Abstract
Emerging evidence suggests that recombinant adeno-associated viral (rAAV) vectors can be used for specific gene targeting in human somatic cells. We have developed an rAAV vector construction procedure employing fusion PCR and a single cloning step that considerably simplifies the knockout process. We demonstrate its utility by disrupting genes at specific positions within human colon cancer cells as well as within immortalized normal epithelial cells. This technology should be broadly applicable to in vitro studies that require the manipulation of the human genome.
INTRODUCTION
Targeted gene inactivation is the most definitive method for evaluating the function of a gene within a specific cell type or organism. Gene disruption through homologous recombination can now be efficiently accomplished in a variety of cell types, including those from bacteria, yeast, chickens and rodents (1). However, the same methods, when applied to human somatic cells, are generally inefficient (2). These difficulties have led to widespread use of ‘knockdowns’ rather than knockout (KO) approaches using either antisense or small interfering RNA technologies (3). Although knockdown approaches can provide important information quickly, the interpretation of such experiments can be difficult because of non-specific effects or incomplete inactivation of the gene product of interest (4–6). This has stimulated efforts to improve the efficiency of KO approaches, particularly in human cells (7–10).
Recombinant adeno-associated viruses (rAAV) can be used to obtain higher frequencies of targeted gene disruptions than generally obtained with conventional plasmid KO vectors (11,12). AAV is a human parvovirus which possesses a single-stranded DNA genome of 4.7 kb. The wild-type virion possesses two open reading frames (ORFs), termed rep and cap, flanked by two inverted terminal repeats (ITRs). The rep ORF encodes proteins involved in viral replication, and the cap ORF encodes proteins necessary for viral packaging. In most rAAV vectors these ORFs are deleted and replaced with a gene expression cassette of interest.
In the current study, we have developed new vectors and cloning approaches that facilitate and simplify the use of rAAV for creating KOs in human cells. We also show, for the first time, that rAAV KO vectors can be used to delete specific sequences at pre-chosen positions. These advances enable the efficient genetic modification of human cells.
MATERIALS AND METHODS
Cells and reagents
The human colon cancer cell line HCT116 (ATCC, Manassas, Virginia) was maintained in HCT116 growth medium [McCoy’s 5A modified medium (HyClone, Logan, UT) supplemented with 10% FBS (HyClone), 100 units/ml penicillin and 100 µg/ml streptomycin (Invitrogen Corp., Carlsbad, CA)]. The human retinal pigment epithelial cell line hTERT-RPE1 (Clontech, Palo Alto, CA) was maintained in RPE growth medium [Dulbecco’s modified Eagle’s medium (DMEM)–Nutrient Mixture F-12 Ham (Sigma-Aldrich Corp., St Louis, MO) supplemented with 10% FBS, 100 units/ml penicillin, 100 µg/ml streptomycin, 2 mM l-glutamine (HyClone) and 0.35% sodium bicarbonate (HyClone)]. HEK 293 cells were obtained from ATCC and cultured in 293 growth medium [DMEM (HyClone) supplemented with 10% FBS (HyClone), 100 units/ml penicillin and 100 µg/ml streptomycin]. For drug selection, the media were supplemented with either geneticin (0.4 mg/ml, Invitrogen) or hygromycin B (100 µg/ml, Calbiochem, San Diego, CA). All cell lines were maintained at 37°C in 5% CO2. All enzymes were purchased from New England Biolabs (Beverly, MA) except where indicated otherwise.
Construction of pNeDaKO plasmid vectors
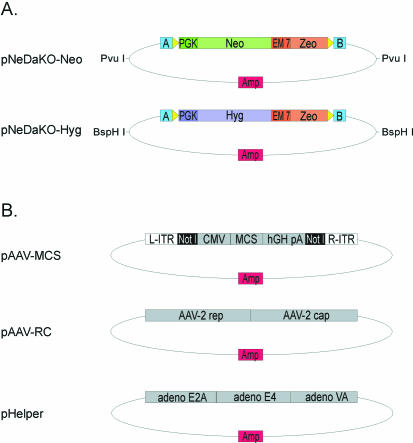
The pNeDaKO (pronounced ‘p-Need a Knockout’) plasmid vectors (Fig. 1) contain either a gene conferring resistance to neomycin (Neo) or hygromycin (Hyg) linked in tandem to a gene conferring zeomycin resistance (Zeo). For the pNeDaKO-Neo plasmid, two PCR fragments were generated and joined via fusion PCR. The first fragment, derived from pKO SelectNeo (Stratagene, La Jolla, CA), was for selection in mammalian cells and contained the Neo gene driven by the phosphoglycerate kinase (PGK) promoter and also contained a polyadenylation signal (pA). An upstream loxP sequence and SacII site were incorporated into the forward primer used to generate this fragment by PCR. The second fragment, derived from pZeoSV (Invitrogen), was for selection in bacterial cells and contained Zeo driven by the EM7 promoter. A downstream loxP sequence and KpnI site were incorporated into the PCR primers used to generate this second fragment. The two fragments were joined via fusion PCR (13). The fusion products were cloned into the SacII and KpnI sites of pBluescript SK+ (Stratagene). PNeDaKO-Hyg was constructed similarly except that the Hyg gene was derived from pIREShyg2 (Clontech) and BstXI replaced SacII. Sequences of ∼20 bp in length that were immediately outside the cloning sites in pBluescript SK+ were surveyed for optimal annealing properties and two were chosen for design of the fusion PCR; these were termed Linker A (5′-CTA AAGGGAACAAAAGCTGGAGC) and Linker B (5′- CGCCCTATAGTGAGTCGTATTAC); see Figure 2 and Results. The electronically compiled sequences of both vectors are provided as Supplementary Material.
