Abstract
In Saccharomyces cerevisiae, entry into mitosis requires activation of the cyclin-dependent kinase Cdc28 in its cyclin B (Clb)-associated form. Clb-bound Cdc28 is susceptible to inhibitory tyrosine phosphorylation by Swe1 protein kinase. Swe1 is itself negatively regulated by Hsl1, a Nim1-related protein kinase, and by Hsl7, a presumptive protein-arginine methyltransferase. In vivo all three proteins localize to the bud neck in a septin-dependent manner, consistent with our previous proposal that formation of Hsl1-Hsl7-Swe1 complexes constitutes a checkpoint that monitors septin assembly. We show here that Hsl7 is phosphorylated by Hsl1 in immune-complex kinase assays and can physically associate in vitro with either Hsl1 or Swe1 in the absence of any other yeast proteins. With the use of both the two-hybrid method and in vitro binding assays, we found that Hsl7 contains distinct binding sites for Hsl1 and Swe1. A differential interaction trap approach was used to isolate four single-site substitution mutations in Hsl7, which cluster within a discrete region of its N-terminal domain, that are specifically defective in binding Hsl1. When expressed in hsl7Δ cells, each of these Hsl7 point mutants is unable to localize at the bud neck and cannot mediate down-regulation of Swe1, but retains other functions of Hsl7, including oligomerization and association with Swe1. GFP-fusions of these Hsl1-binding defective Hsl7 proteins localize as a bright perinuclear dot, but never localize to the bud neck; likewise, in hsl1Δ cells, a GFP-fusion to wild-type Hsl7 or native Hsl7 localizes to this dot. Cell synchronization studies showed that, normally, Hsl7 localizes to the dot, but only in cells in the G1 phase of the cell cycle. Immunofluorescence analysis and immunoelectron microscopy established that the dot corresponds to the outer plaque of the spindle pole body (SPB). These data demonstrate that association between Hsl1 and Hsl7 at the bud neck is required to alleviate Swe1-imposed G2-M delay. Hsl7 localization at the SPB during G1 may play some additional role in fine-tuning the coordination between nuclear and cortical events before mitosis.
INTRODUCTION
Successful division of a eukaryotic cell requires that the events of the cell cycle be properly integrated, both temporally and spatially. To ensure accurate coordination of these processes, eukaryotic cells use specific mechanisms, known as checkpoint pathways, that monitor proper completion of each stage of the cell cycle and can pause cell cycle progression, when necessary, to allow for execution of an unfinished step, correction of errors, or repair of damage. In budding yeast, Saccharomyces cerevisiae, checkpoint pathways have been described that delay or block mitosis in response to defects in DNA replication or damage to DNA (Longhese et al., 1998; Rhind and Russell, 1998) or in response to defects in spindle assembly or dynamics (Amon, 1999; Burke, 2000). In addition to proper replication and segregation of chromosomes, cell division in S. cerevisiae also requires formation and enlargement of the bud (Madden and Snyder, 1998; Chant, 1999). Bud emergence begins early in the cell cycle and bud growth continues until the incipient daughter cell is of the correct size and shape to accept its share of the duplicated chromosomes and its apportionment of organelles and other cellular contents during mitosis. Bud growth, and the cell cycle itself, terminates upon cytokinesis (Balasubramanian et al., 2000; Lippincott and Li, 2000). Although bud formation, bud development, and septation require the actin cytoskeleton, these processes also rely on assembly of a second cytoskeletal structure: the septin filaments (Trimble, 1999). Defects or perturbations in either actin or septin assembly cause a G2 delay (McMillan et al., 1998; Barral et al., 1999), and recent studies provide further evidence for checkpoint mechanisms that monitor assembly of the actin cytoskeleton and the septin filaments (Shulewitz et al., 1999; Lew, 2000).
The septins, a conserved family of GTP-binding proteins (Cooper and Kiehart, 1996; Field and Kellogg, 1999), assemble into cytoplasmic 10-nm filaments that immediately subtend the plasma membrane at the neck between a bud and its mother cell (Byers and Goetsch, 1976) and are required for both cytokinesis and maintenance of proper bud shape (Hartwell, 1971; Longtine et al., 1996; Barral et al., 2000). The septin filaments of mitotic cells are composed of five gene products: Cdc3, Cdc10, Cdc11, Cdc12, and Sep7/Shs1 (Frazier et al., 1998; Mino et al., 1998). As observed by immunofluorescence, the septin filaments appear as an hourglass-like or double-ring structure spanning the isthmus between a mother cell and its bud. The requirement for septin function in cytokinesis is not well understood, but it has been proposed that the septin filaments serve as a scaffold for recruitment and/or organization of other components that play a more direct role in septation (DeMarini et al., 1997; Lippincott and Li, 1998).
In S. cerevisiae, major events of the cell cycle are initiated by the cyclin-dependent protein kinase (CDK) Cdc28. Entry into mitosis requires association of Cdc28 with B-type cyclins (Clb1, Clb2, Clb3, and Clb4) (Fitch et al., 1992; Nasmyth, 1993). Clb-bound Cdc28 is susceptible to inhibitory phosphorylation on a conserved residue (Tyr 19) in its ATP-binding loop by another protein kinase, Swe1. Swe1-mediated Tyr phosphorylation of Cdc28-Clb complexes blocks entry into mitosis (Booher et al., 1993) and must be reversed by the phosphoprotein phosphatase Mih1 (Russell et al., 1989). Because the G2 delay provoked by perturbation of the septin checkpoint is eliminated by the absence of Swe1 (Barral et al., 1999) or by substitution of normal Cdc28 by a Cdc28(Y19F) variant (Shulewitz et al., 1999), the sole cause of the G2 delay appears to be inhibition of Cdc28-Clb complexes by Swe1-dependent phosphorylation at Tyr 19.
Swe1 (819 residues) is subject to negative regulation by two proteins, Hsl1 and Hsl7 (Ma et al., 1996). Hsl1 (1518 residues) is a protein kinase, whose catalytic domain is homologous to Schizosaccharomyces pombe Nim1 (Russell and Nurse, 1987). Hsl7 (827 residues) contains a central region similar to catalytic domains of known S-adenosylmethionine-dependent protein-arginine methyltransferases (Pollack et al., 1999; Ma, 2000) and purportedly possesses this activity (Frankel and Clarke, 2000; Lee et al., 2000). We have shown previously that Hsl7 interacts with both Hsl1 and Swe1, as judged by the two-hybrid method and by coimmunoprecipitation from cell extracts (Shulewitz et al., 1999), suggesting that these proteins function in a complex. This conclusion is supported by the fact that Hsl7 and Hsl1 colocalize at the bud neck during most of the cell cycle and require septin function for this localization (Barral et al., 1999; Shulewitz et al., 1999; Longtine et al., 2000). Septins can be coimmunoprecipitated by Hsl1, but not by Hsl7 (Barral et al., 1999; Shulewitz et al., 1999), suggesting that Hsl1 associates directly with the septin filaments and acts as a tether to localize Hsl7. In agreement with this conclusion, localization of Hsl7 to the bud neck is dependent upon Hsl1, whereas Hsl1 localizes to the bud neck even in the absence of Hsl7 (Barral et al., 1999; Shulewitz et al., 1999; Longtine et al., 2000). In turn, efficient accumulation of Swe1 at the neck is reportedly dependent upon Hsl1 and Hsl7 (Longtine et al., 2000). Targeting of Swe1 for modification (Shulewitz et al., 1999) and its subsequent ubiquitin-mediated degradation (Kaiser et al., 1998) require Hsl1 and Hsl7 (McMillan et al., 1999). Collectively, these observations suggest that assembled septins serve as a platform for formation of Hsl1-Hsl7 complexes, which, in turn, mediate the inactivation and destruction of Swe1, thereby alleviating inhibition of Cdc28-Clb complexes and permitting efficient entry into mitosis (Shulewitz et al., 1999).
Interestingly, it has been reported (Lim et al., 1996) that overexpression of SWE1, or a Cdc28 mutant, Cdc28(Y19E), that presumably mimics permanent Swe1-dependent Tyr phosphorylation prevents separation and/or migration of duplicated spindle pole bodies (SPBs), an event that is a necessary prelude to formation of a short premitotic spindle and that normally occurs in synchrony with the switch from polarized to isotopic bud growth (Lew and Reed, 1993). This observation suggests that, in addition to regulation of Cdc28-Clb function for proper timing of the entry into mitosis, Swe1-dependent control of Cdc28-Clb may also be involved in coordinating spindle dynamics at a premitotic stage.
To gain further understanding into how Hsl1 and Hsl7 action contribute to down-regulation of Swe1, we mapped the region of Hsl7 that mediates its interaction with Hsl1 and examined the effect of mutations in this region on the subcellular localization and function of Hsl7. The behavior of these mutants led to our discovery that Hsl7 localizes to the SPB during early stages of the cell cycle before becoming redistributed to the bud neck. This dynamic movement suggests that Hsl7 may participate in localized depletion of Swe1 (or additional targets) in the vicinity of the SPB well in advance of the Hsl1-Hsl7 promoted- and septin-dependent destruction of Swe1 that precedes the entry into mitosis.
MATERIALS AND METHODS
Strains and Growth Conditions
Yeast strains used in this study are listed in Table 1. Standard rich (YP) and defined minimal (SC) media (Sherman et al., 1986), containing either 2% glucose (Glc), 2% raffinose (Raf), or 2% galactose (Gal) as the carbon source and supplemented with appropriate nutrients to maintain selection for plasmids, were used for yeast cultivation. To impose pheromone-induced G1 arrest, MATa haploids carrying a bar1 allele (sst1–3), which enhances sensitivity to α-factor, were treated with 50 ng/ml (final concentration) α-factor for 3 h at 28°C. Latrunculin A (Molecular Probes, Eugene, OR), cycloheximide (Calbiochem, San Diego, CA), and Benomyl (DuPont, Wilmington, DE) were used at the concentrations indicated.
Table 1.
Saccharomyces cerevisiae strains
| Strain | Genotype | Reference or source |
|---|---|---|
| MJY101 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 lys2Δ∷hisG trp1-1 ura3-1 | Shulewitz et al. (1999) |
| MJY102 | MATa ADE2 can1-100 his3-11,15 leu2-3,112 LYS2 trp1-1 ura3-1 hsl7-Δ20(∷HIS3) | Shulewitz et al. (1999) |
| MJY110 | MATα ADE2 can1-100 his3-11,15 leu2-3,112 lys2Δ∷hisG trp1-1 ura3-1 hsl7-Δ20(∷HIS3) | Shulewitz et al. (1999) |
| MJY112 | MATa ADE2 can1-100 his3-11,15 leu2-3,112 LYS2 trp1-1 ura3-1 | Shulewitz et al. (1999) |
| BJ2168 | MATa leu2 trp1 ura3-52 prb1-1122 pep4-3 prc1-407 gal2 | Jones (1991) |
| MJY153a | BJ2168 hsl1-Δ1(∷URA3) | This study |
| MJY155b | MATa ade2 his3 his6 leu2 lys2 trp1 ura3 sst1-3 | Shulewitz et al. (1999) |
| YD116 | MATa ade2-101oc can1 gal4-542 gal80-538 his3-Δ200 leu2-3,112 lys2-801am trp1-901 ura3-52 ADE2∷GAL1-URA3 LYS2∷UASGAL-lacZ | Durfee et al. (1999) |
| YD119 | MATα (otherwise isogenic to YD116) | Durfee et al. (1999) |
| CWY78 | MATa ade1 his2 leu2 trp1 cdc28-4 | Lorincz and Reed (1986) |
| Y543 | MATα ade2-1 can1-100 his3-11,15 his6 trp1-1 ura3-1 cdc4-1 | T. Durfee |
| YSS19 | MATa ade2-101oc his3-Δ200 leu2Δ1 lys2-801am ura3-52 cdc34-1 | S. Salama, this laboratory |
| VBY610c | MATa ADE2+can1-100 his3-11,15 LEU2+trp1-1 ura3-1 cdc4-1 hsl7-Δ20(∷HIS3) | This study |
| MAY1 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 ura3-1 hsl1-Δ1(∷URA3) | Ma et al. (1996) |
| MJY151d | MATa can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hsl7-Δ20(∷HIS3) swe1-Δ1(∷LEU2) | This study |
| MJY142e | MATα can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hsl1-Δ1(∷URA3) | This study |
| VBY16f | MATα can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hsl7-Δ20(∷HIS3) swe1Δ(∷LEU2) | This study |
| MY3883 | MATa ade2-101oc his3-Δ200 leu2-3,112 ura3-52 cdc31-1 | Sullivan et al. (1998) |
| VBY3113ag | MATa ADE2+his3 leu2-3,112 trp1-1 ura3 hsl7-Δ20(∷HIS3) cdc31-1 | This study |
| HNY21 | MATa ade2 his3 leu2 trp1 ura3 rho1-104 | Y. Takai |
| SLJ139 | MATa ade2 his3 leu2 trp1 ura3 bar1/sst1 cdc16-1 | D. Morgan |
| JC305 | MATa his3 leu2 ura3 bar1/sst1 cdc23-1 | D. Morgan |
| DBY0213 | MATa his4-539 lys2-801am ura3-52 tub2-401 | T. Huffaker |
| VBY4012ah | MATa lys2 trp1-1 ura3 | This study |
| VBY4012ch | MATa lys2 trp1-1 ura3 tub2-401 | This study |
| YSS41 | MATa ade2-101oc leu2-Δ1 ura3-52 cdc15-1 | S. Salama, this laboratory |
| VBY17f | MATa can1-100 his3-11,15 leu2-3-112 trp1-1 ura3-1 hsl1-Δ1(∷URA3) swe1Δ(∷LEU2) | This study |
| VCY1 | MATα can1 his4 leu2-3,112 trp1-1 ura3-52 cdc10-11 | Cid et al. (1998) |
| VBY30i | MATα can1 leu2-3,112 trp1-1 ura3 cdc10-11 | This study |
| VBY31i | MATα can1 leu2-3,112 trp1-1 ura3 cdc10-11 swe1-Δ1(∷LEU2) | This study |
| VBY204j | MATa can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 TRP1∷hsl1(R828A, L831A, N836A)-(HA)3 | This study |
Derived from BJ218 by transformation with DNA containing the hsl1-Δ1(∷URA3) construct (Ma et al., 1996).
In a previous publication (Shulewitz et al., 1999), the fact that this strain is ade2 was omitted from its genotype.
Segregant from MJY102 × Y543.
Derived from MJY102 by transformation with DNA containing the swe1-Δ1(∷LEU2) construct (Booher et al., 1993).
Segregant from MJY110 × MAY1.
Segregant from MJY142 × MJY151.
Segregant from VBY16 × MY3883.
Segregant from DBY0213 × MJY101.
Segregant from VCY1 × VBY17.
Derived from MJY112 by transformation with YIplac204-HSL1-HAmdb (Burton and Solomon, 2000).
Plasmids and Recombinant DNA Methods
Plasmids used in this study are listed in Table 2. These plasmids were constructed according to standard procedures (Sambrook et al., 1989) with the use of Escherichia coli DH5α (Hanahan, 1983) for plasmid propagation. For all DNA amplifications using the polymerase chain reaction (PCR), either PfuI DNA polymerase (Stratagene, La Jolla, CA) or Turbo PfuI DNA polymerase (Stratagene) was used, unless otherwise noted.
Table 2.
