Abstract
In order to create a system in which two independent plasmids can be integrated into a mycobacterial chromosome, a mycobacterial plasmid was constructed containing the phage attachment site attP from the mycobacteriophage L5 genome and additionally containing the bacterial attachment site, attB. This plasmid will integrate into the mycobacterial chromosome via recombination of the plasmid-borne attP site with the chromosomal attB site in the presence of a mycobacterial vector carrying the L5 integrase (int) gene. The integrated plasmid has a plasmid-borne attB site that is preserved and will accept the integration of additional mycobacterial plasmids containing the L5 attP site. This system should be useful in the construction of novel mycobacterial strains. In particular, this system provides a method by which several recombinant antigens or reporter constructs can be sequentially inserted into a mycobacterial strain and subsequently tested.
INTRODUCTION
Integration-proficient vectors have previously been developed for use in mycobacteria (1). Mycobacteriophage L5 contains a phage attachment site, attP, that integrates into the bacterial attachment site, attB, by site-specific recombination via the phage integrase, int, and the mycobacterial integration host factor mIHF (2–9). Vectors containing no mycobacterial origin of replication and having attP and the integrase gene of D29 or L5 readily integrate in single copy into the mycobacterial chromosome (10). Situations may arise where it would be useful to integrate a plasmid into a bacterial chromosome and at a later time have the option to integrate an additional plasmid into the same locus of the chromosome. We describe the development of a system by which the bacterial attachment site, attB, is placed on a mycobacterial plasmid containing attP and is integrated into the Mycobacterium smegmatis chromosome. The integration, via site-specific recombination of attP into the chromosomal attB, destroys the attachment sites by creating attL and attR but results in the persistence of a functional plasmid-borne attB within the integrated plasmid. This site can be used in subsequent studies as the acceptor of additional integrating plasmids containing an attP site and provides a method to sequentially integrate two plasmids within the same locus of the chromosome.
MATERIALS AND METHODS
Bacterial strains and plasmids
Mycobacterium smegmetis strain mc(2)155 and Escherichia coli strain SH288 were used for all experiments (11). The L5 attP and integrase-containing plasmid pBS20 was used as the acceptor for the attB site. pBS20 was cut with EcoRI and XbaI. Eighty-seven base pair primers, BS47 and BS48, were created that contain the L5 attB site. When these primers are hybridized they create overhangs for XbaI and EcoRI. The primers were kinased, hybridized and ligated into the XbaI–EcoRI cut pBS20 resulting in pBS29. pMH94 was a kind gift from Graham Hatfull (1). pBS11 was created by removing the aph-containing HindIII fragment of pMH94 and inserting the hygromycin-resistance-containing PmeI–XbaI fragment from pJG1004 into the SmaI–XbaI sites. pBS33 and pBS37 were created by cutting out the integrase-containing PstI fragment of pBS11 and pBS29 respectively. PBluescriptint was obtained as a kind gift from Burkhard Springer (12).
Assays
Stability of the plasmids was tested by growing M.smegmatis containing either pBS29 and pBS11 or pBS33 and pBS37 in triplicate in 7H9 growth media for 20 generations in the absence of any antibiotic selection. The bacteria were then diluted into fresh 7H9 media and plated onto 7H10 agar plates to obtain single colonies. One hundred individual colonies each were then patched onto 7H10 agar containing hygromycin or 7H10 agar containing kanamycin. Colonies that did not grow had become sensitive to antibiotic.
Southern blot analysis
Chromosomal DNA was purified from M.smegmatis alone or containing pBS37 or pBS37 and pBS33. The chromosomal DNA was cut with MluI or PmlI and electrophoresed on a 0.7% agarose gel. The DNA was transferred to nitrocellulose and was then bound to a 32P-radiolabeled aph gene fragment. The bound aph was then removed and the chromosomal DNA was rebound to a 32P-radiolabeled hygromycin gene fragment.
