Abstract
Branched nucleic acid strands exist as intermediates in certain biological reactions, and bifurcating DNA also presents interesting opportunities in biotechnological applications. We describe here how T4 DNA ligase can be used for efficient construction of DNA molecules having one 5′ end but two distinct 3′ ends that extend from the 2′ and 3′ carbons, respectively, of an internal nucleotide. The nature of the reaction products is investigated, and optimal reaction conditions are reported for the construction of branched oligonucleotides. We discuss the utility of these branched DNA nanostructures for gene detection.
INTRODUCTION
Single DNA strands are typically linear strings of nucleotides with one 5′ and one 3′ extreme end, but there are examples of nucleic acid molecules with segments coupled in parallel from a branching point rather than in series. The well known lariat-shaped intermediate formed in the process of RNA splicing arises through the covalent attachment of the 5′ end of a transcribed and cleaved intron to the 2′ hydroxyl of an adenosine residue, internal to the intron sequence (1). This nucleotide thus represents a branched position in the RNA molecule, connected to downstream sequences via 2′,5′- and 3′,5′-phosphodiester linkages. Also retroelements that exist in some populations of Escherichia coli contain a branched position, an internal G ribonucleotide residue where the 2′ hydroxyl has been used to prime a polymerization reaction using the same RNA strand as a template (2).
Branched nucleic acid strands have been chemically synthesized using branching nucleotides that give rise to fork-like structures (3–5). Such branching oligoribonucleotides have been useful to study messenger RNA splicing and to investigate the biological role of RNA lariats and forks in vivo. Chemically synthesized branched nucleic acid strands have also been used for gene detection by attaching one hybridizing DNA segment to several identical detectable sequences in a forked structure, in order to increase sensitivity in hybridization-based detection of, for example, viral sequences (6). So-called DNA dendrimers have also been used for enhanced detection. These DNA molecules branch dichotomically in several generations and can be constructed by introducing a modified residue having two blocking residues at intervals during solid-phase chemical synthesis for subsequent extension of two separate, but identical 5′ end sequences (7). Braich and Damha developed an interesting strategy to start synthesis of a second 3′ strand during chemical synthesis by deprotecting a 3′ carbon of an internal residue (8). Difficulties of selectively deprotecting this residue, and of performing 5′-to-3′ synthesis from this position, may limit the generality of the procedure.
We were interested in developing enzymatic means to construct DNA strands that branch into two distinct 3′ end sequences from a given sugar residue along a DNA strand. DNA ligases serve the purpose of sealing nicks in double-stranded DNA molecules (9–11), but they have also been employed in a much wider range of reactions, such as the joining of duplex DNA molecules with either cohesive or blunt ends (12), or of ligation substrates where one or more of the strands are RNA (13–15). Under appropriate conditions ligases have also been shown to join oligodeoxyribonucleotides containing abasic sites, gaps or mismatched bases at the ligation junction (16,17). We show here that T4 DNA ligase can also be used to add an extra 3′ strand to an internal nucleotide residue in a single-stranded DNA molecule, connected to the downstream sequence via a 2′,5′ phosphodiester bond. This leaves the 3′ hydroxyl at this internal residue free to be enzymatically joined to the 5′ phosphate of the extra DNA strand if the ligation substrate is positioned through hybridization to a template strand.