Figure 1.
Plasmids required for rAAV-based gene targeting with the pNeDaKO system. (A) Structure of pNeDaKO-Neo and pNeDaKO-Hyg plasmid vectors. Restriction sites for the generation of pNeDaKO fragments used for fusion PCR are shown. A and B refer to linkers A and B (see Materials and Methods); yellow arrows: loxP sites; PGK: phosphoglycerate kinase eukaryotic promoter; Neo: neomycin resistance gene; Hyg: hygromycin resistance gene; EM 7: EM7 prokaryotic promoter; Zeo: zeomycin resistance gene; Amp: ampicillin resistance gene. (B) Associated plasmids necessary for rAAV production. The three plasmids shown were purchased from Stratagene. The fusion PCR products are cloned into the NotI sites in pAAV-MCS. L-ITR: left inverted terminal repeat; R-ITR: right inverted terminal repeat; CMV: cytomegalovirus promoter; MCS: multiple cloning site; hGH pA: human growth hormone poly- adenylation signal; AAV-2 rep and AAV-2 cap: rep and cap sequences required for rAAV replication and packaging; adeno E2A, adeno E4, adeno VA: adenovirus accessory proteins required for efficient rAAV replication.
Figure 2.
Generation of rAAV for gene targeting. Homology arms (HAs) are PCR-amplified from genomic DNA and fused to a selectable cassette via linker sequence homology. NotI restriction sites allow cloning of the fusion product into the AAV plasmid containing the ITR sequences necessary for viral packaging. Dual selection results in a population of plasmids containing the intended targeting cassettes flanked by ITRs. Co-transfection of packaging cells with the targeting plasmid and helper plasmids produces rAAV capable of genomic integration or deletion. A strategy to produce rAAV for the deletion of a generic exon is depicted. PCR primers used for amplification (P1, P2, P3 and P4) and fusion PCR (P1 and P4) are shown. Abbreviations are defined in Figure 1.
Amplification of homology arms
HCT116 genomic DNA was used as the template for generating the left and right homology arms for gene targeting. Regions with minimal numbers of repeated sequences, assessed using RepeatMasker (http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker), were chosen whenever possible. All PCR reactions were performed with Platinum Taq DNA Polymerase High Fidelity (Invitrogen) using the conditions specified by the manufacturer. The homology arms were generated by PCR in 12 separate 10 µl reactions in 96-well plates using the following cycling conditions: 1 cycle of 94°C for 1 min; 4 cycles of 94°C for 10 s, 64°C for 30 s, 68°C for 1 min; 4 cycles of 94°C for 10 s, 61°C for 30 s, 68°C for 1 min; 4 cycles of 94°C for 10 s, 58°C for 30 s, 68°C for 1 min; 22 cycles of 94°C for 10 s, 56.5°C for 30 s, 68°C for 1 min; 1 cycle of 68°C for 5 min. The 12 products were pooled into one tube, extracted with phenol–chloroform, ethanol precipitated and dissolved in 20 µl of water. After electrophoresis on a 0.8% agarose gel, the PCR products were purified using a Qiagen Gel Extraction Kit (Qiagen, Valencia, CA) and eluted into 50 µl of Qiagen Elution Buffer. For the left homology arms, the forward primer contained a NotI site at its 5′ end (Fig. 2 and Table 1). The reverse primer contained the sequence 5′-GCTCCAGCTTTTGTTCCCTTTAG, which is the reverse complement of Linker A engineered into the plasmids described above (Fig. 2 and Table 1). For the right homology arms, the forward primer contained the sequence 5′-CGCCCTATAGTGAGTCGTATTAC of Linker B and the reverse primer contained a NotI site (Table 1).
Table 1. Primer sequences.
