Table 3. Sequence of oligonucleotides used.
| DNA probe | Sequence (5′–3′) | 5′ End | 3′ End |
|---|---|---|---|
| Target-Cy5 | CGT CAT CGG GCT TTG CAT | Cy5 | None |
| Probe-NH2 | ATG CAA AGC CCG ATG ACG | C6-NH2 | Cy3 |
| Probe-SH | ATG CAA AGC CCG ATG ACG | SH | Cy3 |
| Probe-Cy3 | ATG CAA AGC CCG ATG ACG | None | Cy3 |
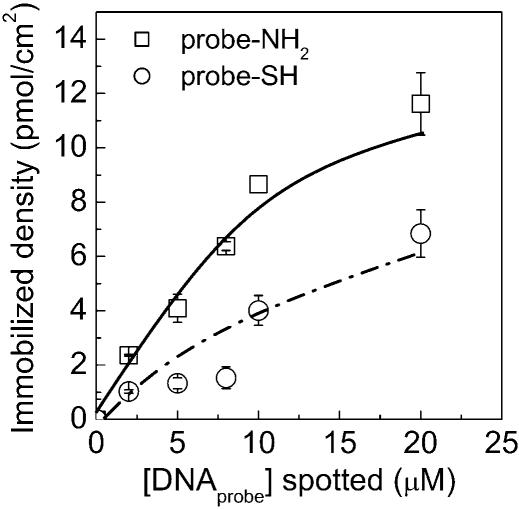
The effect of glutaraldehyde and sulfo-EMCS (Fig. 1D) cross-linkers on the immobilization density was evaluated. The immobilization density obtained was on average 2-fold higher for microarrays prepared with glutaraldehyde (homo-functional) as cross-linker, than with the sulfo-EMCS (hetero-functional) (Fig. 3). The drawback of glutaraldehyde as cross-linker was that the substrates must be used within hours after cross-linking. In contrast, PMMA modified with the sulfo-EMCS has a longer shelf-life (data not shown).
Figure 3.
Immobilized density of oligonucleotides on modified PMMA substrates using different cross-linkers and probe concentrations. The 5′-amino-labeled ssDNA probes (squares) or thiolated DNA probes (circles) were immobilized on PMMA surfaces cross-linked with glutaraldehyde or with sulfo-EMCS, respectively. The aminated PMMA was obtained by the chemistry described in Figure 1A and the probes were immobilized for 2 h.
DNA surface coverage increases with the increasing DNA concentration of the probe solution, reaching a plateau at 10 µM of spotted DNA (Fig. 3). As mentioned above, the theoretical coverage of a close-packed full monolayer of ssDNA is 150 pmol/cm2, considering that the DNA molecules are cylindrical having a 20 Å diameter and oriented perpendicular towards to the plane of the surface. The coverage, presented in Figure 3, corresponds to ∼7% of a DNA monolayer using amino-modified DNA probes and 3% for thiolated DNA probes, assuming that the maximum surface density of ssDNA is 150 pmol/cm2. The chemically aminated PMMA presented here, contains 0.29 nmol/cm2 of NH2 groups [or 0.16 nmol/cm2 for the PMMA modified as Bulmus et al. (26)], which assuming that the surface is flat results in spacing between adjacent amino groups of ∼4.3 Å (or 8 Å). This distance is smaller than the size of the cross-linkers used in this work (10.5 Å, glutaraldehyde and 18 Å, sulfo-EMCS) or the diameter of the DNA helix (∼18–20 Å). Hence, the cross-linker molecules cannot be attached to the surface at the full density of NH2 groups; consequently, the surface density of the immobilized molecules is determined by the dimension of the molecules and not the number of NH2 groups on the surface.