Plasmids used in this study
| Name | Description | Source |
|---|---|---|
| pGEM-5Z | In vitro transcription vector | Promega |
| pBAT4 | In vitro transcription vector | Peranen et al. (1996) |
| pGEX-4T-1 | GST expression vector (E. coli) | Amersham Pharmacia Biotech |
| pGEX-3X | GST expression vector (E. coli) | Amersham Pharmacia Biotech |
| YEpLG-GST | 2 μm LEU2 GAL1 promoter GST expression vector (yeast) | Shulewitz et al. (1999) |
| pACT | Two-hybrid Gal4(TAD) expression vector | Durfee et al. (1993) |
| YCplac111 | CEN LEU2 | Gietz et al. (1988) |
| YCplac22 | CEN TRP1 | Gietz et al. (1988) |
| YCpUG | CEN URA3 GAL1 | Bardwell et al. (1998) |
| YCpLG | CEN LEU2 GAL1 | Bardwell et al. (1998) |
| pGEM-5Z-HSL7 | pGEM-5Z +HSL7 | This study |
| pBAT4-HSL7(1–246) | pBAT4 +HSL7(1–246) | This study |
| pBAT4-HSL7(88–544) | pBAT4 +HSL7(88–544) | This study |
| pBAT4-HSL7(168–345) | pBAT4 +HSL7(168–345) | This study |
| pBAT4-HSL7(316–636) | pBAT4 +HSL7(316–636) | This study |
| pGEX-HSL7 | pGEX-4T-1 +HSL7 | This study |
| YEpLG-GST-HSL7(1–685) | YEpLG-GST +HSL7(1–685) | This study |
| YEpLG-GST-HSL7(1–246) | YEpLG-GST +HSL7(1–246) | This study |
| YEpLG-GST-HSL7(316–636) | YEpLG-GST +HSL7(316–636) | This study |
| pGEX-HSL7(674–827) | pGEX-4T-1 +HSL7(674–827) | This study |
| pGEX-HSL7(674–736) | pGEX-4T-1 +HSL7(674–736) | This study |
| pGEX-HSL7(737–827) | pGEX-4T-1 +HSL7(737–827) | This study |
| pGEX-HSL7(771–827) | pGEX-4T-1 +HSL7(771–827) | This study |
| pGEX-SWE1 | pGEX-3X +SWE1 | This study |
| pGEX-SWE1(284–370) | pGEX-4T-1 +SWE1(284–370) | This study |
| pGEX-HSL1ΔN | pGEX-4T-1 +HSL1(833–1518) | This study |
| pGEX-HSL1ΔNΔC | pGEX-4T-1 +HSL1(1018–1244; 1482–1518) | This study |
| YCpT-ADHp-GAL4(DBD)-HSL7 | Gal4(DBD)-Hsl7 production from ADH1 promoter on a CEN TRP1 plasmid | This study |
| YCpT-ADHp-GAL4(DBD)-HSL7(Δ224–392) | Gal4(DBD)-Hsl7(Δ224–392) production from ADH1 promoter on a CEN TRP1 plasmid | This study |
| YCpT-ADHp-GAL4(DBD)-HSL7(352–827) | Gal4(DBD)-Hsl7(352–827) production from ADH1 promoter on a CEN TRP1 plasmid | This study |
| YCpT-ADHp-GAL4(DBD)-HSL7(224–827) | Gal4(DBD)-Hsl7(224–827) production from ADH1 promoter on a CEN TRP1 plasmid | This study |
| YCpT-ADHp-GAL4(DBD)-HSL7(284–827) | Gal4(DBD)-Hsl7(284–827) production from ADH1 promoter on CEN TRP1 plasmid | This study |
| YCpT-ADHp-GAL4(DBD)-HSL7(1–533) | Gal4(DBD)-Hsl7(1–533) production from ADH1 promoter on CEN TRP1 plasmid | This study |
| pACT-HSL1(987–1518) | pACT +HSL1(987–1518) | Shulewitz et al. (1999) |
| pACT-SWE1(295–819) | pACT +SWE1(295–819) | Shulewitz et al. (1999) |
| YCpL-HSL7 | YCplac111 +HSL7 | This study |
| YCpL-HSL7(V251A) | YCplac111 +HSL7(V251A) | This study |
| YCpL-HSL7(P250Y) | YCplac111 +HSL7(P250Y) | This study |
| YCpL-HSL7(K254E) | YCplac111 +HSL7(K254E) | This study |
| YCpL-HSL7(F242L) | YCplac111 +HSL7(F242L) | This study |
| YCpUG-HSL7 | YCpUG +HSL7 | Shulewitz et al. (1999) |
| YCpUG-HSL7-Myc | YCpUG +HSL7-Myc | Shulewitz et al. (1999) |
| YCpT-GFP-HSL7 | YCplac22 +GFP-HSL7 | Shulewitz et al. (1999) |
| YCpLG-GFP-HSL7 | YCpLG +GFP-HSL7 | Shulewitz et al. (1999) |
| YCpT-GFP-HSL7(V251A) | YCplac22 +GFP-HSL7(V251A) | This study |
| YCpT-GFP-HSL7(P250Y) | YCplac22 +GFP-HSL7(P250Y) | This study |
| YCpT-GFP-HSL7(K254E) | YCplac22 +GFP-HSL7(K254E) | This study |
| YCpT-GFP-HSL7(F242L) | YCplac22 +GFP-HSL7(F242L) | This study |
| YCpT-GFP-HSL7(1–685) | YCplac22 +HSL7(1–685) | This study |
| YCpLG-HSL1(HA)3 | YCpLG +HSL1(HA)3 | Shulewitz et al. (1999) |
| YCpLG-HSL1-K110R(HA)3 | YCpLG +HSL1-K110R(HA)3 | This study |
To express an in-frame fusion of Hsl7 to the Gal4 DNA-binding domain [Gal4(DBD)] from a low copy (CEN) plasmid, a 1.2-kb EcoRV-NsiI fragment was excised from pAS1-HSL7 (Shulewitz et al., 1999) and inserted into YCpT-HSL7 (Shulewitz et al., 1999) that had been cleaved with SmaI and NsiI, yielding YCpT-ADHp-GAL4(DBD)-HSL7.
A C-terminally truncated version of Hsl7 (residues 1–685), fused to the C terminus of the green fluorescent protein (GFP) and expressed from the native HSL7 promoter on a CEN plasmid, was constructed as follows. First, PCR with appropriate primers and GFP(F64L S65T)-Hsl7 DNA (Shulewitz et al., 1999) as the template was used to generate a fragment containing at its 5′ end the sequence 5′-CTG CAG AAA GGA-3′ (PstI site is underlined) and at its 3′ end the sequence 5′-ATG TTG TAA TCT AGA-3′ (XbaI site is underlined; stop codon in bold). Second, this PCR product was cleaved with PstI and XbaI and ligated into plasmid YCpT-HSL7 (Shulewitz et al., 1999) that was cleaved with NsiI and XbaI (in YCpT-HSL7, a naturally occurring NsiI cleavage site is present at codon 2 of the HSL7 coding sequence and a naturally occurring XbaI cleavage site is present between codons 746 and 747). This operation resulted in a NsiI/PstI hybrid junction that reconstructed the initiator codon for GFP, but simultaneously eliminated the NsiI site at this position in GFP. The resulting plasmid, which retains the NsiI site at the junction between the GFP and HSL7 coding sequence, was designated YCpT-GFP-HSL7(1–685). To produce plasmids expressing each of the Hsl1-binding-defective hsl7 alleles as GFP-fusions, the NsiI-XbaI fragment from each mutant DNA was excised from the corresponding YCpT-ADHp-GAL4(DBD)-HSL7 derivative and inserted into YCpT-GFP-HSL7(1–685) in place of the sequence encoding Hsl7(1–685), which had been excised by digestion with NsiI and XbaI. The resulting plasmids (in which the intact HSL7 open reading frame was reconstructed) were designated YCpT-GFP-HSL7(V251A), YCpT-GFP-HSL7(P250Y), YCpT-GFP-HSL7(K254E), and YCpT-GFP-HSL7(F242L). To produce plasmids expressing each of the same hsl7 alleles from the HSL7 promoter on a CEN plasmid, a 0.6-kb BamHI-EcoRV fragment excised from YCpT-HSL7 (Shulewitz et al., 1999) and a 3.0-kb EcoRV-HindIII fragment of each mutant DNA, excised from the corresponding YCpT-ADHp-GAL4(DBD)-HSL7 isolate, were ligated together into YCplac111 (Gietz and Sugnino, 1988) cleaved with BamHI and HindIII. The resulting plasmids were designated YCpL-HSL7(V251A), YCpL-HSL7(P250Y), YCpL-HSL7(K254E), and YCpL-HSL7(F242L). To express wild-type HSL7 from the same plasmid (CEN LEU2), a 3.6-kb BamHI-HindIII fragment excised from YCpT-HSL7 was ligated into the corresponding sites in YCplac111, yielding YCpL-HSL7.
To express residues 833–1518 of Hsl1 as a fusion to glutathione S-transferase (GST) in Escherichia coli, first, a 3.3-kb StuI-SacI fragment was excised from pE14R1 (Ma et al., 1996) and inserted into the vector Litmus28 (New England Biolabs, Beverly, MA) that was cleaved with StuI and SacI. A 2.3-kb EcoRV fragment excised from the resulting plasmid was inserted into pGEX-3X (Amersham Pharmacia Biotech, Piscataway, NJ) that was cleaved with SmaI, yielding pGEX-HSL1(833–1518). To express a derivative of Hsl1(1021–1518)-(HA)3 (Shulewitz et al., 1999) in which a 237–residue segment (1245–1481) was removed by an in-frame deletion, YCpLG-HSL1(1021–1518)-(HA)3 was cleaved with NsiI and religated, yielding YCpLG-HSL1(1021–1244; 1482–1518)-(HA)3. To express essentially the same polypeptide as a GST fusion in E. coli, a 800-bp BamHI fragment excised from YCpLG-HSL1(1021–1244; 1482–1518)-(HA)3 was inserted into the corresponding site of pGEX-4T-1 (Amersham Pharmacia Biotech), generating pGEX-GST-HSL1(1018–1244; 1482–1518).
To express Swe1 as a GST fusion in E. coli, YCpLG-MycSwe1 (Shulewitz et al., 1999) was first cleaved with BamHI and SalI followed by incubation with T4 DNA polymerase to create blunt sites. The resulting flush-ended 2.5-kb fragment was inserted into pGEX-3X (Amersham Pharmacia Biotech) that was cleaved with SmaI, yielding pGEX-SWE1. To express an 86-residue segment (284–369) of Swe1 as a GST fusion in E. coli, PCR was used with appropriate primers to generate a 5′ NsiI site and a 3′ stop codon followed by an XbaI site. The resulting product, containing at its 5′ end the sequence 5′-ATG CAT TCT CCC-3′ (NsiI site underlined) and at its 3′ end the sequence 5′-GAA TAG ATC TAG ACT-3′ (XbaI site underlined; stop codon in bold), was cleaved with NsiI and XbaI and inserted into pGEX-HSL7 from which the HSL7 sequence had been removed by cleavage with NsiI and XbaI, yielding pGEX-SWE1(284–369).
To produce a plasmid for production of radiolabeled full-length Hsl7 by coupled in vitro transcription (from the SP6 promoter) and translation, a 2.5-kb NdeI-PstI fragment from pAS1-HSL7 (Shulewitz et al., 1999) was inserted into pGEM-5Z (Promega, Madison, WI) that was cleaved with NdeI and PstI, yielding pGEM-5Z-HSL7. To produce plasmids for production of radiolabeled fragments of Hsl7 by coupled in vitro transcription (from the T7 promoter) and translation, PCR was used with appropriate primers to generate products in which the desired region of the HSL7 coding sequence was flanked at its 5′ end by an NcoI site and at its 3′ end by either a BamHI or a HindIII site. For Hsl7(1–246), the PCR product contained at its 5′ end the sequence 5′-CCC ATG GAT AGC-3′ (NcoI site underlined; start codon in bold), and at its 3′-end, 5′-CAG TAG GAT CCT-3′ (BamHI site underlined; stop codon in bold). For Hsl7(88–544), the PCR product contained at its 5′ end 5′-CCC ATG GTC-3′ (NcoI site underlined; start codon in bold) and at its 3′ end, 5′-TTC TAG GAT CCC-3′ (BamHI site underlined; stop codon in bold). For Hsl7(1–246) and Hsl7(88–544), the PCR products were cleaved with NcoI and BamHI and inserted into pBAT4 (Peranen et al., 1996) that had been cleaved with NcoI and BamHI, yielding pBAT4HSL7(1–246) and pBAT4-HSL7(88–544), respectively. For Hsl7(168–345), the PCR product was cleaved with NcoI and HindIII and inserted into pBAT4 that had been cleaved with NcoI and HindIII, yielding pBAT4-HSL7(168–345). For Hsl7(316–636), a 1-kb BamHI fragment was excised from YEpLG-GST-HSL7(316–636) (see below) and inserted into the corresponding site of pBAT4, yielding pBAT4-HSL1(316–636).
To express Hsl7(224–827) as a fusion to Gal4(DBD), PCR was used with appropriate primers to generate a fragment containing at its 5′ end the sequence 5′-CG CAT ATG CTG-3′ (NdeI site underlined, start codon in bold) and the naturally occurring XbaI site in HSL7 at its 3′ end. The resulting product was cleaved with NdeI and XbaI and inserted into YCpT-ADHp-GAL4(DBD)-HSL7 that was cleaved with NdeI and XbaI, yielding YCpT-ADHp-GAL4(DBD)-HSL7(224–827). To generate plasmids expressing Hsl7(284–827), Hsl7(352–827), and Hsl7(1–533) as fusions to the Gal4(DBD), a similar PCR strategy was used with appropriate primers, except that the site at the 5′ end was NsiI: Hsl7(284–827), 5′-GGG ATG CAT AAA TAT GCC-3′ (NsiI site underlined; start codon in bold), naturally occurring 3′ XbaI site; Hsl7(352–827), 5′-GAA ATG CAT TTG GTG-3′, naturally occurring 3′ XbaI site; and Hsl7(1–533), naturally occurring 5′ NsiI site, 5′-TGT ATA TAA TCC TCT AGA GAT-3′ (XbaI site underlined, stop codon in bold). To produce Gal4(DBD)-Hsl7(Δ224–392), a similar PCR approach with appropriate primers was used to delete codons 224–392, yielding a fragment with the sequence 5′-TCG TAT GTG GAT CGA ACT-3′ (codon 223 in bold, codon 393 underlined). All four fragments were cleaved with NsiI and XbaI and inserted into YCpT-ADHp-GAL4(DBD)-HSL7 that had been cleaved with NsiI and XbaI, yielding, respectively, YCpT-ADHp-GAL4(DBD)-HSL7(284–827), YCpT-ADHp-GAL4(DBD)-HSL7(352–827), YCpT-ADHp-GAL4(DBD)-HSL7(1–533), and YCpT-ADHp-GAL4(DBD)-HSL7(Δ224–392).
To express a catalytically inactive Hsl1 mutant (K110R) tagged at its C terminus with a triple influenza virus hemagglutinin (HA) epitope from the GAL1 promoter on a CEN plasmid, PCR was used with appropriate primers to substitute the AAA (Lys) at codon 110 with CGT (Arg) and to introduce an SnaBI site at this position. The resulting fragment, containing the sequence 5′-ATA CGT ATT-3′ (SnaBI site underlined, codon 110 in bold), was cleaved with NcoI and inserted into YCpLG-HSL1(HA)3 (Shulewitz et al., 1999) that was cleaved with NcoI, yielding YCpLG-HSL1-K110R(HA)3.
To express Hsl7(1–685) as a GST fusion in yeast, PCR was used with appropriate primers and YCpT-GFP-HSL7(1–685) as the template to generate a fragment with a BamHI site upstream of and immediately adjacent to the initiator codon for the HSL7 codon sequence (5′-GGA TCC ATG CAT-3′; BamHI site underlined, start codon in bold). The resulting product was cleaved with BamHI and XbaI as inserted into YEpLG-GST-HSL7 (Shulewitz et al., 1999) that was cleaved with BamHI and XbaI, yielding YEpLG-GST-HSL7(1–685). To express Hsl7(1–246) as a GST fusion in yeast, a 750-bp NcoI-HindIII fragment excised from pBAT4-HSL7(1–246) was inserted into YEpLG-GST (Shulewitz et al., 1999) that was cleaved with NcoI and HindIII, yielding YEpLG-GST-HSL7(1–246). To express Hsl7(316–636) as a GST fusion in yeast, PCR was used with appropriate primers to generate a fragment containing a BamHI site at its 5′ end and a stop codon after codon 636. The resulting fragment, containing at its 5′ end the sequence 5′-ATT GGA TCC AAT-3′ (BamHI site underlined) and at its 3′ end the sequence 5′-TCG TGA TCT AGA AAT-3′ (stop codon in bold), was inserted into Litmus28 that was cleaved with EcoRV. A 1.0-kb BamHI fragment excised from the resulting plasmid was inserted into YEpLG-GST (Shulewitz et al., 1999) that was cleaved with BamHI and treated with alkaline phosphatase, yielding YEpLG-GST-HSL7(316–636).
To express Hsl7(674–827), Hsl7(674–736), Hsl7(737–827), and Hsl7(771–827) as GST fusions in bacteria, corresponding fragments were generated by PCR with the use of appropriate primers. Each fragment contained a BamHI site at its 5′ end for in-frame fusion to GST. For Hsl7(674–827) and Hsl7(674–736), the sequence at the 5′ end was 5′-GGA TCC TCT TTG GAG-3′; for Hsl7(737–827), 5′-GGA TCC GAA GAA GAA CAG-3′; and, for Hsl7(771–827), 5′-GGA TCC ATC AAT AAG-3′ (BamHI sites underlined). At the 3′ end, three of the fragments contained the naturally occurring stop codon followed by an EcoRI site, whereas the fragment corresponding to Hsl7(674–736) contained an introduced stop codon followed by an EcoRI site, 5′-GAC ATT GAA AAC TAA GAA TTC-3′ (EcoRI site underlined; stop codon in bold). All four fragments were cleaved with BamHI and EcoRI and inserted into pGEX-4T that had been cleaved with BamHI and EcoRI, yielding pGEX-HSL7(674–827), pGEX-HSL7(674–736), pGEX-HSL7(737–827), and pGEX-HSL7(771–827), respectively.
Protein Binding to Immobilized GST Fusions
GST, GST-Hsl1(833–1518), GST-Swe1, or other GST fusions, as indicated, were expressed in E. coli and purified by binding to glutathione-agarose beads (Amersham Pharmacia Biotech), as instructed by the manufacturer. Beads coated with equal amounts of protein were incubated with radiolabeled proteins, prepared by with the use of a commercial kit for coupled in vitro transcription-translation (Promega), at 4°C for 1 h in ice-cold lysis buffer (20 mM Tris-HCl, pH 8.0, 12.5 mM potassium acetate, 4 mM MgCl2, 0.5 mM EDTA, 5 mM sodium bisulfite, 0.1% Tween 20, 12.5% glycerol) followed by three washes (1 ml each) with the same buffer. Bound proteins were recovered from the washed beads by elution with 30 μl of lysis buffer containing freshly prepared 20 mM glutathione (pH 8.0). After incubation for 5 min, the beads were removed by centrifugation in a microfuge at maximum speed for 10 min at room temperature. Proteins in the resulting supernatant fraction were solubilized by addition of 10 μl of 4-times-concentrated SDS-PAGE sample buffer and boiling for 2 min. Samples of the solubilized eluate were resolved by SDS-PAGE and analyzed by autoradiography.
Isolation of Hsl7 Mutants
YCpT-ADHp-GAL4(DBD)-HSL7 was amplified with the use of primers 231–1 (5′-CAA TCA ACT CCA AGC TTG AAG CAA GCC-3′) and HSL7–3 (5′-GTG ACC CAC TGA CCC AGA AGG TTC C-3′) under standard conditions with Taq DNA polymerase (PerkinElmer Cetus, Norwalk, CT), which, in our experience, is sufficiently error-prone to generate mutations at a low frequency. A sample (25 μg) of the resulting PCR product was mixed with a sample of YCpT-ADHp-GAL4(DBD)-HSL7 (5 μg) that had been gapped by prior cleavage with NsiI and XhoI, and the mixture was used for DNA-mediated transformation of strain YD116 (MATa) with the use of the lithium acetate method (Soni et al., 1993). Transformants harboring the resulting library of YCpT-ADHp-GAL4(DBD)-HSL7 derivatives containing potentially mutated HSL7 sequences, generated by gap repair in situ (Muhlrad et al., 1992), were selected by plating on 70 standard (9-cm-diameter) Petri dishes containing SCGlc-Trp medium, which were incubated at 30°C for 3 d. Each of the resulting yeast colonies was transferred, with the use of a sterile toothpick, to a large (14-cm-diameter) Petri dish (260 colonies/dish) containing SCGlc-Trp medium and incubated at 30°C for 3 d. The resulting patches were transferred by replica-plating onto two YPD plates that had been spread with lawns of saturated cultures of YD119 (MATα) that harbored, respectively, either pACT-HSL1(987–1518) or pACT-SWE1(295–819). After overnight incubation at 30°C to allow for mating and propagation of the resulting diploids, the patches from each YPD plate were transferred, by replica plating, to each of two large Petri dishes, one containing SCGlc-Trp-Leu and the other containing SCGlc-Trp-Leu-Ura, to score for expression of the URA3 reporter gene.