| Name | Sequencea | Useb |
|---|---|---|
| P1 | ATACATACGCGGCCGCXXXXXXXXXXXXXXXXXXXX | Forward primer for left HA of YFG; B/U sequence represents a NotI site |
| P2 | GCTCCAGCTTTTGTTCCCTTTAGXXXXXXXXXXXXXXXXXXXX | Reverse primer for left HA of YFG; B/U sequence represents the reverse complement of Linker A |
| P3 | CGCCCTATAGTGAGTCGTATTACXXXXXXXXXXXXXXXXXXXX | Forward primer for right HA of YFG; B/U sequence represents Linker B |
| P4 | ATACATACGCGGCCGCXXXXXXXXXXXXXXXXXXXX | Reverse primer for right HA of YFG; B/U sequence represents a NotI site |
| P1-FHIT | ATACATACGCGGCCGCTGGAATTAAAGAGTGAGTGAAGGC | Forward primer for left HA of FHIT; B/U sequence represents a NotI site |
| P2-FHIT | GCTCCAGCTTTTGTTCCCTTTAGTCTCTAGTCTACTAAAGCACCCAGG | Reverse primer for left HA of FHIT; B/U sequence represents the reverse complement of Linker A |
| P3-FHIT | CGCCCTATAGTGAGTCGTATTACTTACCCTAGATATGGTCTGAAATGG | Forward primer for right HA of FHIT; B/U sequence represents Linker B |
| P4-FHIT | ATACATACGCGGCCGCTCATGTGGTTTATTTGATAGAGCC | Reverse primer for right HA of FHIT; B/U sequence represents a NotI site |
| P1-CCR5 | ATACATACGCGGCCGCTTATTTGGTGAGATGGTGCTTTC | Forward primer for left HA of CCR5; B/U sequence represents a NotI site |
| P2-CCR5 | GCTCCAGCTTTTGTTCCCTTTAGTGATACTGACTGTATGGAAAATGAGA | Reverse primer for left HA of CCR5; B/U sequence represents the reverse complement of Linker A |
| P3-CCR5 | CGCCCTATAGTGAGTCGTATTACGTCCTGCCGCTGCTTGTC | Forward primer for right HA of CCR5; B/U sequence represents Linker B |
| P4-CCR5 | ATACATACGCGGCCGCCCCATAGCAAGACAAAGACCTG | Reverse primer for right HA of CCR5; B/U sequence represents a NotI site |
| FF | GCTGGTGCAGAAATCAATCCC | Forward primer for screening for FHIT targeting events in human cells (see Fig. 4) |
| CF | GCACCATGCTTGACCCAG | Forward primer for screening for CCR5 targeting events in human cells (see Fig. 4) |
| HR | CCACGCCCTCCTACATCG | Reverse primer in Hyg gene for screening targeting events in human cells (see Fig. 4) |
| NR | GTTGTGCCCAGTCATAGCCG | Reverse primer in Neo gene for screening targeting events in human cells (see Fig. 4) |
aAll sequences are shown 5′ → 3′. All primers >30 bp were HPLC purified by the manufacturer.
bYFG: Your Favorite Gene; X20 represents sequences of YFG (see Fig. 2); B/U: bold and underlined; HA: homology arm.
Fusion PCR linking homology arms to selectable markers
The pNeDaKO-Neo plasmid was digested with PvuI, extracted with phenol–chloroform and ethanol precipitated. The 4 kb PvuI fragment containing the drug marker was gel purified with a Qiagen Gel Extraction Kit. When hygromycin selection was desired, the pNeDaKO-Hyg plasmid was digested with BspHI and the 4.5 kb fragment was purified in like fashion. The left and right homology arms were mixed with one of these fragments and the P1 forward primer and the P4 reverse primer were added (Fig. 2 and Table 1). A total reaction volume of 360 µl containing pNeDaKO PvuI fragment (∼400 ng), homology arms (∼350 ng each) and primers (at 0.3 µM) generated sufficient fusion product for subsequent steps. The reaction mix was divided into twelve 30 µl aliquots for PCR, which was performed using the following parameters: 1 cycle of 94°C for 2 min; 20 cycles of 94°C for 30 s, 56°C for 30 s, 68°C for 4 min; 1 cycle of 68°C for 5 min. The 12 PCR reactions were combined into one tube and the primers were removed from the PCR mix using a Qiagen PCR Purification Kit. The fusion PCR products were eluted into 50 µl of Qiagen Elution Buffer and digested with 60 units of NotI in a total volume of 200 µl for 3 h at 37°C. The cleaved products were extracted with phenol and chloroform, precipitated with ethanol and dissolved in 20 µl of water. Following electrophoresis on a 0.8% agarose gel, the ∼4.0 or ∼4.5 kb band (Neo or Hyg, respectively) was purified using a Qiagen Gel Extraction Kit and eluted into 40 µl of Qiagen Elution Buffer. Note that the fusion PCR products were similar in size to the restriction fragments of the pNeDaKO vectors that served as templates and no attempt was made to separate them; the appropriately sized fragments containing both the fusion PCR products and the restriction fragments of pNeDaKO were simply excised from the gel and co-purified.