DNA hybridization
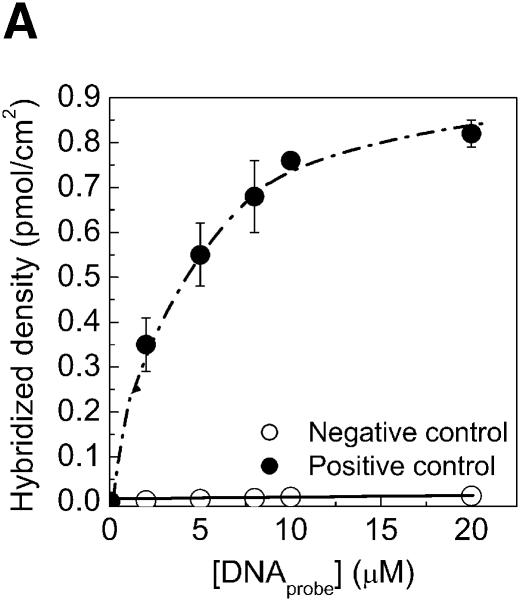
A major concern when preparing DNA microarrays is the accessibility and selectivity of the surface-bound probe for hybridization. The selectivity of duplex formation by the immobilized DNA on aminated PMMA slides, was demonstrated by comparing hybridization of a complementary and a non-complementary DNA strand to an immobilized NH2-DNA probe on the modified PMMA surface. Negligible fluorescence was obtained for hybridization with the non-complementary fluorescent DNA (Fig. 4A), demonstrating that unspecific hybridization did not occur. The signal-to-noise ratio was on average 90, calculated from the signal obtained from complementary hybridized DNA spots and the signal obtained from non-complementary hybridized spots. It should be mentioned that 0.2 µM DNA target (target-Cy5, Table 3) saturated the hybridization signal, since 0.5 and 1 µM DNA target solutions did not increase the hybridization signal.
Figure 4.
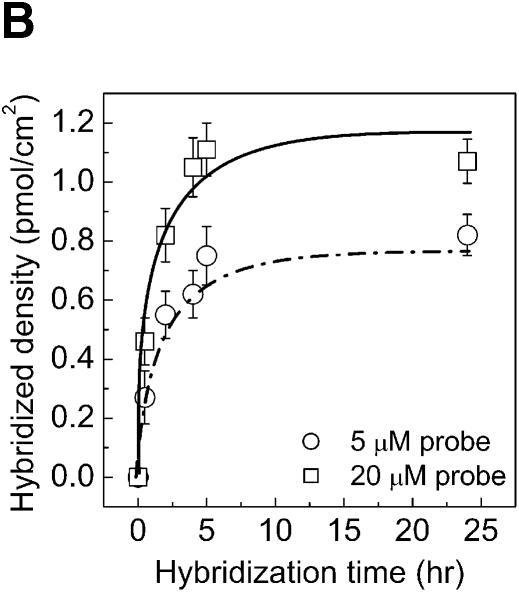
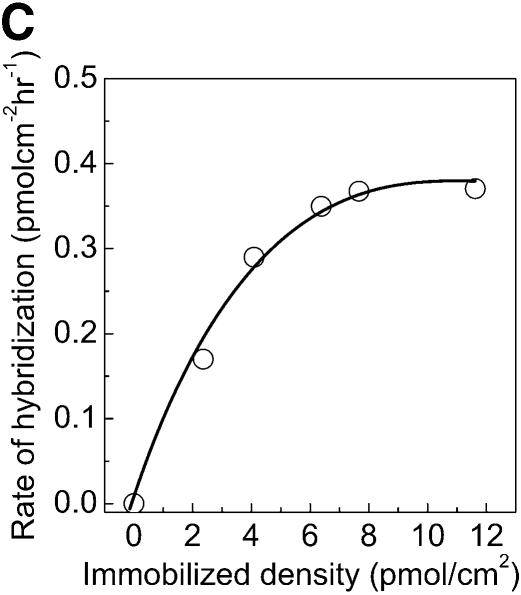
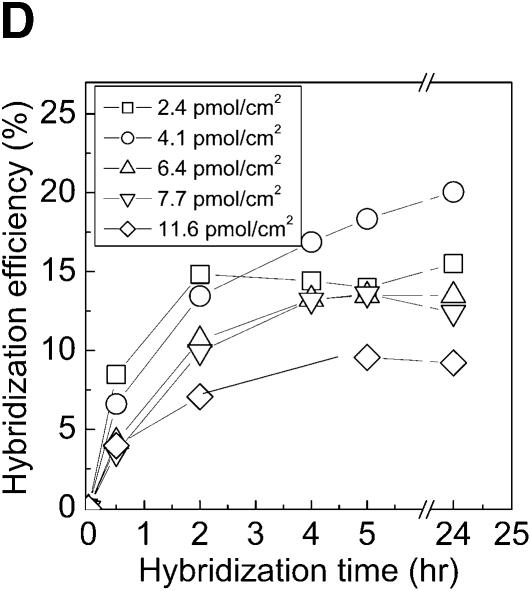
Specificity, kinetics and efficiency of hybridization on PMMA. The aminated PMMA was obtained by the chemistry presented in this report followed by cross-linking with glutaraldehyde. The 5′-aminated probes were immobilized for 2 h, washed and hybridized with 0.2 µM target solution for 2 h. (A) Specificity of DNA probes was tested by hybridization with a complementary (closed circles) and with a non-complementary (open circles) DNA target solution. (B) The kinetics of DNA target hybridization to micorarrays on PMMA. (C) Rate of hybridization for different densities of immobilized DNA probes on aminated PMMA slides. The rate of hybridization is determined from the linear part of the kinetic curves in (B). (D) Hybridization efficiency of amino-labeled DNA probes on PMMA when hybridized to a complementary strand (target-Cy5), varying the hybridization time and the amount of covalently immobilized probes. Efficiency of hybridization was calculated as the fraction of covalently immobilized DNA, which participated in duplex formation with the complementary DNA target.