Immunoprecipitations
Protease-deficient strain BJ2168 (Table 1) carrying plasmids coexpressing GFP-HSL7 with either c-Myc-tagged HSL7, or untagged HSL7, under control of the GAL1 promoter were pregrown under appropriate selective conditions in SCRaf medium to an A600 nm of 0.6, induced by addition of galactose (2% final concentration), and incubated with shaking for 2 h. Cells were harvested, washed with phosphate-buffered saline (PBS), and lysed by vigorous vortex mixing with glass beads in ice-cold lysis buffer (20 mM Tris-HCl, pH 7.2, 12.5 mM potassium acetate, 4 mM MgCl2, 0.5 mM EDTA, 5 mM sodium bisulphite, 0.1% Tween 20, 12.5% glycerol) containing 1 mM dithiothreitol and protease inhibitors (2 μg/ml leupeptin, 2 μg/ml pepstatin A, 1 mM benzamidine, 2 μg/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride). The resulting crude extracts were clarified by centrifugation in a microcentrifuge for 10 min at 4°C, and then by sedimentation at 30,000 × g in a tabletop ultracentrifuge for 30 min at 4°C. Samples (1 mg of total protein) of the extracts were diluted into lysis buffer (200-μl final volume) and mixed with 20 μl of a suspension of protein A + protein G (A/G)-agarose beads (Calbiochem). For preclearing, these samples were incubated for 1 h at 4°C on a roller drum and then the beads were removed by centrifugation in a microcentrifuge at maximum speed for 5 min at 4°C. The resulting supernatant solution was transferred to a fresh tube containing another aliquot (20 μl) of A/G-agarose beads and 1 μl of mouse ascites fluid containing anti-c-Myc monoclonal antibody (mAb), 9E10 (or 1 μl of affinity-purified anti-HA mAb; see below). After incubation on a roller drum for 2 h at 4°C, the bead-bound immune complexes were collected by brief centrifugation, washed three times (1 ml each) with ice-cold lysis buffer, resuspended in SDS-PAGE sample buffer, and solubilized by incubation in a boiling water bath for 10 min. After removal of any residual particulate material by centrifugation for 10 min at room temperature, samples of the resulting supernatant fraction were resolved by SDS-PAGE, transferred to a membrane filter (Immobilon-P; Millipore) with the use of a semidry transfer apparatus (Bio-Rad, Hercules, CA), analyzed by immunoblotting with the use of appropriate primary antibodies, followed by appropriate horseradish peroxidase-conjugated secondary antibodies, and visualized with the use of a commercial chemiluminescence detection system (Renaissance; PerkinElmer Life Science Products, Boston, MA).
Protein Kinase Assay in Immune Complexes
Protease-deficient yeast strain MJY153 (Table 1) carrying either YCpLG-HSL1(HA)3 or YCpLG-HSL1-K110R(HA)3 were pregrown in SCRaf medium lacking leucine to an A600nm of 0.6, induced by addition of galactose (2% final concentration), and incubated with shaking for 2 h. Cells were harvested, washed with PBS, and lysed by vigorous vortex mixing with glass beads in ice-cold lysis buffer (see above) containing 1 mM dithiothreitol, protease inhibitors (2 μg/ml leupeptin, 2 μg/ml pepstatin A, 1 mM benzamidine, 2 μg/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride) and phosphatase inhibitors (10 mM sodium pyrophosphate, 10 mM NaN3, 10 mM NaF, 0.4 mM sodium meta-vanadate, 0.4 mM sodium ortho-vanadate, 0.1 mM β-glycerol-phosphate, and 1 μg/ml phosvitin). The resulting crude extracts were clarified by centrifugation in a microcentrifuge for 10 min at 4°C. The supernatant solution was removed and a sample (1 mg of total protein) was diluted to 200 μl in cold lysis buffer, preadsorbed for 30 min with 20 μl of protein A/G-agarose beads (Calbiochem) at 4°C with rotary agitation. The beads were then removed by sedimentation in a microfuge for 10 min at 4°C. The precleared supernatant fraction was transferred to a fresh tube containing 20 μl of protein A/G-agarose beads and 1 μl of anti-HA mAb HA.11 (Covance Research Products, Richmond, CA). After incubation at 4°C with rotary agitation for 1 h, the bead-bound immune complexes were collected by brief centrifugation, washed three times with 1 ml of cold lysis buffer, and then twice with kinase assay buffer (50 mM HEPES, pH 7.8, 1 mM EGTA, 2 mM MgCl2, 1 mM dithiothreitol, 0.5 mM sodium ortho-vanadate, and 10 mM β-glycerol-phosphate). The suspension of beads was split into two equal portions. One portion was solubilized by boiling in SDS-PAGE sample buffer for 2 min, resolved by SDS-PAGE, transferred electrophoretically to an Immobilon-P membrane (Millipore) with the use of a semidry transfer cell (Bio-Rad), and analyzed by immunoblotting. The other portion was resuspended in kinase buffer (30-μl final volume) containing 20 μM [γ-32P]ATP (1.7 × 107 cpm/nmol), and 1 μg of the appropriate purified GST-Hsl7 derivative and incubated at 30°C for 30 min. To terminate the reaction, 10 μl of 4× SDS-PAGE buffer was added (1× final concentration) followed by boiling for 2 min. Proteins were resolved by SDS-PAGE and analyzed by autoradiography with the use of either x-ray film or a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Preparation of GST-Hsl7 from Yeast
To prepare GST, GST-Hsl7, GST-Hsl7(1–685), and the other derivatives indicated, from yeast, cultures (1 liter) of strain BJ2168 carrying the appropriate plasmid were grown in SCRaf lacking leucine to A600nm of 1.0, induced by addition of galactose (2% final concentration) and incubated with shaking for 12 h at 30°C. Cells were harvested, washed with PBS, and lysed by vigorous vortex mixing with glass beads in ice-cold lysis buffer (see above) containing 1 mM dithiothreitol, protease inhibitors (2 μg/ml leupeptin, 2 μg/ml pepstatin A, 1 mM benzamidine, 2 μg/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride). The resulting crude extracts were clarified by centrifugation in a microcentrifuge for 10 min at 4°C and desalted by passage over a column containing a 10-ml bed of Sephadex G-25 (Amersham Pharmacia Biotech) to remove endogenous glutathione. The column was eluted with 20 ml of lysis buffer and the flow-through fraction was incubated with 500 μl of a slurry of glutathione-agarose beads (Amersham Pharmacia Biotech) at 4°C with rotary agitation for 1 h. The beads were collected by centrifugation, washed three times with 10 ml of wash buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, and 0.1% Tween 20), and placed into an empty glass column. Bead-bound proteins were released by rinsing the beads with 10 ml of elution buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 0.1% Tween 20, and 20 mM freshly prepared glutathione). The eluate was concentrated to a protein concentration of ∼0.2 mg/ml, and the elution buffer was replaced with 50 mM Tris-HCl, pH 8.0, containing 10% glycerol, with the use of a microconcentration device (Microcon-30; Amicon, Beverly, MA) and stored at −70°C.
Indirect Immunofluorescence and Fluorescence Microscopy
For indirect immunofluorescence, exponentially growing cells were fixed with 3.7% formaldehyde in 0.1 M potassium phosphate, pH 6.5, for 30 min at room temperature and washed in 0.1 M potassium phosphate, pH 6.5. Fixed cells were resuspended in 0.2 M Tris-HCl, pH 9.0, containing 20 mM EDTA, pH 8.0, 1 M NaCl, and 80 mM β-mercaptoethanol, incubated at room temperature for 10 min, washed once with potassium phosphate-sodium citrate, pH 5.8, containing 1 M NaCl and twice with potassium phosphate-sodium citrate, pH 5.8, resuspended in 1 ml of solution A (1.2 M sorbitol, 0.1 M potassium phosphate, pH 6.5, 0.5 mM MgCl2) containing 0.14 M β-mercaptoethanol, and digested with 110 μl of Glusulase (PerkinElmer Life Science Products) and 0.6 mg/ml Zymolyase 100T (Seikagaku, Tokyo, Japan). The digested cells were washed twice with solution A, applied to the wells of poly-l-lysine (Ted Pella, Redding, CA)-coated multiwell microscope slides, and permeabilized by treatment at −20°C with, successively, methanol for 6 min and acetone for 30 s. Permeabilized cells were rehydrated in PBS, pH 7.3, blocked in PBS containing 1 mg/ml bovine serum albumin, and incubated overnight at 4°C with an appropriate primary antibody (at the dilution indicated): rat anti-yeast α-tubulin mAb YOL1/34 (1:200) (Kilmartin et al., 1982); affinity-purified mouse polyclonal anti-Hsl7 antibodies (1:500) (Shulewitz et al., 1999); mouse anti-c-Myc mAb 9E10 (1:1000) (Evan et al., 1985); and/or, rabbit polyclonal anti-yeast Tub4 (γ-tubulin) antibodies (1:5000) (generous gift of John Kilmartin, Medical Research Council, Cambridge, United Kingdom). After incubation, cells were washed several times with PBS containing 1 mg/ml bovine serum albumin, and incubated for 2 h in the dark with an appropriate secondary antibody (at the dilution indicated): indocarbocyanine (Cy3)-conjugated goat anti-rat IgG heavy chain (Cappel/Organon Teknika, Malvern, PA) (1:300), Cy3-conjugated donkey anti-mouse immunoglobulin (Jackson ImmmunoResearch, West Grove, PA) (1:500); and/or fluorescein isothiocyanate-conjugated donkey anti-rabbit immunoglobulin (Jackson ImmmunoResearch) (1:200). Finally, stained cells were washed six times with PBS; in some experiments, 4′-6-diamidino-2-phenylindole (DAPI) (1 μg/μl) was added in the fourth wash to counterstain nuclear DNA. For cells expressing GFP, a milder fixation regimen was used: treatment with 3.7% formaldehyde for only 10 min, and the methanol/acetone treatment and rehydration steps were omitted.
For fluorescence microscopy and indirect immunofluorescence, cells were examined with a TE300 microscope (Nikon, Melville, NY) equipped with a 100×/1.4 Plan-Apo objective and a 1.4 numerical aperture condenser. Digital images were acquired with a bottom-ported Orca 100 charge-coupled device camera (Hamamatsu, Bridgewater, NJ) and Phase 3 Imaging Systems software. Samples for time-lapse fluorescence microscopy were embedded in a thin layer of solid SCGlc medium containing purified agarose (instead of standard agar) solidified under sterile conditions on an excavated microscope slide.
Immunoelectron Microscopy
Samples were frozen and subjected to freeze substitution, according to methods described in detail elsewhere (McDonald, 1999). Briefly, yeast cells were cryofixed in a Bal-Tec HPM010 high-pressure freezer, freeze-substituted in 0.2% glutaraldehyde plus 0.1% uranyl acetate for 3 d at −90°C, and then warmed to room temperature over a 12-h period. Cells were rinsed several times in pure acetone and then infiltrated with LR White resin overnight, placed in flat-bottom polypropylene capsules (catalog no. 133–1; Ted Pella) and polymerized at 60°C for 2 d in a nitrogen gas environment. Thin (50-nm) sections were cut on a Reichert UltracutE microtome, floated onto Formvar- and carbon-coated nickel grids, and incubated with affinity-purified mouse polyclonal anti-Hsl7 antibodies (1:50) for 1 h. After rinsing with PBS, the sections were then incubated with 10-nm gold particles decorated with donkey anti-mouse immunoglobulin (1:20) for either 1 h or overnight at 4°C. After rinsing with PBS, the sections were fixed in 0.5% glutaraldehyde for 5 min, rinsed in distilled water, and poststained with uranyl acetate and lead citrate. Sections were examined in a JEOL 100CX electron microscope.
RESULTS
Hsl7 Is Phosphorylated by Hsl1
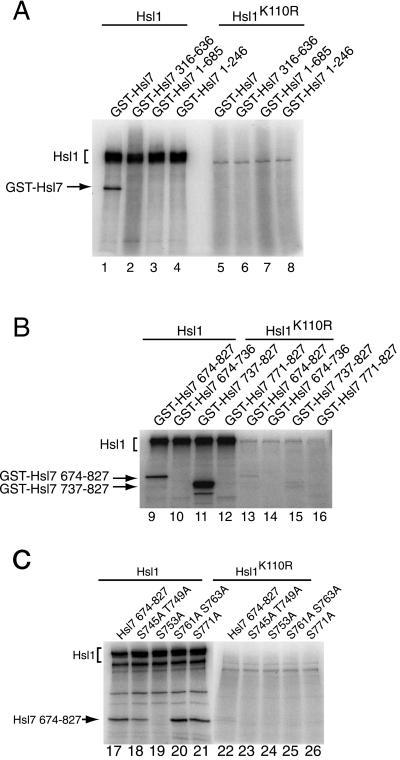
Based on electrophoretic mobility shift, Hsl7 is modified in vivo in an Hsl1-dependent manner (McMillan et al., 1999); however, whether Hsl7 is phosphorylated by Hsl1, or by some other Hsl1-activated protein kinase, was not addressed previously. To examine whether Hsl7 is a direct substrate of Hsl1, wild-type HSL1 and a catalytically inactive (and noncomplementing) mutant, hsl1(K110R), each tagged with a C-terminal triple-HA epitope, were expressed separately in a protease-deficient hsl1Δ strain (MJY153) by brief (2-h) induction from the GAL1 promoter on a CEN plasmid. Extracts were prepared and subjected to immunoprecipitation with the use of anti-HA mAb HA.11. To measure phosphotransferase activity of bead-bound Hsl1, the resulting immune complexes were incubated in a buffer containing Mg2+ and [γ-32P]ATP, along with GST fusions to full-length Hsl7 and various segments of it (purified from yeast by binding to and elution from glutathione-agarose). After quenching the reactions, radiolabeled products were resolved by SDS-PAGE and examined by autoradiography. As anticipated, based on prior observations (Barral et al., 1999), Hsl1 underwent robust autophosphorylation (Figure 1, left); autophosphorylation was abrogated almost completely by the substitution mutation (K110R) in conserved protein kinase domain II (Hanks and Hunter, 1995) (Figure 1, right), as expected. We found that GST-Hsl7 was an efficient substrate for Hsl1, but was not detectably phosphorylated by catalytically inactive Hsl1 (Figure 1A, left versus right), ruling out that the observed phosphorylation was due to a coprecipitating protein kinase. Truncations of Hsl7 (fused to GST), even one that removed just 142 residues from the C terminus, were unable to serve as phosphoacceptors, demonstrating that GST itself is not phosphorylated by Hsl1 and suggesting that this segment contains the site(s) of Hsl1-mediated phosphorylation. Indeed, this same C-terminal region (fused to GST and purified from bacteria) was phosphorylated by Hsl1 at least as efficiently as full-length Hsl7 (Figure 1B); further subdivision of this portion of Hsl7 mapped the Hsl1 phosphorylation site(s) to a 33-residue region (residues 737–770). Site-directed mutagenesis showed that the phosphorylated residue is Ser753 (Figure 1C). Thus, Hsl7 is the first bona fide cellular substrate of Hsl1 to be identified.
Figure 1.
The C terminus of Hsl7 is phosphorylated by Hsl1 protein kinase. Equivalent amounts of protein (1 mg of total) from extracts of a protease-deficient strain (MJY153) expressing either triple-HA-tagged wild-type Hsl1 or a catalytically defective derivative, Hsl1(K110R), from plasmids (YCpLG-HSL1(HA)3 and YCpLG-HLS1-K110R(HA)3, respectively), were subjected to immunoprecipitation with mouse anti-HA mAb HA.11. The resulting immune complexes were resuspended in protein kinase assay buffer, incubated for 10 min at 30°C with Mg2+, [γ-32P]ATP, and a GST-Hsl7 fusion or the indicated C-terminal truncations (purified from yeast) (A), or the indicated C-terminal fragments (purified from bacteria) (B), or the indicated site-directed mutants (purified from bacteria) (C), and then analyzed by SDS-PAGE followed by autoradiography.
Again, as judged by electrophoretic mobility shift, modification of Swe1 in vivo is dependent on Hsl1 (Shulewitz et al., 1999) and on at least one other protein kinase, Elm1 (Sreenivasan and Kellogg, 1999). Moreover, it has been reported that in vitro Nim1/Cdr1, a fission yeast ortholog of Hsl1, can phosphorylate Wee1, the fission yeast relative of Swe1 (Coleman et al., 1993). Hence, we also investigated whether Swe1 is a direct substrate of Hsl1 with the use of the same assay method. To avoid any contribution from the kinase activity of Swe1 itself, we purified a catalytically inactive (and noncomplementing) mutant, Swe1(K472A), both as a GST fusion from bacteria and as an otherwise native protein (tagged with an N-terminal c-Myc epitope) from yeast. When either bacterially expressed GST-Swe1 or Myc-Swe1 produced in yeast were added, no Hsl1-dependent incorporation of radioactivity was observed into either of these purified proteins above the background observed with the catalytically inactive Hsl1 mutant (our unpublished results). Because Hsl7 may help to tether Swe1 to Hsl1 (Shulewitz et al., 1999) and/or possibly methylate Swe1 (Frankel and Clarke, 2000; Lee et al., 2000) and thereby perhaps make it a more efficient substrate for Hsl1, we also performed essentially identical assays in which various amounts of purified GST-Hsl7 was also included; however, no enhancement of Swe1 phosphorylation was observed (our unpublished results). Thus, in marked contrast to Hsl7, and somewhat unexpectedly, Swe1 does not appear to be an efficient substrate for Hsl1.