MATERIALS AND METHODS
Oligonucleotides
Oligonucleotides were synthesized on an Applied Biosystems 380B DNA Synthesizer using phosphoramidite chemistry or ordered from Interactiva or Eurogentech. We purchased the 5′DMT, 3′ TBDMS cytidine (N-Bz) phosphoramidite nucleoside reagent from ChemGenes, catalog number ANP-5682. This reagent is suitable to introduce a ribonucleotide connected via a 2′,5′ phosphodiester bond to the preceding nucleotide. After complete synthesis, deblocking and HPLC purification, this reagent yields a residue with a free 3′ hydroxyl function in the nucleotide chain, referred to herein as a 3′ hydroxy ribonucleotide residue. All sequences are written in the 5′–3′ direction (see also Fig. 1). The sequences of oligonucleotides ON1a, ON1b and ON1c were: GACTGC TCGGACTCAGCCTACACTAGCGACATTGATGGACCTTCCGTCTCGTTCT, where the underlined C represents an unconventional cytosine 3′ hydroxy ribonucleotide in ON1a, a regular cytosine ribonucleotide in ON1b and a cytosine deoxyribonucleotide in ON1c. Oligonucleotide ON1d (GACTGCTCGGACTCAGCCTACACTAGCGAC) was interrupted after the same C residue. Oligonucleotides ON2a, (p-TCATCCAAGGTGCCTTTGCTGAGAC), ON2b (p-TCATCCAAGGTGCCTTTGCTGAGACGTAATAGCG TTGAC) and ON2c (p-TCATCCAAGGTGCCTTTGCTG AGACTTTTTTTTTT-biotin), were used to form 3′ branch sequences. The following reagents were used as templates for branch ligation: ON3a (CAAAGGCACCTTGGATGAGTC GCTAGTGTAGGC), ON3b (GTCTCAGCAAAGGCACC TTGGATGAGTCGCTAGTGTAGGCTGAGTCCGAGCA GTC), ON3c (CAGAACAGTCGTCTCAGCAAAGGCAC CTTGGATGAGTCGCTAGTGTAGGCTGAGTCCGAG), and ON3d (CAGAACAGTCGTCTCAGCAAAGGCA CCTTGGATGAGTCGCTAGTGTAGGCTGAGTCCGAGC AGTCGGCTCAGCAC).
Figure 1.
Structure of a substrate for construction of branched DNA by templated enzymatic ligation. The outer box illustrates that an internal nucleotide, the boxed C residue, is connected to the downstream sequence via the 2′ carbon, and that the 3′ hydroxyl of this nucleotide is juxtaposed to the 5′ phosphate of a separate oligonucleotide by hybridization to a third, template oligonucleotide. H and D indicate cleavage sites for the restriction enzymes HinfI and DdeI, respectively (see Figure 3B).
Finally, oligonucleotides ON4 (AGAACGAGACGGAA GGTCC) and ON5 (GTCTCAGCAAAGGCACCTTG) were used to prime extension reactions from the 2′–5′ and 3′–5′ branches, respectively, of the branched product.
Radiolabeling
Ten picomole oligonucleotide was 5′ radiolabeled by the addition of a 5′ phosphate using 30 units of T4 polynucleotide kinase (Amersham Biosciences) in 50 µl of 10 mM TrisOAc pH 7.5, 10 mM MgAc2, 50 mM KAc and 30 µCi [λ-32P]ATP (3000 mCi/mmol, Amersham Biosciences) at 37°C for 30 min. For 3′ radiolabeling, 10 pmol oligonucleotide was incubated with 18 units of terminal deoxynucleotidyl transferase (Amersham Biosciences) in the same buffer containing 50 µCi [α-32P]dCTP (3000 mCi/mmol, Amersham Biosciences) at 37°C for 30 min. After the reactions the enzymes were heat-inactivated at 65°C for 10 min, and oligonucleotides were purified on MicroSpin™ G-50 columns (Amersham Biosciences).
Branch ligation
Unless otherwise indicated, 1 pmol radiolabeled ON2 and 3 pmol of both ON1 and ON3 were incubated at 65°C for 10 min in 20 µl of reaction buffer containing 10 mM TrisOAc pH 7.5, 10 mM Mg(Ac)2, 50 mM KAc, 0.1 µg/µl bovine serum albumin (BSA), 10 µM ATP, and allowed to cool to room temperature for at least 10 min. Thereafter 2.8 Weiss units of T4 DNA ligase (Amersham Biosciences) was added and the reactions were incubated at 37°C overnight, followed by heat inactivation at 65°C for 10 min.