RESULTS
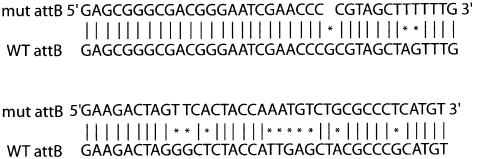
In order to create a system by which multiple plasmids can be integrated into the mycobacterial chromosome, we sought to place a bacterial attachment site, attB, onto the kanamycin-resistant integrating plasmid pBS20, creating pBS29. Two complementary 87 base pair primers were created that contain the attB sequence and hybridized to one another (6). The primers were hybridized, kinased and ligated into pBS20 cut with EcoRI/XbaI. The wild-type attB sequence was unable to be recovered as it was consistently rearranged in the E.coli acceptor cells. A mutated version of attB was obtained (Fig. 1) that can support site-specific recombination with attP. We hypothesize that the wild-type attB sequence of M.smegmatis is toxic for E.coli due to the presence of the mycobacterial tRNAgly within the attB site (3). The mutations in the altered attB site were within the sequence for tRNAgly (6).
Figure 1.
An attB site was cloned into pBS20 creating pBS29. A mutated version of the attB site was obtained. Sequence runs 5′ to 3′ left to right, top to bottom. Vertical bars denote bases that are identical. An asterix denotes bases that have been changed.
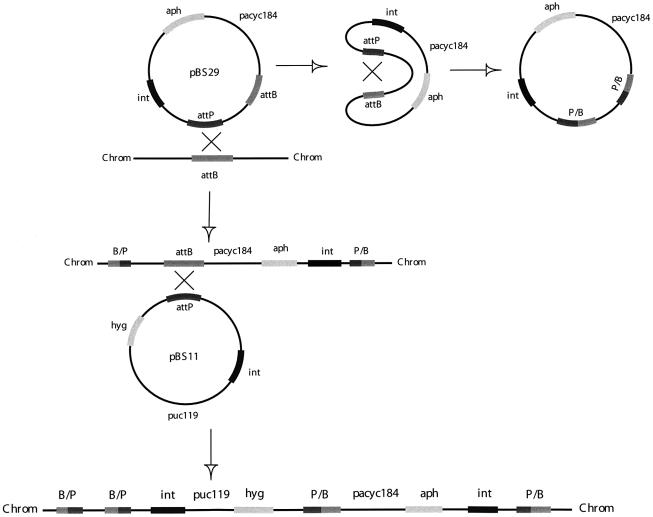
pBS29 was used to transform M.smegmatis. The plasmid attP could either recombine with the plasmid-borne attB, resulting in a vector incapable of integrating into the mycobacterial chromosome thus being lost from the cell, or could recombine with the chromosomal attB site, resulting in integration of the plasmid into the chromosome and preservation of a functional plasmid-borne attB (Fig. 2). pBS29 had a 10× lower transformation efficiency than parental pBS20 (5 × 103 c.f.u./µg versus 5 × 104 c.f.u./µg, respectively). This lower transformation efficiency was probably due to the above described competition between recombination of the attP site with the plasmid attB or chromosomal attB sites. The hygromycin-resistant pBS11, which contains the L5 attP and L5 int was then used to transform M.smegmatis carrying pBS29 integrated into the chromosome (Fig. 2). Transformation efficiency of pBS11 into M.smegmatis carrying pBS29 was only 5-fold less than into wild-type M.smegmatis (1 × 104 c.f.u./µg versus 5 × 104 c.f.u./µg, respectively), indicating the mutant attB serves well as an acceptor for attP integration. As a control, pBS11 and pNBV1 were transformed separately into M.smegmatis carrying pBS20. While pNBV1, an extrachromosomal plasmid carrying the hygromycin gene, had a transformation efficiency of 1 × 104 c.f.u./µg, transformation with pBS11 resulted in no detectable transformants. Therefore the transformation efficiency was <1 c.f.u./0.125 µg of DNA. Thus pBS11 has a low rate of spontaneous integration in the absence of a functional attB site. All transformations were performed in triplicate.
Figure 2.
Schematic representation of the site-specific recombination of the attP site of pBS37 with either the M.smegmatis chromosomal attB site with retention of the plasmid-borne attB site, or the site-specific recombination with the plasmid-borne attB site with the concomitant loss of the plasmid from the bacterium. Once pBS29 is integrated into the mycobacterial chromosome, pBS11 can undergo site-specific recombination of its attP site with the attB site of pBS29.