To understand the dynamics of microarray hybridization, the kinetics of a hybridization reaction on a PMMA array was followed over 24 h. The signal intensity from hybridization proceeds linearly during the first 2 h (Fig. 4B). After this initial stage, the hybridization signal reaches a steady state between 5 and 24 h. Furthermore, the hybridization signal increases with the amount of spotted DNA probes on the surface (Fig. 4B).
The rate of hybridization can be examined as a function of the immobilized DNA density by fitting the data to the equation:
![]()
where V0 is the initial hybridization rate, Vmax is the maximum calculated hybridization rate, I is the density of immobilized DNA probes and K is the concentration of I where the hybridization rate is half of the maximum (33). By plotting the initial rate of hybridization (the linear part of the kinetic curves, Fig. 4B) for different amounts of immobilized capture probes it was possible to observe that the hybridization rate was dependent on the DNA concentration in the spot for low immobilization probe densities, but approaches a maximum above a density of 4–6 pmol/cm2 (Fig. 4C). This density is approximately similar to the 2 pmol/cm2 obtained previously on CMT-GAPS-coated glass slides (33).
However, our result is based on a probe that is 18 bp long while the result on the CMT-GAPS slides was obtained using a 716 bp long probe. The longer probe on CMT-GAPS slides could explain why this slide has a slightly lower density at which the maximum hybridization rate is obtained, since, presumably, a larger probe takes up more space on the surface resulting in a crowding effect at lower densities than a small oligonucleotide.
The surface densities determined for covalently attached DNA and hybridized DNA were used to calculate the percent of covalently attached probes that participated in duplex formation (defined here as hybridization efficiency). There was an optimal immobilized surface DNA density for which the hybridization efficiency reached a maximum 4 pmol/cm2 (which corresponds to a 5 µM spotted solution) and above this density the efficiency decreased with the amount of probes present on the surface (Fig. 4D). This optimal immobilization density is in agreement with the density obtained for achieving the maximum rate of hybridization at the surface. By performing these two types of data analysis (rate and efficiency of hybridization), it can be concluded that above an attachment density of 4 pmol/cm2, there is no gain in the percentage of hybrids formed on the PMMA surface. These results are also in agreement with previous studies (34), that found that the maximum hybridization efficiency on a gold surface was obtained for a 3 pmol/cm2 density of immobilized DNA. This effect is probably due to repulsive electrostatic and steric interactions that increase with the probe density.
The hybridization efficiency, for the aminolysis modified PMMA (according to Fig. 1A), is almost identical for the two types of spacers (15 and 18%, for glutaraldehyde and sulfo-EMCS, respectively) using the same amount of immobilized DNA probes on the surface (2.5 pmol/cm2).
