SUMMARY
Amino acids are required for activation of the mammalian target of rapamycin (mTOR) kinase which regulates protein translation, cell growth, and autophagy. Cell surface transporters that allow amino acids to enter the cell and signal to mTOR are unknown. We show that cellular uptake of L-glutamine and its subsequent rapid efflux in the presence of essential amino acids (EAA) is the rate-limiting step that activates mTOR. L-glutamine uptake is regulated by SLC1A5 and loss of SLC1A5 function inhibits cell growth and activates autophagy. The molecular basis for L-glutamine sensitivity is due to SLC7A5/SLC3A2, a bidirectional transporter that regulates the simultaneous efflux of L-glutamine out of cells and transport of L-leucine/EAA into cells. Certain tumor cell lines with high basal cellular levels of L-glutamine bypass the need for L-glutamine uptake and are primed for mTOR activation. Thus, L-glutamine flux regulates mTOR, translation and autophagy to coordinate cell growth and proliferation.
INTRODUCTION
The target of rapamycin (TOR) Ser/Thr kinase has been conserved throughout eukaryotic evolution to mediate cellular responses to extracellular amino acids and growth factors (Huang and Manning, 2008; Wullschleger et al., 2006). In an amino acid-rich environment TOR is active and regulates protein translation but also inhibits macroautophagy (hereafter referred to as autophagy). When extracellular amino acids are limiting, autophagy recycles intracellular constituents as a way to provide an alternative source of amino acids (Mizushima et al., 2008). Studies using S. cerevisiae provided key insight into the role of TOR as a signal integrator for amino acids. For example, rapamycin, an inhibitor of TOR, mimics the effect that amino acid starvation has on protein translation, cell cycle arrest, ribosome biogenesis and autophagy (Crespo and Hall, 2002). TOR activity is particularly sensitive to availability of preferred nitrogen sources such as L-glutamine or L-asparagine such that in their absence, permeases required for uptake of these amino acids are downregulated and genes required for the utilization of alternative nitrogen sources such as urea and ammonia are upregulated (Cardenas et al., 1999; Hardwick et al., 1999). TOR controls this metabolic switch by regulating the cellular distribution of transcription factors (Gln3 and Rtg1/Rtg2) required for nitrogen-dependent growth (Crespo and Hall, 2002). Nitrogen starvation is also known to activate autophagy in S. cerevisiae (Takeshige et al., 1992) consistent with TOR functioning as a suppressor of autophagy. How L-glutamine regulates TOR activity is currently unclear.
In all eukaryotes studied to date TOR is partitioned into at least two distinct signaling complexes (Wullschleger et al., 2006). In mammalian cells the rapamycin-sensitive mTOR complex (mTORC) 1 is essential for the phosphorylation and activation of the 70 kDa ribosomal protein S6 kinase (S6K) 1 and 2. S6K1 directly impacts cell growth by regulating the pioneer round of protein translation and the eukaryotic initiation factor 3 (eIF3) translation complex (Holz et al., 2005; Ma et al., 2008). mTORC1 also controls the translation of 5' capped mRNAs by directly phosphorylating and inhibiting the eukaryotic translation initiation factor 4E (eIF4E) binding protein (4EBP1) resulting in its dissociation from eIF4E (Gingras et al., 2004). mTORC2 is insensitive to rapamycin, regulates activation of the AKT Ser/Thr kinase, and can alter the actin cytoskeleton (Jacinto et al., 2004; Sarbassov et al., 2004, 2005).
Two key growth factor signals that regulate the mTORC1 pathway emanate from the insulin/insulin-like growth factor (IGF) 1/phosphatidyl inositol-3-OH kinase (PI(3)K) and extra-cellular signal regulated kinase-p90 ribosomal protein S6 kinase (ERK-RSK) pathways (Huang and Manning, 2008). These pathways converge on proteins encoded by the TSC1 and TSC2 tumor suppressor genes which are mutated in the tuberous sclerosis complex (TSC) tumor syndrome (Crino et al., 2006). TSC1 and TSC2 proteins physically associate and suppress the RAS homolog enriched in brain (Rheb), a small G protein required for mTORC1 activation. The TSC1/TSC2 protein complex can be phosphorylated and thereby inactivated by PI(3)K and ERKRSK signaling (Anjum and Blenis, 2008) allowing Rheb to activate mTORC1. In the absence of amino acids, growth factor signals have little or no impact on mTORC1 signaling (Hara et al., 1998) indicating the presence of a key gating mechanism upstream of mTOR. Essential amino acids (EAA) such as L-leucine, L-tryptophan, L-phenylalanine and L-arginine activate the mTORC1 pathway (Blommaart et al., 1995; Hara et al., 1998; Wang et al., 1998). Recent observations indicate that Rag GTPases can physically interact with mTORC1 and regulate its sub-cellular redistribution in response to L-leucine (Sancak et al., 2008). How amino acids enter the cell to regulate mTORC1 activation is currently unknown.
In the present study we identify the mechanism by which L-glutamine exerts control over mTOR signaling. Cellular uptake of L-glutamine is the rate limiting step for EAA- and growth factor-regulation of mTORC1. Following uptake, L-glutamine is effluxed within 1–2 min of adding EAA to cells, leading to reciprocal uptake of L-leucine and rapid activation of S6K1. Solute carrier family 1 member 5 (SLC1A5) is a high affinity L-glutamine transporter and its inhibition blocks uptake of L-glutamine leading to inhibition of mTORC1 signaling and activation of autophagy. Solute carrier family 7 member 5 (SLC7A5)/SLC3A2 is a heterodimeric bidirectional antiporter that regulates the exchange of intracellular L-glutamine for extracellular L-leucine. Together these data show that the counter transport of L-glutamine and EAA controls mTORC1 and autophagy.
RESULTS
L-Glutamine Is Required for Rapamycin-Sensitive mTOR Signaling
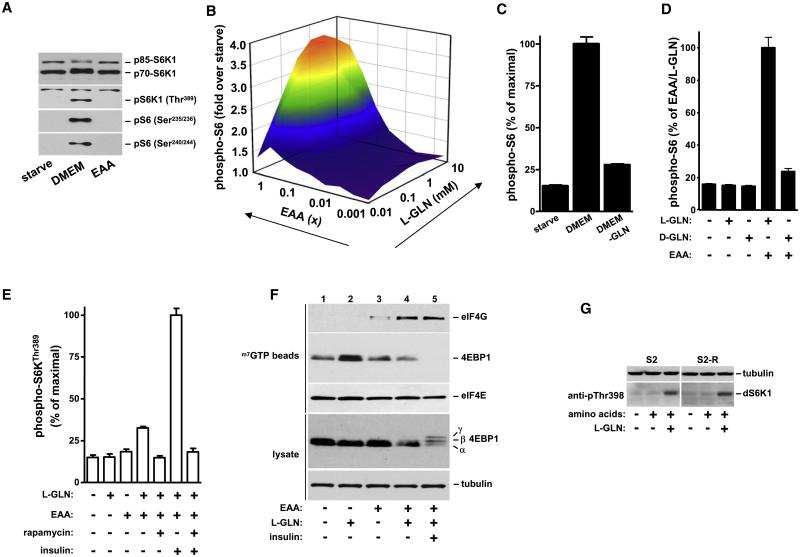
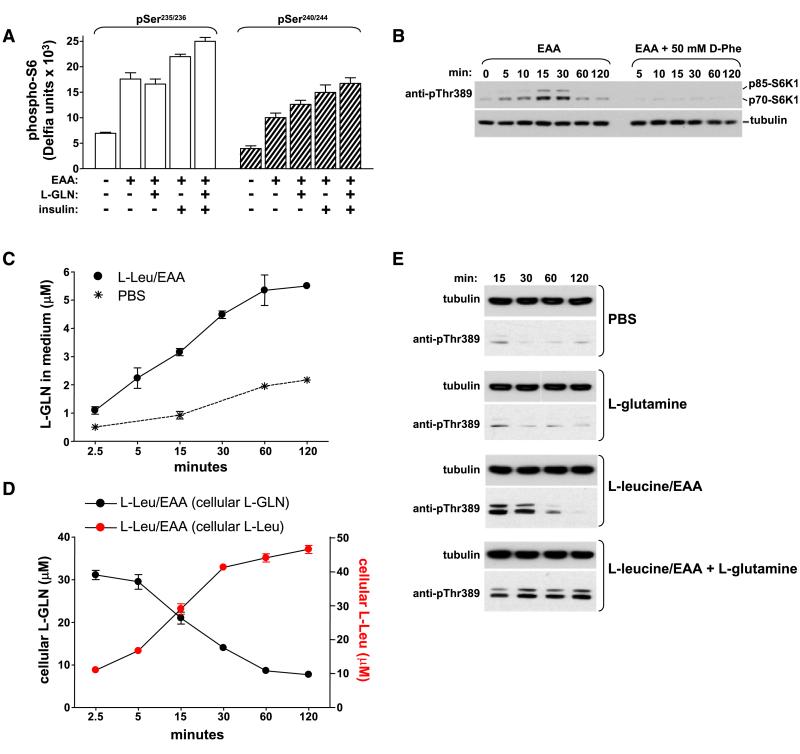
