Abstract
During limb skeletogenesis the cartilaginous long bone anlagen and their growth plates become delimited by perichondrium with which they interact functionally. Yet, little is known about how, despite being so intimately associated with cartilage, perichondrium acquires and maintains its distinct phenotype and exerts its border function. Because perichondrium becomes deranged and interrupted by cartilaginous outgrowths in Hereditary Multiple Exostoses (HME), a pediatric disorder caused by EXT mutations and consequent heparan sulfate (HS) deficiency, we asked whether EXT genes and HS normally have roles in establishing its phenotype and function. Indeed, conditional Ext1 ablation in perichondrium and lateral chondrocytes flanking the epiphyseal region of mouse embryo long bone anlagen – a region encompassing the groove of Ranvier – caused ectopic cartilage formation. A similar response was observed when HS function was disrupted in long bone anlagen explants by genetic, pharmacological or enzymatic means, a response preceded by ectopic BMP signaling within perichondrium. These treatments also triggered excess chondrogenesis and cartilage nodule formation and overexpression of chondrogenic and matrix genes in limb bud mesenchymal cells in micromass culture. Interestingly, the treatments disrupted the peripheral definition and border of the cartilage nodules in such a way that many nodules overgrew and fused with each other into large amorphous cartilaginous masses. Interference with HS function reduced the physical association and interactions of BMP2 with HS and increased the cell responsiveness to endogenous and exogenous BMP proteins. In sum, Ext genes and HS are needed to establish and maintain perichondrium’s phenotype and border function, restrain pro-chondrogenic signaling proteins including BMPs, and restrict chondrogenesis. Alterations in these mechanisms may contribute to exostosis formation in HME, particularly at the expense of regions rich in progenitor cells including the groove of Ranvier.
Keywords: Chondro-perichondrial border, Perichondrium, Heparan sulfate, Ext1, Growth plate, Hereditary multiple exostoses, Groove of Ranvier
Introduction
Long bones begin to form at mid stages of embryogenesis in many vertebrate species, including mice and chicks (Hinchliffe and Johnson, 1980; Ota and Kuratani, 2009). Onset of their development becomes apparent with the formation of uninterrupted Y-shaped preskeletal mesenchymal condensations that correspond to the proximal stylopod elements (humerus and femur) and medial zeugopod elements (radius/ulna and tibia/fibula). Soon after, the condensed cells differentiate into chondrocytes that produce cartilage matrix, establish the cartilaginous skeletal primordia and become organized into growth plates (Kronenberg, 2003). The cells adjacent to the cartilaginous elements remain mesenchymal, forming the perichondrium that surrounds the elements all along their longitudinal length as well as at their epiphyseal ends where the synovial joints begin to form (Eames et al., 2003; Hall and Miyake, 2000; Holder, 1977). The mesenchymal cells located at those joint sites and now interrupting the elements at the prospective elbow and knee joints are collectively called the interzone (Pacifici et al., 2005). While initially loose and non-descript, the perichondrial cells become more organized over developmental time eventually giving rise to the distinct layers of perichondrium. Specifically, the inner layer becomes composed of cuboidal progenitor cells and the outer layer contains elongated fibrogenic and mechanically-endowed cells (Bairati et al., 1996; Bandyopadhyay et al., 2008; Gigante et al., 1996; Scott-Savage and Hall, 1980). Starting in diaphysis, the growth plate chondrocytes undergo maturation and hypertrophy and produce the signaling protein Indian hedgehog (Ihh) that diffuses into the adjacent perichondrium and induces formation of the intra-membranous bone collar (Koyama et al., 1996a; Vortkamp et al., 1996). This elicits a local transition from perichondrium to periosteum characterized by marked changes in cell phenotype and gene expression (Koyama et al., 1996b). Ihh is also part of another regulatory loop with periarticular cells that produce parathyroid hormone-related protein (PTHrP) and regulate the overall rates of chondrocyte maturation in the growth plate (Vortkamp et al., 1996). In addition, several members of the bone morphogenetic protein (BMP) and fibroblast growth factor (FGF) families are expressed in growth plate and/or perichondrium and have been shown to be part of interactive loops regulating Ihh and PTHrP expression and overall growth plate activities (Pathi et al., 1999; Zou et al., 1997). These and several other studies have provided compelling evidence that the growth plate and perichondrium are engaged in multiple interactions critical for skeletal development and growth (Alvarez et al., 2001, 2002; Colnot et al., 2004; Long and Linsenmayer, 1998). However, what has remained less understood is how, though adjacent and so intimately interconnected to cartilage, perichondrium is actually able to acquire and maintain its phenotype and exert its boundary functions.
Heparan sulfate (HS) constitutes the glycosaminoglycan moiety of cell surface and matrix proteoglycans that include syndecans, glypicans and perlecan (Bernfield et al., 1999). The HS chains are composed of repeating d-glucuronic acid (GlcA) and N-acetyl-D-glucosamine (GlcNAc) residues that are assembled into linear polysaccharides by the Golgi-associated heterodimer of Ext1 and Ext2 glycosyltransferases (Esko and Selleck, 2002). During assembly, the nascent chains undergo extensive modifications that include N-deacetylation/N-sulfation, epimerization, and O-sulfation. These multiple and systematic reactions result in the creation of patterns of specific negatively-charged sulfated segments of modified sugars within the chains of HS-proteoglycans expressed in different tissues and organs (Bulow and Hobert, 2006). Thus endowed, the resultant heparan sulfate proteoglycans can influence cell determination and differentiation, cell-matrix interactions, and cell-growth factor interactions (Bishop et al., 2007; Hacker et al., 2005). Cell-growth factor interactions are of particular relevance since several signaling proteins and growth factors critical for skeletogenesis including BMPs, FGFs and hedgehog family members, are HS-binding factors (Bernfield et al., 1999; Lin, 2004). Previously we and other groups showed that deficiency in Ext1 and/or Ext2 expression alters the organization and functioning of the growth plate and can lead to growth aberrations (Hilton et al., 2005; Koziel et al., 2004; Stickens et al., 2005; Zak et al., 2011). Growth plate and skeletal aberrations were also observed in mutant mice of HS modifying genes such as 6-O-sulfotransferase-1 and N-sulfotransferase 1. Ablation of Perlecan or Glypican-3 genes, both of which encode HSPG core proteins and are expressed in the growth plate, also results in growth plate and skeletal abnormalities (Arikawa-Hirasawa et al., 1999; Habuchi et al., 2007; Viviano et al., 2005; Yasuda et al., 2010). These observations suggest that HS-dependent mechanisms are major regulators of skeletogenesis and growth plate function, but far less is known about whether similar mechanisms operate in perichondrium to regulate its phenotype and roles.
Hereditary multiple exostoses (HME; also called Multiple Osteochondroma or Multiple Hereditary Exostoses) is a congenital pediatric skeletal disorder caused by heterozygous loss-of-function mutations in EXT1 or EXT2 and consequent HS deficiency (Ahn et al., 1995; Lind et al., 1998). HME is characterized by cartilaginous and bony outgrowths (exostoses) that form next to, but never within, the growth plates. Given their location, the exostoses interrupt the continuity of the perichondrium, violate the chondro-perichondrial border and protrude into surrounding tissues causing multiple complications, and it has been suggested that they arise from perichondrial cells (Hecht et al., 2005; Porter and Simpson, 1999). Thus, we reasoned that Ext genes and HS could normally be part of mechanisms by which the perichondrium acquires and maintains its fibrogenic phenotype and exerts its boundary roles along the chondro-perichondrial border. The genetic, pharmacologic and biochemical-biophysical data we report here do support such a possibility.
Materials and methods
Mouse lines, mating and genotyping
Gdf-5-Cre transgenic mice were described previously (Rountree et al., 2004) and line B was used in the present study. Creation of loxP-modified Ext1 allele and establishment of the Ext1 floxed mouse line (Ext1flox/flox) were described previously (Inatani et al., 2003). eYFP mice were obtained from Jackson labs (stock number 006148). Pregnant mice and postnatal mice were sacrificed by IACUC approved methods. Genotyping was carried out with DNA isolated from tail clips.