Alkaline treatment
One picomol branched 3′-labeled oligonucleotide in 6.5 µl of ligation reaction was incubated at 37°C overnight after the addition of 6.5 µl of 0.6 M NaOH, 2 mM EDTA (18). Thereafter 2.2 µl of 2 M HAc was added for neutralization, and the nucleic acid was precipitated with ethanol and dissolved in 6.5 µl of H2O. An equal volume of loading buffer was added [25 mM Tris-borate pH 8.3, 10 mM EDTA, 75% formamide, 0.05% (w/v) xylene cyanol FF, 0.05% (w/v) bromophenol blue], and the samples were separated in a denaturing 10% polyacrylamide gel and analyzed on a PhosphorImager (Amersham Biosciences).
Restriction enzyme digestion
Radiolabeled ligation product (25 fmol) was digested with 5 units of HinfI or 4 units of DdeI restriction enzyme (Amersham Biosciences) in 5 µl of 20 mM TrisOAc pH 7.5, 20 mM Mg(Ac)2, 100 mM KAc, at 37°C overnight.
Solid-phase replication of branched oligonucleotides
Ten microliter branch ligation reactions with 0.25 pmol 5′ radiolabeled and 3′ biotinylated ON2c, were immobilized to 20 µl of streptavidin-coated Dynabeads (Dynal), denatured, and washed according to recommended procedures. The extension primers, 0.25 pmol of ON4 or ON5, were incubated together with the immobilized branched DNA at 65°C for 10 min in 5 µl of extension buffer. They were allowed to cool to room temperature for at least 10 min. The 10 µl extension reactions contained either 200 ng φ29 DNA polymerase (a kind gift of Dr Margarita Salas) in 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 20 mM (NH4)2SO4, 1 mM of 200 µM dNTP, 0.15 µg/µl BSA, 1 mM dithiothreitol, or 7.5 units of T7 Sequenase version 2.0 DNA polymerase (Amersham Biosciences) in 40 mM Tris–HCl pH 7.5, 20 mM MgCl2, 50 mM NaCl, 200 µM dNTP, 0.15 µg/µl BSA. The reactions were incubated at 37°C for 1 h, and stopped by adding an equal volume of loading buffer. The samples were analyzed on a denaturing 10% polyacrylamide gel.
RESULTS AND DISCUSSION
Construction of branched DNA by ligation
Ligation of DNA strands by DNA ligases typically involves the joining of 5′ phosphates and 3′ hydroxyls of adjoining DNA ends, hybridizing to a common template DNA strand (9–11,19). In order to attempt ligase-mediated construction of branched DNA, we synthesized an oligonucleotide with an internal 3′ hydroxy ribonucleotide residue. This nucleotide was thus connected to the downstream sequence via a 2′,5′ phosphodiester linkage, leaving a free 3′ hydroxyl at this internal position. The 3′ hydroxyl group was brought close to the 5′ phosphate of a separate oligonucleotide by hybridizing the strands to be joined to a common template oligonucleotide. The question we asked was whether a DNA ligase could join a 5′ phosphate of one strand to an internal 3′ hydroxyl along a different DNA strand in a DNA-templated reaction, despite the DNA strand extending from the adjacent 2′ carbon. Figure 1 illustrates the structure of the oligonucleotide reagents and of the complex serving as substrate for the ligation reaction.
Characterization of the ligation product
We investigated the structure of the ligation products by denaturing polyacrylamide gel electrophoresis. Of the three oligonucleotides in the ligation substrates, only the branch oligonucleotides, ON2a or ON2b, had 5′ phosphates—the 32P added for labeling. Therefore only ligation to the 5′ end of ON2 oligonucleotides could be detected. Figure 2A demonstrates that the migration of the ligation products was dependent on the sizes of the labeled 3′ branch oligonucleotides, confirming that these reagents participated in the ligation products. Western and Rose have shown that under certain conditions T4 DNA ligase is able to form an intramolecular loop by joining the end of a single DNA strand to that of an oligonucleotide hybridizing to the DNA strand, without apparent Watson–Crick basepairing on one side of the ligation site (20). A similar reaction could have joined the branch oligonucleotides (ON2) to the 3′ ends of the template oligonucleotides (ON3). No such effect was seen under the conditions used here, however, as the different sizes of the three different ON3 oligonucleotides that were used to template the ligation reactions clearly did not influence the migration of the ligation products (Fig. 2A).