mTORC1 signaling is inhibited after growth factor and amino acid deprivation. Addition of serum-free DMEM to starved HeLa cells re-activates S6K1 and results in the phosphorylation of S6K1 (causing an SDS-PAGE mobility shift) as well as ribosomal protein S6, a key downstream target of S6K1 (Figure 1A). In contrast, incubating cells with EAA at the same concentration found in DMEM does not regulate S6K1 indicating that a component present in DMEM is required for mTORC1 activation. The remaining components found in DMEM include sodium pyruvate, sodium bicarbonate, ferric nitrate, nonessential amino acids (NEAA) and L-glutamine. Using an in-cell western phospho-S6 assay (Figure S1A available with this article online) we observed that adding sodium pyruvate, sodium bicarbonate, ferric nitrate or NEAA to EAA does not reconstitute the effect of DMEM on mTORC1 activation (data not shown and see below). In contrast, the addition of L-glutamine to EAA upregulates rapamycin-sensitive mTOR-S6K-S6 signaling (Figures 1B, 1E, and S1). Importantly, L-glutamine and EAA alone have little effect but together they synergize to activate mTOR (Figure 1B). Maximal effects occur with 1 mM L-glutamine, comparable to circulating levels which range from 0.5 to 0.7 mM (Dechelotte et al., 1991; Mittendorfer et al., 2001). To determine if L-glutamine in DMEM is responsible for activating the mTOR pathway, DMEM lacking this amino acid was added to starved cells. Remarkably, despite the presence of EAA and other nutrients in L-glutamine-free medium, regulated S6 phosphorylation is reduced by 85% (Figure 1C). The effect of L-glutamine on mTORC1 is also stereoselective (Figure 1D). Since DMEM contains 4.5 g/L D-glucose and the starvation and stimulation solutions contain 1 g/L, we confirmed that this variation was not contributing to the inability of EAA to regulate mTORC1-S6K1 activation (Figure S1B). The phosphorylation of 4EBP1 and subsequent recruitment of the eukaryotic translation initiation factor 4G (eIF4G) to the eIF4E cap complex is also sensitive to L-glutamine (Figure 1F, compare lanes 3 and 4). These observations in HeLa cells extend to other mammalian cell lines in which rapamycin-sensitive mTOR activity depends on L-glutamine (Figure S2) as well as in S2 Drosophila cells (Figure 1G) indicating that this is an evolutionarily conserved process.
Figure 1.
Glutamine-Dependent Regulation of mTORC1 Effector Signaling
(A) HeLa cells deprived of growth factors and nutrients (starve) were treated with DMEM or EAA for 60 min. The phosphorylation of S6K1Thr389 and S6Ser235/236 and S6240/244, and total levels of S6K1 (70 kDa cytoplasmic and 85 kDa nuclear isoforms are indicated) were assessed using western blotting.
(B) HeLa cells were starved as in (A) and then treated with the indicated concentrations of EAA and/or L-glutamine for 60 min. EAA concentration (x-fold) was based on final concentration in DMEM (1×). Phosphorylation of S6Ser235/236 was quantitated using the in-cell western assay and expressed as fold-increase compared to starved cells.
(C) Starved HeLa cells were incubated with DMEM or L-glutamine-deficient DMEM (DMEM-GLN) for 60 min and the phosphorylation of S6Ser235/236 was quantitated.
(D) Starved HeLa cells were treated for 60 min as indicated and the phosphorylation of S6Ser235/236 was quantitated. 1 mM L- or D-glutamine was used.
(E) Starved HeLa cells were treated as indicated and the phosphorylation of S6K1Thr389 was quantitated as described in the supplement.
(F) Starved HeLa cells were treated as indicated for 60 min and levels of 4EBP1 and eIF4G associated with m7 GTP beads analyzed using western blotting.
(G) S2 or S2-R Drosophila cells starved of serum and amino acids were then treated with amino acids in the presence or absence of L-glutamine.
Data shown are representative of three independent experiments. Data with error bars represent mean ± SD.
L-Glutamine Is Rate Limiting for mTORC1 Activation
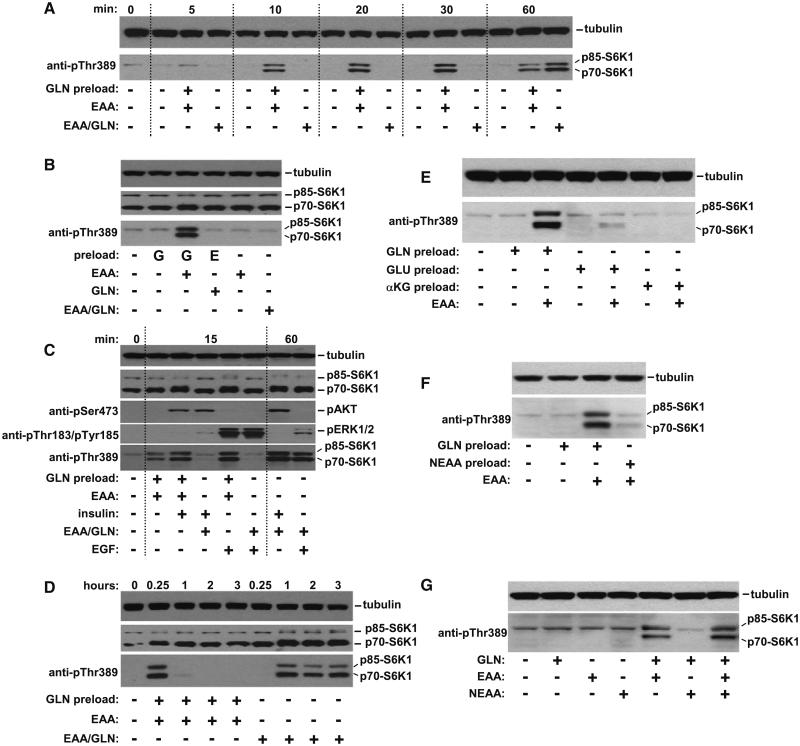
To understand the relationship between EAA and L-glutamine, starved HeLa cells were treated with L-glutamine for 60 min (preload), extracellular L-glutamine washed away and then EAA was added for different times (Figure 2A). Under these conditions S6K1 activation kinetics are significantly accelerated compared to the simultaneous addition of EAA and L-glutamine where phosphorylation of S6K1 is observed after 60 min (Figure 2A). Pretreating starved cells with EAA and then L-glutamine does not activate S6K1 (Figure 2B), confirming that L-glutamine is upstream of the EAA input. These observations reveal that L-glutamine is rate limiting for EAA-mediated activation of S6K1. We next evaluated whether L-glutamine is required for integrating growth factor signals upstream of mTORC1. Insulin-regulated activation of AKT and EGF-regulated activation of ERK1/2 is not affected by L-glutamine pretreatment (Figure 2C). However, pretreating with L-glutamine sensitizes S6K1 to insulin and EGF indicating an important role in signal integration. (Figure 2C).
Figure 2.
L-Glutamine Integrates EAA and Growth Factor Signaling to mTORC1 and Regulates the Temporal Dynamics of S6K1
(A) Starved HeLa cells were preincubated with L-glutamine (GLN preload) or starve medium (–) for 1 hr prior to adding EAA or EAA/GLN for the times indicated. Phosphorylation of S6K1 was analyzed by western blotting.
(B) Cells were pretreated with either L-glutamine (G) or EAA (E) for 1 hr and incubated with EAA, L-glutamine or EAA/GLN for 15 min prior to lysis.
(C) Cells were treated as in (A) except that insulin (100 nM) or EGF (25 ng/mL) were included as indicated.
(D) Cells pretreated with either L-glutamine or starve medium for 1 hr were then incubated with EAA or EAA/GLN, respectively, for the indicated times.
(E) Cells pretreated for 1 hr with 10 mM L-glutamine, L-glutamic acid (GLU) or α-ketoglutaric acid (αKG), were washed and then incubated with starve or EAA-containing medium for 15 min.
(F) Cells pretreated with either 10 mM L-glutamine or 13 nonessential amino acids (NEAA) for 1 hr, were washed and then incubated with starve or EAA-containing medium for 15 min.