Ligation of fusion product into pAAV-MCS
The pAAV-MCS vector carries the ITR sequences necessary for AAV packaging plus NotI sites useful for cloning the fusion PCR product (Fig. 1). Two micrograms of pAAV-MCS plasmid (Stratagene) was digested with 40 units NotI for 2 h at 37°C in 200 µl. To this tube, 2 µl of 10 000 units/ml calf intestinal alkaline phosphatase was added and incubated at 37°C for an additional 15 min. The digest was extracted with phenol and chloroform, precipitated with ethanol and dissolved in 20 µl of water. Following electrophoresis on a 0.8% agarose gel, the 3.0 kb fragment containing the plasmid backbone and ITR sequences was purified using a Qiagen Gel Extraction Kit and eluted into 50 µl of Qiagen Elution Buffer. Ligation was performed with the Rapid DNA Ligation Kit (Roche, Basel, Switzerland) by adding 1 µl (∼10 ng) of the pAAV-MCS NotI fragment and 7 µl (∼70 ng) of the fusion PCR product to 2 µl of 5× DNA dilution buffer, 10 µl of 2× T4 DNA ligase buffer and 1 µl of T4 DNA ligase and incubating at room temperature for 1 h. The reaction was extracted with phenol and chloroform, precipitated with ethanol and dissolved in 40 µl of water. Half of the DNA (20 µl) was electroporated into 10 µl of DH10B electro-competent Escherichia coli cells (catalog #18290015, Invitrogen). Seven hundred microliters of Luria Bertani (LB) broth was added to the electroporation cuvette, and the contents were transferred to a 1.5 ml tube and incubated for 15 min at 37°C. Half of this (350 µl) was then transferred to a 50 ml conical tube containing 10 ml of LB containing 40 µg/ml zeomycin and 100 µg/ml ampicillin. The tube was incubated at 37°C shaking at 250 r.p.m. until log phase growth was evident (16–36 h, depending on efficiency of ligation). Other aliquots from the electroporated bacteria were spread on agar plates when individual clones were desired. Plasmid DNA was purified from E.coli cells using a Qiagen Mini Prep Kit, and proper ligation was confirmed with a NotI digest (Fig. 3C).
Figure 3.
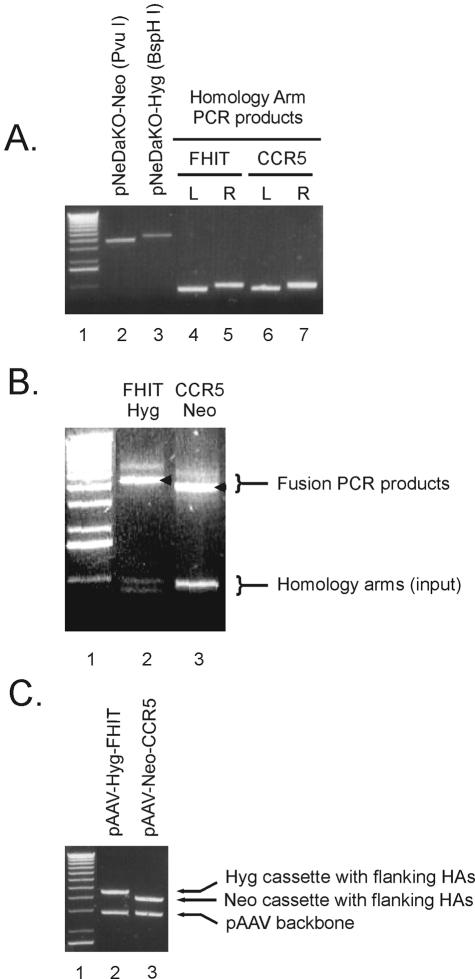
Analytical agarose gels representing various steps in the generation of rAAV targeting constructs. The first lane in each gel contains ∼500 ng of 1 kb + DNA ladder (Invitrogen). (A) DNA components used in fusion PCR. Lane 2: pNeDaKO-Neo PvuI fragment; lane 3: pNeDaKO-Hyg BspHI fragment; lanes 4 and 5: left (L) and right (R) homology arm PCR products for FHIT; lanes 6 and 7: left (L) and right (R) homology arm PCR products for CCR5. (B) Fusion PCR products for FHIT and CCR5. The arrows indicate the desired PCR products while the less prominent products that migrate more slowly are PCR artifacts. (C) Confirmation of gene targeting vector construction. FHIT and CCR5 targeting constructs were digested with NotI and separated by electrophoresis on a 0.8% agarose gel. HAs: homology arms
Packaging of rAAV targeting constructs
The targeting construct made above (2.5 µg) was mixed with pAAV-RC and pHelper plasmids (2.5 µg of each) from the AAV Helper-Free System (Stratagene) and transfected into HEK 293 cells (ATCC) using Lipofectamine (Invitrogen). This DNA was dissolved in Opti-MEM reduced-serum media (Invitrogen) to a total volume of 750 µl (i.e. if volume of DNA was 50 µl, volume of Opti-MEM was 700 µl). Similarly, 54 µl of Lipofectamine was dissolved in Opti-MEM to a total volume of 750 µl. The two tubes were combined and the DNA–Lipofectamine mix was incubated at room temperature for 15 min. HEK 293 cells at 70–80% confluence in a 75 cm2 flask were washed with Hank’s Balanced Salt Solution (HBSS, HyClone) and then 7.5 ml Opti-MEM was added. To this, the 1.5 ml DNA–Lipofectamine mixture was added dropwise, and the cells were incubated at 37°C for 3–4 h. The Opti-MEM was replaced with 293 growth medium and the cells were allowed to grow for 48 h prior to harvesting virus. Virus was harvested according to the AAV Helper-Free System instructions with minor modifications. Briefly, the media was aspirated from the flask and the 293 cells were scraped into 1 ml of phosphate-buffered saline (Invitrogen), transferred to a 2 ml microfuge tube, and subjected to three cycles of freeze–thaw. Each cycle consisted of 10 min freeze in a dry ice–ethanol bath, and 10 min thaw in a 37°C water bath, vortexing after each thaw. The lysate was then clarified by centrifugation at 12 000 r.p.m. in a microfuge to remove cell debris and the supernatant containing rAAV was divided into three aliquots of ∼330 µl each and frozen at –80°C.