Figure 2.

Formation of ligation products involving an oligonucleotide with a 2′,5′ linkage. (A) Dependence of ligation product size on the length of the labeled 5′ phosphorylated ON2a (25 nt) and ON2b (39 nt) and independence of template lengths. M, molecular weight markers. (B) Oligonucleotides with a 2′,5′ linkage, a regular 3′–5′ DNA linkage or a 3′–5′ linkage connecting a single ribonucleotide residue to the downstream sequence were combined with templates of the indicated lengths. Reaction products were analyzed on a denaturing 10% polyacrylamide gel.
Ligation products were obtained in high yields, ranging from 70–78% of the labeled strands in the illustrated reactions. Only strands containing an internal 2′,5′ linkage with a free 3′ hydroxyl were substrates for the ligation reaction. No ligation was observed with oligonucleotides having a regular 3′–5′ linkage in the corresponding position, whether or not there was a free 2′ hydroxyl present (Fig. 2B).
DNA strands containing internal RNA residues are susceptible to cleavage by treatment with alkali. The 3′ hydroxy ribonucleotide residue, having an internal 3′ hydroxyl, is also expected to render the strands sensitive to alkaline hydrolysis. By contrast, successful ligation products that have this internal 3′ carbon attached to an extraneous DNA strand should no longer be sensitive to alkali. In the experiment shown in Figure 3A the 3′ radiolabeled oligonucleotide with a 3′ hydroxy ribonucleotide residue was shown to be sensitive to alkaline cleavage as expected, whereas the ligation product was not. The pattern of cleavage of the ligation products with restriction enzymes HinfI and DdeI is also consistent with a branched structure of the ligation products. We compared restriction cleavage of a conventional nick-ligated and a branch-ligated product (Fig. 3B). The branched products migrated distinctly from the corresponding linear fragments of the same size, and the migration of the restriction digested products was consistent with the expected cleavage pattern. A HinfI site situated at the branch point could be cleaved in the nick-ligated duplex but not in the branch-ligation reaction product, indicating that the branched structure inhibited the restriction enzyme. We conclude that the ligation products indeed represent branched structures, created by ligation of the 5′ phosphorylated strands to other strands having internal 2′,5′ linkages.
Figure 3.
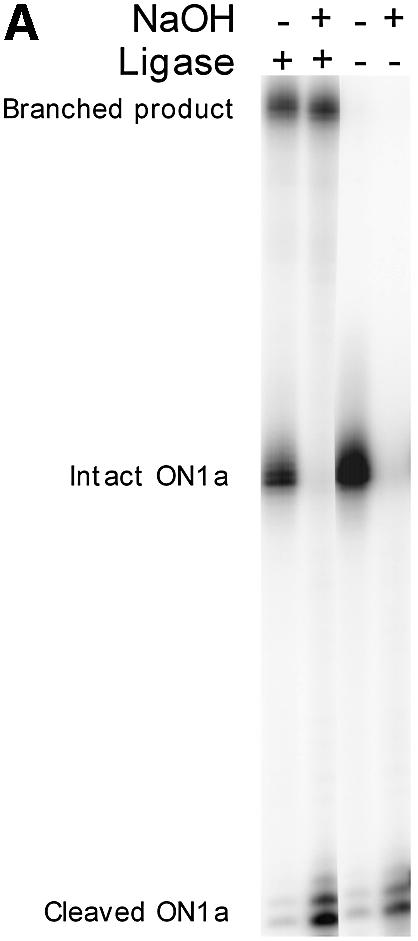

Analysis of the structure of the ligation products formed in branch-ligation reactions. (A) Investigation of the susceptibility of 3′-labeled molecules with a 2′,5′ linkage to alkali treatment before and after branch ligation. (B) Regular nick- and branch-ligated products, radiolabeled via a 5′ phosphate, were investigated for their susceptibility to cleavage by the restriction enzymes HinfI and DdeI. Cleavage sites are indicated in Figure 1. The samples were analyzed on a denaturing 10% polyacrylamide gel. M, molecular weight markers.