Hsl7 Binds Directly to Hsl1 and Swe1
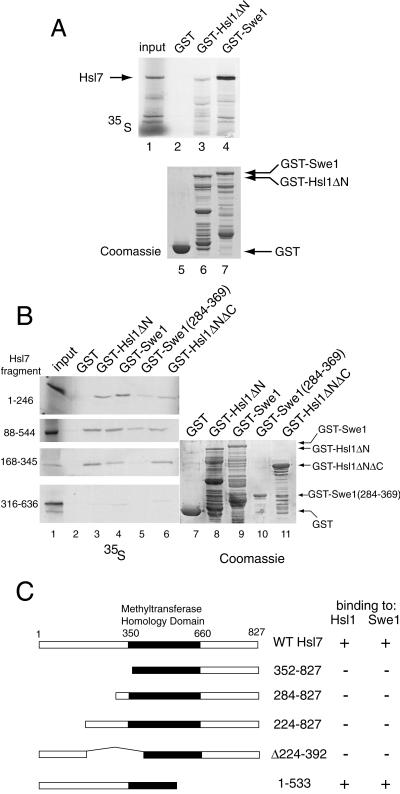
We have shown previously that Hsl7 can associate with either Hsl1 or Swe1, both in vivo (with the use of the two-hybrid method) and in cell extracts (as judged by coimmunoprecipitation) (Shulewitz et al., 1999). Prior studies suggested that the C terminus of Hsl1 is responsible for its association with Hsl7, whereas most of Swe1 appeared to be necessary for its association with Hsl7 (Shulewitz et al., 1999). To determine whether these associations represent direct physical interactions between Hsl7 and each of these partners, we prepared radiolabeled Hsl7 by in vitro translation in a rabbit reticulocyte lysate and examined its ability to bind to GST alone or to GST-Hsl1(833–1518) and GST-Swe1, which had been expressed in and purified from bacteria and then immobilized on glutathione-agarose beads. Hsl7 bound reproducibly to both GST-Hsl1(833–1518) and GST-Swe1, whereas no detectable binding to GST was ever observed (Figure 2A, top). In most experiments, Hsl7 appeared to bind more strongly to GST-Swe1 (≥5% of input) and less strongly to Hsl1 (∼1% of input); however, due to nonspecific degradation, the fraction of the bead-bound GST-Hsl1(833–1518) fusion that was intact was always less than the fraction of bead-bound GST-Swe1 that was intact (Figure 2A, bottom). These results demonstrate that the interactions between Hsl7 and Hsl1, and between Hsl7 and Swe1, are direct and do not require bridging by any other yeast protein.
Figure 2.
Hsl1- and Swe1-binding sites reside in the N-terminal domain of Hsl7. (A) Radiolabeled Hsl7 (35S), prepared by in vitro translation, was incubated with equivalent amounts of glutathione-agarose beads preadsorbed with equivalents amounts of bacterially expressed GST (lane 2), GST-Hsl1(833–1518) (GST-Hsl1ΔN) (lane 3), or GST-Swe1 (lane 4). After washing, the beads were eluted with SDS-PAGE sample buffer and the bound protein analyzed by SDS-PAGE and autoradiography, along with a portion of the 35S-labeled Hsl7 (10% of the amount added in the binding reactions; “input”) (lane 1). Equivalent samples of the beads containing each bacterially produced protein (GST, lane 5; GST-Hsl1(833–1518), lane 6; GST-Swe1, lane 7) were analyzed by SDS-PAGE and staining with Coomassie brilliant blue to demonstrate equal loading. (B) Various fragments of Hsl7 indicated (1–246, 88–544, 168–345, 316–636) were each prepared in radiolabeled form by in vitro translation and incubated, as described in A, with beads carrying GST (lane 2), GST-Hsl1(833–1518) (lane 3), GST-Swe1 (lane 4), GST-Swe1(284–369) (lane 5), and GST-Hsl1(1018–1244; 1482–1518) (GST-Hsl1ΔNΔC). After washing, the beads were eluted with excess glutathione, and the released protein was analyzed by SDS-PAGE and autoradiography, as in A (left panels). Equivalent samples of the beads containing each bacterially produced protein, as indicated, were analyzed by SDS-PAGE and staining with Coomassie brilliant blue to demonstrate equal loading. (C) A ura3 reporter strain (YD116) carrying a GAL1 promoter-dependent URA3 gene was cotransformed with plasmids expressing either Gal4(TAD)-Hsl1(987–1518) or Gal4(TAD)-Swe1(295–819) (Shulewitz et al., 1999) and plasmids expressing Gal4(DBD) fused to the indicated portions of Hsl7. Methyltransferase homology domain denotes the segment of Hsl7 homologous to known protein-arginine methyltransferases (Pollack et al., 1999; Ma, 2000). The ability (+) or inability (−) of each pair of fusions to activate transcription of the GAL1-dependent URA3 reporter gene, as judged by growth in the absence of uracil, is summarized in the right-hand columns.
Delineation of the Hsl1- and Swe1-binding Domains of Hsl7
To gain more insight about the nature of the interactions between Hsl7 and both Hsl1 and Swe1, we identified the segments of Hsl7 that mediate its direct physical association with these proteins by using two independent methods. First, as an extension of the binding assays that demonstrated that radiolabeled full-length Hsl7 can bind specifically to both GST-Hsl1(833–1518) and GST-Swe1 (Figure 2A), we prepared various subfragments of Hsl7 in radiolabeled form by in vitro translation and tested their ability to bind to GST-Hsl1(833–1518) and to GST-Swe1, and also to smaller segments of both Swe1 and Hsl1 that appear to harbor their minimal Hsl7-binding domains (Shulewitz, 2000). Indeed, we found that three overlapping regions of the N terminus of Hsl7 [Hsl7(1–246), Hsl7(88–544), and Hsl7(168–345)] were able to associate specifically with immobilized, bacterially expressed GST-Hsl1(833–1518) and GST-Hsl1(1018–1244; 1482–1518), but not with GST alone (Figure 2B). As judged by the fraction of the input retained, Hsl7(168–345) bound with the highest apparent affinity. In contrast, a more C-terminal segment, Hsl7(316–636), showed little or no binding to either Hsl1(833–1518) or GST-Swe1 above the nonspecific background (Figure 2B). Likewise, a more N-terminal segment, Hsl7(1–226), displayed no binding above background (our unpublished results). Therefore, the minimal region of Hsl7 sufficient for binding to Hsl1 in vitro is contained in the region spanned by residues 168–246. The results for Hsl7 binding to Swe1 were somewhat more ambiguous; however, sequences in Hsl7 N-terminal to residue 168 appear to make important contributions to its association with Swe1 (Figure 2B).
To confirm these conclusions in vivo, we used the two-hybrid method (Fields and Sternglanz, 1994). As we previously reported, a Gal4(DBD)-Hsl7 fusion interacts with Gal4(TAD)-Hsl1(987–1518) and Gal4(TAD)-Swe1(295–819) fusions in the two-hybrid assay (Shulewitz et al., 1999). Therefore, we generated a modest collection of deletions (mainly N-terminal truncations) of Gal4(DBD)-Hsl7 and tested whether they retained the ability to interact with Gal4(TAD)-Hsl1(987–1518) and Gal4(TAD)-Swe1(295–819). The only construct that preserved Hsl1- and Swe1-binding was one that included the intact N-terminal domain of Hsl7 (residues 1–533) (Figure 2C), which includes the regions identified in the biochemical studies (Figure 2, A and B). Indeed, removal of residues 1–223 eliminated the ability of Hsl7 to associate with Hsl1 and Swe1 (Figure 2C). Likewise, removal of the region between residues 224 and 391 was sufficient to abrogate interaction with Hsl1 and Swe1. Taken together, the binding studies and the two-hybrid analysis, delineate an ∼160-residue segment of Hsl7 (residues 88–246) that is necessary and sufficient for its interaction with Hsl1 and Swe1 both in vivo and in vitro.
Point Mutants of Hsl7 Specifically Defective for Association with Hsl1
To pinpoint individual residues in Hsl7 important for its interaction with Hsl1 and Swe1, we devised a variation on the differential interaction trap method (White, 1996; Inouye et al., 1997) to screen for mutations in Hsl7 that interfere with its association with Hsl1, but not its association with Swe1, and vice-versa. The method we developed is described in detail in MATERIALS AND METHODS. In brief, to achieve the uniform expression and plasmid maintenance required for a large-scale screen, instead of expressing Gal4(DBD)-Hsl7 from the ADH1 promoter on the 2 μm DNA plasmid used previously (pAS1-HSL7) (Shulewitz et al., 1999), we constructed a low-copy (CEN) plasmid expressing Gal4(DBD)-Hsl7 from the ADH1 promoter (YCpT-ADHp-GAL4(DBD)-HSL7). DNA containing the entire HSL7 gene was randomly mutagenized with the use of error-prone PCR and the resulting amplification products were recombined into YCpT-ADHp-GAL4(DBD)-HSL7 by in vivo gap repair (Muhlrad et al., 1992) in the MATa reporter strain YD116 (Durfee et al., 1999). The transformants obtained, carrying the collection of mutagenized Gal4(DBD)-Hsl7, were individually mated to two MATα reporter strains otherwise isogenic to YD116, one carrying plasmid pACT-HSL1(987–1518) and the other carrying plasmid pACT-SWE1(295–819). Therefore, two diploids were generated from each original transformant: one containing mutagenized Gal4(DBD)-Hsl7 and Gal4(TAD)-Hsl1(987–1518), and the other containing the same Gal4(DBD)-Hsl7 variant and Gal4(TAD)-Swe1(295–819). Successful interaction was scored by expression of the reporter gene (URA3 under upstream activation sequenceGAL control), as judged by ability to grow on −Ura plates.
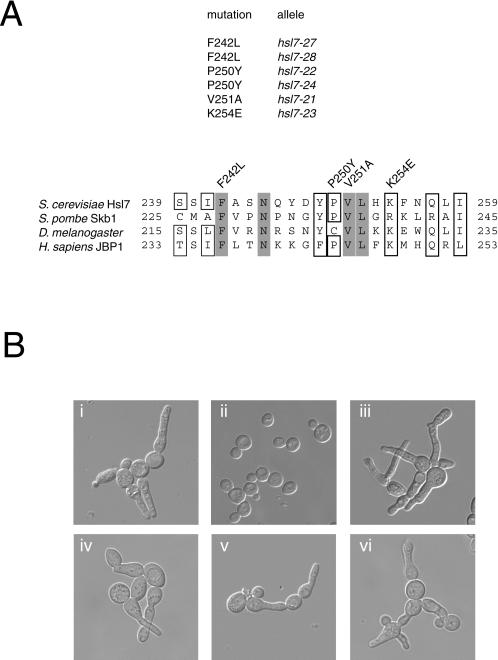
As expected, most of the Gal4(DBD)-Hsl7-containing transformants yielded Ura+ diploids with both Gal4(TAD)-Hsl1(987–1518) and Gal4(TAD)-Swe1(295–819), indicating that neither protein-protein interaction had been perturbed. Some yielded diploids that were both Ura−, indicating that both interactions had been destroyed. However, of 7000 transformants initially screened, 30 reproducibly yielded Ura+ diploids when mated to the cells expressing Gal4(TAD)-Swe1(295–819), but Ura− diploids when mated to cells expressing Gal4(TAD)-Hsl1(987–1518). This class of mutants was designated “Hbd” (for Hsl1-binding-defective). For reasons we do not yet understand, no mutants of the opposite class, so-called “Sbd” (for Swe1-binding-defective), were obtained. Of the 30 Hbd mutants, six alleles (hsl7–21, hsl7–22, hsl7–23, hsl7–24, hsl7–27, and hsl7–28) were chosen for further detailed study because immunoblot analysis indicated that each mutant Gal4(DBD)-Hsl7 was apparently full-length (our unpublished results). The entire HSL7 open reading frame in each of these six alleles was determined by automated nucleotide sequence analysis of the corresponding DNA. All six alleles contained only a single base pair change that resulted in a single amino acid substitution mutation (Figure 3A, top). The hsl7–27 and hsl7–28 alleles, although causing the same amino acid change, resulted from different base pair changes and, hence, were not siblings; however, the hsl7–22 and hsl7–24 alleles were identical. Thus, four different single-substitution mutations were isolated: F242L (hsl7–27 and hsl7–28); P250Y (hsl7–22/hsl7–24); V251A (hsl7–21); and, K254E (hsl7–23). Reassuringly, all four point mutations fell within or immediately adjacent to the C-terminal boundary of the region of Hsl7 (residues 168–246) identified as important for interaction with Hsl1 by the two-hybrid and in vitro binding experiments. Moreover, all four mutations fall within a segment of Hsl7 that has high homology to its known orthologs in other organisms (Figure 3A, bottom) and alter residues that are highly conserved, suggesting that these mutations fall within a conserved Hsl1-binding motif.
Figure 3.
Point mutations in Hsl7 define its Hsl1-binding site and prevent function. (A) Positions of the different single-residue substitution mutations (and their corresponding allele numbers) that prevent association of Hsl7 with Hsl1, as assessed by the two-hybrid method, are indicated within the primary structure of Hsl7 (and several of its homologues from the organisms indicated). Gray boxes indicate absolutely conserved residues; open boxes indicated highly conserved residues. (B) An hsl7Δ strain (MJY102) was transformed with an empty vector (i) or the same plasmid expressing either normal Hsl7 (ii), Hsl7(F242L) (iii), Hsl7(P250Y) (iv), Hsl7(V251A) (v), or Hsl7(K254E) (vi), grown in liquid medium to midexponential phase, and photographed with the use of differential interference contrast microscopy.
Association of Hsl7 with Hsl1 Is Required for Down-Regulation of Swe1 at G2-M
Our isolation of alleles of Hsl7 that perturb only its interaction with Hsl1 provided a means to determine whether stable association with Hsl1 is required for Hsl7 function. To do so, we first examined whether each of the four mutants was able to complement an hsl7Δ mutation. As has been amply demonstrated previously (Ma et al., 1996; Shulewitz et al., 1999), hsl7Δ cells display a dramatically elongated bud (Figure 3B, i). Wild-type HSL7 expressed from its own promoter on a low copy (CEN) plasmid completely restored normal morphology (Figure 3B, ii). In marked contrast, when expressed in the same manner, none of the hsl7 mutants [Hsl7(F242L), Hsl7(P250Y), Hsl7(V251A), or Hsl7(K254E)] was able to complement (Figure 3B, iii-vi), despite the fact that each of the mutant proteins was expressed at a level indistinguishable from wild-type Hsl7, as judged by immunoblotting (our unpublished results). None of the four mutant proteins had any detectable effect when expressed in an otherwise isogenic HSL7+ strain (our unpublished results), indicating that they do not exert any toxic, dominant-negative effect on cell morphology. Because it has been amply demonstrated that the elongated buds observed in hsl7Δ cells are indicative of hyperactive Swe1 (Ma et al., 1996; McMillan et al., 1999; Shulewitz et al., 1999), the fact that each of the Hsl1-binding–defective Hsl7 mutants was unable to rescue this phenotype shows that stable association of Hsl7 with Hsl1 is required for down-regulation of Swe1.
Association of Hsl7 with Hsl1 Is Required for Hsl7 Localization at Bud Neck
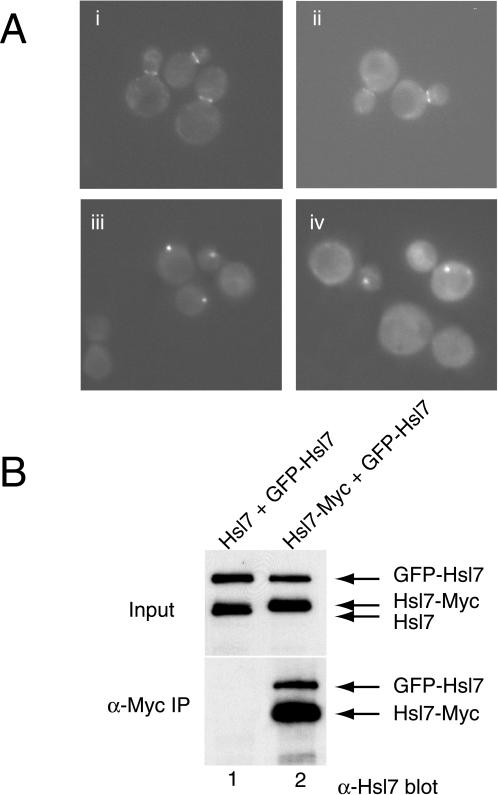
We have recently demonstrated that Hsl7 fails to localize to the bud neck in an hsl1Δ cell, but is still localized to the bud neck in cells expressing catalytically inactive Hsl1(K110R) (Shulewitz et al., 1999), suggesting that binding of Hsl7 to Hsl1 (rather than modification by Hsl1) is required for recruitment of Hsl7 to this location. The availability of the Hsl1-binding–defective Hsl7 alleles allowed us to test directly whether association of Hsl7 with Hsl1 is required for accumulation of Hsl7 at the bud neck, even in cells expressing normal levels of wild-type Hsl1. We have shown previously that a GFP-Hsl7 fusion is fully functional and, like native Hsl7, targets to the bud neck in a septin- and Hsl1-dependent manner (Shulewitz et al., 1999). Hence, each Hsl7 mutant was fused to GFP to permit examination of its subcellular distribution. Each of the four mutants behaved identically; illustrative data are shown for two of them, Hsl7(F242L) and Hsl7(P250Y), in Figure 4A. To make examination of the bud neck unambiguous, cells with a swe1Δ mutation were used so that even the hsl7Δ cells would have a normal morphology. When expressed in wild-type cells (Figure 4A, i and ii), or in swe1Δ cells (our unpublished results) GFP-Hsl7(F242L) and GFP-Hsl7(P250Y) were efficiently targeted to the bud neck, as observed previously for wild-type Hsl7 fused to GFP. However, in cells lacking endogenous Hsl7, namely, hsl7Δ cells (our unpublished results) or hsl7Δ swe1Δ cells (Figure 4A, iii and iv), neither GFP-Hsl7(F242L) nor GFP-Hsl7(P250Y) were present detectably at the bud neck, but were present as a cytoplasmic “dot.” In contrast, in hsl7Δ or hsl7Δ swe1Δ cells, wild-type GFP-Hsl7 decorates the bud neck exclusively (Shulewitz et al., 1999). These data demonstrated, first, that interaction with Hsl1 is indeed required for stable accumulation of Hsl7 at the bud neck. However, as noted, when endogenous wild-type Hsl7 was present, each of the GFP-tagged Hsl7 mutants was able to localize to the bud neck, suggesting that Hsl7 is normally an oligomeric protein and that each of the Hsl7 mutants retained the capacity to multimerize with wild-type Hsl7.