(G) Starved HeLa cells were treated as indicated for 1 hr and lysed.
All data is representative of at least three independent experiments.
Although pretreating cells with L-glutamine supports robust S6K1 phosphorylation within minutes of adding EAA (Figures 2A-2C), S6K1 activation subsides around 60 min (Figure 2A). Cells treated in this way exhibit transient S6K1 activation compared to cells treated with EAA and L-glutamine together where S6K1 phosphorylation persists for several hours (Figures 2D and S3A). Interestingly, addition of L-glutamine after 3 hr reactivates S6K1 (Figure S3A) indicating that the transient activation kinetics are due to L-glutamine limitation rather than consumption of EAA or pathway desensitization. In certain tumor cell lines L-glutamine can enhance metabolic flux via the process of glutaminolysis which generates tricarboxylic acid (TCA) cycle intermediates (DeBerardinis et al., 2007; Kovacevic and Morris, 1972). In order to determine if the L-glutamine effect on S6K1 is due to increasing cellular energy we asked if L-glutamic acid and α-ketoglutarate (products of glutaminolysis) can sensitize mTOR to EAA. In contrast to L-glutamine, L-glutamic acid and α-ketoglutarate are unable to regulate S6K1 activation (Figure 2E). L-glutamine can also contribute to the biosynthesis of certain NEAAs such as L-asparagine and L-arginine. NEAAs are unable to regulate the activation of S6K1 either in pretreatment mode (Figure 2F) or when added in combination with EAA and/ or L-glutamine (Figure 2G). Taken together, these data show that L-glutamine-regulated activation of mTOR-S6K1-S6 is a proximal event and is not derived from increased glutaminolysis and/or NEAA synthesis.
Inhibition of SLC1A5 and SLC7A5/SLC3A2 Antagonizes mTORC1
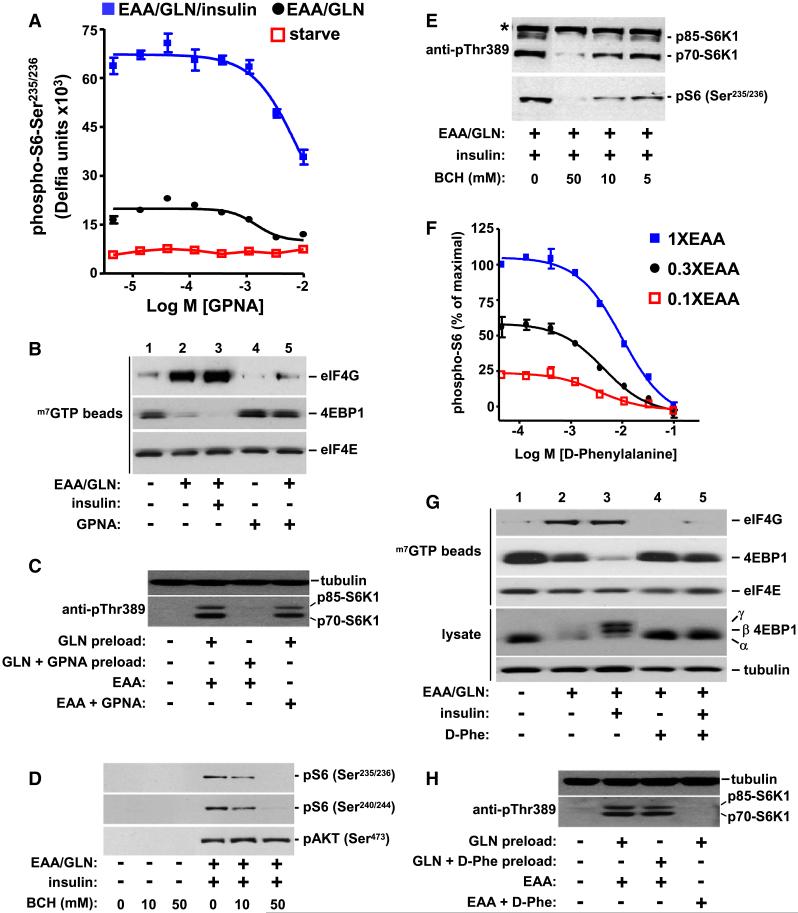
We sought to identify a molecular mechanism that could explain why L-glutamine is rate-limiting for mTORC1 activation. L-glutamine influx into mammalian cells can be regulated by at least four different transporters; Na+-dependent systems A, ASC, N, and the Na+-independent system L, comprised of low affinity L-glutamine transporters (McGivan and Bungard, 2007). Intriguingly, a relationship between SLC1A5 (also known as ASCT2) and SLC7A5 (also known as LAT1) has been proposed (Verrey, 2003) which explains the interplay between L-glutamine and EAA upstream of mTORC1 (Figures 2A-2C). SLC1A5 is a highaffinity, Na+-dependent transporter for L-glutamine (Utsunomiya-Tate et al., 1996). SLC7A5 transports branched side-chain amino acids such as L-leucine into cells in exchange for the efflux of intracellular amino acids such as L-glutamine (Meier et al., 2002; Yanagida et al., 2001). SLC7A5 functions as part of a disulfide bonded heterodimeric complex with SLC3A2 and resides at the plasma membrane (Mastroberardino et al., 1998). Thus, we hypothesized that intracellular L-glutamine, transported into the cell by SLC1A5, serves as an efflux substrate for SLC7A5/SLC3A2 allowing for the counter transport of EAA. L-γ-glutamyl-p-nitroanilide (GPNA), an inhibitor of SLC1A5-regulated transport (Esslinger et al., 2005) dose-dependently inhibits S6 phosphorylation and the eIF4E cap complex (Figures 3A and 3B). To evaluate the specificity of GPNA in suppressing the L-glutamine input, cells were pretreated with Lglutamine in the presence or absence of GPNA for 60 min, and then exposed to EAA in the presence or absence of GPNA for 15 min (Figure 3C). The presence of GPNA during the pretreatment period, but not with EAA, completely inhibits S6K1 activation (Figure 3C). Collectively, these data indicate that the SLC1A5 inhibitor GPNA antagonizes mTORC1 signaling by limiting cellular uptake of L-glutamine.
Figure 3.
Pharmacological Modulation of SLC1A5 and SLC7A5 Inhibits mTOR Regulation
(A) Starved HeLa cells were treated as indicated in the presence of increasing concentrations of GPNA (1 hr).
(B) Starved cells were treated for 60 min and the cap translation complex analyzed. 10 mM GPNA was used.
(C) Cells were pretreated (preload) for 60 min with L-glutamine in the presence or absence of GPNA prior to treatment with EAA for 15 min in the presence or absence of GPNA.
(D and E) Starved HeLa cells were treated with EAA containing 1 mML-glutamine (EAA/GLN) and 100 nM insulin in the absence or presence of BCH for 60 min. A nonspecific reactive band (*) is indicated.
(F) The phosphorylation of S6Ser235/236 was quantitated following treatment with the indicated concentrations (X) of EAA in the presence of 1mML-glutamine and D-phenylalanine.
(G) Starved HeLa cells were treated as indicated in the presence and absence of 50 mM D-phenylalanine (D-Phe).
(H) Cells were pretreated for 60 min as indicated and then treated with EAA for 15 min in the presence or absence of D-Phe.
Data with error bars represent mean ± SD.
To interrogate the requirement of SLC7A5/SLC3A2, a candidate transporter for EAA, 2-aminobicyclo-(2,2,1)heptanecarboxylic acid (BCH) was used. BCH, an inhibitor of SLC7A5 (Kanai et al., 1998; Mastroberardino et al., 1998; Yanagida et al., 2001), suppresses S6K-S6 phosphorylation (Figures 3D, 3E, S3B, and S3C). BCH does not inhibit AKT phosphorylation (Figure 3D) or phorbol 12-myristate 13-acetate (PMA) regulation of S6 phosphorylation in starved HeLa cells (Figure S3D) consistent with the ability of RSK to directly phosphorylate S6 in a mTORC1-independent manner (Anjum and Blenis, 2008). Thus, BCH specifically inhibits the amino acid input to mTORC1. As an alternative way to modulate SLC7A5/SLC3A2 function, D-phenylalanine was used. Unlike L-phenylalanine, a high affinity SLC7A5 substrate which regulates S6K1 activation (Hara et al., 1998; Kanai et al., 1998; Mastroberardino et al., 1998; Yanagida et al., 2001), D-phenylalanine inhibits SLC7A5/ SLC3A2-mediated transport (Kanai et al., 1998). D-phenylala-nine inhibits rapamycin-sensitive S6 phosphorylation (Figure 3F) and the cap translation complex (Figure 3G). In contrast to the mode of action of GPNA, D-phenylalanine specifically inhibits the EAA rather than the L-glutamine input (Figure 3H). Collectively these results show that a BCH- and D-phenylalanine sensitive activity is responsible for EAA regulation of mTORC1 and are consistent with previous studies showing that they inhibit the bidirectional transporter SLC7A5/SLC3A2.