The hybridization signals obtained on chemically modified PMMA slides were compared, under the same experimental conditions, to the hybridization signals obtained on PMMA modified according to Bulmus et al. (26), silanized glass slides, commercial silylated glass and commercial plastic Euray™ slides (Fig. 5A). The hybridization signals obtained were similar for hybridization to silanized glass and chemically modified PMMA. The percentage of immobilized probes involved in hybridization was strongly dependent on the amount of capture probes present at the surface, for all the substrates studied (Fig. 5B). Above 2–4 pmol/cm2 the yield of hybridization did not increase even if the density of immobilized probes increased. High surface coverage of ssDNA probes did not consequently lead to the formation of more hybrids, since overloading the surface with probes may cause a crowding effect that can lower the accessibility of the surface-bound probes (34). However, the maximum surface coverage for dsDNA on a surface has been determined to be 50 pmol/cm2 (35), which is 5-fold higher than the maximum number of probes immobilized on PMMA slides and on glass slides (10 pmol/cm2). The crowding effect should thus be small, but still the efficiency of hybridized probes was <20%, for all the substrates analyzed, meaning that besides the density of probes, other parameters should be taken into account, like the thermodynamic equilibrium between the immobilized probes and the complementary target DNA in solution, and the spatial orientation of the probes on the surface, after the immobilization.
Figure 5.
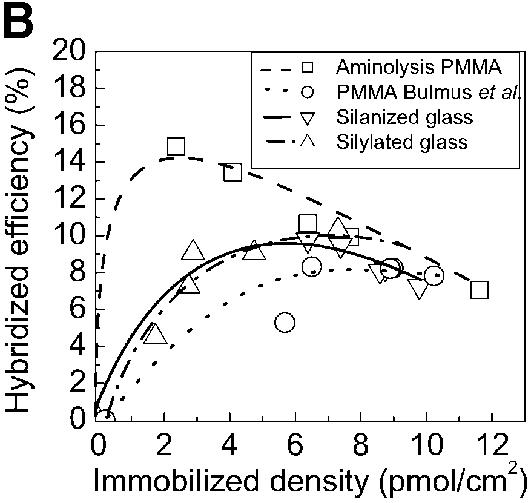
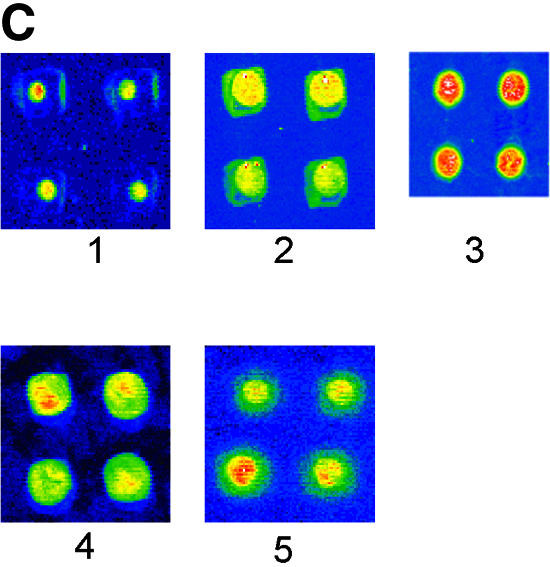
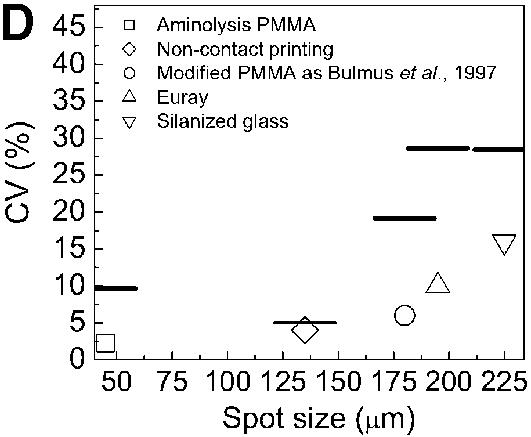
Comparison of hybridization signals obtained on different substrates and chemistries. The amino-labeled DNA probes were immobilized on glutaraldehyde-modified slides in the case of PMMA and silanized glass. On commercial slides (silylated glass and Euray™) the probes were directly reacted with the surface. Each slide type was spotted with 0, 2, 5, 10 and 20 µM of probe-NH2 and incubated for 2 h to immobilize the probes. The slides were subsequently hybridized with 0.2 µM of target-Cy5, for 2 h. (A) Hybridization densities and (B) efficiencies achieved on different substrates and with different chemical strategies to yield aminated reactive surfaces. (C) Spot morphology obtained after hybridization on PMMA modified by the method presented in this paper (1), on PMMA modified as in Bulmus et al. (26) (2), on silanized glass (3), on commercial plastic slides (Euray™) (4) and on a non-contact printed PMMA aminolysed substrate (5). (D) CV of spot sizes (symbols) and fluorescence intensity (lines) observed after hybridization on the substrates used in 5C (n = 3, 30 spots total).