Figure 4.
Differential subcellular localization of Hsl7 mutants defective for binding to Hsl1. (A) Two of the Hsl1-binding-defective Hsl7 mutants, Hsl7(P250Y) (i and iii) and Hsl7(F242L) (ii and iv), expressed as GFP fusions from the HSL7 promoter on low copy number TRP1-marked (CEN) plasmids, were introduced into either a wild-type HSL7+ strain (MJY112) (i and ii) or an hsl7Δ strain (MJY151) (iii and iv). The transformants were grown to mid-exponential phase at 30°Cin SCGlc-Trp, and samples of each culture were viewed directly in a fluorescence microscope. (B) Protease-deficient strain (BJ2168) was transformed with plasmids expressing either untagged Hsl7 or a c-Myc-epitope tagged derivative, as indicated, and also cotransformed with a plasmid expressing GFP-Hsl7. Equivalent amounts of protein (1 mg of total) from extracts of these cells (input) were subjected to immunoprecipitation with mouse anti-Myc mAb 9E10, as described in MATERIALS AND METHODS. The resulting immunoprecipitates (α-Myc IP) were resolved by SDS-PAGE and analyzed by immunoblotting with mouse polyclonal anti-Hsl7 (bottom), along with samples (respresenting ∼1% of the amount of extract protein that was subjected to immunoprecipitation) (top).
Hsl7 Oligomerization
To test directly the capacity of Hsl7 to self-associate, differentially tagged versions of Hsl7 were coexpressed in the same strain (BJ2168) and their ability to interact was assessed by coimmunoprecipitation. For this purpose, we used a plasmid expressing GFP-Hsl7 and a plasmid expressing either Hsl7-myc or untagged Hsl7 (as a control), each expressed from the GAL1 promoter on a CEN plasmid. Lysates were prepared from these cells after brief (2-h) induction with galactose, and the resulting clarified extracts were subjected to immunoprecipitation with anti-Myc mAb 9E10. The immune complexes were resolved by SDS-PAGE and analyzed by immunoblotting with appropriate antibodies. As expected, in the extracts containing untagged Hsl7 and GFP-Hsl7, neither protein was immunoprecipitated by the anti-c-Myc mAb (Figure 4B, lane 1). In contrast, Hsl7-myc was efficiently immunoprecipitated by the anti-c-Myc mAb and GFP-Hsl7 was coimmunoprecipitated nearly stoichiometrically (Figure 4B, lane 2). These results demonstrate that in vivo Hsl7 does indeed self-associate to form a multimeric species.
Hsl7 Mutants Reveal a New Subcellular Location for Hsl7
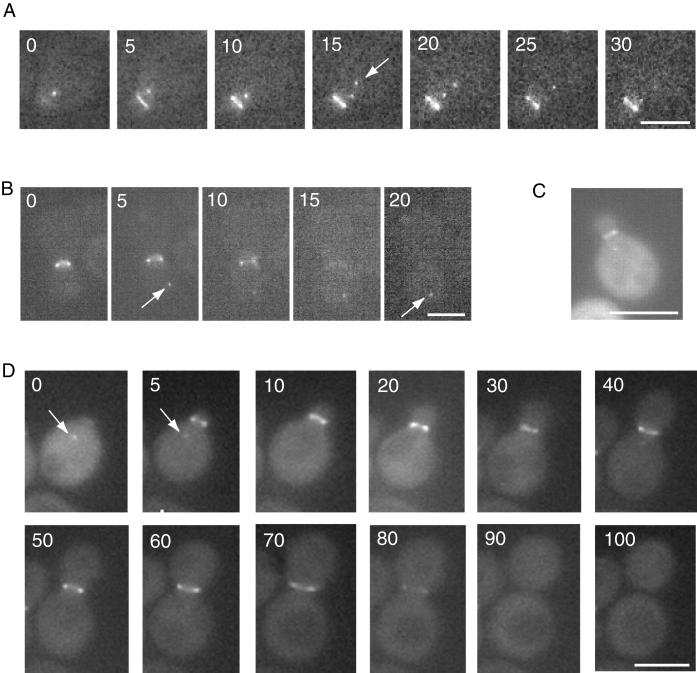
As mentioned immediately above, all four of the Hsl7 point mutants unable to interact stably with Hsl1 localized as a bright cytoplasmic dot when present as the sole source of Hsl7 in the cell (Figure 4A, iii and iv). Likewise, in hsl1Δ cells, wherein interaction of even wild-type Hsl7 with Hsl1 cannot occur, we found that a GFP fusion to normal Hsl7 also localized exclusively as a dot in the cytosol (see below). This cytoplasmic dot was observed under other circumstances as well. For example, Hsl7(1–685), which lacks 142 C-terminal residues (Figure 1), is nonetheless able to complement a hsl7Δ mutation when expressed from the HSL7 promoter on a low-copy (CEN) plasmid, restoring round cell morphology and showing no G2 delay (our unpublished results). Correspondingly, when expressed from the HSL7 promoter on a CEN plasmid, GFP-Hsl7(1–685) localized strongly to the bud neck in hsl7Δ cells; however, in many unbudded cells, in cells with small buds, and even in some cells with medium-sized buds, we also observed a very bright fluorescent dot (or, occasionally, 2 dots) (Figure 5, A and B). Similarly, when a GFP fusion to wild-type Hsl7 was overexpressed from the GAL1 promoter on a CEN plasmid (YCpLG-GFP-HSL7), in addition to bright decoration of the bud neck, the presence of a prominent cytoplasmic dot could be frequently visualized, even although the background fluorescence of the cytoplasm was elevated due to the overproduction (Figure 5C). Unlike Hsl7(1–685), in the case of GFP fused to full-length Hsl7, only a single cytoplasmic dot was observed reproducibly.
Figure 5.
Dynamic relocalization of Hsl7 during the cell division cycle. Time-lapse fluorescence microscopy of live hsl7Δ cells (MJY102) harboring YCpT-GFP-Hsl7(1–685) during bud emergence and early stages of the cell cycle (A) or during cytokinesis (B). Arrows indicate appearance of a cytoplasmic dot. (C) Wild-type strain (MJY112) harboring YCpLG-GFP-HSL7 was grown on SCRaf-Leu medium at 30°C to mid-exponential phase and then galactose was added (2% final concentration). After 5 h of induction, the live cells were viewed in the fluorescence microscope. (D) Time-lapse fluorescence mincroscopy of a live wild-type cell (MJY112) expressing GFP-Hsl7 (full length) from the HSL7 promoter on a CEN plasmid (YCpT-GFP-HSL7) throughout an entire cell cycle, showing cytoplasmic dot (arrow) of GFP-Hsl7 in an unbudded cell whose disappearance and redeposition at the bud neck is concomitant with bud emergence.
Dynamic Localization of Hsl7 During the Cell Cycle
Collectively, the observations described immediately above suggested that accumulation of Hsl7 as a cytoplasmic dot was not an artifact of mutant forms of Hsl7. To confirm that localization to the cytosolic dot occurred normally and was physiologically relevant, we first carefully examined cells expressing GFP-Hsl7 at near-normal levels from the HSL7 promoter on a CEN plasmid (YCpT-GFP-HSL7) in asynchronous culture. In a total population of 393 cells examined, 65% showed exclusive staining at the bud neck, as we have reported previously (Shulewitz et al., 1999), whereas 28% showed Hsl7 concentrated exclusively as a small intracellular dot and 7% displayed both staining patterns simultaneously (our unpublished results). We noted that cells with a bud (ranging from small to large) always displayed staining at the neck, whereas all of the cells that displayed the cytosolic dot and lacked a signal at the neck were unbudded and cells with both staining patterns always had only a small bud. These data suggested that Hsl7 localization changes throughout the cell cycle in a regular manner. To obtain additional and direct support for this conclusion, and to avoid artificial means for synchronizing the cells, localization of GFP-Hsl7 was followed by time-lapse fluorescence microscopy in individual cells embedded in thin agar slabs. Newborn (unbudded) cells always displayed a cytoplasmic dot, invariably closer to the incipient bud site than to any other part of the cell surface; and, just after bud emergence, the signal at the dot faded as the fluorescent ring at the bud neck appeared (Figure 5D). The signal at the neck remained through most of the cell cycle, but disappeared rather abruptly, concomitant with the onset of anaphase but before cytokinesis (Figure 5D). The loss of signal at late mitosis was not due to cell cycle-dependent proteolysis of Hsl7 because immunoblotting of extracts prepared from synchronized cultures indicates that Hsl7 is quite a stable protein (McMillan et al., 1999; Shulewitz, 2000). Alternatively, since localization of Hsl7 to the bud neck requires stable association with Hsl1, as we have demonstrated here, the loss of the GFP-Hsl7 signal at the bud neck could be due to cell cycle-dependent degradation of Hsl1. Indeed, it has recently been shown that Hsl1 is targeted for destruction by the cell cycle-regulated ubiquitin ligase known as the “anaphase-promoting complex” (or APC) (Burton and Solomon, 2000).
Relocalization of Hsl7 from the cytoplasmic dot to the bud neck follows the same kinetics as the appearance of Hsl1, which is detected at the daughter-side septin ring from bud emergence until anaphase (Barral et al., 1999). However, in G1 cells, Hsl1 cannot be detected and certainly does not localize as a cytoplasmic dot, even when an Hsl1-GFP fusion is overexpressed from the GAL1 promoter (Cid, unpublished results). Presumably, Hsl1 cannot accumulate until the APC is inactivated, which does not occur until late G1 or the G1-S transition (Amon et al., 1994).
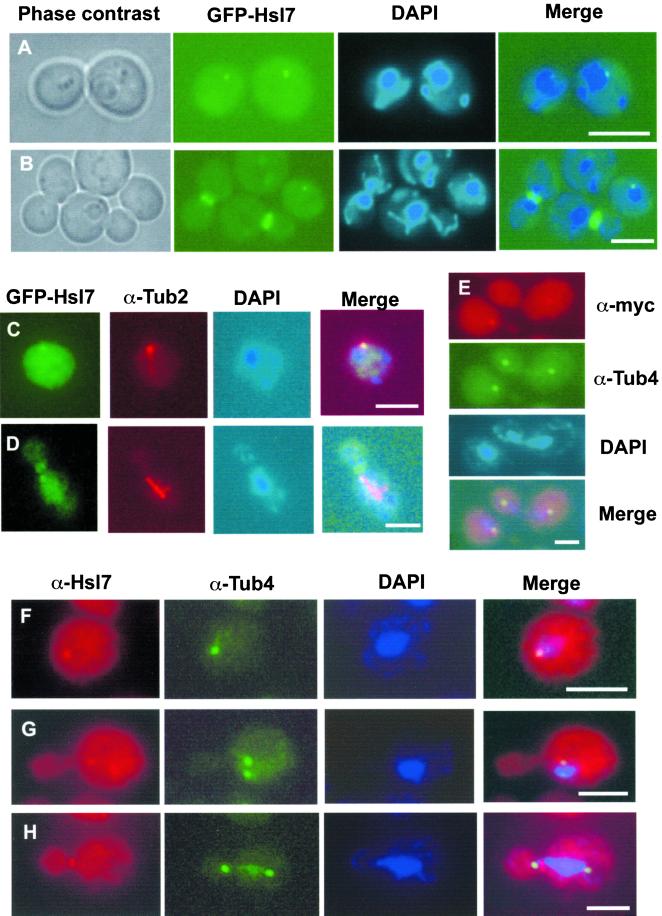
Hsl7 Localizes to the Cytoplasmic Face of the Spindle Pole Body in G1 Cells
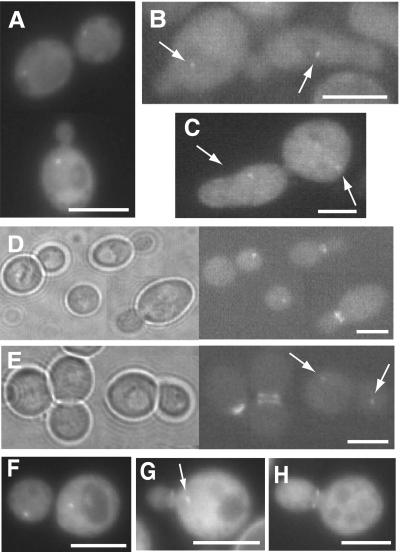
To determine whether the cytoplasmic dot corresponded to a known subcellular structure or compartment, several complementary approaches were taken. First, costaining of cells expressing GFP-Hsl7 with the DNA dye DAPI demonstrated that, in every cell containing a dot, the spot was always immediately juxtaposed to the nucleus (Figure 6, A and B). Second, cells expressing GFP-Hsl7 were costained with a rat monoclonal anti-yeast α-tubulin antibody to determine the position of the dot with respect to microtubules (under mild fixation conditions to preserve GFP fluorescence). The GFP signal always coincided with the single astral microtubule array in G1 cells (Figure 6C) and, occasionally, with one end of the short spindle formed in S/G2 cells (Figure 6D), suggestive of a protein that colocalizes with the SPB. Third, with the use of indirect immunofluorescence with mouse monoclonal anti-Myc antibodies to examine Hsl7 tagged with a C-terminal Myc epitope (Figure 6E) or affinity-purified mouse polyclonal anti-Hsl7 antibodies to examine native Hsl7 (Figure 6F), and rabbit polyclonal antibodies against Tub4 (yeast γ-tubulin), a specific marker for the SPB (Sobel and Snyder, 1995; Geissler et al., 1996; Marschall et al., 1996), the same dot was observed in G1 cells as was seen with the use of GFP-Hsl7, and its position was congruent with Tub4. In cells with a short spindle (presumably S phase), Hsl7 decorated only one of the two SPBs (Figure 6G). However, as observed before for GFP-Hsl7 (Shulewitz et al., 1999), in M phase cells (marked by an elongated spindle), all of the Hsl7 was localized to the bud neck and no detectable Hsl7 was present at either SPB (Figure 6H).
Figure 6.
Cytoplasmic Hsl7 dot corresponds to the SPB. (A and B) Perinuclear localization of the cytoplasmic dot in G1 cells. Fluorescence microscopy was performed on wild-type cells (MJY112) expressing GFP-Hsl7 (green) from the HSL7 promoter on a CEN plasmid (YCpT-GFP-HSL7) that were grown to mid-exponential phase in SCGlc-Trp at 30°C and stained with a DNA-specific dye (DAPI) to discern the nucleus (blue). (C and D) Wild-type cells (MJY112) expressing GFP-Hsl7 from the GAL1 promoter on a CEN plasmid (YCpLG-GFP-HSL7) were grown in SCRaf-Leu at 30°C to mid-exponential phase and then induced by addition of galactose (2% final concentration). After 6 h, the cells were lightly fixed, permeabilized, stained with rat anti-α-tubulin mAb YOL134 (and an appropriate Cy3-labeled secondary antibody), counterstained with DAPI, and viewed in a fluorescence microscope, as described in MATERIALS AND METHODS. Diffuse greencytoplasmic and nuclear signal is nonspecific autofluorescence, whereas in a G1 cell the prominent green cytoplasmic dot (GFP-Hsl7) is always coincident with a bundle of astral microtubules (C), and in a preanaphase cell, GFP-Hsl7 is always at the bud neck, but can be found occasionally decorating one end of the spindle (D). (E) Wild-type cells (MJY112) expressing C-terminally c-Myc-tagged Hsl7 (Hsl7-Myc) from the GAL1 promoter on a CEN plasmid (YCpUG-HSL7-Myc) were grown in SCRaf-Ura medium at 30°C to mid-exponential phase and then induced with galactose (2% final concentration). After 6 h, cells were fixed, permeabilized, stained with mouse anti-c-Myc mAb 9E10 (visualized with an appropriate Cy3-conjugated secondary antibody) and with rabbit anti-Tub4 polyclonal antibodies (visualized with an appropriate fluorescein isothiocyanate-conjugated secondary antibody), counterstained with DAPI, and visualized in a fluorescence microscope. The cytoplasmic dot of Hsl7-myc is congruent with the γ-tubulin signal in the early G1 cell (lower left) and, in a cell at late stage of mitosis, Hsl7-myc is deposited at one of the developing SPBs (upper right). (F–H) Same cells as in E, except that Hsl7 (red) was stained with affinity-purified mouse polyclonal anti-Hsl7 antibodies (rather than with mouse anti-c-Myc mAb) before costaining with anti-Tub4 antibodies (green) and counterstaining with DAPI. Unbudded (G1) cells always show a single dot of Hsl7 congruent with Tub4 (F), whereas, characteristically, early after SPB duplication and separation Hsl7 associates assymetrically with only one SPB (G). By the time a cell has initiated mitosis, Hsl7 is found exclusively at the bud neck and is never observed at either SPB (H). Bars, 5 μm.