In situ hybridization and proliferation analysis
In situ hybridization was carried out as described (Koyama et al., 1999). Limbs fixed with 4% paraformaldehyde overnight were embedded in paraffin and sectioned. Serial 5-μm-thick sections from wild type and mutants mounted on the same slide were pretreated with 10 μg/ml proteinase K (Sigma, St. Louis, MO) in 50 mM Tris–HCl, 5 mM EDTA pH 7.5 for 1 min at room temperature, immediately post-fixed in 4% paraformaldehyde buffer for 10 min, and then washed twice in 1 × PBS containing 2 mg/ml glycine for 10 min/wash. Sections were treated for 15 min with a freshly prepared solution of 0.25% acetic anhydride in triethanolamine buffer and were hybridized with antisense or sense 35S-labeled riboprobes (approximately 1 × 106 DPM/section) at 50 °C for 16 h. cDNA clones used as templates for probes included: a 121 bp mouse collagen IIA (284–404, NM_008109); a 356 bp mouse collagen IIB (2409–2764, NM_008109); a 254 bp mouse histone 4C (H4C) (546–799; AY158963); a 515 bp mouse collagen X (1302–1816); a 741 bp mouse Sox9 (116–856; NM_011448) and a 591 bp mouse Tenascin-C (544–1124, NM_011607.3). After hybridization, slides were washed three times with 2 × SSC containing 50% formamide at 50 °C for 20 min/wash, treated with 20 μg/ml RNase A for 30 min at 37 °C, and washed three times with 0.1 × SSC at 50 °C for 10 min/wash. Sections were dehydrated by immersion in 70, 90, and 100% ethanol for 5 min/step, coated with Kodak NTB-3 emulsion diluted 1:1 with water, and exposed for 10–14 days. Slides were developed with Kodak D-19 at 20 °C for 3 min, and stained with hematoxylin and eosin. Bright and dark-field images of in situ hybridization were taken with a SPOT insight camera (Diagnostic Instruments, Inc.) operated with SPOT 4.0 software. No further processing of images was performed apart from assembling montages in Photoshop (version 8, Adobe).
Explant cultures
Forelimb elements were isolated from P1 Ext1flox/flox neonates and cultured on filter membranes in DMEM supplemented with 1% FBS. Adeno-CMV-Cre (Vector Biolabs) was added to the media at a concentration of 109 virus particles per ml of media. Control samples were treated with Ad-CMV-GFP (Vector Biolabs) at the same concentration. Samples were incubated for 6 days. Similar experiments were performed using P1 WT forelimbs with the addition of 10 μM Surfen (Drug Synthesis and Chemistry Branch, Division of Cancer Treatment and Diagnosis, National Cancer Institute, Bethesda, MD).
Immunohistochemistry
Immunostaining for phospho-Smad1/5/8 was carried out with paraffin sections that were first deparaffinized and then treated with 1 mg/ml pepsin for 10 min at 37 °C to de-mask the tissue. Sections were incubated with anti-phospho-Smad1/5/8 polyclonal antibody (Cell Signaling 9511) at 1:200 dilution in 3% NGS in PBS overnight at 4 °C. Following rinsing, sections were then incubated with biotinylated anti-rabbit secondary antibody and the signal was visualized using a HRP/DAB detection IHC kit according to the manufacturer’s instructions (Abcam). Bright-field images were taken with a SPOT insight camera (Diagnostic Instruments, Inc.) operated with SPOT 4.0 software.
Preparation, treatment and analysis of micromass cultures
Micromass cultures were prepared from the mesenchymal cells of E11.5 mouse embryo limb buds (Ahrens et al., 1979). Dissociated cells were suspended at a concentration of 5 × 106 cells/ml in DMEM containing 3% fetal bovine serum and antibiotics. Micromass cultures were initiated by spotting 20 μl of the cell suspensions (1.5 × 105 cells) onto the surface of 12-well tissue culture dishes. After a 90-minute incubation at 37 °C in a humidified CO2 incubator to allow for cell attachment, the cultures were supplied with 0.25 ml of medium. After 24 h, medium supplemented with the indicated concentrations of Surfen, heparitinase III (Sigma), recombinant human BMP2 (R&D Systems), recombinant mouse Noggin (R&D Systems), or combinations of Surfen/rhBMP2 or Surfen/Noggin were added to the cultures. Fresh reagents (drug and/or protein) were given with medium change every other day. Equivalent amounts of vehicle were added to control cultures. Cultures were stained with Alcian blue (pH 1.0) after 5 days to monitor chondrogenic cell differentiation. Micromass analysis was performed using ImageJ. Images were made binary under an RGB threshold, and “Particle Analysis” was utilized to measure nodule size, number, and Alcian blue positive area.
Timed matings were used to collect E11.5 Ext1flox/flox embryonic limb buds for micromass cultures. Cells were plated using the above procedure with the addition of adeno-CMV-Cre (Vector Biolabs, MOI 1000) after cell attachment.
Semiquantitative and real time PCR analyses
Total RNA was extracted from cells by the guanidine phenol method using TRIzol reagent (Invitrogen) according to the manufacturer’s protocols. One microgram of total RNA was reverse transcribed using the SuperScript III First-Strand Synthesis System (Life Technologies). Quantitative real-time PCR was conducted using SYBR Green PCR Master Mix in an Applied Biosystems 7900HT according to the manufacturer’s protocols. The housekeeping gene GAPDH was used as an internal control for quantification. The following primer sets were used: Gapdh forward primer (5′-CGTCCCGTAGACAAAATGGT-3′) and reverse primer (5′-TTGATGGCAACAATCTCCAC-3′); Sox9 forward primer (5′-GAGCTCAGCAAGACTCTGGG-3′) and reverse primer (5′-CGGGGCTGGTACTTGTAATC-3′); Runx2 forward primer (5′-CGCACGACAACCGCACCAT-3′) and reverse primer 5′-AACTTCCTGTGCTCCGTGCTG-3′); Agg forward primer (5′-GGAGCGAGTCCAACTCTTCA-3′) and reverse primer (5′-CGCTCAGTGAGTTGTCATGG-3′); Col2a1 forward primer (5′-CTACGGTGTCAGGGCCAG-3′) and reverse primer (5′-GTGTCACACACACAGATGCG-3′); Col10a1 forward primer (5′-CATAAAGGGCCCACTTGCTA-3′) and reverse primer (5′ACCAGGAATGCCTTGTTCTC-3′). Band intensities for semiquantitative PCR were normalized to GAPDH and compared while the band intensities and cycle numbers are linear. The following primer sets were used: BMP2 forward primer (5′-TCTTCCGGGAACAGATACAGG-3′) and reverse primer (5′-TCTCCTCTAAATGGGCCACTT-3′); BMPRI forward primer (5′-GCTTGCGGCAAtCGTGTCTAA-3′) and reverse primer (5′-GCAGCCTGTGAAGATGTAGAGG-3′); BMPRII forward primer (5′-CACACCAGCCTTATACTCTAGATA-3′) and reverse primer (5′-CACATATCTGTTATGAAACTTGAG-3′).
Reporter assays
Mouse mesenchymal C3H10T1/2 cells were seeded in monolayer culture in a 96-well plate and serum-starved overnight. Cultures were co-transfected with 100 ng per well of canonical BMP signaling reporter Id1-Luc plasmid (Korchynsky and ten Dijke, 2002) and 1 ng/well of phRG-TK (Promega) using 0.2 μl of Lipofectamine LTX (Invitrogen) according to the manufacturer’s protocol. After 6 h, media was replaced with medium supplemented by indicated concentrations of Surfen, rhBMP2, recombinant mouse Noggin, or combinations of Surfen/BMP2 or Surfen/Noggin. Control wells received equivalent amounts of vehicle (0.1% BSA/PBS or DMSO). Twenty-four hours later, cells were harvested and subjected to dual luciferase assay (Promega); transfection efficiency was normalized to Renilla luciferase activity generated by phRG-TK.
Protein analysis
C3H10T1/2 cells were grown to 70% confluence in 6-well plates and treated with indicated concentrations of rhBMP2 or Surfen. Total cellular proteins harvested in SDS-PAGE sample buffer were electrophoresed on 4–15% SDS-Bis-Tris gels (40 μg per lane) and transferred to PVDF membranes (Invitrogen). Membranes were incubated overnight at 4 °C with dilutions of antibodies against phospho-Smad1/5/8 (Cell Signaling Technology 9511, 1:1000) or Smad1 (Abcam 63439, 1:500). Enhanced chemiluminescent immunoblotting detection system (Pierce) was used to detect the antigen-antibody complexes. The membranes were re-blotted with antibodies to α-tubulin (Sigma T-5168, 1:2000) for normalization, and band intensities were quantified by computer-assisted image analysis.
Surface plasmon resonance (SPR) analysis
SPR experiments were performed on a Biacore 3000 (GE Healthcare). Flow channel 2 on a SA chip was loaded with roughly 500 RU biotinylated heparin (Sigma, 5 μg/ml) with a 1 min injection at a flow rate of 5 μl/min. Flow channels 1 and 2 were then capped with a 1 min injection of biotin (10 mM) at 5 μl/min. Kinetic studies of BMP2 binding to heparin was done with 2 fold dilutions from 75 to 0 nM over 10 samples, 50 μl protein was injected followed by 4 min of dissociation, with and without a 30 μl injection of 15 μM Surfen. All kinetics were done at 25 °C in PBS+0.1% BSA at a flow rate of 50 μl/min. Binding data was analyzed using the BIAevaluation software and fitted to a 1:1 Langmuir model.