The spot-to-spot variation was compared and evaluated with respect to spot size and fluorescence intensity after hybridization (Fig. 5C and D). On all the polymeric plastic substrates a ‘foot-print’ or halo was present around each spot (Fig. 5C), representing an outline of the pin printing tool, and probably due to differences in surface properties between the printing pin and the substrate surface. The spot size variation for the PMMA substrate modified by aminolysis was low [45 µm, coefficient of variance (CV) 2%], when compared with the variation observed for the other chemically treated substrates (180–225 µm, CV 5–17%), as shown in Figure 5D. Using a non-contact printing method (Packard Biochip Arrayer) uniform (135 µm, CV 5%) rounded spots were obtained (Fig. 5C), appearing without shrinking.
The fluorescence signal variation observed on aminolysed PMMA was 10%, while PMMA modified as described by Bulmus et al. (26), Euray™ slides and silanized glass substrates had an average variation between 20 and 30% (Fig. 5D). The PMMA background signal was similar to the glass substrates, but lower than the other modified PMMA and commercial plastic slides (data not shown).
The chemically aminated PMMA surfaces (protocol according to Fig. 1A) resulted in the highest hybridization efficiency after 5 h of hybridization for a probe DNA density of 4 pmol/cm2. For these conditions the hybridized density and the yield of hybridization were 0.75 pmol/cm2 and 18%, respectively. Considering that 0.29 nmol/cm2 of NH2 groups are present on the aminated surface after chemical modification, only 0.26% of these groups were used in hybridization. The efficiency of hybridization depends on (i) the number of methyl ester groups accessible in the PMMA, (ii) the efficiency of obtaining primary amino groups, (iii) the degree of derivatization with the cross-linker, (iv) the number of accessible aldehyde or maleimide groups to the modified DNA probes, (v) the attachment efficiency of oligonucleotides to the modified PMMA surfaces and (vi) the efficiency in the reaction of hybrid formation between immobilized ssDNA and target DNA. All these steps can lead to the low yields of hybridization observed. The values reported here for immobilization (9.75 pmol/cm2) of DNA probes on chemically modified PMMA sheets (according to Fig. 1A), is 300-fold higher than the values previously reported on PMMA (33.1 fmol/cm2) (29). Apparently, the procedure described by Waddell et al. (29) resulted in high amounts of NH2 on the surfaces (6.85 nmol/cm2); however, very few of these were involved in immobilization of DNA probes.
Heat stability
The heat stability of the immobilized DNA probes and the aminated PMMA slides was evaluated by subjecting the immobilized DNA probes to thermal cycling under PCR-like conditions for 2, 5, 10 and 20 cycles (as described in Materials and Methods). Afterwards, the slides were hybridized with the complementary DNA target and even after 20 cycles of PCR, the hybridization signal was only decreased by 4.3%. These results indicate that the probes are covalently immobilized to the plastic surface and that this type of chemistry and plastic polymer can be very useful in the development of new devices for different kinds of bioassays involving heat, e.g. PCR techniques.
CONCLUSIONS
In this paper a detailed study of immobilization and hybridization reactions on chemically modified PMMA surfaces was reported. The chemical reaction described in this paper to modify PMMA was simpler and faster than the procedures already described for PMMA modification. The density of immobilized and hybridized DNA to microarrays on chemically modified PMMA was similar to densities obtained on silanized glass and on PMMA chemically modified by others. Immobilized and hybridized densities of 10 and 0.75 pmol/cm2 were obtained for 2 h of immobilization (with 20 µM of spotted DNA probes) and 5 h of hybridization (with 0.2 µM of DNA target), respectively. Furthermore, the covalently immobilized oligonucleotides were stable to heat cycling suggesting that functions like PCR could be integrated in the same biochip.
The ability to robustly immobilize DNA and other biomolecules on plastic polymers, presents new possibilities in the development and fabrication of specialized lab- on-a-chip systems, including both sample preparation, amplification and detection.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Dr Weimin Rong for his assistance in surface chemistry and Jo Lazar for her assistance with the spotting robots. F. Fixe acknowledges a PhD grant from FCT (SFRH/BD/2000/3001). This work was supported by The Danish Research Council (grant 2014-00-0003, DABIC).