Optimization of ligation conditions
Ligation reactions proceed in four steps. First the enzyme is activated by covalent coupling of ATP, followed by binding to the substrate. The adenylyl group is then transferred to the 5′ phosphate on the nicked DNA, and finally the activated phosphate is attacked by a nearby 3′ hydroxyl group, forming a phosphodiester bond between the two adjacent strands and releasing AMP (10,11,19). Attempts to use T4 DNA ligase to join difficult substrates, such as blunt ends of duplex DNA molecules or DNA strands hybridized to an RNA template, tend to result in the interruption of the ligation process after the cofactor has been transferred to the 5′ end of the ligation substrate. This leads to the accumulation of 5′-adenylated DNA strands. This tendency can be reduced, however, and ligation can be allowed to proceed by using substantially lower amounts of the cofactor ATP (21,22). Adenylated intermediates accumulate also in branch ligation. At 1 mM ATP, the concentration commonly used in nick ligation, adenylation dominated over ligation (Fig. 4A and B). By lowering ATP concentrations to 1–10 µM, the ligation product yield increased to levels similar to those of nick ligation. Similar yields of branched products were observed at temperatures of 5, 16 or 37°C (results not shown).
Figure 4.
Comparison of the effect of ATP concentration on the yield of (A) ligation and (B) adenylation products of regular nick-ligated (open symbols) and branch-ligated (filled symbols) products as a function of time. The large and small symbols represent reactions performed with 1 mM and 1 µM ATP, respectively.
Compared to the joining of conventional nicked ligation substrates, considerably more ligase is required for efficient branch ligation, and branched ligation products accumulate at a far slower rate, reaching yields of ∼70–80% only after overnight incubation (Fig. 4A). Maximal ligation yields were observed at the highest concentration of ligase used, corresponding to an estimated four ligase molecules per substrate (Fig. 5). Blunt-end ligation requires 10–100× more ligase than cohesive end ligation, and both RNA templated ligation as well as the looped ligation investigated by Western and Rose require a molar excess of ligase over substrate molecules. The reason for the large requirement of ligase in the branch-ligation reaction is not understood. It is possible that the enzyme remains bound to the substrate after the ligase has covalently joined the oligonucleotides, unable to go on to ligate new substrates. However, no band shift was obtained when the ligation reaction products were analyzed for electrophoretic mobility shift according to Rossi et al. (21), indicating that the ligase leaves the substrate after ligation (data not shown).
Figure 5.
The yield of branched product was investigated as a function of ligase concentration. Circles represent branched ligation products and squares represent adenylation products.
The branched site blocks replication
It was of interest to determine if the branched oligonucleotides formed by enzymatic ligation could serve as templates for DNA polymerases. Lorsch et al. have shown that reverse transcriptase is able to read through a 2′,5′ linkage in a template (23). We tested several DNA polymerases and found that most were able to read through the 3′ hydroxy ribonucleotide residue in the linear ON1a and the 3′–5′ ribonucleotide in ON1b, although paused products were observed (results not shown). By contrast, both φ29 DNA polymerase and T7 Sequenase generated products interrupted around the branch-point of branched templates, although a faint band, somewhat smaller than the expected full-length product, can also be seen on the gel when extension was primed from the 3′ branch (see arrows in Fig. 6). We conclude that the branched site served as a block for replication.
Figure 6.

Attempted replication of immobilized branched 5′-radiolabeled oligonucleotides across the 2′–5 and the 3′–5′ linkages at the branch point. Comparison between φ29 DNA polymerase and T7 Sequenase versus 2.0 DNA polymerase. Reaction products were analyzed on a denaturing 10% polyacrylamide gel. The extension reactions were primed from the 2′ branch in lanes 1, 2, 5 and 6, and from the 3′–5′ branch in lanes 3, 4, 7 and 8. M, molecular weight markers.