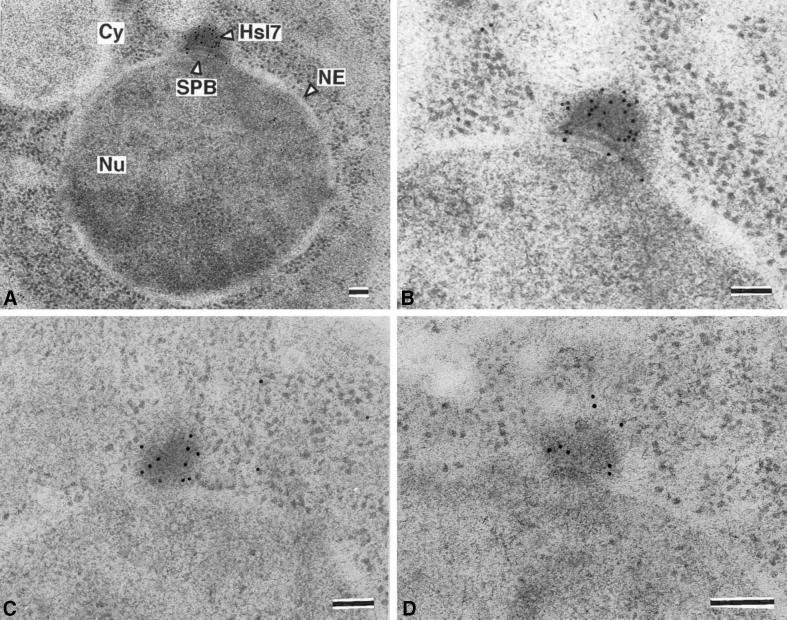
Careful inspection of the merged images revealed that, although Hsl7 largely colocalized with Tub4 at the SPB, the Hsl7 staining always extended slightly beyond the Tub4 staining and always toward the cytoplasm and away from the nucleus. This observation suggested that Hsl7 localizes specifically to the cytoplasmic side of the SPB. To verify this conclusion, frozen thin sections of wild-type yeast cells expressing Hs17-myc were stained with affinity-purified mouse polyclonal anti-Hs17 antibodies and gold-labeled secondary antibodies, and examined in the electron microscope. In every section where a single (nonduplicated) SPB was observed, the gold particles clustered prominently at the SPB on the cytoplasmic side of the nuclear envelope, coincident with an electron-dense “cloud” of SPB-associated material (Figure 7), and nowhere else. Control cells (hsl7Δ) showed no gold particles at this or any other location (our unpublished results). The prominence of the electron-dense material on the outer (cytosolic) face of the SPB is due, in part, to its better preservation with the use of the cryofixation method required for immunoelectron microscopy (compared with standard aldehyde-based fixation procedures) and, in part, to overproduction of Hsl7-myc. However, compensating for ambiguities with regard to the plane of sectioning when comparing these images and those for normal control cells, the cytoplasmic density on the SPB in the Hsl7-myc overproducers is only modestly enlarged (no more than 1.5–2 times its “normal” size) (McDonald, unpublished results). Taken together, these results demonstrate that endogenous Hsl7 localizes to the cytoplasmic side of the SPB during G1 phase, before its total redistribution to the septin ring later in the cell cycle.
Figure 7.
Hsl7 is specifically localized to the cytosolic face of the SPB. Strain MJY112 harboring YCpUG-HSL7-Myc was induced with galactose for 5 h and then prepared for immunoelectron microscopy, as described in MATERIALS AND METHODS. Frozen thin-sections were stained with affinity-purified mouse polyclonal anti-Hsl7 antibodies then with gold particles coated with donkey anti-mouse immunoglobulin, and examined by transmission electron microscopy. In sections of G1 cells, staining was specific for the SPB and no detectable staining of any other place or structure was observed (A). Staining of the SPB was reproducibly observed in every G1 cell examined (B–D). Cy, cytoplasm; NE, nuclear envelope; Nu, nucleus. Bars, 100 nm.
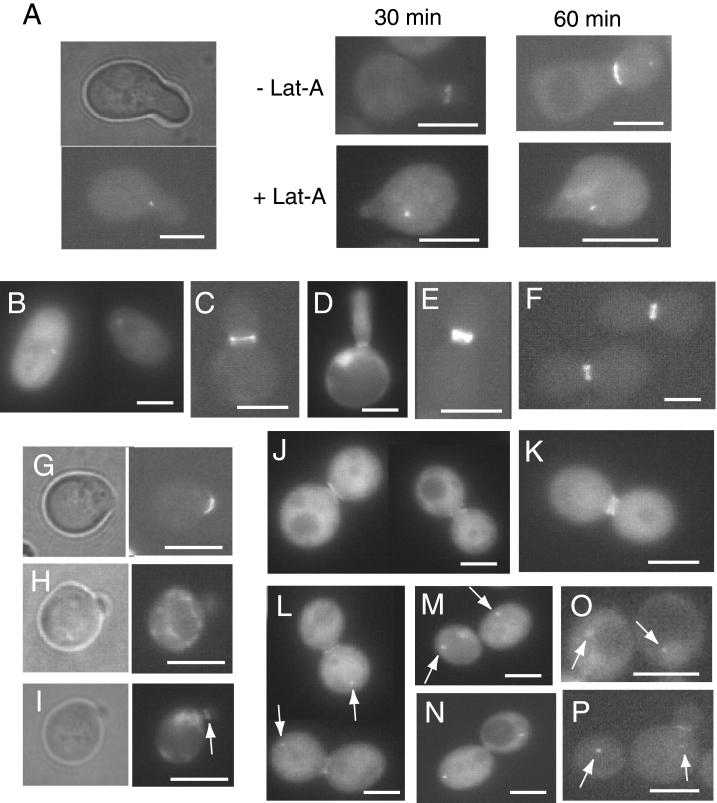
Displacement of Hsl7 from the SPB Occurs Concomitant with Bud Emergence
Redistribution of Hsl7 from the SPB to the septins seems to occur at the time of bud emergence, or shortly thereafter (Figure 5D). To further delineate the timing of Hsl7 translocation from the SPB to the bud neck, several approaches were taken. First, cultures of cells expressing GFP-Hsl7 at a near normal level (from the HSL7 promoter on a CEN plasmid) were synchronized in G1 by treatment with the mating pheromone α-factor. After 3 h, cells exhibited the pear-shaped morphology indicative of a pheromone-induced G1 arrest and projection (“shmoo tip”) formation. Under these conditions, and consistent with arrest during G1 phase of the cell cycle, GFP-Hsl7 was localized as a single cytoplasmic dot in every cell (Figure 8A, left), which colocalized with markers of the SPB (our unpublished results). The pheromone-treated cells were then collected, washed to remove the α-factor, and resuspended in fresh medium in two equal portions, one of which was treated with latrunculin A (200 μM), an effective actin polymerization inhibitor in yeast (Ayscough et al., 1997), and the other received an equivalent volume of solvent (dimethyl sulfoxide) alone as a control. In the absence of latrunculin, the washed cells proceeded synchronously through the cell cycle, and Hsl7 began to redistribute from the SPB to the neck of the incipient bud by 20–30 min after removal from α-factor (Figure 8A, right). This timing seemed to correspond to events that just precede bud emergence because most of the cells (≥80%) had a distinct bud projecting from the former shmoo tip with GFP-Hsl7 present at the neck by 40 min after release from pheromone arrest. By 50–60 min after release, GFP-Hsl7 was completely redistributed to the bud neck (Figure 8A, right). In contrast, in the latrunculin A-treated cells, where bud emergence was prevented, Hsl7 remained at the SPB (Figure 8A, right).
Figure 8.
Cell-cycle stage dependence and functional requirements for relocalization of Hsl7. (A) Hsl7 resides at the SPB in cells arrested in G1 by mating pheromone treatment. Strain MJY155 harboring YCpT-GFP-HSL7, expressing GFP-Hsl7 from the HSL7 promoter on a CEN plasmid, was synchronized by exposure to α-factor and viewed in the fluoresence microscope to confirm formation of the characteristic “shmoo” morphology (left). The arrested cells were then washed to release them from the pheromone-imposed block and allowed to resume growth for the indicated times in the absence (−) or presence (+) of latrunculin-A (right). (B) Strain CWY78 (cdc28–4ts) harboring YCpLG-GFP-HSL7 was induced with galactose and shifted to restrictive temperature (37°C) for 4 h. (C) Strain Y543 (cdc4–1ts) harboring YCpT-GFP-HSL7 was shifted to 37°C for 3 h. (D) Strain YSS19 (cdc34–1ts) harboring YCpLG-GFP-HSL7 was induced with galactose and shifted to 37°C for 9 h. (E) Strain VBY610 (cdc4–1 hsl7Δ) harboring YCpT-GFP-Hsl7(1–685) and shifted to 37°C for 3 h. Note that, although this C-terminally truncated version of Hsl7 can be found at one or both SPBs during S phase (as well as at the bud neck) (Figure 5A), Hsl7(1–685) nonetheless relocates quantitatively to the septin rings in cdc4–1 cells arrested at restrictive temperature in which initiation of DNA replication is blocked. (F) Strain VBY3113a (cdc31–1ts hsl7Δ) harboring YCpT-GFP-Hsl7(1–685) and shifted to 37°C for 3 h. (G) Strain HVY21 (rho1–104ts) expressing Cdc10-GFP from plasmid pLA10 (Cid et al., 1998) shifted to 37°C for 90 min. (H) Same strain as in G harboring YCpT-HSL1-GFP under identical conditions; in most cells (91%), like the one shown here, Hsl1-GFP did not localize to the neck of the tiny buds. (I) Same strain as in G harboring YCpT-GFP-HSL7 under the same conditions; in most cells (87%), GFP-Hsl7 did not localize to the neck of the tiny bud, whereas in a minority of the population (13%), like the one shown here, some GFP-Hsl7 could be detected at the bud neck(arrow). (J) Strain SLJ139 (cdc16–1ts) harboring YCpLG-GFP-HSL7 was induced with galactose and shifted to 37°C for 6 h. Background is high here because cells were pre-stained with DAPI for visualization of nuclei. (K) Strain JC305 (cdc23–1ts) harboring YCpLG-GFP-HSL7 was induced with galactose and shifted to 37°C for 6 h. (L) Strain YSS41 (cdc15–1ts) harboring YCpLG-GFP-HSL7 was induced with galactose and shifted to 37°C for 3 h; both the bud neck and an SPB (arrows) were faintly stained. The arrested cells were shifted into fresh medium at 26°C and allowed to resume growth for 45 min in either the absence (M) or the presence (N) of latrunculin-A. Wild-type strain VBY4012a (O) and its otherwise isogenic derivative VBY4012c (tub2–401cs) (P), each harboring a CEN plasmid expressing GFP-Hsl7(1–685) from the HSL7 promoter, were shifted to nonpermissive temperature (16°C) for 8 h. Bars, 5 μm.
Next, localization of GFP-Hsl7 in cells carrying various cdc mutations that block progression at early stages of the cell cycle was examined. In cells carrying the cdc28–4 allele, which arrests specifically in G1 when shifted to nonpermissive temperature (Segal et al., 1998), GFP-Hsl7 was localized exclusively to the SPB (Figure 8B). In contrast, in cells carrying the cdc4–1 allele, which blocks initiation of DNA replication, but not bud emergence (Pringle and Hartwell, 1981), due to the failure of a ubiquitin ligase (SCFCdc4) to target a specific Cdk inhibitor (Sic1) for destruction (Feldman et al., 1997), GFP-Hsl7 was located exclusively at the bud neck at restrictive temperature (Figure 8C). Likewise, in cells carrying the cdc34–1 allele, which also blocks initiation of DNA replication, but not bud emergence (Pringle and Hartwell, 1981), because it inactivates the ubiquitin-conjugating enzyme required for the function of multiple SCF complexes (Mathias et al., 1998), including that required for Swe1 destruction (Kaiser et al., 1998), GFP-Hsl7 was localized at the base of the markedly elongated buds that formed in these mutant cells at nonpermissive temperature (Figure 8D). For these experiments, we also constructed a cdc4–1 hsl7Δ double mutant to avoid the possibility that endogenous Hsl7 might compete with GFP-Hsl7 expressed from the plasmid and thereby obscure localization of GFP-Hsl7 to the SPB. Even under these conditions, GFP-Hsl7 was located at the bud neck and no detectable SPB staining was observed (Figure 8E). To try to pinpoint when during bud emergence Hsl7 relocates from the SPB to the bud neck, the distributions of GFP-Hsl7, Hsl1-GFP, and a septin (Cdc10-GFP) were examined in cells expressing each of these proteins and carrying a specific temperature-sensitive rho1 allele (rho1–104), which supports formation of very small abortive buds at the restrictive temperature that are then susceptible to lysis (Yamochi et al., 1994). Complete loss of Rho1 function compromises both cell wall biosynthesis and proper polarization of the actin cytoskeleton (Cabib et al., 1998). After 90 min at restrictive temperature, 50 intact (unlysed) cells of each culture were examined. In 100% of the cells, the septin (Cdc10-GFP) accumulated at the base of the button-like buds (Figure 8G), whereas only 9 and 13% of the cells, respectively, had Hsl1-GFP (Figure 8H) or GFP-Hsl7 (Figure 8I) at the same location. Given that we have amply demonstrated above that association with Hsl1 is required for recruitment of Hsl7 to the bud neck, the slightly lower value observed for Hsl1-GFP probably reflects a limitation in our ability to detect Hsl1-GFP because it decorates the septin ring less brightly than GFP-Hsl7. None of the cells expressing GFP-Hsl7 displayed a cytoplasmic dot. We conclude, therefore, that bud emergence is necessary, but not sufficient, for efficient recruitment of Hsl7 (mediated by Hsl1) to the septins at the bud neck, and that displacement of Hsl7 from the SPB precedes, and can occur in the absence of, its deposition at the bud neck.
Displacement of Hsl7 from the SPB Is Independent of SPB Dynamics
Among the events that occur before or concomitant with bud emergence are duplication of the SPB and separation/migration of the resulting two SPBs to form the poles of the short spindle. Hence, we examined whether ejection of Hsl7 from the SPB and its localization at the bud neck was dependent on any of these aspects of SPB dynamics. As mentioned above, GFP-Hsl7 was able to relocate from the SPB to the bud neck in both cdc4 and cdc34 mutants, in which SPB separation does not occur (Pringle and Hartwell, 1981). Thus, SPB separation is not necessary for displacement of Hsl7 from the SPB. To examine whether duplication of the SPB is required for removal of Hsl7, we constructed a hsl7Δ cdc31–1 double mutant. The cdc31–1 mutation causes cells to arrest with a single SPB, although bud development proceeds normally (Pringle and Hartwell, 1981). At restrictive temperature, GFP-Hsl7 localized exclusively to the septin ring at the bud neck in cdc31–1 hsl7Δ cells (Figure 8F). Thus, both displacement of Hsl7 from the SPB and its relocalization to the bud neck appeared to be independent of both SPB duplication and separation. However, the latter results should perhaps be interpreted with caution because Cdc31 is a component of the SPB (Spang et al., 1993; Biggins and Rose, 1994). Hence, Cdc31 itself could be involved in anchoring Hsl7 to the SPB; and, perturbing Cdc31 function might allow Hsl7 to detach from the SPB. However, we could detect no obvious exacerbation of phenotypes when the cdc31–1 hsl7Δ double mutant was compared with its parental single mutants, nor did we observe any effect of HSL7 overexpression in cdc31–1 cells (our unpublished results). Although indirect, the lack of obvious genetic interaction is at least consistent with the view that Hsl7 and Cdc31 do not associate physically. Moreover, Cdc31 is a component of the so-called “half-bridge” substructure of the SPB (Spang et al., 1993; Biggins and Rose, 1994), whereas our ultrastructural analysis by immunoelectron microscopy places Hsl7 mainly at the periphery of the SPB (Figure 7).
Redistribution of Hsl7 from Septin Ring to SPB
Based upon our time-lapse photomicroscopy (Figure 5, B and D), Hsl7 is displaced from the bud neck in late anaphase, concomitant with the destruction of Hsl1, and becomes associated with the SPB by the completion of cytokinesis. To examine this aspect of the relocation of Hsl7 in more detail, we first examined localization of GFP-Hsl7 in cells carrying temperature-sensitive mutations in two core subunits of the APC (Page and Hieter, 1999). On shift to nonpermissive conditions, cdc16 and cdc23 mutants undergo mitotic arrest (Lamb et al., 1994). In cells arrested at this stage, GFP-Hsl7 localized exclusively to the bud neck, and SPB localization could not be observed (Figure 8, J and K). This observation suggests that APC-dependent degradation of Hsl1 is required to free Hsl7 from the bud neck, as a prelude to its relocalization to the SPB. Normally, GFP-Hsl7 and native Hsl7 at the bud neck reside exclusively on the septin ring facing the daughter cell (Shulewitz et al., 1999). Interestingly, when either cdc16 or cdc23 cells were left at restrictive temperature for >2 h, this pronounced asymmetry was lost (i.e., the mother-side septin ring became stained nearly as brightly as the bud-side septin ring).
We examined GFP-Hsl7 localization in cells arrested at a slightly later stage of mitosis with the use of the cdc15–1 mutation, which inactivates a protein kinase necessary for exit from mitosis (Jaspersen et al., 1998; Xu et al., 2000), causing a blockade of the anaphase-to-telophase transition. At the nonpermissive temperature, GFP-Hsl7 at the bud neck was visible, but faint, in nearly all of the cells, but in a large fraction of them (∼33%) a very faint signal at the SPB could also be observed in both the mother and/or the incipient daughter cell (Figure 8L). The reduction in GFP-Hsl7 fluorescence at the neck suggests that removal of Hsl7 from the septin scaffold has commenced at this stage. Because the cdc15–1 block is reversible upon temperature down-shift (Surana et al., 1993), the arrested cells were collected and resuspended in fresh medium at the permissive temperature, allowing the cells to enter telophase and continue through the cell cycle. As the cells progressed through cytokinesis, the GFP-Hsl7 signal disappeared from the bud neck, whereas fluorescence at the SPB became distinctly more intense. By 60 min after down-shift, most cells had completed cytokinesis, and in the great majority (≥80%) of the resulting single unbudded cells the SPB was brightly stained (Figure 8M). In contrast to the movement of GFP-Hsl7 from the SPB to the bud neck, addition of 200 μM latrunculin A did not affect the redistribution of GFP-Hsl7 from the bud neck to the SPB upon downshift of cdc15–1 cells (Figure 8N). Thus, when cells are allowed to complete mitosis, Hsl7 is released from the septin rings and translocates to the SPB in an actin-independent manner.