The stability of the vectors was investigated. Mycobacterium smegmatis containing both pBS29 and pBS11 was grown in 7H9 media without antibiotic selection for 20 generations and was then plated onto 7H10 plates without antibiotics. Bacteria were patched onto 7H10 plates containing kanamycin or 7H10 plates containing hygromycin to test for maintenance of the plasmids as evidenced by antibiotic resistance. Plasmids pBS29 and pBS11 were unstable in M.smegmatis as 28% of the bacteria became kanamycin sensitive and 8% became hygromycin sensitive. The bacteria that had become sensitive to kanamycin were analyzed for the presence of the kanamycin gene within the chromosome. Twenty colonies were tested by PCR using primers which amplify the aph gene. None of the colonies that were kanamycin sensitive had the aph gene amplify while those that were kanamycin resistant had the aph gene amplify (data not shown), indicating that the aph-containing pBS29 had been lost from the chromosome and that sensitivity to kanamycin was not due to a mutation in aph.
It has recently been found that the L5 integrase can act to excise integrated vectors at a low rate and contribute to plasmid instability (12–14). As first demonstrated by Lewis and Hatfull, placement of the integrase gene on a separate unlinked non-replicating plasmid could induce integration of plasmids carrying only the attP site while preventing low-level plasmid loss. To accomplish this int and attP can be placed on separate vectors. AttP can be placed on an integrating vector while int on a plasmid without a mycobacterial origin of replication. Co-transformation leads to extremely stable integration of the attP integration vector with subsequent loss of the int-containing vector (3,14). We removed the int-containing PstI fragments of pBS29 and pBS11 creating pBS37 and pBS33 respectively. pBS37 was subsequently co-transformed into M.smegmatis with a vector devoid of a mycobacterial origin of replication and containing the L5 integrase (pBluescriptint). The transformants were obtained and subsequently used for transformation with pBS33 and pBluescriptint. The resultant bacteria were tested for the stability of pBS33 and pBS37 in M.smegmatis by growth for 20 generations in 7H9 in the absence of antibiotic selection. In contrast to the previous work with pBS29 and pBS11, pBS33 and pBS37 were stably maintained with no detectable (<1%) loss of kanamycin or hygromycin resistance.
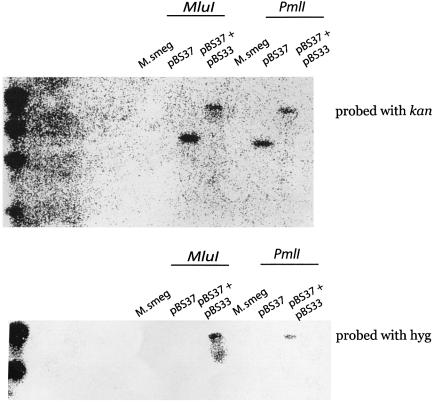
Further Southern blot analysis revealed that pBS33 is integrated into the same locus as pBS37 (Fig. 3). Chromosomal DNA was purified from M.smegmatis containing no plasmid, pBS37, or pBS33 and pBS37, and then cut with the restriction enzymes MluI or PmlI which do not cut within pBS33 or pBS37. The cut chromosomal DNA was electrophoresed on a 0.7% agarose gel, transferred to nitrocellulose, and bound to 32P-labeled DNA containing aph or hyg. Mycobacterium smegmatis does not contain the genes aph or hyg and hence does not bind the radiolabeled DNA fragments (Fig. 3). MluI and PmlI cut chromosomal DNA from M.smegmatis containing pBS37 had a fragment of ∼8.5 and 8.0 kb respectively that bound to the 32P-labeled aph fragment (Fig. 3). MluI and PmlI cut chromosomal DNA from M.smegmatis containing both pBS37 and pBS33 had chromosomal fragments that bound to both 32P-labeled aph and hyg that were significantly higher in molecular weight presumably because they contained the integrated pBS33 (Fig. 3).
Figure 3.
Mycobacterium smegmatis chromosomal DNA was cut with MluI or PmlI and run on a 0.7% agarose gel and transferred to nitrocellulose. The chromosomal DNA was then bound to 32P-labeled kan or hyg gene fragments.
Transformation frequencies of M.smegmatis with pBS37 and subsequently pBS33 are low due to the obligate use of the int on a second vector thus requiring two plasmids to enter the bacterium at once. This procedure, however, results in the extremely stable sequential integration of pBS37 and pBS33 into the mycobacterial chromosome.