Efflux of L-Glutamine and Uptake of L-Leucine Precede S6K1 Activation
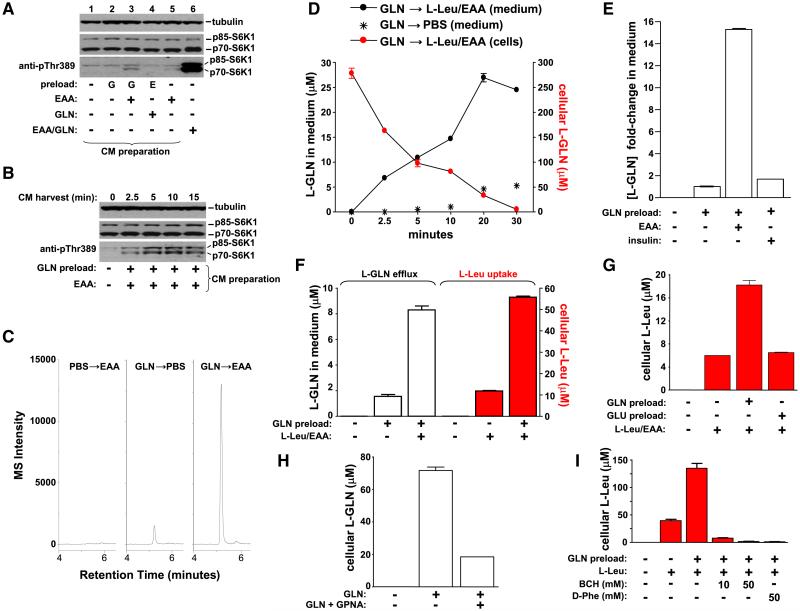
The above data supports a model in which SLC1A5-mediated uptake of L-glutamine provides a signal for EAA to regulate mTOR signaling. Since SLC7A5/SLC3A2 is capable of using L-glutamine as an efflux substrate (Meier et al., 2002; Yanagida et al., 2001), we tested the hypothesis that EAA regulates the efflux of intracellular L-glutamine by quantifying its presence in medium after EAA treatment. Starved cells were pretreated with L-glutamine for 1 hr and washed as described above (Figure 2A). After treating with EAA for 15 min, conditioned medium was collected and transferred to naive starved cells (Figure 4A, lane 3). This conditioned medium supports activation of S6K1 though not to the extent as direct addition of EAA to L-glutamine-preloaded cells (Figure 4A, compare lanes 3 and 6). Conditioned medium taken from EAA-treated, nonprimed cells does not activate S6K1 (Figure 4A, lane 5) proving that the effect of conditioned medium is dependent on L-glutamine. These results indicate that a cellular factor, likely L-glutamine, is released in the presence of EAA and which is capable of regulating mTOR-S6K1 activity. This factor is released after only 2.5 min in the presence of EAA (Figure 4B).
Figure 4.
Bidirectional Transport of L-Glutamine and EAA Is Upstream of mTOR Activation
(A) Starved cells were preincubated with L-glutamine or starve medium (–) for 1 hr prior to adding EAA or starve medium (15 min). Conditioned medium (CM) was removed and added to naive starved cells for 1 hr (lanes 1-5) and S6K1 phosphorylation analyzed. Phosphorylation of S6K1 in an extract from EAA-treated L-glutamine primed cells is shown (lane 6).
(B) Conditioned medium was prepared as in (A) except collection was 2.5, 5, 10, and 15 min after adding EAA.
(C) LC/MS/MS ion chromatograms of L-glutamine in conditioned medium taken at 2.5 min from cells pretreated with starve medium (PBS) prior to EAA (left trace), pretreated with L-glutamine prior to PBS (middle trace) or pretreated with L-glutamine prior to EAA (right trace).
(D) Starved HeLa cells were pretreated with labeled L-glutamine for 1 hr, washed, and then treated with EAA containing 0.4 mM labeled L-leucine (L-Leu/EAA) for various times (minutes). The absolute levels of labeled L-glutamine in conditioned medium (black symbols) and cell extracts (red symbols) was quantitated using LC/MS/MS.
(E) Starved cells were treated as in (C) and the L-glutamine efflux (fold-change in medium concentration) after treatment with PBS, EAA or insulin for 2.5 min quantitated.
(F) Cells were treated as in (D) and after 2.5 min the absolute levels of effluxed labeled L-glutamine (black) and labeled L-leucine in cells (red) was quantitated.
(G) Cells pretreated with 10 mM labeled L-glutamine or labeled L-glutamic acid for 1 hr were washed, treated with EAA containing labeled L-leucine for 2.5 min and extracted.
(H) Starved cells were treated with labeled L-glutamine for 1 hr in the absence or presence of 10 mM GPNA. The absolute levels of L-glutamine in cell extracts were quantitated.
(I) Cells were treated with labeled L-glutamine for 1 hr, washed and incubated with 0.4 mM labeled L-leucine in the absence and presence of BCH or D-Phe for 2.5 min.
Data with error bars represent mean ± SD.
To prove that L-glutamine is effluxed out of the cell in response to EAA treatment, conditioned medium was analyzed using HPLC-triple quadruple mass spectrometry (LC/MS/MS). L-glutamine is undetectable in conditioned medium taken from cells treated with EAA for 2.5 min (Figure 4C, left trace) however, a 10-fold increase in L-glutamine concentration is detected in conditioned medium taken from L-glutamine primed-EAA treated cells compared to control cells (Figure 4C and 4E). The absolute concentration of L-glutamine in this medium was approximately 100-fold lower than that required to maximally regulate mTORC1 pathway activity (Figure 1) consistent with the submaximal effect associated with conditioned medium in naive cells (Figure 4A). Using a stable-isotope labeled L-glutamine analog we observed that efflux of L-glutamine with time correlates with depletion of cellular L-glutamine (Figure 4D). Importantly, insulin treatment does not lead to L-glutamine efflux indicating that the above effects are specific to EAA (Figure 4E). These observations prove that L-glutamine is rapidly effluxed out of the cell in response to EAA. If SLC7A5/SLC3A2 regulates exchange of L-glutamine for EAA then L-glutamine should enhance the uptake of L-leucine. Indeed, by using labeled L-glutamine and L-leucine analogs, reciprocal exchange of these amino acids takes place (Figure 4F). L-glutamic acid is not capable of regulating L-leucine uptake (Figure 4G) and unlike L-glutamine, its cellular levels are not depleted following treatment with EAA (Fig. S5A and B). This provides clear evidence that L-glutamine metabolites are not involved in mTORC1 activation. Finally, uptake of labeled L-glutamine into cells is inhibited by GPNA (Figure 4H) and both basal and L-glutamine-regulated L-leucine uptake is inhibited by BCH and D-phenylalanine (Figure 4I).