Hsl7 Relocation Does Not Require New Protein Synthesis or Microtubules
The experiments presented above did not distinguish between two possibilities: 1) Hsl7 molecules at the SPB redistribute to the bud neck, or 2) Hsl7 molecules are degraded after their release from the SPB and only newly synthesized Hsl7 localizes at the bud neck. To address this issue, cells were arrested in G1 by treatment with α-factor, and then pheromone was removed to allow cells to progress synchronously through the cell cycle in either the absence or presence of cycloheximide (10 μg/ml), a concentration of this inhibitor sufficient to abrogate yeast protein synthesis. In the samples treated with cycloheximide, bud emergence was considerably delayed (but proceeds via utilization of pools of preformed components). At 1 h after release from pheromone, only 14% of the cells (n = 100) displayed buds, and by 2 h after release only 30% of the population displayed buds (and bud size was very heterogeneous). Nonetheless, in every cell that contained a bud, Hsl7 localized as a ring to the bud neck (our unpublished results), suggesting that de novo synthesis of Hsl7 is not required for its translocation from the SPB to the septins. To determine whether Hsl7 recruitment to the bud neck depended on microtubules, G1-arrested cells were released from pheromone in medium lacking or containing the mictrotubule-depolymerizing drug benomyl (10 μg/ml). In the pheromone-treated cultures, 100% of cells displayed the single SPB-associated dot (Figure 8A). After release, the untreated cells all showed bright staining exclusively at the bud neck within 30 min, as observed before (Figure 8A). Treatment with benomyl delayed budding by ∼10 min and caused the cells to become slightly asynchronous; nevertheless, by 1 h after release, that majority (55%) of the cells (n = 100) displayed GFP-Hsl7 exclusively at the septin rings, only 9% retained obvious dots, and the remainder were ambiguous (our unpublished results). This observation suggests that microtubule assembly is also not required for relocation of Hsl7 to the bud neck.
The effects of cycloheximide treatment were also examined in cells released from cdc15-induced mitotic arrest. On decreasing the temperature, the presence of cycloheximide did not strongly affect movement of GFP-Hsl7 from the septin rings to the SPB; at 1 h after downshift, the majority (70%) of the cells (n = 100) displayed bright dots exclusively and the remainder of the cells were similar, but showed some residual staining at the bud neck. Thus, like redistribution of Hsl7 from the SPB to the septins, the return of Hsl7 from the bud neck to the SPB does not require new protein synthesis, suggesting that the same population of Hsl7 is able to shuttle between the two locations. In contrast, when cdc15-arrested cells were downshifted in the presence of benomyl, the cells were unable to resume cell cycle progression even after 1 h, and not unexpectedly GFP-Hsl7 remained at the neck. To clarify whether this observation implied a role for astral microtubules in Hsl7 movement from the bud neck to the SPB, GFP-Hsl7(1–685) (which marks the SPB especially brightly; Figure 5A) was introduced into a strain that contains the cold-sensitive tub2–401 mutation. This tub2 allele specifically disrupts astral, but not spindle, microtubules at the nonpermissive temperature (Sullivan and Huffaker, 1992). After 8 h at restrictive temperature (16°C), the mutant formed abnormally large cells, but the vast majority of the unbudded cells displayed the characteristic SPB-associated dot (Figure 8, O and P). Moreover, no difference was observed at this low temperature between the tub4–401 strain and its otherwise isogenic parent (n = 100 each) when the proportion of cells showing GFP-Hsl7(1–685)-decorated SPBs in the population was counted, suggesting that astral microtubules are not necessary for Hsl7 movement from the bud neck to the SPB. Thus, the inhibition of Hsl7 relocalization by benomyl in cdc15 cells released from temperature-induced arrest may reflect a blockade of mitotic exit by the drug itself, perhaps as a consequence of reactivation of the spindle assembly checkpoint (Rudner and Murray, 1996).
Displacement of Hsl7 from SPB Is Independent of Hsl1
Hsl7 associates with the SPB at a cell cycle stage when Hsl1 is not present in the cell (Barral et al., 1999; McMillan et al., 1999) due to its APC-dependent degradation (Burton and Solomon, 2000). Conversely, as we have shown here, interaction of Hsl7 with Hsl1 is required for Hsl7 localization to the bud neck. Because redistribution of Hsl7 from the SPB to the septins occurs when Hsl1 is resynthesized and itself localizes to the bud neck (Barral et al., 1999; McMillan et al., 1999; Shulewitz et al., 1999), it seemed possible that, in addition to playing a role in anchoring Hsl7 at the bud neck, Hsl1 might also play a role in displacing Hsl7 from the SPB. However, several lines of evidence indicate that release of Hsl7 from the SPB occurs independently of Hsl1. First, as expected based on our findings presented above, GFP-Hsl7 localized to the SPB in an hsl1Δ swe1Δ double mutant (elimination of Swe1 prevents cell elongation resulting from absence of Hsl1; Ma et al., 1996) (Figure 9A) and in a septin mutant, cdc10–11, at the restrictive temperature, in either the presence (Figure 9B) or absence (Figure 9C) of a functional SWE1 gene. Strikingly, when individual hsl1Δ (or hsl1Δ swe1Δ) cells were followed by time-lapse fluorescence microscopy (of the sort shown in Figure 5D), the GFP-Hsl7 dot disappears shortly after bud emergence, just as in wild-type cells (our unpublished results). Conversely, overproduction of Hsl1 from the GAL1 promoter on a CEN plasmid (YCpLG-HSL1-HA3) (Figure 9, D and E) or elevation of Hsl1 level by expression of an integrated nondegradable version (Burton and Solomon, 2000) that persists in G1 (Figure 9F) had no dramatic effects on the distribution of GFP-Hsl7 between the SPB and the septins (Figure 9, F–H), did not prevent GFP-Hsl7 from targeting to the SPB in G1 cells, and did not cause premature recruitment of GFP-Hsl7 to the bud neck. Thus, regardless of whether Hsl1 was absent, overproduced, or stabilized by mutation, there was no effect on the dynamics of Hsl7 association with and release from the SPB in unbudded (G1) cells.
Figure 9.
Hsl7 localization to the SPB, and its release, are independent of Hsl1. (A) Live cells of strain VBY17 (hsl1Δ swe1Δ) harboring YCpT-GFP-HSL7 display GFP-Hsl7 staining of the SPB in G1 cells (bottom) or in cells at telophase (top), but not at any other cell cycle stage. Presence of swe1Δ mutation prevents formation of elongated buds due to loss of Hsl1. Live cells of strain VBY30 (cdc10–11ts) (B) or strain VBY31 (cdc10–11 swe1Δ) (C), each harboring YCpT-GFP-HSL7, shifted to 37°C for 3 h; due to septin delocalization, GFP-Hsl7 staining of the neck of the elongated buds is ablated, whereas SPB staining (arrows) is unaffected (arrows). Strain MJY112 harboring both YCpT-GFP-HSL7 and YCpLG-HSL1-(HA)3 (Shulewitz et al., 1999) was grown in Raf medium (D) or shifted to galactose medium for 3 h (E); although overproduction of Hsl1 causes abnormal GFP-Hsl7 staining of both the mother- and daughter-side septin rings (E) (Cid, Shulewitz, and Thorner, unpublished results), SPBs (arrows) still recruit GFP-Hsl7 normally in unbudded (G1) cells. (F–H) Strain VBY204 strain, in which the normal HSL1 locus has been substituted by a triple mutant (HSL1R828A L831A N836A) that renders Hsl1 more stable during G1 (Burton and Solomon, 2000), and also harboring YCpLG-GFP-HSL7, was induced with galactose for 6 h; typical cells in G1 (F), after bud emergence (G), and at mitosis (H) show the same distribution of GFP-Hsl7 to the SPB or the neck as do wild-type cells (Figure 5D).
DISCUSSION
We have recently proposed that, in budding yeast, Hsl7 serves as an adapter linking the Hsl1 protein kinase and its putative substrate, Swe1, allowing Hsl1 to inactivate Swe1 in response to assembly of the septin filaments at the bud neck (Shulewitz et al., 1999). To better understand the role of Hsl7, we explored in this study several features of Hsl7 in greater detail. Consistent with the proposal that Hsl7 has an adapter function, we were able to demonstrate direct physical association between Hsl7 and Hsl1, and between Hsl7 and Swe1, and that stable Hsl7-Hsl1 interaction is required for down-regulation of Swe1. We mapped the site in Hsl7 required for its interaction with Hsl1 by several independent methods (in vitro binding, two-hybrid interaction, random mutagenesis) and were able to pinpoint individual residues in Hsl7 essential for Hsl1 binding, which appear to define a conserved Hsl1-binding site. We found that Hsl7 oligomerizes in vivo and serves as a direct substrate for Hsl1 in vitro. Somewhat surprisingly, under the same conditions, Swe1 is not phosphorylated by Hsl1, regardless of the presence of Hsl7. Finally, we discovered a new subcellular location for Hsl7. We found that Hsl7 associates with the SPB in a cell cycle-dependent manner, decorating the cytoplasmic face of the SPB in G1 cells before relocating to the septin rings as the bud develops. This new finding raises the possibility that Hsl7 has other functions in addition to its role at the bud neck.
Formation of Hsl7-Hsl1 Complexes Is Required for Inactivation of Swe1
Both Hsl1 (Barral et al., 1999) and Hsl7 (Shulewitz et al., 1999) localize to the daughter-side septin ring in budded cells. Deposition of Hsl7 at this location requires the presence of Hsl1, whereas Hsl1 can localize to the bud neck in the absence of Hsl7 (Barral et al., 1999; Shulewitz et al., 1999). Moreover, Hsl7 and Hsl1 associate which each other, as indicated by our previous, more circumstantial evidence (Shulewitz et al., 1999) and as demonstrated conclusively in this study. We showed here that radiolabeled Hsl7 prepared by in vitro translation can associate with the C terminus of Hsl1 (purified as a GST fusion from bacteria), indicating that this association is direct and does not require the participation of any other yeast proteins, consistent with our previous evidence that Hsl1 recruits Hsl7 to the bud neck and not vice versa (Shulewitz et al., 1999). Furthermore, single-residue substitution mutations in Hsl7 that prevent Hsl1 binding (but do not perturb any other property of Hsl7 tested), no longer localize to the septins and cannot complement the elongated bud morphology of an hsl7Δ cell, in strong support of the model that catastrophic down-regulation of Swe1 occurs at the bud neck and requires formation of stable Hsl7-Hsl1 complexes at that site. We showed previously that the state of modification of Swe1, as judged by a gel mobility shift assay, is dramatically affected by the absence of either Hsl1 or Hsl7 (Shulewitz et al., 1999). Our current work shows that just preventing Hsl7-Hsl1 association blocks Swe1 inactivation. Presumably, therefore, when Hsl7-Hsl1 complexes do not assemble, Swe1 cannot be appropriately modified to become a target for its SCFMet30-mediated ubiquitinylation and subsequent proteasome-dependent destruction (Kaiser et al., 1998). Although Swe1 is a phosphoprotein (Sia et al., 1998; Sreenivasan and Kellogg, 1999), it is not yet clear whether the modification required for destruction of Swe1 is phosphorylation or methylation (or both).
With the use of an immune-complex kinase assay in which Hsl1 displayed both potent autophosphorylation and phosphotransferase activity (with the use of GST-Hsl7 as an exogenous substrate), Hsl1 was unable to phosphorylate Swe1. Although this negative result does not rule out that Hsl1 is able to phosphorylate Swe1 under some conditions, it raises the possibility that Swe1 may be regulated by posttranslational modifications not mediated by Hsl1. In this regard, it is intriguing that Hsl7 has a domain with readily detectable homology to S-adenosyl-methionine-dependent protein-arginine methyltransferases (PRMTs) (Ma, 2000) and that both Hsl7 itself (Frankel and Clarke, 2000; Lee et al., 2000) and its human ortholog, JBP1 (Pollack et al., 1999), reportedly display PRMT activity in vitro. Hence, it is tempting to speculate that Hsl7 may convert specific Arg residues in Swe1 to their ω-NG-monomethyl-Arg, ω-NG,NG-bismethyl-Arg, and/or ω-NG,N′G-dimethyl-Arg derivatives and that such modifications are important for marking Swe1 as a substrate for Hsl1 (or some other protein kinase) and/or as a target for ubiquitinylation by the SCFMet30. Consistent with this possibility, we found that Hsl7 can associate with Swe1 in the absence of any other yeast protein, as might be expected for an enzyme-substrate complex. However, the portion of Hsl7 responsible for its interaction with Swe1 seems to lie in the amino-terminal region of Hsl7, well upstream of the PRMT homolog domain (residues 350–660). Moreover, site-directed mutagenesis of seven different conserved residues in the PRMT domain of Hsl7 fail to ablate its ability to complement the elongated bud phenotype of an hsl7Δ mutant (Shulewitz, 2000). Furthermore, in our hands (Shulewitz and S.E. Crown, unpublished results), GST-Hsl7 or Hsl7-myc expressed in and purified from yeast or bacterial cells (in any of four different buffers), with or without prephosphorylation by Hsl1, fails to display detectable methyltransferase activity (with the use of any of three different standard methyl-acceptor substrates) under conditions where another yeast PRMT, Hmt1/Rmt1 (Gary et al., 1996; McBride et al., 2000), displays potent activity. Finally, even in the presence of Hsl7, Hsl1 failed to phosphorylate Swe1. Thus, at the present time, the most reasonable conclusion is that the Hsl1-Hsl7 module recruits Swe1 to the septin ring at the bud neck so that Swe1 can be phosphorylated by another protein kinase resident at this location. Candidates for this function include the following: one (or both) of the other two Nim1-related protein kinases in S. cerevisiae, Kcc4 and Gin4, which also localize to the bud neck and appear to be involved in promoting and/or monitoring septin assembly (Carroll et al., 1998; Barral et al., 1999; Longtine et al., 2000); another bud neck-associated protein kinase, Elm1, that also contributes to proper mitotic regulation (Sreenivasan and Kellogg, 1999; Bouquin et al., 2000); and the polo kinase homolog Cdc5, which also localizes, in part, to the bud neck (Song et al., 2000).
Hsl1-independent Functions of Hsl7
Our finding that Hsl7 localizes to the SPB in G1 cells suggests that it may have a discrete role at this site. Hsl1 does not appear to localize to the SPB, suggesting that Hsl7 function at the SPB is independent of Hsl1. Indeed, release of Hsl7 from the SPB at G1-S and association of Hsl7 with the SPB in late telophase-early G1 occur normally in the complete absence of Hsl1, even though Hsl7 cannot localize to the bud neck in the same cells. Thus, other components must be responsible for recruitment of Hsl7 to, and displacement of Hsl7 from, the SPB.
A truncation of Hsl7, Hsl7(1–685), lacking its C-terminal 142 amino acids, is no longer an in vitro substrate for Hsl1, whereas this C-terminal fragment alone is an efficient substrate for Hsl1, and the phosphorylated residue lies in this short segment. However, we found that the amino-terminal region of Hsl7 was required for its tight binding to Hsl1. The simplest explanation for these observations is that the amino-terminal region of Hsl7 contains a “docking site” that mediates its high-affinity interaction with Hsl1 and is widely separated (at least in terms of primary structure) from the phosphoacceptor residues in Hsl7, a situation that has been observed for many other protein kinase-substrate pairs (Bardwell et al., 1996). Hsl7(1–685) restores normal morphology to an hsl7Δ mutant just as well as full-length Hsl7. One interpretation of this result is that phosphorylation of Hsl7 by Hsl1 is dispensible for its function in the down-regulation of Swe1. Alternatively, however, the missing C-terminal segment of Hsl7 could normally be a negative regulatory domain and phosphorylation by Hsl1 (or removal by the truncation mutation) could alleviate its inhibitory constraint. In this regard, it is noteworthy that GFP-Hsl7(1–685) decorates the SPB more brightly than an otherwise identical GFP fusion to full-length Hsl7. Also, GFP-Hsl7(1–685) more than occasionally decorates both SPBs (as well as the bud neck) well into S phase, whereas GFP-Hsl7 is removed from the SPB shortly after bud emergence and is localized thereafter exclusively at the bud neck. Even when GFP-Hsl7 is rampantly overproduced, causing some of it to remain at the SPB during S phase, it localizes asymmetrically, i.e., to only one end of the premitotic spindle, unlike Hsl7(1–685). Thus, the C-terminal segment of Hsl7 (and/or its phosphorylation by Hsl1) may have some role in regulating or restricting the degree of Hsl7 association with SPBs.
Sequential Localization of Key Cell Cycle Regulatory Components at the SPB
We have shown here that Hsl7 is displaced from the bud neck during late anaphase, relocates to the outer plaque of the SPB at late telophase-early G1, and remains at the SPB until G1/S (or until S/G2, in the case where Hsl7 is overexpressed or deprived of its C-terminal segment). To our knowledge, this G1-specific pattern of SPB decoration is unique to Hsl7. Other proteins that play a role in cell cycle regulation, like certain components of the mitotic exit network (such as Cdc15 and Tem1), also have been shown recently to localize transiently to the outer plaque of the SPB (Cenamor et al., 1999; Bardin et al., 2000; Pereira et al., 2000; Xu et al., 2000), but this association occurs specifically in M phase. Appearance of Tem1, a small GTPase, at the SPB (Bardin et al., 2000) seems to correspond in timing to the disappearance of Hsl7 from the SPB in our experiments. Given that Hsl7 is a premitotic marker and Tem1 is mitotic marker of the SPB, it is tempting to speculate that displacement of Hsl7 might be required before efficient recruitment of Tem1 and, thus, that sequential localization of proteins to the SPB plays a role in optimizing coordination of nuclear dynamics with events (like assembly of the septin-based cytokinesis apparatus) occurring at the cell cortex. Indeed, two regulators involved in the exit from mitosis, the Cdc5 and Cdc15 protein kinases, as well as the type I phosphoprotein phosphatase, Glc7, localize sequentially during mitosis, first to the SPB and, late in mitosis, to the site of cytokinesis (Bloecher and Tatchell, 2000; Song et al., 2000; Xu et al., 2000). Our experiments with the use of a temperature-sensitive cdc15 mutant suggest that Cdc15 function per se is not required for localization of Hsl7 to the SPB, but rather that release of Hsl7 from the septin ring and its relocation to SPB requires efficient mitotic exit. Nonetheless, it is noteworthy that disappearance of Cdc15 from the SPB and its appearance at the bud neck (Xu et al., 2000) are correlated with the appearance of Hsl7 at the SPB and its disappearance from the septin scaffold before cytokinesis. Degradation of Hsl1 by the APC (Burton and Solomon, 2000) is sufficient to explain displacement of Hsl7 from the bud neck during anaphase. Consistent with this conclusion, in temperature-sensitive mutants that inactivate the APC, GFP-Hsl7 remained anchored at the septin rings and did not associate detectably with the SPB. Our studies and those of others cited immediately above provide substantial evidence that successive movements of signaling proteins from the SPB to the bud neck constitute part of the control mechanisms that properly integrate nuclear dynamics with cortical events during the cell division cycle.