REFERENCES
- 1.Marshall A. and Hodgson,J. (1998) DNA chips: an array of possibilities. Nat. Biotechnol., 16, 27–31. [DOI] [PubMed] [Google Scholar]
- 2.Ramsay G. (1998) DNA chips: state-of-the art. Nat. Biotechnol., 16, 40–44. [DOI] [PubMed] [Google Scholar]
- 3.Gerhold D., Rushmore,T. and Caskey,C.T. (1999) DNA chips: promising toys become powerful tools. Trends Biochem. Sci., 24, 168–173. [DOI] [PubMed] [Google Scholar]
- 4.Southern E., Mir,K. and Shchepinov,M. (1999) Molecular interactions on micorarrays. Nature Genet., 21 (Suppl. 1), 5–9. [DOI] [PubMed] [Google Scholar]
- 5.Pease A.C., Solas,D., Sullivan,E.J., Cronin,M.T., Holmes,C.P. and Fodor,S.P. (1994) Light generated oligonucleotide arrays for rapid DNA sequence analysis. Proc. Natl Acad. Sci. USA, 91, 5022–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maskos U. and Southern,E.M. (1992) Oligonucleotide hybridisations on glass supports: a novel linker for oligonucleotide synthesis and hybridisation properties of oligonucleotides synthesised in situ. Nucleic Acids Res., 20, 1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Southern E.M., Case-Green,S.C., Elder,J.K., Johnson,M., Mir,K.U., Wang,L. and Williams,J.C. (1994) Arrays of complementary oligonucleotides for analysing the hybridisation behaviour of nucleic acids. Nucleic Acids Res., 22, 1368–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamoto T., Suzuki,T. and Yamamoto,N. (2000) Microarray fabrication with covalent attachment of DNA using bubble jet technology. Nat. Biotechnol., 18, 438–441. [DOI] [PubMed] [Google Scholar]
- 9.Gingeras T.R., Kwoh,D.Y. and Davis,G.R. (1987) Hybridization properties of immobilized nucleic acids. Nucleic Acids Res., 15, 5373–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Britcher L.G., Kehoe,D.C., Matisons,J.G., Smart,R.S.C. and Swincer,A.G. (1993) Silicones on glass surfaces, coupling agent analogs. Langmuir, 9, 1609–1613. [Google Scholar]
- 11.Beier M. and Hoheisel,J.D. (1999) Versatile derivatisation of solid support media for covalent bonding on DNA-microchips. Nucleic Acids Res., 27, 1970–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halliwell C. and Cass,A. (2001) A factorial analysis of silanisation conditions for the immobilisation of oligonucleotides on glass surfaces. Anal. Chem., 73, 2476–2483. [DOI] [PubMed] [Google Scholar]
- 13.Janolino V. and He,S. (1982) Analysis and optimisation of methods using water-soluble carbodiimide for immobilization of biochemicals to porous glass. Biotechnol. Bioeng., 24, 1069–1080. [DOI] [PubMed] [Google Scholar]
- 14.Joos B., Kuster,H. and Cone,R. (1997) Covalent attachment of hybridisable oligonucleotides to glass supports. Anal. Biochem., 247, 96–101. [DOI] [PubMed] [Google Scholar]
- 15.Rogers Y.H., Jiang-Baucom,P., Huang,Z.J., Bogdanov,V., Anderson,S. and Boyce-Jacino,M.T. (1999) Immobilization of oligonucleotides onto a glass support via disulfide bonds: a method for preparation of DNA microarrays. Anal. Biochem., 266, 23–30. [DOI] [PubMed] [Google Scholar]
- 16.Chrisey L.A., Lee,G.U. and O’Ferrall,C.E. (1996) Covalent attachment of synthetic DNA self-assembled monolayer films. Nucleic Acids Res., 24, 3031–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chrisey L.A., O’Ferrall,C.E., Spargo,B.J., Dulcey,C.S. and Calvert,J.M. (1996) Fabrication of patterned DNA surfaces. Nucleic Acids Res., 24, 3040–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strother T., Cai,W., Zhao,X., Hamers,R.J. and Smith,L.M. (2000) Synthesis and characterisation of DNA-modified silicon (111) surfaces. J. Am. Chem. Soc., 122, 1205–1209. [Google Scholar]