BMP2/heparin in vitro binding assay
Heparin-agarose beads (Sigma) were added to individual wells of a 96-well plate. Non-specific binding was blocked overnight at 4 °C with 0.1% BSA in 1 PBS. 0–20 μM Surfen was added to wells in triplicate and allowed to bind for 1 h. rhBMP was added at the indicated concentrations and incubated for 1 h. Samples of supernatant were harvested in SDS-PAGE sample buffer and electrophoresed on 12% Bis-Tris gels. Gels were stained with silver nitrate according to the manufacturer’s instructions (Biorad) and quantified by computer-assisted image analysis.
Results
Ectopic cartilage forms in Ext1-deficient mouse embryo long bones
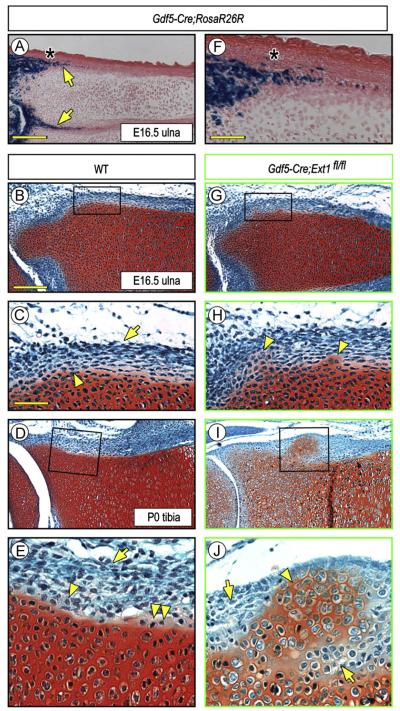
In a recent study, we crossed Ext1flox/flox and Gdf5Cre mice to conditionally ablate Ext1 expression in the mesenchymal interzone cells that separate contiguous cartilaginous long bone anlagen in mouse embryo limbs, resulting in a major drop in local HS levels (Mundy et al., 2011). We found that the interzone’s function was markedly disrupted and that the adjacent anlagen often fused together, suggesting that the interzone’s ability to serve as a border and boundary had been compromised or even lost. Interestingly, we noted in companion Gdf5-Cre;R26R-LacZ embryos that Cre was expressed not only in interzone cells as expected (Koyama et al., 2008; Rountree et al., 2004), but also in inner layer perichondrial cells present in the epiphyseal region of the anlagen (Fig. 1A and F, arrows), a region anatomically encompassing the developing groove of Ranvier (Fig. 1A and F, asterisk) (Burkus and Ogden, 1984). Thus, we created additional Gdf5Cre; Ext1flox/flox mice to determine whether and how Ext1 deficiency would affect the phenotype and function of perichondrial cells in that region over developmental time. In control E16.5 embryos and P0 neonates (Gdf5 or Gdf5Cre; Ext1flox/+), the perichondrial cells had morphological characteristics typical for those stages and the border between the cells and the adjacent Safranin O-positive epiphyseal and growth plate cartilage was obvious, continuous, smooth and well defined (Fig. 1B and D). The cells in the inner perichondrial layer were looser in density and cuboidal-round in shape (Fig. 1C and E, arrowhead), while those in the outer layer were denser and elongated (Fig. 1C and E, arrow). Occasionally, a few Safranin O-slightly positive cells were seen along the otherwise smooth border (Fig. 1E, double arrowhead), likely representing newly-apposed and newly-differentiated chondrocytes involved in normal appositional growth and lateral expansion of the epiphysis (Burkus and Ogden, 1984; Solomon, 1966). In E16.5 Ext1-deficient embryos (Gdf5Cre; Ext1flox/flox), however, the chondro-perichondrial border appeared to be compromised and uneven and had become clearly so by P0 (Fig. 1G and I). In particular, ectopic Safranin-O-positive cartilaginous tissue masses had formed within it, were close to the cartilaginous anlagen at early stages, and had grown larger and further outwards by P0 (Fig. 1H and J, arrowheads). Penetrance of this phenotype was 100% (11/11). Notably, most of the now numerous cells surrounding the cartilaginous outgrowth at P0 were round in shape (Fig. 1J, arrows), hinting to the possibility that there had been an expansion of inner layer progenitor cells around the ectopic cartilage.
Fig. 1.
Conditional deletion of Ext1 causes ectopic cartilage formation in long bones. (A and F) Histochemical staining of E16.5 Gdf5-Cre;R26R-LacZ limb sections showing that reporter activity is present in inner perichondrial layer (arrows) in addition to its expected presence in developing joints. Underlying epiphyseal cartilage and growth plate are both negative. The asterisk points to the prospective position of the groove of Ranvier. (B–E) Safranin O/Fast green-stained sections of E16.5 and P0 wild type (WT) long bone sections showing that inner and outer perichondrial layers contain typical cuboidal (arrowheads) and elongated cells (arrows), respectively. A few Safranin O-mildly positive round cells are present at the border (E, double arrowhead) likely representing newly-apposed chondrocytes. (G–J) Sections from Gdf5-Cre; Ext1fl/fl mutant limbs displaying ectopic cartilage formation. At E16, the ectopic Safranin O-positive tissue is small and close to native cartilage (G, H, arrowheads), but becomes more conspicuous by P0 (I, J, arrowheads) and is surrounded by round perichondrial cells (J, arrows). Scale bars, 500 μm in A, 250 μm in B, 100 mμ in C and F.
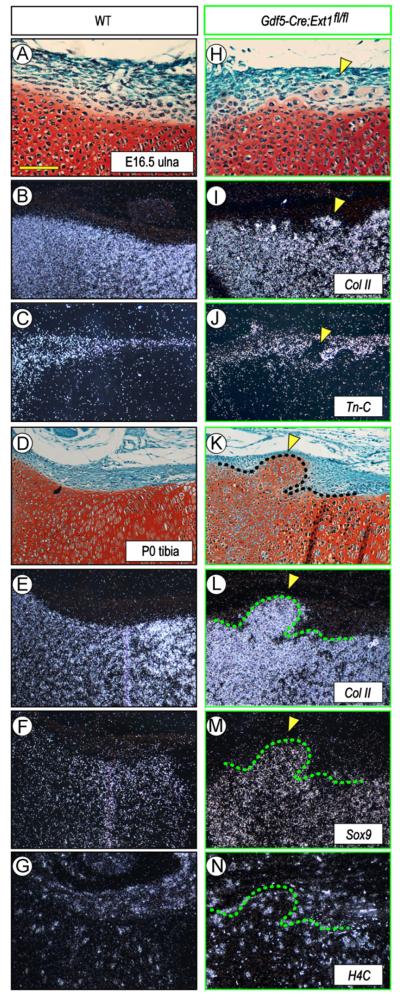
In situ hybridization confirmed that the ectopic tissue was in fact cartilaginous and expressed such typical cartilage master and matrix genes as Sox9 and collagen type IIB (Fig. 2H, I, K, L, and M, arrowheads), while these genes were restricted to the cartilaginous anlage and were undetectable in perichondrium in wild types (Fig. 2A, B, D, E, and F). Expression analysis of histone 4C, a marker of proliferating cells, showed that the ectopic chondrocytes and surrounding cells were all actively proliferating, thus possibly contributing to overall tissue expansion and outgrowth (Fig. 2N). Interestingly, analysis of tenascin-C expression (Fig. 2C), a marker of articular tissue and epiphyseal perichondrium (Koyama et al., 1995), showed that the ectopic cartilage was largely negative as was the adjacent epiphyseal and growth plate cartilage (Fig. 2J, arrowheads), indicating that the ectopic cartilage resembled phenotypically transient growth plate cartilage rather than permanent articular cartilage. To further analyze the nature and origin of the ectopic tissue, we analyzed P0 Gdf5Cre; Ext1flox/flox; eYFP mice in which the Ext1-deficient cells would also be eYFP-positive. As to be expected, there was strong reporter signal within the joints and flanking perichondrial region, but also within the ectopic tissue (Fig. S1, arrowheads). Expression did not characterize every cell present, suggesting that the ectopic tissue consisted of mutant and non-mutant cells. Growth plate cartilage was YFP-negative (Fig. S1).
Fig. 2.
Gene expression is altered along mutant chondro-perichondrial border in vivo. (A–G) WT long bone sections at E16.5 and PO (A and D) displaying typical expression patterns of the cartilage markers collagen IIB (B,E) and Sox9 (F) and the perichondrial marker tenascin-C (C). Proliferative H4C-positive cells are present in both perichondrium and cartilage (G). (H–N) Sections from Gdf5-Cre; Ext1fl/fl mutant limbs (H and K) show that the ectopic cartilage within perichondrium expresses collagen IIB (I and L) and Sox9 (M) but not tenascin-C (J) and contains, and is surrounded by, H4C-positive cells (N). Arrowheads indicate the location of ectopic cartilage. Scale bar, 100 μm.