We have demonstrated that Hsl7 accumulates at two different subcellular locations: on the daughter cell-side of the bud neck during cell division, where it physically associates with Hsl1; and on the cytoplasmic face of the SPB before mitosis in an Hsl1-independent manner. When situated at the bud neck, Hsl7 participates in processes that lead to the timely and catastrophic destruction of Swe1 (McMillan et al., 1999; Shulewitz et al., 1999). Conversely, cells that maintain high Swe1 levels are unable to enter mitosis and arrest in G2 as a consequence of sustained tyrosine phosphorylation of the B-cyclin/CDK (Clb/Cdc28) complex (Lew, 2000). Inhibiting Clb/Cdc28 activity by overexpression of Swe1 (Booher et al., 1993), by depletion of B-type cyclins (Fitch et al., 1992; Nasmyth, 1993), or by failure to degrade the Clb/Cdc28-specific CDK inhibitor Sic1 (Verma et al., 1997) arrests cells before the onset of mitosis and has two strikingly diagnostic consequences: 1) blockade of SPB separation, arresting nuclear dynamics with side-by-side SPBs and preventing the formation of the short (premitotic) spindle (Lim et al., 1996); and 2) a pronounced delay in shifting from polarized to isotropic growth of the bud, leading to the appearance of markedly elongated buds (Lew and Reed, 1993). Thus, active Clb/Cdc28 is essential both for SPB separation and for the switch from polarized to isotropic growth, suggesting that specific targets for Clb/Cdc28 are found at those subcellular locations. Our earlier work (Shulewitz et al., 1999) and the data presented here demonstrate that assembly of Hsl7-Hsl1 complexes at the bud neck is required for timely and efficient inactivation of the majority of the cellular content of Swe1 to promote the G2-M transition. Likewise, the role of Hsl7 at the SPB (perhaps in conjunction with an SPB-associated protein kinase) may be to bring about the early and highly localized inactivation of a small pool of Swe1 (or some other target), thereby allowing a commensurately small and localized pool of Clb/Cdc28 to act. If correct, this kind of highly localized action many be one of the molecular mechanisms that cells use to ensure that the events of the cell cycle are optimally coordinated both spatially and temporally.
ACKNOWLEDGMENTS
We thank John Kilmartin, Tim Huffaker, Dave Morgan, Waheeda Khalfan, Becky Davis, Mark Rose, Sofie Salama, Curt Wittenberg, Yoshimi Takai, Tim Durfee, Janet Burton, and Mark Solomon for plasmids, strains, and/or antibodies; and Carla J. Inouye for constructing plasmid pGEM-5Z-HSL7. This work was supported by postdoctoral fellowships from the Madrid Educational Exchange Commission/Fulbright Foundation and the Del Amo Foundation (to V.J.C.), by predoctoral traineeships GM-07032 and CA-09041 and a William V. Power Graduate Fellowship (to M.J.S.); and by National Institutes of Health Research Grant GM-21841 and facilities provided by the Berkeley campus Cancer Research Laboratory (to J.T.).
Abbreviations used:
- CDK
cyclin-dependent protein kinase
- Cy3
indocarbocyanine dye
- Gal
galactose
- GFP
Aequorea victoria green fluorescent protein
- GST
Schistosoma japonicum glutathione S-transferase
- HA
influenza virus hemagglutinin
- mAb
monoclonal antibody
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PRMT
S-adenosylmethionine-dependent protein-arginine methyltransferase
- Raf
raffinose
- SPB
spindle pole body
REFERENCES
- Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, McCollum D, Surana U. Tying the knot: linking cytokinesis to the nuclear cycle. J Cell Sci. 2000;113:1503–1513. doi: 10.1242/jcs.113.9.1503. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Bardwell L, Cook JG, Chang EC, Cairns BR, Thorner J. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol Cell Biol. 1996;16:3637–3650. doi: 10.1128/mcb.16.7.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell L, Cook JG, Zhu-Shimoni JX, Voora D, Thorner J. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc Natl Acad Sci USA. 1998;95:15400–15405. doi: 10.1073/pnas.95.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y, Mermali V, Mooseker MS, Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol Cell. 2000;5:841–851. doi: 10.1016/s1097-2765(00)80324-x. [DOI] [PubMed] [Google Scholar]
- Barral Y, Parra M, Bidlingmaier S, Snyder M. Nim1-related kinases coordinate cell-cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Rose MD. Direct interaction between yeast spindle pole body components: Kar1p is required for Cdc31p localization to the spindle pole body. J Cell Biol. 1994;125:843–852. doi: 10.1083/jcb.125.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloecher A, Tatchell K. Dynamic localization of protein phosphatase type 1 in the mitotic cell cycle of Saccharomyces cerevisiae. J Cell Biol. 2000;149:125–140. doi: 10.1083/jcb.149.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher RN, Deshaies RJ, Kirschner MW. Properties of the Saccharomyces cerevisiae wee1 and its differential regulation of p34cdc28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquin N, Barral Y, Courbeyrette R, Blondel M, Snyder M, Mann C. Regulation of cytokinesis by the Elm1 protein kinase in Saccharomyces cerevisiae. J Cell Sci. 2000;113:1435–1445. doi: 10.1242/jcs.113.8.1435. [DOI] [PubMed] [Google Scholar]
- Burke DJ. Complexity in the spindle checkpoint. Curr Opin Genet Dev. 2000;10:26–31. doi: 10.1016/s0959-437x(99)00040-4. [DOI] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. Hsl1p, a Swe1p inhibitor, is degraded via the anaphase-promoting complex. Mol Cell Biol. 2000;20:4614–4625. doi: 10.1128/mcb.20.13.4614-4625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976;69:717–721. doi: 10.1083/jcb.69.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E, Drgonova J, Drgon T. Role of small G proteins in yeast cell polarization and wall biosynthesis. Annu Rev Biochem. 1998;67:307–333. doi: 10.1146/annurev.biochem.67.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Altman R, Schieltz D, Yates JR, III, Kellogg D. The septins are required for the mitosis-specific activation of the Gin4 kinase. J Cell Biol. 1998;143:709–717. doi: 10.1083/jcb.143.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenamor R, Jimenez J, Cid VJ, Nombela C, Sanchez M. The budding yeast Cdc15 localizes to the spindle pole body in a cell-cycle-dependent manner. Mol Cell Biol Res Commun. 1999;2:178–184. doi: 10.1006/mcbr.1999.0173. [DOI] [PubMed] [Google Scholar]
- Chant J. Cell polarity in yeast. Annu Rev Cell Dev Biol. 1999;15:365–391. doi: 10.1146/annurev.cellbio.15.1.365. [DOI] [PubMed] [Google Scholar]
- Cid VJ, Adamikova L, Cenamor R, Molina M, Sanchez M, Nombela C. Cell integrity and morphogenesis in a budding yeast septin mutant. Microbiology. 1998;144:3463–3474. doi: 10.1099/00221287-144-12-3463. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Tang Z, Dunphy WG. Negative regulation of the wee1 protein kinase by direct action of the nim1/cdr1 mitotic inducer. Cell. 1993;72:919–929. doi: 10.1016/0092-8674(93)90580-j. [DOI] [PubMed] [Google Scholar]
- Cooper JA, Kiehart DP. Septins may form a ubiquitous family of cytoskeletal filaments. J Cell Biol. 1996;134:1345–1348. doi: 10.1083/jcb.134.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini DJ, Adams AEM, Fares H, DeVirgilio C, Valle G, Chuang JS, Pringle JR. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Durfee T, Draper O, Zupan J, Conklin DS, Zambryski PC. New tools for protein linkage mapping and general two-hybrid screening. Yeast. 1999;15:1761–1768. doi: 10.1002/(SICI)1097-0061(199912)15:16<1761::AID-YEA494>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsey G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Field CM, Kellogg D. Septins: cytoskeletal polymers or signaling GTPases? Trends Cell Biol. 1999;9:387–394. doi: 10.1016/s0962-8924(99)01632-3. [DOI] [PubMed] [Google Scholar]
- Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- Fitch I, Dahmann C, Surana U, Amon A, Nasmyth K, Goetsch L, Byers B, Futcher B. Characterization of four B-type cyclin genes of the budding yeast Sachharomyces cerevisiae. Mol Biol Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A, Clarke S. PRMT3 is a distinct member of the protein arginine N-methyltransferase family: conferral of substrate specificity by a zinc-finger domain. J Biol Chem. 2000;275:32974–32982. doi: 10.1074/jbc.M006445200. [DOI] [PubMed] [Google Scholar]
- Frazier JA, Wong ML, Longtine MS, Pringle JR, Mann M, Mitchison TJ, Field C. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J Cell Biol. 1998;143:737–749. doi: 10.1083/jcb.143.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Lin WJ, Yang MC, Herschman HR, Clarke S. The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. J Biol Chem. 1996;271:12585–12594. doi: 10.1074/jbc.271.21.12585. [DOI] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soues S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the gamma-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugnino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. FASEB J. 9, 576–596. 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. [PubMed] [Google Scholar]
- Hartwell LH. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69,:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Inouye C, Dhillon N, Durfee T, Zambryski PC, Thorner J. Mutational analysis of STE5 in the yeast Saccharomyces cerevisiae: application of a differential interaction trap assay for examining protein-protein interactions. Genetics. 1997;147:479–492. doi: 10.1093/genetics/147.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Tinker-Kulberg RL, Morgan DO. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EW. Tackling the protease problem in yeast. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- Kaiser P, Sia RAL, Bardes EGS, Lew DJ, Reed SI. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes Dev. 1998;12:2587–2597. doi: 10.1101/gad.12.16.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Wright B, Milstein C. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JR, Michaud WA, Sikorski RS, Hieter PA. Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J. 1994;13:4321–4328. doi: 10.1002/j.1460-2075.1994.tb06752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Cook JR, Pollack BP, Kinzy TG, Norris D, Pestka S. Hsl7p, the yeast homologoue of human JBP1, is a protein methyltransferase. Biochem Biophys Res Commun. 2000;274:105–111. doi: 10.1006/bbrc.2000.3049. [DOI] [PubMed] [Google Scholar]
- Lew D. Cell-cycle checkpoints that ensure coordination between nuclear and cytoplasmic events in Saccharomyces cerevisiae. Curr Opin Genet Dev. 2000;10:47–53. doi: 10.1016/s0959-437x(99)00051-9. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and the cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HH, Goh PY, Surana U. Spindle pole body separation in Saccharomyces cerevisiae requires dephosphorylation of the tyrosine 19 residue of Cdc28. Mol Cell Biol. 1996;16:6385–6397. doi: 10.1128/mcb.16.11.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott M, Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol. 1998;143:1947–1960. doi: 10.1083/jcb.143.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. Involvement of PCH family proteins in cytokinesis and actin distribution. Microsc Res Tech. 2000;49:168–172. doi: 10.1002/(SICI)1097-0029(20000415)49:2<168::AID-JEMT9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Longhese MP, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P. DNA damage checkpoint in budding yeast. EMBO J. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, DeMarini DJ, Valencik ML, Al-War OS, Fares H, De Virgilio C, Pringle JR. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Theesfeld CL, McMillan JN, Weaver E, Pringle JR, Lew DJ. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:4049–4061. doi: 10.1128/mcb.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz AT, Reed SI. Sequence analysis of temperature-sensitive mutations in the Saccharomyces cerevisiae gene CDC28. Mol Cell Biol. 1986;6:4099–4103. doi: 10.1128/mcb.6.11.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XJ. Cell-cycle regulatory proteins Hsl7p/Skb1p belong to the protein methyltransferase superfamily. Trends Biochem Sci. 2000;25:11–12. doi: 10.1016/s0968-0004(99)01509-1. [DOI] [PubMed] [Google Scholar]
- Ma X-J, Lu Q, Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Sacchaaromyces cerevisiae. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- Madden K, Snyder M. Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast gamma-tubulin-like protein: implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias M, Steussy CN, Goebl MG. An essential domain within Cdc34p is required for binding to a complex containing Cdc4p and Cdc53p in Saccharomyces cerevisiae. J Biol Chem. 1998;273:4040–4045. doi: 10.1074/jbc.273.7.4040. [DOI] [PubMed] [Google Scholar]
- McBride, A.E., Weiss, V.H., Kim, H.K., Hogle, J.M., and Silver, P.A. (2000). Analysis of the yeast arginine methyltransferase Hmt1p/Rmt1p and its in vivo function. Cofactor binding and substrate interactions. 275, 3128–3136. [DOI] [PubMed]
- McDonald KL. High pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabeling. Methods Mol Biol. 1999;1:77–97. doi: 10.1385/1-59259-201-5:77. [DOI] [PubMed] [Google Scholar]
- McMillan JN, Longtine MS, Sia RA, Theesfeld CL, Bardes ES, Pringle JR, Lew DJ. The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol Cell Biol. 1999;19:6929–6939. doi: 10.1128/mcb.19.10.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan JN, Sia RAL, Lew DJ. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J Cell Biol. 1998;142:1487–1499. doi: 10.1083/jcb.142.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino A, Tanaka K, Kamei T, Umikawa M, Fujiwara T, Takai Y. Shs1p: a novel member of septin that interacts with Spa2p, involved in polarized growth in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1998;251:732–736. doi: 10.1006/bbrc.1998.9541. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr Opin Cell Biol. 1993;5:166–179. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- Page AM, Hieter P. The anaphase-promoting complex: new subunits and regulators. Annu Rev Biochem. 1999;68:583–609. doi: 10.1146/annurev.biochem.68.1.583. [DOI] [PubMed] [Google Scholar]
- Peranen J, Rikkonen M, Hyvonen M, Kaariainenen L. T7 vectors with a modified T7lac promoter for expression of proteins in Escherichia coli. Anal Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E. The Bub2 spindle checkpoint links nuclear migration with mitotic exit. Mol Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- Pollack BP, Kotenko SV, He W, Izotova LS, Barnoski BL, Pestka S. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J Biol Chem. 1999;274:31531–31542. doi: 10.1074/jbc.274.44.31531. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Hartwell LH. The Saccharomyces cerevisiae cell cycle. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1981. pp. 97–142. [Google Scholar]
- Rhind N, Russell P. Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- Russell P, Moreno S, Reed SI. Conservation of mitotic controls in fission and budding yeast. Cell. 1989;57:295–303. doi: 10.1016/0092-8674(89)90967-7. [DOI] [PubMed] [Google Scholar]
- Russell P, Nurse P. The mitotic inducer nim1+ functions in a regulatory network of protein kinase homologs controlling the initiation of mitosis. Cell. 1987;49:569–576. doi: 10.1016/0092-8674(87)90459-4. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Segal M, Clarke DJ, Reed SI. Clb5-associated kinase activity is required early in the spindle pathway for correct preanaphase nuclear positioning in Saccharomyces cerevisiae. J Cell Biol. 1998;143:135–145. doi: 10.1083/jcb.143.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Shulewitz MJ. Septin assembly regulates cell cycle progression through activation of a protein kinase signaling pathway. Ph.D. Dissertation. Berkeley, CA: University of California; 2000. [Google Scholar]
- Shulewitz MJ, Inouye CJ, Thorner J. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia RA, Bardes ES, Lew DJ. Control of Swe1p degradation by the morphogenetic checkpoint. EMBO J. 1998;17:6678–6688. doi: 10.1093/emboj/17.22.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel SG, Snyder M. A highly divergent gamma-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. J Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Grenfell TZ, Garfield S, Erikson RL, Lee KS. Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol Cell Biol. 2000;20:286–298. doi: 10.1128/mcb.20.1.286-298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni R, Carmichael JP, Murray JA. Parameters affecting lithium acetate-mediated transformation of Saccharomyces cerevisiae and development of a rapid and simplified procedure. Curr Genet. 1993;24:455–459. doi: 10.1007/BF00351857. [DOI] [PubMed] [Google Scholar]
- Spang A, Courtney I, Fackler U, Matzner M, Schiebel E. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J Cell Biol. 1993;123:405–416. doi: 10.1083/jcb.123.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan A, Kellogg D. The Elm1 kinase functions in a mitotic signaling network in budding yeast. Mol Cell Biol. 1999;19:7983–7994. doi: 10.1128/mcb.19.12.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DS, Biggins S, Rose MD. The yeast centrin, Cdc31p, and the interacting protein kinase, Kic1p, are required for cell integrity. J Cell Biol. 1998;143:751–765. doi: 10.1083/jcb.143.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble WS. Septins: a highly conserved family of membrane-associated GTPases with functions in cell division and beyond. J Membr Biol. 1999;169:75–81. doi: 10.1007/s002329900519. [DOI] [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- White MA. The yeast two-hybrid system: forward and reverse. Proc Natl Acad Sci USA. 1996;93:10001–10003. doi: 10.1073/pnas.93.19.10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Huang HK, Kaiser P, Latterich M, Hunter T. Phosphorylation and spindle pole body localization of the Cdc15p mitotic regulatory protein kinase in budding yeast. Curr Biol. 2000;10:329–332. doi: 10.1016/s0960-9822(00)00382-1. [DOI] [PubMed] [Google Scholar]
- Yamochi W, Tanaka K, Nonaka H, Maeda A, Musha T, Takai Y. Growth site localization of Rho1 small GTP-binding protein and its involvement in bud formation in Saccharomyces cerevisiae. J Cell Biol. 1994;125:1077–1093. doi: 10.1083/jcb.125.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]