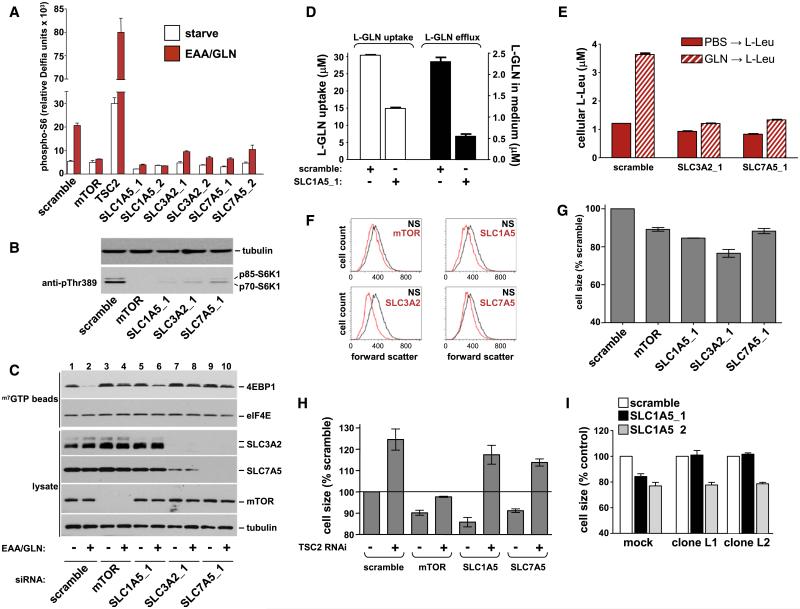
SLC1A5 and SLC7A5/SLC3A2 Are Required for Amino Acid Exchange, mTORC1 Signaling and Cell Growth
To determine if SLC1A5 and SLC7A5/SLC3A2 are required for amino acid-regulation of mTORC1, we identified two siRNAs for each gene that knocked-down the target mRNA by 80%–90% (Figures S4A-S4C). As expected, amino acid regulation of S6 phosphorylation is inhibited with mTOR siRNA and increased with TSC2 siRNA (Figure 5A). Each of the SLC-targeted siRNAs inhibit S6-S6K1 phosphorylation similarly to the mTOR siRNA (Figures 5A and 5B). Next, we examined the effect of these RNAi reagents on regulation of the cap complex. As expected, downregulation of mTOR prevents dissociation of 4EBP1 from the eIF4E cap complex and even enhances basal 4EBP1 association (Figure 5C, compare lanes 2 and 4, 1 and 3). Knockdown of either SLC1A5, SLC7A5 or SLC3A2 also enhances basal association of 4EBP1 with the cap complex and restores 4EBP1 to the complex in the presence of L-glutamine and EAA (Figure 5C, compare lanes 2, 6, 8, and 10). Similar effects occur in asynchronously cycling, serum starved or insulin-treated cells (Figure S4D). Following downregulation of SLC3A2, eIF4G recruitment was not inhibited by BCH (Figure S3E) confirming that BCH targets SLC7A5/SLC3A2. Note that expression of SLC3A2 and SLC7A5 is codependent (Mastroberardino et al., 1998) explaining the decreased expression of SLC3A2 and SLC7A5 when either is targeted with RNAi (Figure 5C and S4D). L-glutamine uptake is significantly reduced when SLC1A5 is downregulated resulting in decreased efflux after adding EAA (Figure 5D). Downregulation of SLC3A2 or SLC7A5 blocks L-glutamine-regulated uptake of L-leucine (Figure 5E). These data prove that SLC1A5 and SLC7A5/SLC3A2 support amino acid exchange in HeLa cells and that this is required for activation of mTORC1.
Figure 5.
RNAi-Mediated Downregulation of SLC1A5, SLC3A2, or SLC7A5 Inhibits Amino Acid Transport and mTOR Pathway Activity
(A) HeLa cells transfected with the indicated siRNAs were amino acid starved (open bars) and treated for 60 min with EAA containing 1 mM L-glutamine (red bars). Phosphorylation of S6Ser235/236 was quantitated using the Delfia readout.
(B) After siRNA transfection (96 hr) S6K1 phosphorylation was analyzed.
(C) Cells transfected with siRNAs were treated as indicated (1 hr) and the cap translation complex analyzed.
(D) Cells transfected with scramble or SLC1A5_1 siRNA were starved and preincubated with labeled L-glutamine (1 hr). Cells were extracted for quantitation of labeled L-glutamine uptake (open bars) or treated with labeled L-leucine/EAA (2.5 min) for quantitation of effluxed labeled L-glutamine (filled bars).
(E) Cells were transfected with the indicated siRNAs, starved and pretreated with labeled L-glutamine (hatched bars) or starve medium (filled bars) for 1 hr, washed and then treated with 0.4 mM labeled L-leucine for 2.5 min. Cell extracts were processed for absolute quantitation of L-leucine.
(F) HeLa cells were transfected with siRNAs (nonsilencing, NS) for 72 hr and collected for cell size quantitation.
(G) Cell size data from three independent experiments (mean ± SEM) is shown.
(H) HeLa cells were transfected with scramble, mTOR, SLC1A5 or SLC7A5 siRNAs in the presence or absence of a TSC2 siRNA and processed for cell size quantitation.
(I) Vector-expressing (mock) or RNAi-insensitive GFP-SLC1A5-expressing HeLa cells (clones L1 and L2) were transfected with siRNAs and processed for cell size quantitation.
The data shown (mean ± SD) represent three independent experiments.
The ability of mTOR to regulate translation is responsible for cellular and organismal growth (Arsham and Neufeld, 2006). We therefore quantitated relative cell size after downregulation of SLC1A5, SLC7A5 or SLC3A2. Compared to a nonsilencing siRNA, downregulation of mTOR results in approximately 10% reduction in cell size (Figures 5F and 5G) consistent with previous reports in mammalian cells (Fingar et al., 2002; Kim et al., 2002). For control purposes TSC2 downregulation was used to show that under these experimental conditions HeLa cells can also grow in size (Figure S6A). Transfection with SLC1A5, SLC3A2 or SLC7A5 siRNAs reduces cell size 10%-20% below that of control transfected cells (Figures 5F, 5G, and S6A). Two separate approaches were taken to demonstrate that the cell size phenotypes were due to (1) mTOR pathway-specific inhibition and (2) on-target RNAi. We first determined if downregulation of TSC2 would restore cell growth in the presence of SLC1A5 or SLC7A5 siRNA. Under this experimental paradigm downregulation of mTOR suppresses the TSC2 cell growth phenotype and further reduces cell size when compared to mock transfected cells (Figure 5H) consistent with mTOR acting downstream of TSC2. In contrast, downregulation of either SLC1A5 or SLC7A5 in the presence of TSC2 siRNA results in increased cell growth (Figure 5H). This indicates that SLC1A5 and SLC7A5 may act upstream of TSC2 to mediate an amino acid signal to mTOR. It should be noted however that in these experiments cell growth was not rescued to the same level as TSC2 downregulation alone (i.e., 75% rescue for SLC1A5 and 50% for SLC7A5). This could be due to incomplete knockdown of TSC2 or that SLC1A5 and SLC7A5/SLC3A2 can signal to mTOR independently of TSC2. The later possibility is supported by other reports (Kim et al., 2008; Sancak et al., 2008; Smith et al., 2005) as well as present observations that L-glutamine and EAA significantly increase S6 phosphorylation in cells transfected with TSC2 siRNA (Figure 5A). Finally, cell size phenotypic rescue experiments were performed to prove that the SLC1A5 RNAi phenotype is on-target. HeLa cells stably expressing a GFP-SLC1A5 allele insensitive to SLC1A5_1 but not SLC1A5_2 siRNA were generated (Figure S6B). The reduction in cell size associated with SLC1A5_1 was completely suppressed in these cells (Figure 5I) proving that the observed phenotypes are due to SLC1A5. Note however that the stable HeLa clones are still sensitive to the SLC1A5_2 siRNA (Figure 5I) which downregulates both the exogenous and endogenous SLC1A5 (Figure S6B).
High Levels of Endogenous L-Glutamine Prime mTORC1 Activation
Upon screening cell lines for L-glutamine-dependent mTOR signaling, lines were identified that showed indifference to exogenous L-glutamine. For example, in MCF7 cells EAA alone is sufficient to regulate D-phenylalanine-sensitive phosphorylation of S6K1 and S6 (Figures 6A and 6B). Intriguingly, the S6K1 activation kinetics (Figure 6B) mirror what takes place in L-glutamine preloaded HeLa cells (Figure 2A) suggesting that MCF7 cells are primed for the EAA input. Adding EAA to MCF7 cells increases the medium concentration of L-glutamine 3- to 4-fold (Figure 6C) indicating that these cells synthesize L-glutamine which can regulate uptake of EAA. L-glutamine is readily detected in MCF7 cells and this rapidly decreases upon treatment with L-leucine/EAA consistent with its efflux from the cell (Figure 6D). Bidirectional transport of L-glutamine and labeled L-leucine in MCF7 cells subsides after 30-60 min (Figures 6C and 6D) in line with S6K1 inactivation (Figure 6B). To determine if the cellular concentration of L-glutamine is limiting sustained S6K1 activity, exogenous L-glutamine was added in combination with EAA. Under these conditions total cellular L-glutamine levels are elevated (data not shown) and S6K1 is continually activated for up to 2 hr (Figure 6E). These data illustrate that bidirectional flux of L-glutamine and L-leucine takes place in multiple cells types and reveal the possibility that certain tumor lines with elevated cellular L-glutamine levels are primed for EAA-regulation of mTORC1.
Figure 6.
Efflux of Endogenous L-Glutamine Regulates S6K1 Activation in MCF7 Cells
(A) MCF7 cells were amino acid starved, treated as indicated (1 hr) and processed for quantitation of S6 phosphorylation.
(B) Starved MCF7 cells were treated with EAA in the absence or presence of 50 mM D-Phe for various times.
(C) Starved MCF7 cells were treated with labeled L-leucine/EAA (filled circles) or starve medium (stars) for various times and conditioned medium processed for quantitation of L-glutamine.