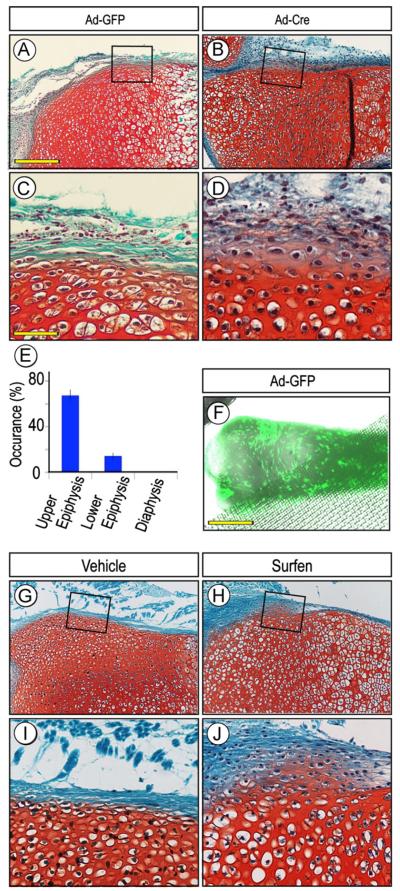
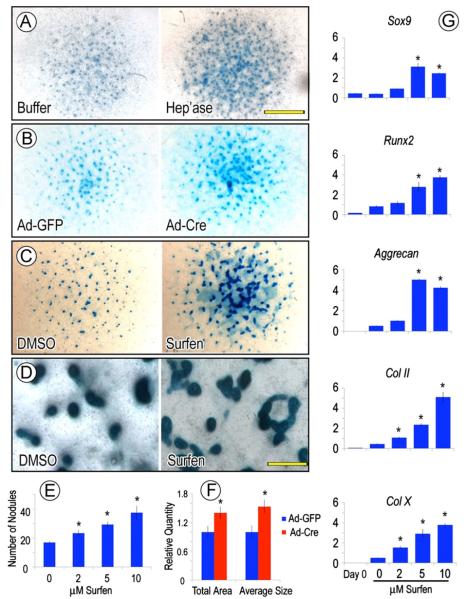
Perichondrium surrounds the cartilaginous anlagen along their entire periphery, but the approach above using Gdf5-Cre mice allowed us to examine only the portion flanking the epiphysis. Thus, to determine whether other perichondrial portions were susceptible to ectopic cartilage formation, we isolated limbs from P1 Ext1flox/flox neonates, removed the ectodermal tissue layer and reared the resulting explants in organ culture in medium containing control GFP-encoding adenovirus (Ad-GFP) or Cre-encoding adenovirus (Ad-Cre). Cultures were monitored over time and processed for histochemical analysis of cartilage formation and location over time. These experiments showed that ectopic cartilage had in fact formed by 3–5 days of incubation in the majority of the Ad-Cre-treated explants (13 out of 20 in three independent experiments) (Fig. 3B and D), while no cartilage had formed in companion explants receiving adeno-GFP and their entire border was intact and their perichondrium was wholly fibrogenic (Fig. 3A and C). Most interestingly, when we compared the frequency of ectopic cartilage formation with respect to the long bone longitudinal axis, it was clear that the incidence was higher in the epiphyseal than diaphyseal region by a ratio of about 4 to 1 (Fig. 3E, p < 0.01). Such differential incidence did not appear to reflect a differential distribution/action of adenovirus given that companion specimens incubated with adeno-GFP exhibited strong signal all along their surface (Fig. 3F). Similar results were obtained when wild type P1 explants were exposed to medium containing the HS antagonist Surfen (Schuksz et al., 2008). Ectopic cartilage had clearly formed in the epiphyseal region of Surfen-treated explants (5/7) but less frequently in the diaphyseal region (Fig. 3H and J); companion explants treated with vehicle showed no ectopic cartilage (Fig. 3G and I).
Fig. 3.
Induction and location of ectopic cartilage vary along perichondrium. (A–F) Sections of P0 Ext1flox/flox limb explants incubated with adenovirus encoding Cre recombinase (Ad-Cre) for 5 days and exhibiting ectopic Safranin O-positive tissue within perichondrium (B and D) but is absent in companion control adeno-GFP (Ad-GFP) specimens (A and C). Incidence of ectopic cartilage formation is much higher in epiphyseal than diaphyseal regions (E), but this does not appear to reflect differential distribution of virus that was fairly uniform (F). (G-J) Sections of P0 Ext1flox/flox limb explants treated with Surfen for 4 days and showing ectopic cartilage (H and J) that, however, was absent in vehicle-treated controls (G and I). Note that in all controls (A, C, G, and I), the chondro-perichondrial border is intact and continuous. Scale bars, 250 μm in A, 75 μm in C, and 500 μm in F.
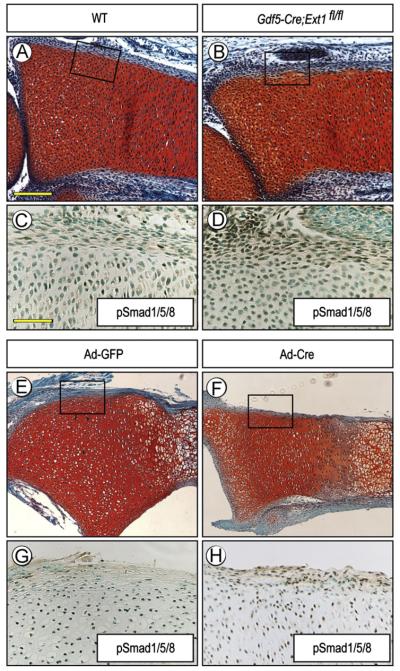
HS mediates the interaction of chondrogenic factors with target cells, but is also critical to limit their availability, distribution and action (Bernfield et al., 1992; Lin, 2004). Thus, we asked whether the formation of ectopic cartilage in Ext1-deficient long bones was preceded by, and possibly due to, ectopic activation of pro-chondrogenic pathways. For proof-of-principle, we chose BMP signaling since it has strong pro-chondrogenic activity (Weston et al., 2000). Longitudinal sections of long bone anlagen from E15.5 control and Gdf5Cre; Ext1flox/flox embryos were processed for immunostaining with Smad1/5/8 antibodies. Indeed, in Ext1-deficient specimens, the perichondrial cells displayed clear and positive nuclear staining (85.8±6.6%, n=3) (Fig. 4B and D), while most perichondrial cells in controls were negative (17.3±10.9%, n=4) (Fig. 4A and C). A similar ectopic activation of BMP signaling was observed in the perichondrium of Ext1flox/flox long bone explants exposed to adeno-Cre (78.6±5.5%, n=5) (Fig. 4F and H) but not in those exposed to control adeno-GFP (23.3±4.5%, n=3) (Fig. 4E and G). Student’s t-test showed that these changes were statistically significant in mutant (p=6 × 10−6) and adeno-Cre treated samples (p=2 × 10−4) as compared to controls.
Fig. 4.
Ectopic BMP signaling in perichondrium. (A–D) Serial sections of E15.5 WT and Gdf5-Cre; Ext1fl/fl radii processed for Safranin-O staining (A and B) or pSmad1/5/8 immunostaining (C and D). Note the strong ectopic immunostaining in the nuclei of perichondrial cells in mutant (D) but only weaker staining in WT (C). The proliferative zone of growth plate in all specimens exhibits positive staining that acted as an internal positive control. (E and H) Nuclear immunostaining is also prevalent in perichondrium of P1 Ext1flox/flox explants exposed to adeno-Cre (F and H), while very few cells are positive in control explants treated with adeno-GFP (E and G). Scale bars, 250 μm in A, 100 μm in C.