DISCUSSION
It may be desirable to create recombinant mycobacterial strains that overexpress various antigens. In some cases it may be desirable to express more than one antigen. We have created a method to stably integrate more than one plasmid into the Mycobacterium tuberculosis chromosome. The first plasmid, containing an attP and an additional attB, was integrated into the chromosomal attB site with the aid of an int gene on a non-replicating vector. An additional plasmid containing an attP site was then integrated into the chromosome using the integrated plasmid-borne attB site and the int gene on a non-replicating vector. This system would make it feasible to construct and compare a pair of mycobacterial strains expressing one and two antigens, or one and two reporter constructs. We propose that this new method will be useful to researchers and aid in vaccine development.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Graham Hatfull for providing us with pMH94 and Burkhart Springer for providing us with pBluescriptint. This work was supported by a postdoctoral fellowship support to B.S. from the Heiser Foundation, a research grant to B.S. from the Potts Foundation, NIH grants to W.B AI37856 and AI43846 and an NIH grant to B.S. AI054794.
REFERENCES
- 1.Lee M.H., Pascopella,L., Jacobs,W.R. and Hatfull,G.F. (1991) Site specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis,Mycobacterium tuberculosis, and bacilli Calmette–Guerin. Proc. Natl Acad. Sci. USA, 88, 3111–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snapper S.B., Lugosi,L., Jekkel,A., Melton,R., Kieser,T., Bloom,B.R.and Jacobs,W.R.,Jr (1988) Lysogeny and transformation of Mycobacteria: stable expression of foreign genes. P roc. Natl Acad. Sci. USA, 85, 6987–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee M.L. and Hatfull,G.F. (1993) Mycobacteriophage L5 integrase-mediated site-specific integration in vitro. J. Bact., 175, 6836–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pena C.E.A., Kahlenburg,J.M. and Hatfull,G.F. (1998) The role of supercoiling in mycobacteriophage L5 integrative recombination. Nucleic Acids Res., 26, 4012–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pena C.E.A., Lee,M.H., Pedulla,M.L. and Hatfull,G.F. (1997) Characterization of the mycobacteriophage L5 attachment site, attP. J. Mol. Biol., 266, 76–92. [DOI] [PubMed] [Google Scholar]
- 6.Pena C.E.A., Stoner,J.E. and Hatfull,G.F. (1996) Positions of strand exchange in mycobacteriophage L5 integration and characterization of the attB site. J. Bact., 178, 5533–5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pena C.A., Kahlenberg,M. and Hatfull,G.F. (1999) Protein–DNA complexes in mycobacteriophage L5 integrative recombination. J. Bact., 181, 454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pedulla M.L., Lee,M.H., Lever,D.C. and Hatfull,G.F. (1996) A novel host factor for integration of mycobacteriophage L5. Proc. Natl Acad. Sci. USA, 93, 15411–15416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pena C.E.A., Kahlenberg,M. and Hatfull,G.F. (2000) Assembly and activation of site-specific recombination complexes. Proc. Natl Acad. Sci. USA, 97, 7760–7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribeiro G., Viveiros,M., David,H. and Costa,J.V. (1997) Mycobacteriophage D29 contains an integration system similar to that of the temperate mycobacteriophage L5. Microbiology, 143, 2701–2708. [DOI] [PubMed] [Google Scholar]
- 11.Snapper S.B., Melton,R.E., Mustafa,S., Kieser,T. and Jacobs,W.R.,Jr (1990) Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol., 4, 1911–1919. [DOI] [PubMed] [Google Scholar]
- 12.Springer B., Sander,P., Sedlacek,L., Ellrott,K. and Bottger,E.C. (2001) Instability and site-specific excision of integration-proficient mycobacteriophage L5 plasmids: development of stably maintained integrative vectors. Int. J. Med. Microbiol., 290, 669–675. [DOI] [PubMed] [Google Scholar]
- 13.Lewis J.A. and Hatfull,G.F. (2000) Identification and characterization of mycobacteriophage L5 excisionase. Mol. Microbiol., 35, 350–360. [DOI] [PubMed] [Google Scholar]
- 14.Pena C.E.A., Stoner,J. and Hatfull,G.F. (1998) Mycobacteriophage D29 integrase mediated recombination: specificity of mycobacteriophage integration. Gene, 225, 143–151. [DOI] [PubMed] [Google Scholar]