Concluding remarks
Among the DNA ligases, the T4 DNA ligase enzyme is unusually adept at joining modified nucleic acid substrates. To our knowledge it is the only ligase capable of efficient blunt-end ligation (11). By using high concentration of T4 DNA ligase and low ATP concentration the yield of the branched ligation product can reach levels similar to those for regular nick ligation, although the reaction is quite slow.
Templated branch ligation allows the construction of DNA strands having two different 3′ ends. This further extends the already rich scope for DNA nano-scale structures, composed of rigid duplex and flexible single-stranded segments (24). In gene detection applications, branched DNA strands could extend the range of functionalities of probe molecules. For example, it is possible to construct padlock probes (25) that after circularization and firm binding to a target molecule can still extend a 3′ end that can be used to prime rolling-circle replication of a separately added circular DNA strand for enhanced detection (26). Branched oligonucleotides could also be used as bi-allelic padlock probes having two different 3′ ends, specific for alternate alleles at a given locus, and separately detectable.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by Amersham Biosciences, the Swedish Medical and Technological Research Councils of Sweden, the Wallenberg Foundation, and Polysaccharide Research AB Uppsala. A.K. was supported by a stipend from the Swedish Institute. Dr Margarita Salas kindly provided the φ29 polymerase. We thank Drs Marek Kwiatkowski, Mats Gullberg and Mats Nilsson for providing valuable comments on the manuscript.
REFERENCES
- 1.Padgett R.A., Konarska,M.M., Grabowski,P.J., Hardy,S.F. and Sharp,P.A. (1984) Lariat RNAs as intermediates and products in the splicing of messenger RNA precursors. Science, 225, 898–903. [DOI] [PubMed] [Google Scholar]
- 2.Yee T., Furuichi,T., Inouye,S. and Inouye,M. (1984) Multicopy single-stranded DNA isolated from a Gram-negative bacterium, Myxococcus xanthus. Cell, 38, 203–209. [DOI] [PubMed] [Google Scholar]
- 3.Horn T. and Urdea,M.S. (1989) Forks and combs and DNA: the synthesis of branched oligodeoxyribonucleotides. Nucleic Acids Res., 17, 6959–6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sund C., Foldesi,A., Yamakage,S. and Chattopadhyaya,J. (1991) Synthesis of branched nona- and deca-RNA modelling the lariat formed in pre-mRNA processing reaction (splicing). Nucleic Acids Symp. Ser., 24, 9–12. [PubMed] [Google Scholar]
- 5.Damha M.J., Ganeshan,K., Hudson,R.H.E. and Zabarylo,S.V. (1992) Solid-phase synthesis of branched oligoribonucleotides related to messenger RNA splicing intermediates. Nucleic Acids Res., 20, 6565–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urdea M.S. (1994) Branched DNA signal amplification. Bio/Technol., 12, 926–928. [DOI] [PubMed] [Google Scholar]
- 7.Horn T., Chang,C.-A., Fultz,T.J., Ahle,D., Hamren,S.J. and Urdea,M.S. (1991) Branched oligodeoxyribonucleotides for use as amplification multimers in bioassays: chemical synthesis and characterization. Nucleic Acids Symp. Ser., 24, 201–202. [PubMed] [Google Scholar]
- 8.Braich R.S. and Damha,M.J. (1997) Regiospecific solid-phase synthesis of branched oligonucleotides. Effect of vicinal 2′,5′- (or 2′,3′-) and 3′,5′-phosphodiester linkages on the formation of hairpin DNA. Bioconjugate Chem., 8, 370–377. [DOI] [PubMed] [Google Scholar]