Interference with Ext1 expression and HS function stimulates chondrogenesis in vitro
The in vivo data above indicate that Ext1 deficiency stimulates cartilage formation. To verify this notion and gain additional insights on mechanisms, we resorted to high density micromass cultures of early limb bud mesenchymal cells (Ahrens et al., 1979). Upon seeding in culture, the cells attach and form a tissue-like multilayer and, over time, give rise to cartilaginous nodules each round in shape and fully separated from neighboring nodules by fibroblastic and muscle cells (Pacifici et al., 1980). Thus, we isolated limb bud mesenchymal cells from E11.5 wild type and Ext1flox/flox mouse embryos and seeded them in micromass culture overnight. To interfere with Ext1 expression and HS function, the Ext1flox/flox cultures were exposed to adeno-Cre (or adeno-GFP as control), whereas the wild type cultures were treated with different doses of Surfen or heparitinase I added daily to the medium (or vehicle as control). Chondrogenesis and nodule formation were monitored over the following 6–8 days by microscopic inspection and histochemical staining. As to be expected, all control cultures displayed a sizable number of Alcian blue-positive nodules by day 5 or 6 (Fig. 5A, B, and C, left panels), but the number and size of the nodules were clearly higher in all the treated cultures (Fig. 5A, B, and C, right panels). Imaging analysis showed that the number of nodules was nearly doubled by treatment with 10 μM Surfen (Fig. 5E), while both the total cartilage area and average nodule size were increased in adeno-Cre cultures (Fig. 5F) (n=5, p < 0.01). An excess number of nodules was also described in heparitinase-treated chick micromass cultures in a previous study (Fisher et al., 2006). Of much interest was the finding that while the majority of nodules in control cultures were well defined, round, evenly dispersed and fully distinct from neighboring nodules (Fig. 5D, left panel), many of the nodules present in treated cultures were ill shaped and had fused into large amorphous cartilaginous masses (Fig. 5D, right panel). Furthermore, quantitative PCR revealed increases in transcript levels of several key chondrogenic gene regulators, including Sox9 and Runx2, in a dose-dependent manner (Fig. 5G). Transcripts encoding the cartilage matrix constituents aggrecan, collagen type IIB and collagen type X were also upregulated.
Fig. 5.
HS antagonism stimulates chondrogenesis in vitro. (A–D) Representative micromass cultures of E11.5 mouse embryo limb mesenchymal cells showing that formation of Alcian blue-positive cartilage nodules is stimulated by treatment with heparitinase I (A), adeno-Cre (B) or Surfen (C and D) for 6 days compared to respective vehicle-treated controls. Note that, interestingly, nodules are round and distinct in controls (D, left panel), while they are fused into large amorphous masses in Surfen-treated cultures (D, right panel). (E and F) Histograms showing that nodule number increases in a dose-dependent manner in Surfen-treated cultures (E); similarly, total nodule area and average size are higher in adeno-Cre-infected than control adeno-GFP cultures (F). Values are means+S.E. of five wells. (G) Quantitative PCR analysis showing that expression of chondrogenic genes increases proportionally to Surfen dose, in line with the dose-dependent increases in nodule number (E). Values are representative of three independent experiments. Scale bars, 3 mm in A, 300 μm in D. *, p < 0.05 compared to control.
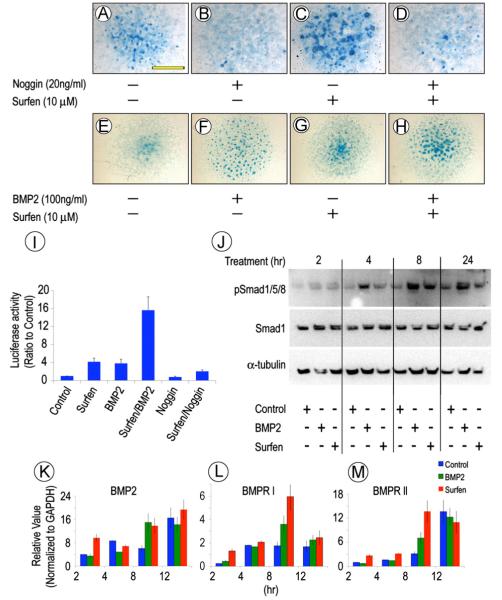
To determine whether the increased chondrogenesis in treated cultures was associated with, and likely caused by, changes in BMP signaling, similar E11.5 mouse embryo limb bud micromass cultures were treated with Surfen in absence or presence of the BMP inhibitor Noggin. While Surfen treatment had led to excess Alcian blue-positive nodule formation (Fig. 6C), this response was fully counteracted by Noggin co-treatment (Fig. 6D). In good correlation, the increased chondrogenesis induced by Surfen was further enhanced by co-treatment with rhBMP2 (Fig. 6E and H). To determine whether BMP signaling was activated early during the chondrogenic response, freshly-isolated limb bud cells were transfected with the BMP reporter plasmid Id1-Luc (Korchynsky and ten Dijke, 2002), seeded in micromass culture, and treated with: vehicle; Surfen; rhBMP2; Surfen plus BMP2; Noggin; or Surfen plus Noggin. Twenty-four hours later, whole cell homogenates were processed for luciferase assays normalized with renilla luciferase. When given singly, Surfen (10 μM) or rhBMP2 (0.1 mg/ml) stimulated reporter activity about 3-fold above controls (Fig. 6I). Co-treatment with Surfen plus rhBMP2 had strong additive effects and boosted reporter activity by over 14 fold (Fig. 6I). Noggin treatment by itself reduced basal endogenous Id1-Luc activity and reduced the stimulation by Surfen (Fig. 6I). Potency of BMP antagonism by Noggin has been shown to be comparable in the absence of HS-binding (Paine-Saunders et al., 2002).
Fig. 6.
HS antagonism modulates chondrogenic response to BMP2. (A–H) Representative mouse limb bud cell micromass cultures stained with Alcian blue on day 6 showing that cartilage nodule formation is stimulated by Surfen (C) or rhBMP2 (F) over respective controls (A and E). The pro-chondrogenic effects of Surfen are prevented by Noggin co-treatment (D), but are further enhanced by rhBMP2 co-treatment (H). (I) Id1-luc reporter assays indicating that treatment with either rhBMP2 or Surfen increases BMP signaling while rhBMP2/Surfen co-treatment stimulates it over 10-fold; Noggin treatment reduces it. Values are means+S.E. of three experiments. (J) Immunoblots showing that the levels of phosphorylated Smad1/5/8 are increased by treatment with rhBMP2 or Surfen by 4 h and remain high, while total Smad1 levels remain steady. (K–M) RT-PCR analysis showing that BMP2, BMPRI and BMPRII expression increases over time after initiating rhBMP2 or Surfen treatment. Values are means±S.E. of three experiments. Scale bar, 3 mm in A.
Whole cell homogenates from the above cultures were processed for immunoblot analysis of pSmad1/5/8 and total Smad1 levels. Clearly, Surfen and rhBMP2 greatly increased pSmad1/5/8 levels as early as 4 h of treatment and, interestingly, the increases persisted at least until 24 h, while overall Smad1 levels did not change significantly (Fig. 6J). Such persistent signaling likely reflected the high density tissue-like (and thus more physiologic) organization of the micromass cultures, given that when tested in cells in monolayer (a more artificial condition), BMP signaling usually peaks at 2 h and then decreases (Kuo et al., 2010). RT-PCR analysis with companion samples showed that endogenous BMP2 gene expression was increased as well by treatment with Surfen or rhBMP2 over control values (p < 0.05) as was expression of both BMPR-I and BMPR-II, possibly contributing to signal amplification and persistence (Fig. 6K and M).
Modulation of BMP2/HS interactions by Surfen
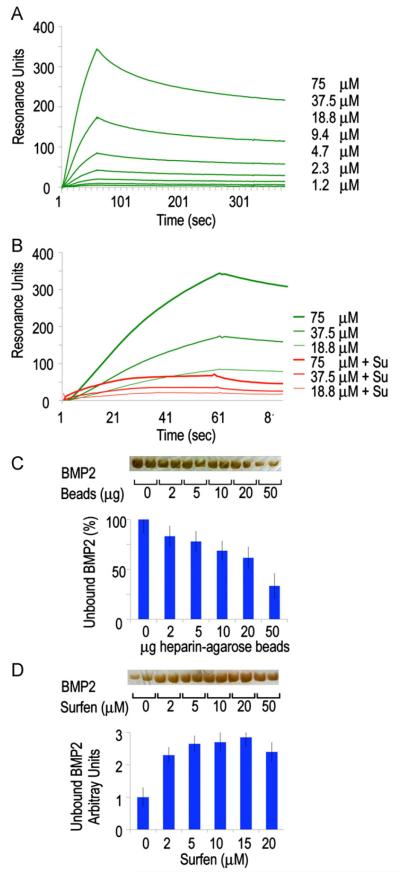
The enhanced chondrogenesis and ectopic cartilage formation seen after interference with HS function (as well as the early stimulation of BMP reporter activity and Smad1/5/8 phosphorylation) may reflect increased levels of “free” BMP protein able to interact with its receptors and elicit biologic responses (Umulis et al., 2009). The binding kinetics of the BMP/HS interaction and the effect Surfen has on this interaction was investigated using surface plasmon resonance. We coupled biotinylated heparin chains to streptavidin-derivatized sensor chip and injected rhBMP2 over the surface in concentrations ranging between 0.3 and 75 μM (Fig. 7A; Table 1) (Bramono et al., 2012; Kuo et al., 2010). Pretreating the heparin with Surfen (15 μM) drastically decreases the amount of rhBMP2 that can bind to the surface (Fig. 7B). Association and dissociation rates as well as constants for the rhBMP2/HS interaction were calculated by fitting the data to a 1 to 1 binding model. To calculate the kinetic parameters for rhBMP2 binding to Surfen-treated HS, it was assumed that the association rate of the rhBMP2 to the available HS was the same as for the wild-type interaction. The data shows that pre-treating heparin with Surfen reduces rhBMP2 binding (Fig. 7B) and that the rhBMP2 bound to Surfen treated heparin dissociates faster (Table 1).