- 19.Csaki A., Moller,R., Straube,W., Kohler,J.M. and Fritzsche,W. (2001) DNA monolayer on gold substrates characterized by nanoparticle labeling and scanning force microscopy. Nucleic Acids Res., 29, e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steel A.B., Levicky,R.L., Herne,T.M. and Tarlov,M.J. (2000) Immobilization of nucleic acids at solid surfaces: effect of oligonucleotide length on layer assembly. Biophys. J., 79, 975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Ness J., Kalbfleisch,S., Petrie,C.R., Reed,M.W., Tabone,J.C. and Vermeulen,N.M. (1991) A versatile solid support system for oligodeoxynucleotide probe-based hybridisation assays. Nucleic Acids Res., 19, 3345–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Coyne,M.Y., Will,S.G., Levenson,C.H. and Kawasaki,E.S. (1991) Single-base mutational analysis of cancer and genetic diseases using membrane bound modified oligonucleotides. Nucleic Acids Res., 19, 3929–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Healey B.G., Matson,R.S. and Walt,D.R. (1997) Fiberoptic DNA sensor array capable of detecting point mutations. Anal. Biochem., 251, 270–279. [DOI] [PubMed] [Google Scholar]
- 24.Proudnikov D., Timofeev,E. and Mirzabekov,A. (1998) Immobilization of DNA in polyacrylamide gel for the manufacture of DNA and DNA-oligonucleotide microchips. Anal. Biochem., 259, 34–41. [DOI] [PubMed] [Google Scholar]
- 25.Nahar P., Wali,N.M. and Gandhi,R.P. (2001) Light induced activation of an inert surface for covalent immobilization of a protein ligand. Anal. Biochem., 294, 148–153. [DOI] [PubMed] [Google Scholar]
- 26.Bulmus V., Ayhan,H. and Piskin,E. (1997) Modified PMMA monosize microbeads for glucose oxidase immobilization. Chem. Eng. J., 65, 71–76. [Google Scholar]
- 27.Henry A.C. and McCarley,R.L. (2001) Selective deposition of metals on plastics used in the construction of microanalytical devices: photo-directed formation of metal features on PMMA. J. Phys. Chem. B, 105, 8755–8761. [Google Scholar]
- 28.Henry A.C., Tutt,T.J., Galloway,M., Davidson,Y.Y., McWhorter,C.S., Soper,S.A. and McCarley,R.L. (2000) Surface modification of poly(methyl methacrylate) used in the fabrication of microanalytical devices. Anal. Chem., 72, 5331–5337. [DOI] [PubMed] [Google Scholar]
- 29.Waddell E., Wang,Y., Stryjewski,W., McWhorter,S., Henry,A.C., Evans,D., McCarley,R.L. and Soper,S.A. (2000) High-resolution near-infrared imaging of DNA microarrays with time resolved acquisition of fluorescence lifetimes. Anal. Chem., 72, 5907–5917. [DOI] [PubMed] [Google Scholar]
- 30.Brown L., Plant,A., Durst,R. and Brizgys,M. (1990) Radiometric and fluorimetric determination of aminosilanes and protein covalently bound to thermally pretreated glass substrates. Anal. Chem. Acta, 228, 107–116. [Google Scholar]
- 31.Fixe F., Faber,A., Goncalves,D., Prazeres,D., Cabeca,R., Chu,V., Ferriera,G. and Conde,J.P. (2002) Thin film micro arrays with immobilized DNA for hybridization analysis. Mat. Res. Soc. Symp. Proc., 73, O2.3.1. [Google Scholar]
- 32.Moon J.H., Shin,J.W., Kim,S.Y. and Park,J.W. (1996) Formation of uniform aminosilane thin layers: an imine formation to measure relative surface density of the amine group. Langmuir, 12, 4621–4625. [Google Scholar]
- 33.Stillman B.A. and Tonkinson,J.L. (2001) Expression microarray hybridisation kinetics depend on length of the immobilized DNA but are independent of immobilization substrate. Anal. Biochem., 295, 149–157. [DOI] [PubMed] [Google Scholar]
- 34.Peterson A.W., Heaton,R.J. and Georgiadis,R.M. (2001) The effect of surface probe density on DNA hybridisation. Nucleic Acids Res., 29, 5163–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georgiadis R., Peterlinz,K.P. and Peterson,A.W. (2000) Quantitative measurements and modeling of kinetics in nucleic acid monolayer films using SPR spectroscopy. J. Am. Chem. Soc., 122, 3166–3173. [Google Scholar]