- 9.Lehman I.R. (1974) DNA ligase: structure, mechanism, and function. Science, 186, 790–797. [DOI] [PubMed] [Google Scholar]
- 10.Higgins N.P. and Cozzarelli,N.R. (1979) DNA-joining enzymes: a review. Methods Enzymol., 68, 50–71. [DOI] [PubMed] [Google Scholar]
- 11.Engler M.J. and Richardson,C.C. (1982) DNA ligases. The Enzymes, XV, 3–29. [Google Scholar]
- 12.Sgaramella V. (1972) The covalent joining of DNA molecules at their base-paired ends. Farmaco [Sci.], 27, 809–817. [PubMed] [Google Scholar]
- 13.Kleppe K., van de Sande,J.H. and Khorona,H.G. (1970) Polynucleotide ligase-catalyzed joining of deoxyribo-oligonucleotides on ribopolynucleotide templates and of ribo-oligonucleotides on deoxyribopolynucleotide templates. Proc. Natl Acad. Sci. USA, 67, 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fareed G.C., Wilt,E.M. and Richardson,C.C. (1971) Enzymatic breakage and joining of deoxyribonucleic acid. 8. Hybrids of ribo- and deoxyribonucleotide homopolymers as substrates for polynucleotide ligase of bacteriophage T4. J. Biol. Chem., 246, 925–932. [PubMed] [Google Scholar]
- 15.Nilsson M., Barbany,G., Antson,D.O., Gertow,K. and Landegren,U. (2000) Enhanced detection and distinction of RNA by enzymatic probe ligation. Nat. Biotechnol., 18, 791–793. [DOI] [PubMed] [Google Scholar]
- 16.Wu D.Y. and Wallace,R.B. (1989) The ligation amplification reaction (LAR)–amplification of specific DNA sequences using sequential rounds of template-dependent ligation. Genomics, 4, 560–569. [DOI] [PubMed] [Google Scholar]
- 17.Landegren U., Kaiser,R., Sanders,J. and Hood,L. (1988) A ligase-mediated gene detection technique. Science, 241, 1077–1080. [DOI] [PubMed] [Google Scholar]
- 18.Sriskanda V. and Shuman,S. (1998) Specificity and fidelity of strand joining by Chlorella virus DNA ligase. Nucleic Acids Res., 26, 3536–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss B., Jacquemin-Sablon,A., Live,T.R., Fareed,G.C. and Richardson,C.C. (1968) Enzymatic breakage and joining of deoxyribonucleic acid. VI. Further purification and properties of polynucleotide ligase from Escherichia coli infected with bacteriophage T4. J. Biol. Chem., 243, 4543–4555. [PubMed] [Google Scholar]
- 20.Western L.M. and Rose,S.J. (1991) A novel DNA joining activity catalyzed by T4 DNA ligase. Nucleic Acids Res., 19, 809–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossi R., Montecucco,A., Ciarrocchi,G. and Biamonti,G. (1997) Functional characterization of the T4 DNA ligase: a new insight into the mechanism of action. Nucleic Acids Res., 25, 2106–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson M., Antson,D.O., Barbany,G. and Landegren,U. (2001) DNA-templated DNA ligation for transcript analysis. Nucleic Acids Res., 29, 578–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorsch J.R., Bartel,D.P. and Szostak,J.W. (1995) Reverse transcriptase reads through a 2′-5′ linkage and a 2′-thiophosphate in a template. Nucleic Acids Res., 23, 2811–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seeman N.C. (2003) DNA in a material world. Nature, 421, 427–431. [DOI] [PubMed] [Google Scholar]
- 25.Nilsson M., Malmgren,H., Samiotaki,M., Kwiatkowski,M., Chowdhary,B.P. and Landegren,U. (1994) Padlock probes: circularizing oligonucleotides for localized DNA detection. Science, 265, 2085–2088. [DOI] [PubMed] [Google Scholar]
- 26.Lizardi P.M., Huang,X., Zhu,Z., Bray-Ward,P., Thomas,D.C. and Ward,D.C. (1998) Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature Genet., 19, 225–232. [DOI] [PubMed] [Google Scholar]