Fig. 7.
Binding interactions between BMP2 and heparin are reversed by HS antagonism. (A) Association–dissociation curves of rhBMP2 injected on a streptavidin chip coated with biotinylated heparin. Binding constants were calculated and are shown in Table 1. (B) Binding of rhBMP2 is substantially decreased when the heparin is pre-treated with Surfen (red lines). (C and D) Solid phase assays (bottom histograms) and gel electrophoresis analysis (top gel bands) showing that the fraction of unbound rhBMP2 decreases with increasing amounts of heparin-coated beads (C), while this trend is reversed in the presence of increasing doses of Surfen (D). Values are means±S.E. of four wells.
Table 1.
Rate and equilibrium constants for rhBMP2/heparin binding.
| Vehicle | Surfen | |
|---|---|---|
| kon (M−1s−1) | 1.8 (±0.4)×103 | 1.8 (±0.4)×103 |
| koff (s−1) | 8.6 (±0.9)×10−4 | 4.6 (±0.8)×10−3 |
| Ka (M−1) | 2.0 (±0.1)×106 | 3.9 (±0.8)×105 |
| Kd (M) | 4.9 (±0.2)×10−7 | 2.6 (±0.5)×10−6 |
Rate constants (koff and kon) and association (Ka) and dissociation (Kd) constants for rhBMP2 binding to heparin, in absence and presence of Surfen, as measured by surface plasmon resonance. Constants are a mean of three separate experiments.
To verify the dynamicism of BMP/HS interactions, we used a solid phase assay with heparin-coupled agarose beads. Increasing amounts of beads were first incubated with a fixed amount of rhBMP2 (100 ng) for 1 h; beads were then centrifuged to separate bound and unbound (“free”) protein and the latter was resolved by gel electrophoresis. Optical measurements after silver staining showed that the amount of unbound rhBMP2 decreased linearly with increasing amounts of heparin beads as expected (Fig. 7C and D). Control experiments indicated that BMP2 has no affinity for unconjugated agarose beads (Fig. S2). We selected the bead amount producing the lowest levels of unbound rhBMP2 (50 μg), and carried out additional binding assays using such amount of beads and a fixed amount of rhBMP2 (100 ng). Beads and protein were incubated for 1 h after which increasing concentrations of Surfen were added. One hour later, beads were centrifuged and the unbound protein was resolved and quantified by electrophoresis as above. Clearly, Surfen was able to displace rhBMP2 from the heparin beads and increased the levels of unbound protein by 2–3 fold in a dose-dependent manner (Fig. 7E and F). The levels of unbound protein were proportional to biological activity as indicated by reporter assays (not shown).
Discussion
The results of the study provide evidence that perichondrium does in fact rely on Ext1 expression and HS to acquire and maintain its phenotype, organization and roles. When such endogenous mechanisms are experimentally altered, the chondro-perichondrial border becomes uneven and ectopic cartilage forms and grows within it. Overt formation of ectopic cartilage is first appreciable around E16.5 which corresponds to approximately 3 days from when Gdf5-Cre expression becomes detectable in the developing epiphyseal areas (Koyama et al., 2008). Similarly, ectopic cartilage first becomes visible within 3–4 days from when the long bone explants were exposed to adeno-Cre, heparitinase or Surfen. Interestingly, however, ectopic BMP signaling within perichondrium is appreciable sooner and precedes overt cartilage formation. These data signify that perichondrium must possess a number of mechanisms to maintain the phenotype of its cells and resist their phenotypic reprogramming. It may also be that the tissue possesses an adequate reservoir of HS at the moment of experimental intervention that needs to undergo turnover and/or structural and functional destabilization before pro-chondrogenic pathways and mechanisms can take over and initiate ectopic cartilage formation. Whatever the explanation, what is interesting is that ectopic cartilage eventually forms and not other tissues, implying that the lineage options for the cells residing along the border are not open-ended but rather limited and seemingly pre-specified.
Our finding that the incidence of ectopic cartilage formation is higher in the epiphysis than diaphysis has important implications given that epiphysis and diaphysis undergo markedly different developmental processes during development. The major process occurring in the early diaphysis is the formation of the primary ossification center consequent to cartilage hypertrophy and blood vessel invasion (Kronenberg, 2003). In comparison, the epiphysis is a more complex and dynamic structure in which the joints form and acquire their distinct morphologies and unique tissues, the growth plate is present and sustains longitudinal growth, and a secondary ossification center will eventually form. As importantly, the epiphysis needs to enlarge and expand laterally by appositional growth and achieve a much larger diameter compared to the diaphysis (Burkus and Ogden, 1984; Porter and Simpson, 1999; Solomon, 1966); such expansion is needed to accommodate the joints and other tissues and sustain the overall epiphysis mechanically. Indeed, we do often observe the presence of chondrocytes intimately associated to the surface of the control cartilaginous anlagen that likely represent newly-apposed and newly-differentiated cells originating from perichondrial inner layer (see Fig. 1E). Thus, it is conceivable and possible that the ectopic cartilage formation occurring in Ext1-deficient long bone anlagen in vivo or explant culture is an amplification of that natural process propelled by ectopic pro-chondrogenic signaling activity, increased availability of “free” chondrogenic factors, and precocious recruitment of progenitor cells into the chondrogenic lineage. The pro-chondrogenic pathways involved could include BMP signaling as our data suggest, but also others. For example, we showed previously that hedgehog proteins greatly stimulate chondrogenesis (Enomoto-Iwamoto et al., 2000) and that a re-distribution of Ihh from growth plate to perichondrium leads to ectopic cartilage formation (Koyama et al., 2007).
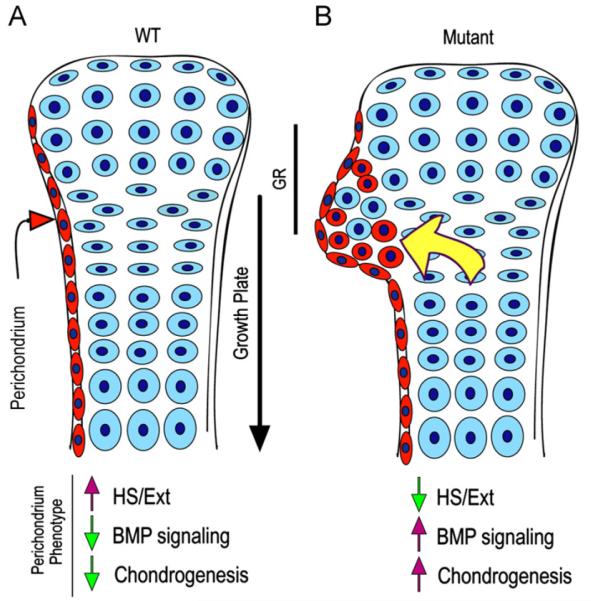
Recruitment of progenitor cells from perichondrium and/or the groove of Ranvier was previously invoked to explain the origin of exostosis-forming cells in children with HME and to account for the fact that the exostoses form next to, but never within, the growth plate itself (Hecht et al., 2005; Porter and Simpson, 1999). A possible role of cells from the groove of Ranvier is particularly attractive since the groove does contain both fast- and slow-proliferating progenitor cells, the latter typical of a stem cell niche (Karlsson et al., 2009). Studies we and others have conducted in mouse models of HME have indicated that conditional ablation of Ext1 in growth plate chondrocytes – using Col2-Cre or Col-CreER mice – also leads to formation of ectopic cartilage around the epiphyses (Jones et al., 2010; Matsumoto et al., 2010a;Zak et al., 2011). These and other studies (Clement et al., 2008) have raised the alternative possibility that the exostoses are actually produced by growth plate chondrocytes themselves and in particular by chondrocytes located on the periphery of the growth plate abutting the chondro-perichondrial border. As in the case of progenitor cells above, the mutant growth plate chondrocytes would overgrow into and within perichondrium and act as benign cartilage tumor cells because they are deficient in Ext expression and HS production. Indeed, when they were initially studied, Ext genes were defined as tumor suppressors, a definition they still have (McCormick et al., 1998). It is interesting to note also that ectopic cartilage in the mouse models and exostosis cartilage from HME patients appear to contain a mixed population of both mutant and wild type cells, suggesting that the mutant cells may “recruit” normal cells to spur further ectopic growth and that relatively few mutant cells are sufficient for exostosis formation (de Andrea et al., 2012; Jones et al., 2010;Matsumoto et al., 2010a). This concept is also supported by our data demonstrating a combination of both YFP-positive and negative cells (representing Ext1-deficient and WT cells, respectively) within ectopic cartilage (Fig. S1). Thus, in view of the data in the present study, we propose here a model that aims to reconcile previous data and disparate conclusions regarding the origin of exostosis-forming cells (model in Fig. 8). It is possible that the initial cells triggering the onset of ectopic cartilage formation in mice (or exostoses in patients) could be growth plate or perichondrial in origin. Once this process initiates, it would self-propel because: the mutant cells have higher responsiveness to growth factors such as BMPs and higher chondrogenic capacity; the growth factors themselves would be more broadly distributed and freer to interact and act; and the mutant cells would recruit wild type (perichondrial or growth plate) cells to further propel the overgrowth process.