(D) MCF7 cells were treated as in (C) and extracted at the times indicated for quantitation of L-glutamine (black circles) or labeled L-leucine (red circles).
(E) Starved MCF7 cells were treated as indicated and S6K1 activation analyzed using western blotting.
Data with error bars represent mean ± SD.
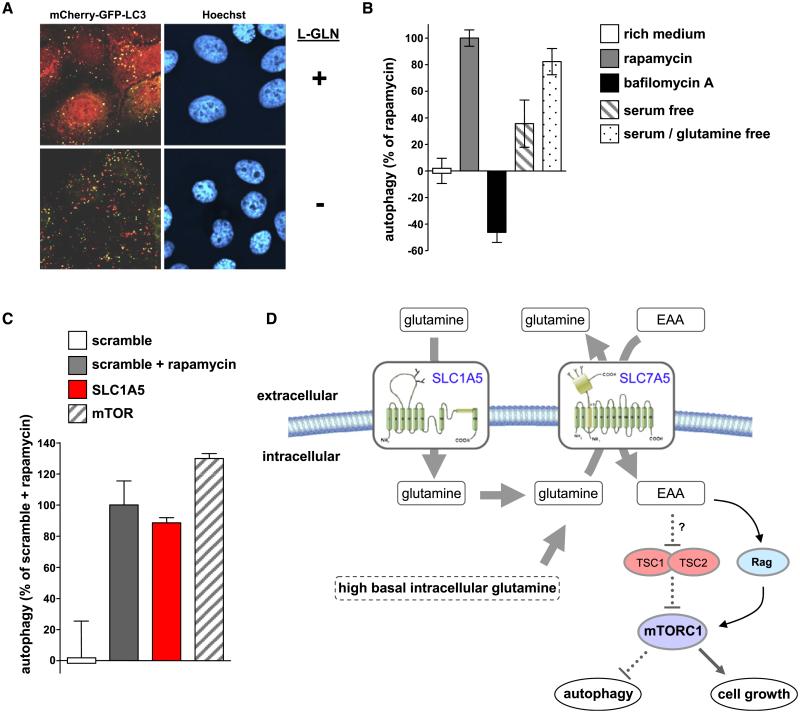
L-Glutamine and SLC1A5 Suppress Autophagy
Inhibition of mTOR signaling by withdrawing amino acids or adding rapamycin activates autophagy (Blommaart et al., 1995). We hypothesized that the absence of L-glutamine or downregulation of SLC1A5 would also activate autophagy. To test this we developed a tandem fluorescent image-based system (Kimura et al., 2007) to quantitate the redistribution of the autophagosomal membrane protein LC3. RT112 cells exhibit L-glutamine-dependent activation of S6K1 (Figure S6C) and were engineered to express mCherry-eGFP-LC3. Since eGFP is unstable in cellular compartments pH < 6.0, an increase in the mCherry:eGFP puncta ratio indicates enhanced autophagy flux and allows discrimination between the distribution of LC3 before and after lysosomal fusion. In serum-free medium LC3 codistributes into mCherry- and eGFP-positive puncta (Figure 7A, yellow puncta) but cytoplasmic codistribution of mCherry- and eGFP-LC3 is also observed indicating that autophagy is not maximally activated. After removing extracellular L-glutamine, the cytoplasmic localization disappears and LC3 codistribution into mCherry- and eGFP-positive puncta is inhibited (Figure 7A). This indicates a shift of LC3 into acidic, GFP-sensitive compartments such as lysosomes. These effects were quantitated using high content image analysis of individual mCherry and eGFP LC3 puncta (Figure 7B). As controls, rapamycin and bafilomycin A were used as activators and inhibitors of autophagy, respectively (Figure 7B). Incubation of cells in serum-free (L-glutamine-containing) DMEM increases the basal level of autophagy, consistent with the ability of growth factors to regulate mTOR (Martin and Blenis, 2002). Importantly, incubation with serum- and L-glutamine-free DMEM results in a much larger activation of autophagy, almost reaching the level achieved by rapamycin (Figure 7B). Since EAA are still present in the L-glutamine-free medium this observation proves that L-glutamine is essential for autophagy suppression and is consistent with the inability of EAA alone to activate mTOR and suppress autophagy. Finally, downregulation of SLC1A5 using RNAi enhances autophagy flux to the same extent as rapamycin or mTOR inhibition (Figures 7C and S6D). These data show that L-glutamine transport into cells via SLC1A5 is required for suppression of autophagy.
Figure 7.
L-Glutamine and SLC1A5 Regulate Autophagy
(A) Confocal images of mCherry-eGFP-LC3a distribution before (+) and after (−) L-glutamine starvation are shown.
(B) Quantitation of autophagy flux in RT112 cells.
(C) RT112 cells transfected with the individual siRNAs for 72 hr were serum starved for 24 hr and processed for autophagy flux quantitation. Rapamycin was added to scrambled siRNA transfected cells during the starvation period.
(D) EAA regulation of mTOR signaling, cell growth and suppression of autophagy requires intracellular uptake and subsequent efflux of L-glutamine (SLC3A2 dimerized with SLC7A5 is shown in yellow).
All data (mean ± SD) are representative of at least three independent experiments.
DISCUSSION
The ability of amino acids to regulate TOR signaling is functionally conserved across eukaryotes and is essential for mTORC1 to integrate growth factor signals (Dann and Thomas, 2006). Presumably amino acid transporters function upstream of mTORC1 to allow the cell to sense amino acid availability and launch an anabolic response (increased translation and growth) or when amino acids are limiting, a catabolic response such as autophagy. Amino acid transporters that perform such a role have not been identified. Here we have shown that (1) L-glutamine is an essential and rate-limiting sensitizing factor that allows EAA and growth factors to activate mTOR, (2) the high affinity L-glutamine transporter SLC1A5 and the heterodimeric SLC7A5/SLC3A2 bidirectional transporter are required for amino acid transport, mTORC1 pathway activity and cell growth, and (3) L-glutamine and SLC1A5 function are required to suppress autophagy.
A Two-Step Amino Acid Transport Mechanism Regulates mTORC1
The findings presented here are consistent with a two-step process that defines how amino acids activate mTOR (Figure 7D). First, SLC1A5 regulates an increase in the intracellular concentration of L-glutamine. Second, SLC7A5/SLC3A2 uses intracellular L-glutamine as an efflux substrate to regulate the uptake of extracellular L-leucine which subsequently leads to activation of mTORC1. This model is supported by the fact that SLC7A5/SLC3A2 is an obligate exchanger (Meier et al., 2002) and amino acid uptake requires an intracellular efflux substrate such as L-glutamine (Meier et al., 2002; Yanagida et al., 2001). The finding that reactivation of mTORC1 in starved cells depends on cellular uptake of L-glutamine is consistent with its role as an efflux substrate for SLC7A5/SLC3A2. Furthermore, LC/MS/MS analysis identified reciprocal exchange of intracellular L-glutamine for extracellular L-leucine in mammalian cells minutes before the activation of S6K1.
Some of the preferred SLC7A5 substrates are known to contribute to the activation of S6K1 (Hara et al., 1998) consistent with the observation that BCH and RNAi-mediated suppression of SLC7A5 inhibited mTORC1 activity and cell growth. Downregulation of SLC3A2 destabilized SLC7A5 which is in line with reports that cell surface expression of the heterodimer requires both subunits (Kanai et al., 1998; Mastroberardino et al., 1998). SLC3A2 can also heterodimerize with other transporters related to SLC7A5 (Verrey, 2003) such as SLC7A8 (also known as LAT2). Like SLC7A5, SLC7A8 requires SLC3A2 for proper cellular distribution and so it is possible that the observations associated with SLC7A5 or SLC3A2 RNAi could be indirectly mediated via effects on SLC7A8 expression and/or localization. This is unlikely since D-phenylalanine, which potently antagonizes mTORC1, inhibits transport via SLC7A5 but not SLC7A8 (Kanai et al., 1998; Segawa et al., 1999). Thus, SLC7A5/SLC3A2 is the key EAA transporter upstream of mTORC1. SLC7A5 is upregulated in many tumors and cancer cell lines (Fuchs and Bode, 2005; Wolf et al., 1996; Yanagida et al., 2001) indicating that it may be involved in supporting the increased anabolic needs of tumors. Furthermore, SLC7A5 expression is induced by phorbol esters and PDGF (Liu et al., 2004; Nii et al., 2001) and its 3' UTR contains destabilizing AU-rich elements (Boado et al., 1999; Gaugitsch et al., 1992), a feature associated with several protooncogene products.