Gene targeting of human cells
The cells to be targeted were grown in a 75 cm2 flask until 60–80% confluence (6–8 × 106 cells/flask). The cells were washed 1× with HBSS, the HBSS was removed, and then 330 µl of rAAV lysate and 4 ml of the appropriate growth media were added to the flask. The virus was allowed to infect cells at 37°C for 2–3 h. Afterwards, 8 ml of growth media was added to the flask and the cells were allowed to grow for 48 h. After 48 h, the cells in each 75 cm2 flask were harvested by trypsinization and distributed into ten 96-well plates with media containing either geneticin or hygromycin B. The plates were wrapped in Saran Wrap to minimize evaporation and incubated at 37°C for 2–3 weeks prior to harvest.
Screening for targeting events
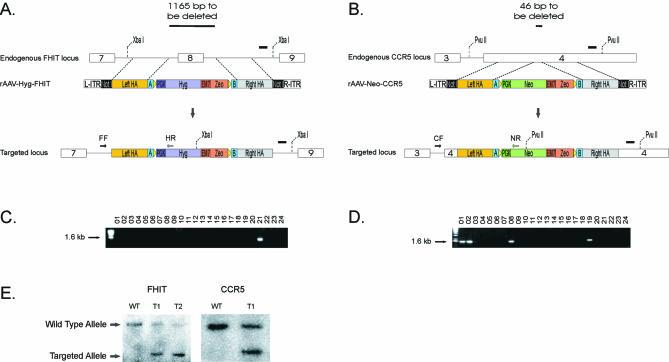
Genomic DNA was extracted from colonies grown in 96-well plates using the Qiagen 96 Well Blood Kit (Qiagen), and DNA was eluted in 100 µl of Elution Buffer according to the manufacturer’s instructions. Locus-specific targeting events were screened by PCR using a forward primer that was situated outside of the left homology arm and a reverse primer that was situated within the Neo (or Hyg) gene. A forward primer within the drug resistance gene and a reverse primer outside of the right homology arm could also be used to screen for recombinants. Sequences of the primers used in the experiments presented below are listed in Table 1. PCR screening was carried out in 10 µl reaction volumes using the same conditions, cycling times and temperatures noted above for amplification of the homology arms except that all the extension times were 130 s instead of 1 min, and 30 cycles instead of 22 cycles were used for 56.5°C step. The presence of a PCR product of the correct size (∼1.6 kb) was indicative of a specific targeting event at the locus of interest. Southern blot analysis was performed as described in (9). Genomic DNA was digested with XbaI (FHIT) or PvuII (CCR5) and the blots probed with ∼600 bp probes derived from sequences downstream of the right homology arms (Fig. 4A, B and E).
Figure 4.
Screen for targeting events. (A) rAAV-Hyg-FHIT was used to generate a 1165 bp deletion of FHIT that included exon 8 and surrounding intronic sequences. Following rAAV-Hyg-FHIT infection, cells were selected for hygromycin resistance. Targeted integration removed exon 8 and surrounding sequences. FF: FHIT forward screening primer; HR: hygromycin reverse screening primer (Table 1). (B) rAAV-Neo-CCR5 was used to generate a 46 bp deletion within exon 4 of CCR5. Following rAAV infection, cells were selected for geneticin resistance. Targeted integration removes a small portion of exon 4. CF: CCR5 forward screening primer; NR: neomycin reverse screening primer (Table 1). (C) Representative gel showing PCR products of 24 HCT116 hygromycin-resistant clones after infection with rAAV-Hyg-FHIT obtained with primers FF and HR. The FHIT gene in clone 21 was correctly targeted. (D) Representative gel showing PCR products of 24 hTERT-immortalized RPE geneticin-resistant clones after infection with rAAV-Neo-CCR5 obtained with primers CF and NR. The CCR5 gene in clones 1, 2, 8 and 19 was correctly targeted. (E) Southern blot of wild-type (WT) and targeted (T) clones demonstrating the restriction digest patterns expected from targeted homologous integration in HCT116 cells. FHIT wild-type (5.4 kb) and targeted (2.8 kb) alleles and CCR5 wild-type (3.6 kb) and targeted (2.2 kb) alleles are shown. In both cases, the probes were derived from sequences downstream of the right homology arms (black rectangles in A and B). The 5.4 kb FHIT wild-type allele was not transferred as well as the smaller targeted allele, explaining the difference in signal strength in FHIT lanes T1 and T2. Abbreviations are defined in Figures 1 and 2.
Cre-mediated excision of the drug resistance marker in targeted cells
When it is desirable to remove the antibiotic resistance cassettes from the targeted locus in human cells, such cells can be transfected with supercoiled pΔE1-creHA plasmid (9). The protocol for transfection with Lipofectamine is the same as that described above. Alternatively, we have used an adenovirus-expressing Cre for the same purpose (14).