Fig. 8.
Model of ectopic cartilage/exostosis formation. (A) In WT developing long bones, perichondrium (in red) and cartilage (in blue) acquire and maintain their distinct phenotypes and would interact in a variety of ways to sustain skeletal growth and morphogenesis. Normal phenotypic traits of perichondrium would include Ext/HS expression, low protein signaling (such as BMP signaling) and low chondrogenic activity. (B) In conditional Ext mutants (or following treatment with Surfen or heparitinase I), perichondrium would lose its normal phenotypic properties and become depleted of Ext/HS, but would exhibit higher signaling protein and chondrogenic activities. These changes would induce progenitor cells, including those from the groove of Ranvier (GR), to undergo chondrogenesis and form local ectopic cartilage/exostosis (red round cells). The outgrowth process could be aided by recruitment of wild type growth plate cells and also by increased diffusion of BMPs, hedgehogs and growth plate-derived factors (yellow arrow) as we suggested previously (Koyama et al., 2007). Similar overall mechanisms could operate when conditional Ext ablation is directed to growth plate chondrocytes as seen in recent studies (see text). Exostosis formation in HME patients could follow a similar pathogenic cascade or may be more complex given that their EXT mutations are systemic.
The data from the limb mesenchymal cells in micromass cultures sustain these possibilities and provide additional mechanistic clues. The levels of Id1-luc reporter activity, pSmad1/5/8 and BMP2/BMPR expression all increase rapidly within 4–8 h of HS interference but are sustained for at least 24 h, and the timing and degrees of these responses are similar to those elicited by treatment with exogenous rhBMP2. We show also by surface plasmon resonance and solid phase assays that the HS/rhBMP2 physical association is highly dynamic and can be reversed or even prevented. The data suggest that the overall HS-dependent regulatory mechanisms are dynamic as well and can be quickly altered to increase (and maintain) responsiveness to BMPs and possibly other factors. Due to elevated ligand and receptor expression as well as physical association of the two, such increased and long-lasting responsiveness could be instrumental to prolong active signaling. In turn, this could lead to the increases in chondrogenesis and cartilage nodule formation occurring over time in these cultures (or ectopic cartilage formation in vivo and explant culture). Why then do the nodules fuse and what does this phenomenon mean? It is possible that the fusion of the nodules is merely due to the fact that they had increased in size and number and there was insufficient space left for them to remain individually. An alternative and more likely possibility is that the treated or mutant nodules had lost their innate boundary control mechanisms, fusing into large but amorphous and irregular cartilaginous masses. In fact, it was previously shown that each nodule is normally surrounded by a multilayered tissue that closely resembles perichondrium in cell arrangement and configuration and gene expression patterns (Solursh et al., 1982), and we recently showed that there was extensive intermingling of pSmad1/5/8-positive chondrocytes and fibronectin-positive perichondrial-like cells within/around the nodules in micromass cultures prepared with mutant Prx1Cre; Ext1flox/flox mouse embryo limb bud cells (Matsumoto et al., 2010b).
In addition to chondro-perichondrial cell intermingling, we observed an overall decrease in nodule formation in the Prx1Cre; Ext1flox/flox micromass cultures rather than an increase we observe here following treatment of Ext1flox/flox micromass cultures with adeno-Cre, heparitinase or Surfen. Similarly, the mutant Prx1Cre; Ext1flox/flox limbs displayed growth retardation and significant decreases in chondrogenesis and cartilage development rather than a stimulation of ectopic cartilage formation we observe in our Gdf5Cre; Ext1flox/flox embryos. There are several explanations and interesting implications for these seemingly contradictory observations. One possibility is that because Prx1Cre targets the entire limb mesenchyme from very early stages, the resulting generalized deficiency in Ext1 could have hampered overall cell survival and proliferation, leading to a hypomorphic cartilage phenotype in vivo and reduced nodule formation in vitro. Mouse embryogenesis is in fact halted around E8.5 when both Ext1 alleles are deleted (Lin et al., 2000). In the present study, we used adenovirus infection or enzymatic or small molecule treatment to reduce (but not eliminate) Ext1 expression or HS levels/function in cultured cells possibly accounting for the stimulation of chondrogenesis we observe. A second possibility is that Ext1 may have different roles at different stages of chondrogenic cell commitment and differentiation and/or different skeletal sites and contexts, thus eliciting different phenotypic outcomes if it is deleted early versus late or broadly versus locally (Bishop et al., 2007; Lin, 2004). While more work is needed to sort out these and other possibilities, our data do indicate that Ext deficiency and HS interference render the perichondrium unable to maintain its phenotype and remain close to, but distinct from, growth plate cartilage. Further understanding of these mechanisms should illuminate the intricacies of this essential tissue–tissue interaction and should also pave the way toward effective therapies for HME and related conditions.
Supplementary Material
Acknowledgments
We are grateful to Dr. David Kingsley for providing the Gdf5-Cre mice and to Dr. Joel Bennett for help with the SPR analyses. This work was supported by NIH grants RC1AR058382 and R01AR061758 (to M.P. and E.K.) and R01AR055670 (to YY). J.H. is the recipient of the NRSA predoctoral award F31DE022204 from the NIH. We dedicate this paper to the Multiple Hereditary Exostoses Research Foundation (Brooklyn, NY) for their continuous efforts on behalf of the MHE community.
Footnotes
Appendix A. Supporting information Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2013.02.008.