In Drosophila melanogaster, distinct amino acid transporters known as minidiscs and slimfast orchestrate cellular and organismal growth (Colombani et al., 2003; Martin et al., 2000). mini-discs and slimfast function in the fat body to sense nutrient availability and control body size, the later doing so via dTOR (Colombani et al., 2003). Interestingly, both transporters have high homology with members of the SLC7 family with minidiscs sharing 48% sequence identity specifically with SLC7A5 (Martin et al., 2000). Together with the present finding that L-glutamine is required for activation of dS6K in S2 cells, it is likely that bidirectional flux of amino acids has been conserved throughout eukaryotic evolution to regulate TOR and cell growth.
L-Glutamine Regulates the Temporal Dynamics of mTORC1-S6K1 Signaling
L-glutamine-mediated sensitization of mTORC1 activation was sharply dose-dependent suggesting that a rise in intracellular L-glutamine concentration provides a switch for uptake of EAA leading to mTORC1 signaling. Preloading cells with L-glutamine prior to treatment with EAA or growth factors accelerated S6K1 activation indicating that SLC1A5-regulated uptake of L-glutamine is rate-limiting for mTORC1 pathway activation. This striking observation likely explains the 30-60 min lag that typically precedes mTOR-S6K1 activation in response to amino acids and growth factors. Importantly, upon depletion of intracellular L-glutamine, L-leucine uptake stops and mTORC1 is inactivated. These observations, which were made in several cell types, lead to the prediction that L-glutamine is required to sustain mTORC1-S6K1 signals during cell cycle progression. This is likely to be of fundamental importance since normal proliferative responses require coordination of cell division and mTOR-dependent control of cell size (Fingar et al., 2002). Moreover, observations made in MCF7 cells raise the possibility that tumor cell lines with elevated L-glutamine levels are primed for EAA-regulation of mTORC1 (Figure 7D). One possible mechanism explaining the elevated levels of L-glutamine could be aberrant upregulation of glutamine synthetase. Although further studies will be required to determine under what conditions this might occur, overexpression of glutamine synthetase in tumors has been reported (Christa et al., 1994; Gebhardt and Williams, 1995).
L-Glutamine: At the Center of Cell Growth and Metabolism
L-glutamine is the most abundant nonessential amino acid and regulates the proliferation of diverse cell types (Eagle et al., 1956; Reitzer et al., 1979). Interestingly, skeletal muscle is one of the richest stores of L-glutamine in the body with intramuscular concentrations around 20 mM (Mittendorfer et al., 2001) and is a tissue type that is dependent on mTORC1 for cell growth (Bodine et al., 2001; Ohanna et al., 2005). The demand for L-glutamine can often exceed the ability of individual cells to synthesize it de novo rendering it a “conditionally essential” amino acid (Fuchs and Bode, 2006). This is particularly important in tumor cells where it has been shown that glutaminolysis is used to replenish intermediates for the TCA cycle and to generate reductive power in the form of NADPH (DeBerardinis et al., 2007). Importantly, the observations made using L-glutamic acid and α-ketoglutarate provide evidence that glutaminolysis is not contributing to mTORC1 activation. Instead, our data are consistent with a model in which intracellular L-glutamine acts upstream of mTORC1 to regulate EAA uptake by acting as an efflux substrate for SLC7A5/SLC3A2.
Current models describing how mTORC1 is activated by nutrients show that EAA are required for integration of growth factor signals (Arsham and Neufeld, 2006; Jacinto and Lorberg, 2008). The present data suggests an update to these models in recognition of the fact that L-glutamine is rate limiting for mTORC1 activation by EAA and growth factors. Why does L-glutamine have this critical role? Tissue homeostasis depends on the coordination of protein translation, metabolic flux and nitrogen balance, processes that depend on L-glutamine. Not surprisingly, tumor cells have an increased demand for L-glutamine (DeBerardinis et al., 2007) and during cachexia circulating and skeletal muscle levels of L-glutamine are greatly reduced (Holecek, 2002). As mTORC1 is a major regulator of cell size and tissue mass in both normal and diseased states (Bodine et al., 2001; Ohanna et al., 2005; Shaw and Cantley, 2006) it is perhaps appropriate that its activity is sensitive to both intra-cellular and circulating levels of L-glutamine.
EXPERIMENTAL PROCEDURES
Mammalian Cell Culture, Treatments, and Extract Preparation
HeLa and MCF7 cells were cultured as described in the supplementary information. Amino acid starvation was performed by depriving cells of FBS for 20 hr and then incubation with D-PBS containing CaCL2, MgCl2, 15 mg/L phenol red, 20 mM HEPES and 1 g/L D-glucose (starve medium) for 3 hr. Starved cells were cultured with a mixture of EAA at the concentration found in DMEM (MEM amino acids) in the presence or absence of 1 or 10 mM L-glutamine. L-glutamic acid, α-ketoglutarate and nonessential amino acids (NEAA) solution (Sigma, St. Louis, MO) were diluted in starve medium and the resulting solutions adjusted to pH 7.2. GPNA (gamma-L-glutamyl-p-nitroanilide hydrochloride, (MP Biomedicals, Solon, OH) and BCH (2-aminobicyclo-(2,2,1)-heptane-2-carboxyclic acid, (Sigma, St. Louis, MO) were dissolved in stimulation medium and pH adjusted to 7.2. For L-glutamine preincubation experiments, amino acid starved cells were cultured in the presence of 10 mM L-glutamine for 1 hr at 37°C. The medium was then removed and cell monolayers washed twice with starve medium before adding test medium for the times indicated. Details of cell lysis buffer, Drosophila cell culture and stimulations, antibodies, eIF4E-cap complex assay and the in-cell western assay are provided in the supplementary information.
LC/MS/MS Measurement of L-Glutamine and Stable-Isotope Labeled Amino Acid Analogs
The L-glutamine concentration in culture medium was quantified using LC/MS/MS with an API4000 triple quadruple MS system (Applied Biosystems Inc, Framingham, MA) coupled to an Agilent 1200 HPLC (Santa Clara, CA) and a Leap CTC Autosampler (Carrboro, NC). LC conditions and stable-isotope labeled amino acid details are provided in the supplementary information. Calibration standards were prepared in acetonitrile/water solvent mixture (50/50 v/ v) containing 0.5% blank culture medium. Samples of conditioned culture medium, collected at different time points after incubation with cells, were diluted 1:200 (v/v) with the acetonitrile/water solvent mixture containing the internal standard. Diluted samples (5 μL) were injected and analyzed by LC/ MS/MS. For quantitation of L-glutamine and labeled amino acids in cell extracts, cell monolayers were washed twice with ice-cold PBS, scraped into PBS (200 μL), snap frozen three times and centrifuged (14,000 rpm, 10 min) to remove cell debris. After sample normalization based on total protein levels, protein in lysates was removed by precipitation and centrifugation after the addition of two equivalent aliquots of acetonitrile. Absolute levels of amino acids in the supernatants were determined as described above for the medium samples.
RNAi and Cell Size Assays
siRNA sequence information and design of the RNAi-insensitive SLC1A5 cDNA are described in the supplementary information. Final concentration of siRNA in each experiment was 25 nM. TSC2 or scramble siRNAs were transfected 24 hr prior to transfection with scramble, mTOR, SLC1A5 or SLC7A5 siRNA (Figure 5H). FACS based cell size quantitation was performed as described previously (Edinger and Thompson, 2002) with specific details described in the supplementary information.
Quantitation of Autophagy
RT112 cells stably expressing mCherry-eGFP-LC3a were cultured in clear bottom 384-well plates (24 hr). Cells were re-fed with rich medium (DMEM/ 10% FBS), 30 nM rapamycin, 100 nM bafilomycin A, serum free DMEM (containing 4 mM L-glutamine) or serum free DMEM lacking L-glutamine. After 18 hr cells were fixed, stained with Hoechst and LC3a puncta analyzed using an IN Cell Analyzer 1000 (GE Healthcare, Piscataway, NJ). Each data point shown in Figures 7B and 7C represents the average of 8 fields of view with 50-100 cells/field. Autophagy flux is calculated based on the mCherry-GFP puncta intensity ratio and expressed as a percentage of rapamycin-induced autophagy. Representative images (Figure 7A) were collected using a LSM 510 META confocal microscope (Carl Zeiss MicroImaging, Inc, Thornwood, NY).
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Stephen Helliwell, Dan Garza, Dmitri Wiederschain, Greg Michaud, Alex Huang, Leslie Pond, and John Westwick for support, comments, and advice; and to Alan Abrams for help with graphics; and Akos Szilvasi for help with confocal imaging. All authors (except L.C.C.) are employees of Novartis Pharmaceuticals.