RESULTS AND DISCUSSION
Plasmid vectors
We designed two plasmids, one bearing the Neo resistance cassette (pNeDaKO-Neo), and the other bearing the Hyg resistance cassette (pNeDaKO-Hyg), both driven by the PGK promoter (Fig. 1). The same homology arms could thereby be used to create different rAAV vectors, one conferring resistance to geneticin and the other to hygromycin. The availability of two different vectors facilitated KOs in cells which already contain one drug resistance gene (such as the hTERT-RPE1 cells described below). Additionally, the two vectors could be used sequentially to knock out both alleles of a target gene without the need for an intervening Cre-mediated excision of the drug resistance marker. The flanking loxP sites make it possible to remove all drug resistance genes from the targeted cells when desirable. The plasmids also contained the bacterial EM7 promoter driving expression of the Zeo gene. This component was useful for the generation of the targeting construct, as described below.
Generation of rAAV gene targeting constructs by fusion PCR
The steps described below are diagrammed in Figure 2 and methodological details are provided in Materials and Methods.
Step 1: PCR-amplification of homology arms. The optimal packaging size of rAAV is between 4.1 and 4.9 kb (15). As detailed in Table 2, the Hyg cassette plus other features of pNeDaKO-Hyg that were incorporated into the rAAV added up to 2.7 kb. The combined length of the ITRs was 282 bp, leaving up to ∼1.9 kb for the homology arms. We therefore generally used ∼900 bp for each arm. Although the pNeDaKO-Neo allowed an additional 500 bp to be incorporated into the homology arms, we found it convenient to use the same homology arms to create both Hyg- and Neo-based targeting vectors, and 900 bp homology arms proved to be large enough to generate KOs at acceptable efficiencies.
Table 2. rAAV component sizes.
| rAAV-Neo-YFG | rAAV-Hyg-YFG | ||
|---|---|---|---|
| Fragment | Size (bp) | Fragment | Size (bp) |
| L-ITR | 141 | L-ITR | 141 |
| Left homology arm | 900 | Left homology arm | 900 |
| Linker A | 23 | Linker A | 23 |
| 5′ loxP to Linker A | 11 | 5′ loxP to Linker A | 13 |
| 5′ loxP | 34 | 5′ loxP | 34 |
| PGK | 529 | PGK | 513 |
| Neo + pA | 1091 | Hyg B + pA | 1603 |
| EM7 + Zeo | 453 | EM7 + Zeo | 453 |
| 3′ loxP | 34 | 3′ loxP | 34 |
| 3′ loxP to Linker B | 10 | 3′ loxP to Linker B | 10 |
| Linker B | 23 | Linker B | 23 |
| Right homology arm | 900 | Right homology arm | 900 |
| R-ITR | 141 | R-ITR | 141 |
| Total | 4290 | Total | 4788 |
The forward primer used to PCR-amplify the left homology arm and the reverse primer used to amplify the right homology arm both contained NotI restriction sites so that the final constructs could be cloned into the pAAV-MCS vector (Fig. 1) for subsequent viral packaging. The reverse primer used to PCR-amplify the left homology arm also contained a 23 bp linker sequence (reverse complement of linker ‘A’ in Figure 1). Analogously, the forward primer used to generate the right homology arm contained the sequence (not the reverse complement) of linker ‘B’. These linkers provided sequence overlap with the plasmid fragment from pNeDaKO-Hyg or pNeDaKO-Neo; this overlap was required for the fusion PCR in step 2.
Step 2. Fusion PCR. The amplified homology arms were mixed with pNeDaKO-Hyg or pNeDaKO-Neo restriction fragments in equimolar ratios and PCR was performed with primers P1 and P4 (Fig. 2). This generated a fusion PCR product containing the drug resistance cassette flanked by the left and right homology arms.
Step 3. Ligation. The fusion PCR product was digested with NotI and ligated into the pAAV-MCS NotI fragment. The ligation product was transformed into electro-competent E.coli and double selected with ampicillin and zeomycin. The AAV-MCS component provided the Amp element and the fusion PCR product provided the Zeo element. The double selection ensured that all recovered plasmids contained the desired sequences. For the electroporation, we included control ligations using pAAV-MCS NotI fragment alone (without PCR product) and PCR product alone (without pAAV-MCS fragment) to ensure the absence of contaminating DNAs in the ligation components. These controls should yield no colonies resistant to both ampicillin and zeomycin. DNA from the pooled population of doubly resistant clones grown in liquid culture was used for the generation of rAAV. If desired, random clones from this pooled DNA can be sequenced to ensure that the desired sequences are present and that mutations were not introduced during the PCR or cloning steps. However, it has previously been shown that low levels of mutations in targeting constructs do not preclude homologous integration in human cells (2). Therefore, cloning is not generally necessary and the pooled DNA can be directly used for rAAV production.
Step 4. Viral packaging. DNA from the targeting construct plus pHelper and pAAV-RC DNAs were co-transfected into HEK 293 cells using Lipofectamine. As shown in Figure 1, the pHelper plasmid contained the adeno E2A, E4 and VA genes while the pAAV-RC contained the rep and cap genes necessary for efficient packaging. Time-course experiments showed that the optimum time to recover rAAV from the transfected cells was 48–72 h following transfection (16 and unpublished data). The rAAV preparations generally contained ∼3 × 108 rAAV particles/ml, as determined by real-time PCR (17), and 330 µl aliquots generated ∼2000 (Neo) or ∼200 (Hyg) drug-resistant colonies upon infection of one 75 cm2 flask of HCT116 cells. In general, one third of the virus generated from one 75 cm2 flask was sufficient for infection of one 75 cm2 flask containing the cells to be targeted.
Generation of targeted deletions
In the seminal paper by Hirata et al. (11), two different strategies for generating KOs with rAAV were described. The first used endogenous promoter elements and intron–exon junctions to drive expression of the Neo gene, and the second used an exogenous promoter. Both strategies resulted in the introduction of the Neo gene within an exon, with no deletion of adjacent exonic sequences. The targeting frequency was ∼5-fold higher when endogenous promoters were used, and we have confirmed this observation with other rAAV constructs (unpublished data). However, we found it more convenient to use an exogenous promoter, facilitating the construction of multiple targeting vectors in a uniform manner. The exogenous promoter strategy also permitted targeting of genes that are expressed at low levels in the cell type of interest under the conditions used for transfection.
During the course of this work, we discovered that rAAV targeting could be used to generate deletions of endogenous sequences, even though rAAV targeting has historically resulted in the insertion of rAAV sequences between two adjacent nucleotides. We have obtained reasonable targeting efficiencies with deletions up to 1165 bp in length, as described below. Such deletions can be very useful for certain purposes, such as study of the null phenotype or delineation of the role of a specific exon within a given gene (following cre-mediated excision of the inserted Neo or Hyg sequences).
Among several vector designs and procedures attempted during our study, we found that the ones described in Figures 1 and 2, along with the accompanying details presented in the Materials and Methods, were preferred because of their efficiency, generality and ease of use and are therefore described in full. Representative results obtained with these procedures are presented in Figures 3 and 4. Figure 3A shows an ethidium bromide-stained agarose gel containing pNeDaKO plasmid fragments and PCR products of homology arms from the FHIT and CCR5 loci (Step 1). Figure 3B shows fusion PCR products generated with these homology arms plus the restriction fragments from pNeDaKO-Hyg and pNeDaKO-Neo for FHIT and CCR5, respectively (Step 2). Figure 3C shows NotI digests of plasmids obtained after cloning the fusion PCR products into pAAV-MCS and selecting with ampicillin and zeomycin (Step 3).
Cartoons depicting the FHIT and CCR5 loci are shown in Figure 4A and B, respectively. For FHIT targeting, we chose to delete an entire exon and surrounding intronic sequences, while for CCR5, 46 bp from exon 4 were deleted when homologous integration occurred. Figure 4C and D show a typical PCR-based screen for successful targeting events. Because the homology arms used for targeting were generally only 900 bp (as noted above), the sizes of the PCR products generated from successful targeting events were relatively small (∼1600 bp). We generally screened with one primer representing a sequence upstream of the left homology arm and another primer representing Neo or Hyg gene sequences. Targeting events can be confirmed with an independent PCR using a primer containing a sequence downstream of the right homology arm and another primer containing a sequence from the Neo or Hyg genes.
Figure 4C and D depict typical results obtained upon such screening. The FHIT gene was found to be correctly targeted in two of 92 HCT116 clones resistant to hygromycin after infection with rAAV generated with a pNeDaKO-Hyg-derived vector (Fig. 4C). The CCR5 gene was found to be correctly targeted in 12 of 173 hTERT-immortalized RPE clones resistant to geneticin after infection with rAAV generated from pNeDaKO-Neo-derived vectors (Fig. 4D). For comparison, 13 of 244 HCT116 Neo-resistant clones obtained after infection with the rAAV for CCR5 were properly targeted. Southern blots were used to confirm the validity of the PCR-based screen with randomly chosen clones (Fig. 4E).
Although rAAV-based KOs are now routinely performed in our lab, there is still some variability in the frequency with which such KOs are obtained. We have to date successfully disrupted 10 different human genes in five different human cell lines using variations of the approaches described above at efficiencies ranging from 0.4–13% (data not shown). Though we have not systematically investigated the basis for this 30-fold variability, we believe that part of it is due to sequence- and chromatin-specific factors. Indeed, previous studies in murine ES cells as well as human somatic cells with conventional KO vectors have revealed similar unexplained variability (1). When targeting efficiency is low, anecdotal experiments in our laboratory suggest a few approaches that have sometimes proved useful. Firstly, the sequence of the final targeting vector (in pAAV-MCS) should be determined, along with the sequence of the endogenous locus in the cells to be targeted. Though a few mismatches are tolerable, clones with the least number of mismatches are preferable. Secondly, the targeting frequency may be higher when the stretch of endogenous sequences that are to be deleted is relatively small, so that deletions of one or a few base pairs is preferable over deletions encompassing 1000 bp (Fig. 4). Thirdly, it is preferable to choose homology arms that have no repeats, as defined by RepeatMasker. If this is impossible, then arms with the minimal number of repeats should be chosen; the maximum fraction of repeated sequences in any of our homology arms was ∼50%. Fourthly, the efficiency can sometimes be improved by simply targeting a different exon. As the homology arms are each <1 kb in size, and the vectors are so easy to generate, it is generally not difficult to construct several targeting vectors for different regions of the same gene.