Footnotes
Supplemental Data include Supplemental Experimental Procedures and six figures and can be found with this article online at http://www.cell.com/supplemental/S0092-8674(08)01519-5.
REFERENCES
- Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 2008;9:747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- Arsham AM, Neufeld TP. Thinking globally and acting locally with TOR. Curr. Opin. Cell Biol. 2006;18:589–597. doi: 10.1016/j.ceb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- Boado RJ, Li JY, Nagaya M, Zhang C, Pardridge WM. Selective expression of the large neutral amino acid transporter at the blood-brain barrier. Proc. Natl. Acad. Sci. USA. 1999;96:12079–12084. doi: 10.1073/pnas.96.21.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christa L, Simon MT, Flinois JP, Gebhardt R, Brechot C, Lasserre C. Overexpression of glutamine synthetase in human primary liver cancer. Gastroenterology. 1994;106:1312–1320. doi: 10.1016/0016-5085(94)90024-8. [DOI] [PubMed] [Google Scholar]
- Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- Crespo JL, Hall MN. Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2002;66:579–591. doi: 10.1128/MMBR.66.4.579-591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N. Engl. J. Med. 2006;355:1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- Dann SG, Thomas G. The amino acid sensitive TOR pathway from yeast to mammals. FEBS Lett. 2006;580:2821–2829. doi: 10.1016/j.febslet.2006.04.068. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechelotte P, Darmaun D, Rongier M, Hecketsweiler B, Rigal O, Desjeux JF. Absorption and metabolic effects of enterally administered glutamine in humans. Am. J. Physiol. 1991;260:G677–G682. doi: 10.1152/ajpgi.1991.260.5.G677. [DOI] [PubMed] [Google Scholar]
- Eagle H, Oyama VI, Levy M, Horton CL, Fleischman R. The growth response of mammalian cells in tissue culture to L-glutamine and L-glutamic acid. J. Biol. Chem. 1956;218:607–616. [PubMed] [Google Scholar]
- Edinger AL, Thompson CB. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol. Biol. Cell. 2002;13:2276–2288. doi: 10.1091/mbc.01-12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esslinger CS, Cybulski KA, Rhoderick JF. Ngamma-aryl glutamine analogues as probes of the ASCT2 neutral amino acid transporter binding site. Bioorg. Med. Chem. 2005;13:1111–1118. doi: 10.1016/j.bmc.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs BC, Bode BP. Amino acid transporters ASCT2 and LAT1 in cancer: partners in crime? Semin. Cancer Biol. 2005;15:254–266. doi: 10.1016/j.semcancer.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Fuchs BC, Bode BP. Stressing out over survival: glutamine as an apoptotic modulator. J. Surg. Res. 2006;131:26–40. doi: 10.1016/j.jss.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Gaugitsch HW, Prieschl EE, Kalthoff F, Huber NE, Baumruker T. A novel transiently expressed, integral membrane protein linked to cell activation. Molecular cloning via the rapid degradation signal AUUUA. J. Biol. Chem. 1992;267:11267–11273. [PubMed] [Google Scholar]
- Gebhardt R, Williams GM. Glutamine synthetase and hepatocarcinogenesis. Carcinogenesis. 1995;16:1673–1681. doi: 10.1093/carcin/16.8.1673. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. mTOR signaling to translation. Curr. Top. Microbiol. Immunol. 2004;279:169–197. doi: 10.1007/978-3-642-18930-2_11. [DOI] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holecek M. Relation between glutamine, branched-chain amino acids, and protein metabolism. Nutrition. 2002;18:130–133. doi: 10.1016/s0899-9007(01)00767-5. [DOI] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Huang J, Manning BD. The TSC1–TSC2 complex: a molecular switchboard controlling cell growth. Biochem. J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Lorberg A. TOR regulation of AGC kinases in yeast and mammals. Biochem. J. 2008;410:19–37. doi: 10.1042/BJ20071518. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Segawa H, Miyamoto K, Uchino H, Takeda E, Endou H. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98) J. Biol. Chem. 1998;273:23629–23632. doi: 10.1074/jbc.273.37.23629. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Kovacevic Z, Morris HP. The role of glutamine in the oxidative metabolism of malignant cells. Cancer Res. 1972;32:326–333. [PubMed] [Google Scholar]
- Liu XM, Reyna SV, Ensenat D, Peyton KJ, Wang H, Schafer AI, Durante W. Platelet-derived growth factor stimulates LAT1 gene expression in vascular smooth muscle: role in cell growth. FASEB J. 2004;18:768–770. doi: 10.1096/fj.03-0886fje. [DOI] [PubMed] [Google Scholar]
- Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Martin JF, Hersperger E, Simcox A, Shearn A. minidiscs encodes a putative amino acid transporter subunit required non-autonomously for imaginal cell proliferation. Mech. Dev. 2000;92:155–167. doi: 10.1016/s0925-4773(99)00338-x. [DOI] [PubMed] [Google Scholar]
- Martin KA, Blenis J. Coordinate regulation of translation by the PI 3-kinase and mTOR pathways. Adv. Cancer Res. 2002;86:1–39. doi: 10.1016/s0065-230x(02)86001-8. [DOI] [PubMed] [Google Scholar]
- Mastroberardino L, Spindler B, Pfeiffer R, Skelly PJ, Loffing J, Shoemaker CB, Verrey F. Amino-acid transport by heterodimers of 4F2hc/CD98 and members of a permease family. Nature. 1998;395:288–291. doi: 10.1038/26246. [DOI] [PubMed] [Google Scholar]
- McGivan JD, Bungard CI. The transport of glutamine into mammalian cells. Front. Biosci. 2007;12:874–882. doi: 10.2741/2109. [DOI] [PubMed] [Google Scholar]
- Meier C, Ristic Z, Klauser S, Verrey F. Activation of system L heterodimeric amino acid exchangers by intracellular substrates. EMBO J. 2002;21:580–589. doi: 10.1093/emboj/21.4.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittendorfer B, Volpi E, Wolfe RR. Whole body and skeletal muscle glutamine metabolism in healthy subjects. Am. J. Physiol. Endocrinol. Metab. 2001;280:E323–E333. doi: 10.1152/ajpendo.2001.280.2.E323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nii T, Segawa H, Taketani Y, Tani Y, Ohkido M, Kishida S, Ito M, Endou H, Kanai Y, Takeda E, et al. Molecular events involved in up-regulating human Na+-independent neutral amino acid transporter LAT1 during T-cell activation. Biochem. J. 2001;358:693–704. doi: 10.1042/0264-6021:3580693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohanna M, Sobering AK, Lapointe T, Lorenzo L, Praud C, Petroulakis E, Sonenberg N, Kelly PA, Sotiropoulos A, Pende M. Atrophy of S6K1(-/-) skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. Nat. Cell Biol. 2005;7:286–294. doi: 10.1038/ncb1231. [DOI] [PubMed] [Google Scholar]
- Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J. Biol. Chem. 1979;254:2669–2676. [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, BarPeled L, Sabatini DM. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, ErdjumentBromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Segawa H, Fukasawa Y, Miyamoto K, Takeda E, Endou H, Kanai Y. Identification and functional characterization of a Na+-independent neutral amino acid transporter with broad substrate selectivity. J. Biol. Chem. 1999;274:19745–19751. doi: 10.1074/jbc.274.28.19745. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J. Biol. Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsunomiya-Tate N, Endou H, Kanai Y. Cloning and functional characterization of a system ASC-like Na+-dependent neutral amino acid transporter. J. Biol. Chem. 1996;271:14883–14890. doi: 10.1074/jbc.271.25.14883. [DOI] [PubMed] [Google Scholar]
- Verrey F. System L: heteromeric exchangers of large, neutral amino acids involved in directional transport. Pflugers Arch. 2003;445:529–533. doi: 10.1007/s00424-002-0973-z. [DOI] [PubMed] [Google Scholar]
- Wang X, Campbell LE, Miller CM, Proud CG. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem. J. 1998;334:261–267. doi: 10.1042/bj3340261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DA, Wang S, Panzica MA, Bassily NH, Thompson NL. Expression of a highly conserved oncofetal gene, TA1/E16, in human colon carcinoma and other primary cancers: homology to Schistosoma mansoni amino acid permease and Caenorhabditis elegans gene products. Cancer Res. 1996;56:5012–5022. [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yanagida O, Kanai Y, Chairoungdua A, Kim DK, Segawa H, Nii T, Cha SH, Matsuo H, Fukushima J, Fukasawa Y, et al. Human L-type amino acid transporter 1 (LAT1): characterization of function and expression in tumor cell lines. Biochim. Biophys. Acta. 2001;1514:291–302. doi: 10.1016/s0005-2736(01)00384-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.