The techniques described above should greatly facilitate the manipulation of endogenous sequences in human cells. Preparation of the homology arms and final pNeDaKO vectors are considerably simpler and faster than is possible with conventional systems, in part due to the small size of the homology arms required with this approach. Moreover, the targeting efficiency obtained with rAAV-based targeting is often 10–100× higher than obtained with methods that employ plasmid DNA transfection rather than rAAV infection (18). As noted in the Introduction, gene disruption is currently the most direct and interpretable approach to assess gene function. As it is clear that the function of many genes is both cell-type- and species-dependent (19), it is essential to assess a gene’s function in the cell type that most closely reflects the biological question of interest. For human disease genes in particular, gene disruptions in model systems such as Drosophila, Caenorhabditis elegans and Mus musculus can provide essential information about function but do not always reflect the gene’s functional role in specific types of human cells. In addition to basic research questions, KO cell lines and their parental controls provide excellent tools for drug discovery (20).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank S. E. Kern, E. Gallmeier and K. E. Bachman for critical review of the manuscript, and S. Szabo for sequencing support. This work was supported by the Clayton Fund and NIH grants CA 43460, CA 57345 and CA 62924.
REFERENCES
- 1.Bunz F. (2002) Human cell knockouts. Curr. Opin. Oncol., 14, 73–78. [DOI] [PubMed] [Google Scholar]
- 2.Sedivy J.M. and Dutriaux,A. (1999) Gene targeting and somatic cell genetics—a rebirth or a coming of age? Trends Genet., 15, 88–90. [DOI] [PubMed] [Google Scholar]
- 3.Hannon G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 4.Jackson A.L., Bartz,S.R., Schelter,J., Kobayashi,S.V., Burchard,J., Mao,M., Li,B., Cavet,G. and Linsley,P.S. (2003) Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol., 21, 635–637. [DOI] [PubMed] [Google Scholar]
- 5.Sledz C.A., Holko,M., de Veer,M.J., Silverman,R.H. and Williams,B.R. (2003) Activation of the interferon system by short-interfering RNAs. Nature Cell Biol., 5, 834–839. [DOI] [PubMed] [Google Scholar]
- 6.Bridge A.J., Pebernard,S., Ducraux,A., Nicoulaz,A.L. and Iggo,R. (2003) Induction of an interferon response by RNAi vectors in mammalian cells. Nature Genet., 34, 263–264. [DOI] [PubMed] [Google Scholar]
- 7.Mansour S.L., Thomas,K.R. and Capecchi,M.R. (1988) Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature, 336, 348–352. [DOI] [PubMed] [Google Scholar]
- 8.Hanson K.D. and Sedivy,J.M. (1995) Analysis of biological selections for high-efficiency gene targeting. Mol. Cell. Biol., 15, 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jallepalli P.V., Waizenegger,I.C., Bunz,F., Langer,S., Speicher,M.R., Peters,J., Kinzler,K.W., Vogelstein,B. and Lengauer,C. (2001) Securin is required for chromosomal stability in human cells. Cell, 105, 445–457. [DOI] [PubMed] [Google Scholar]
- 10.Porteus M.H. and Baltimore,D. (2003) Chimeric nucleases stimulate gene targeting in human cells. Science, 300, 763. [DOI] [PubMed] [Google Scholar]
- 11.Hirata R., Chamberlain,J., Dong,R. and Russell,D.W. (2002) Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat. Biotechnol., 20, 735–738. [DOI] [PubMed] [Google Scholar]
- 12.Porteus M.H., Cathomen,T., Weitzman,M.D. and Baltimore,D. (2003) Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol. Cell. Biol., 23, 3558–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- 14.Park B.H., Vogelstein,B. and Kinzler,K.W. (2001) Genetic disruption of PPARdelta decreases the tumorigenicity of human colon cancer cells. Proc. Natl Acad. Sci. USA, 98, 2598–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong J.Y., Fan,P.D. and Frizzell,R.A. (1996) Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum. Gene Ther., 7, 2101–2112. [DOI] [PubMed] [Google Scholar]
- 16.Xiao X., Li,J. and Samulski,R.J. (1998) Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol., 72, 2224–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veldwijk M.R., Topaly,J., Laufs,S., Hengge,U.R., Wenz,F., Zeller,W.J. and Fruehauf,S. (2002) Development and optimization of a real-time quantitative PCR-based method for the titration of AAV-2 vector stocks. Mol. Ther., 6, 272–278. [DOI] [PubMed] [Google Scholar]
- 18.Yanez R.J. and Porter,A.C. (1998) Therapeutic gene targeting. Gene Ther., 5, 149–159. [DOI] [PubMed] [Google Scholar]
- 19.Cassman M. (2003) Computational biology: counting on the neuron. Science, 300, 756–757. [DOI] [PubMed] [Google Scholar]
- 20.Torrance C.J., Agrawal,V., Vogelstein,B. and Kinzler,K.W. (2001) Use of isogenic human cancer cells for high-throughput screening and drug discovery. Nat. Biotechnol., 19, 940–945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.