References
- Ahn J, Ludecke HJ, Lindow S, Horton WA, Lee B, Wagner MJ, Horsthemke B, Wells DE. Cloning of the putative tumour suppressor gene for hereditary multiple exostoses (EXT1) Nat. Genet. 1995;11:137–143. doi: 10.1038/ng1095-137. [DOI] [PubMed] [Google Scholar]
- Ahrens PB, Solursh M, Reiter RS, Singley CT. Position-related capacity for differentiation of limb mesenchyme in cell culture. Dev. Biol. 1979;69:436–450. doi: 10.1016/0012-1606(79)90303-8. [DOI] [PubMed] [Google Scholar]
- Alvarez J, Horton J, Sohn P, Serra R. The perichondrium plays an important role in mediating the effects of TGF-β1 on endochondral bone formation. Dev. Dyn. 2001;221:311–321. doi: 10.1002/dvdy.1141. [DOI] [PubMed] [Google Scholar]
- Alvarez J, Sohn P, Zeng X, Doetschman T, Robbins DJ, Serra R. TGFβ2 mediates the effects of Hedgehog on hypertrophic differentiation and PTHrP expression. Development. 2002;129:1913–1924. doi: 10.1242/dev.129.8.1913. [DOI] [PubMed] [Google Scholar]
- Arikawa-Hirasawa E, Watanabe H, Takami H, Hassell JR, Yamada Y. Perlecan is essential for cartilage and cephalic development. Nat. Genet. 1999;23:354–358. doi: 10.1038/15537. [DOI] [PubMed] [Google Scholar]
- Bairati A, Comazzi M, Gloria M. An ultrastructural study of the perichondrium in cartilages of the chick embryo. Acta Embryol. 1996;194:155–167. doi: 10.1007/BF00195009. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Kubilus JK, Crochiere ML, Linsenmayer TF, Tabin CJ. Identification of unique molecular subdomains in the perichodnrium and periosteum and their role in regulating gene expression in the underlying chondrocytes. Dev. Biol. 2008;321:162–174. doi: 10.1016/j.ydbio.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- Bernfield M, Kokenyesi R, Kato M, Hinkes MT, Spring J, Gallo RL, Lose EJ. Biology of the syndecans: a family of transmembrane heparan sulfate proteoglycans. Annu. Rev. Cell Biol. 1992;8:365–393. doi: 10.1146/annurev.cb.08.110192.002053. [DOI] [PubMed] [Google Scholar]
- Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans finetune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- Bramono DS, Murali S, Rai B, Ling L, Poh WT, Lim ZX, Stein GS, Nurcombe v., Van Wijnen AJ, Cool SM. Bone marrow derived heparan sulfate potentiates the osteogenic activity of bone morphogenetic protein-2 (BMP2) Bone. 2012;50:954–964. doi: 10.1016/j.bone.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulow HE, Hobert O. The molecular diversity of glycosaminoglycans shapes animal development. Annu. Rev. Cell Dev. Biol. 2006;22:375–407. doi: 10.1146/annurev.cellbio.22.010605.093433. [DOI] [PubMed] [Google Scholar]
- Burkus JK, Ogden JA. Development of the distal femoral epiphysis; a microscopic morphological investigation of the zone of ranvier. J. Pediatr. Orthop. 1984;4:661–668. doi: 10.1097/01241398-198411000-00001. [DOI] [PubMed] [Google Scholar]
- Clement A, Wiweger M, von der Hardt S, Rusch MA, Selleck S, Chien C-B, Roehl HH. Regulation of zebrafish skeletogenesis by ext2/dackel and papst1/pinscher. PLoS Genet. 2008;4:e1000136. doi: 10.1371/journal.pgen.1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colnot C, Lu C, Hu D, Helms JA. Distinguishing the contributions of the perichondrium, cartilage, and vascular endothelium to skeletal development. Dev. Biol. 2004;269:55–69. doi: 10.1016/j.ydbio.2004.01.011. [DOI] [PubMed] [Google Scholar]
- de Andrea CE, Reijnders CM, Kroon HM, de Jong D, Hogendoorn PC, Szuhai K, Bovee JV. Secondary peripheral chondrosarcoma evolving from osteochondroma as a result of outgrowth of cells with functional EXT. Oncogene. 2012;9:1095–1104. doi: 10.1038/onc.2011.311. [DOI] [PubMed] [Google Scholar]
- Eames BF, de la Fuente L, Helms JA. Molecular ontogeny of the skeleton. Birth Defects Res. (Part C) 2003;69:93–101. doi: 10.1002/bdrc.10016. [DOI] [PubMed] [Google Scholar]
- Enomoto-Iwamoto M, Nakamura T, Aikawa T, Higuchi Y, Yuasa T, Yamaguchi A, Nohno T, Noji S, Matsuya T, Kurisu K, Koyama E, Pacifici M, Iwamoto M. Hedgehog proteins stimulate chondrogenic cell differentiation and cartilage formation. J. Bone Min. Res. 2000;15:1659–1668. doi: 10.1359/jbmr.2000.15.9.1659. [DOI] [PubMed] [Google Scholar]
- Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- Fisher MC, Li Y, Seghatoleslami MR, Dealy CN, Kosher RA. Heparan sulfate proteoglycans including syndecan-3 modulate BMP activity during limb cartilage differentiation. Matrix Biol. 2006;25:27–39. doi: 10.1016/j.matbio.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Gigante A, Specchia N, Nori S, Greco F. Distribution of elastic fiber types in the epiphyseal region. J. Orthop. Res. 1996;14:810–817. doi: 10.1002/jor.1100140519. [DOI] [PubMed] [Google Scholar]
- Habuchi H, Nagai N, Sugaya N, Atsumi F, Stevens RL, Kimata K. Mice deficient in heparan sulfate 6-O-sulfotransferase-1 exhibit defective heparan sulfate biosynthesis, abnormal placentation, and late embryonic lethality. J. Biol. Chem. 2007;282:15578–15588. doi: 10.1074/jbc.M607434200. [DOI] [PubMed] [Google Scholar]
- Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nat. Rev. Mol. Cell Biol. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- Hall BK, Miyake T. All for one and one for all: condensations and the initiation of skeletal development. BioEssays. 2000;22:138–147. doi: 10.1002/(SICI)1521-1878(200002)22:2<138::AID-BIES5>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Hecht JT, Hayes E, Haynes R, Cole GC, Long RJ, Farach-Carson MC, Carson DD. Differentiation-induced loss of heparan sulfate in human exostosis derived chondrocytes. Differentiation. 2005;73:212–221. doi: 10.1111/j.1432-0436.2005.00025.x. [DOI] [PubMed] [Google Scholar]
- Hilton MJ, Gutierrez L, Martinez DA, Wells DE. EXT1 regulates chondrocyte proliferation and differentiation during endochondral bone development. Bone. 2005;36:379–386. doi: 10.1016/j.bone.2004.09.025. [DOI] [PubMed] [Google Scholar]
- Hinchliffe JR, Johnson DR. The Development of the Vertebrate Limb. Oxford University Press; New York: 1980. pp. 72–83. [Google Scholar]
- Holder N. An experimental investigation into the early development of the chick elbow joint. J. Embryol. Exp. Morphol. 1977;39:115–127. [PubMed] [Google Scholar]
- Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 2003;302:1044–1046. doi: 10.1126/science.1090497. [DOI] [PubMed] [Google Scholar]
- Jones KB, Piombo V, Searby C, Kurriger G, Yang B, Grabellus F, Roughley PJ, Morcuende JA, Buckwalter JA, Capecchi MR, Vortkamp A, Sheffield VC. A mouse model of osteochondromagenesis from clonal inactivation of Ext1 in chondrocytes. Proc. Natl. Acad. Sci. USA. 2010;107:2054–2059. doi: 10.1073/pnas.0910875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson C, Thornemo M, Barreto Henriksson H, Lindahl A. Identification of a stem cell niche in the zone of Ranvier within the knee joint. J. Anat. 2009;215:355–363. doi: 10.1111/j.1469-7580.2009.01115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korchynsky O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- Koyama E, Golden EB, Kirsch T, Adams SL, Chandraratna RAS, Michaille J-J, Pacifici M. Retinoid signaling is required for chondrocyte maturation and endochondral bone formation during limb skeletogenesis. Dev. Biol. 1999;208:375–391. doi: 10.1006/dbio.1999.9207. [DOI] [PubMed] [Google Scholar]
- Koyama E, Leatherman JL, Noji S, Pacifici M. Early chick limb cartilaginous elements possess polarizing activity and express Hedgehog-related morphogenetic factors. Dev. Dyn. 1996a;207:344–354. doi: 10.1002/(SICI)1097-0177(199611)207:3<344::AID-AJA11>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Koyama E, Shimazu A, Leatherman JL, Golden EB, Nah H-D, Pacifici M. Expression of syndecan-3 and tenascin-C: possible involvement in periosteum development. J. Orthop. Res. 1996b;14:403–412. doi: 10.1002/jor.1100140310. [DOI] [PubMed] [Google Scholar]
- Koyama E, Leatherman JL, Shimazu A, Nah H-D, Pacifici M. Syndecan-3, tenascin-C and the development of cartilaginous skeletal elements and joints in chick limbs. Dev. Dyn. 1995;203:152–162. doi: 10.1002/aja.1002030204. [DOI] [PubMed] [Google Scholar]
- Koyama E, Shibukawa Y, Nagayama M, Sugito H, Young B, Yuasa T, Okabe T, Ochiai T, Kamiya N, Rountree RB, Kingsley DM, Iwamoto M, Enomoto-Iwamoto M, Pacifici M. A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev. Biol. 2008;316:62–73. doi: 10.1016/j.ydbio.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama E, Young B, Nagayama M, Shibukawa Y, Enomoto-Iwamoto M, Iwamoto M, Maeda Y, Lanske B, Song B, Serra R, Pacifici M. Conditional Kif3a ablation causes abnormal hedgehog signaling topography, growth plate dysfunction, and excessive bone and cartilage formation during mouse skeletogenesis. Development. 2007;134:2159–2169. doi: 10.1242/dev.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziel L, Kunath M, Kelly OG, Vortkamp A. Ext1-dependent heparan sulfate regulates the range of Ihh signaling during endochondral ossification. Dev. Cell. 2004;6:801–813. doi: 10.1016/j.devcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- Kuo W-J, Digman MA, Lander AD. Heparan sulfate acts as a bone morphogenetic protein co-receptor by facilitating ligand-induced receptor hetero-oligodimerization. Mol. Biol. Cell. 2010;21:4028–4041. doi: 10.1091/mbc.E10-04-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- Lin X, Wei G, Shi Z, Dryer L, Esko JD, Wells DE, Matzuk MM. Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice. Dev. Biol. 2000;224:299–311. doi: 10.1006/dbio.2000.9798. [DOI] [PubMed] [Google Scholar]
- Lind T, Tufaro F, McCormick C, Lindahl U, Lidholt K. The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J. Biol. Chem. 1998;273:26265–26268. doi: 10.1074/jbc.273.41.26265. [DOI] [PubMed] [Google Scholar]
- Long F, Linsenmayer TF. Regulation of growth region cartilage proliferation and differentiation by perichondrium. Development. 1998;125:1067–1073. doi: 10.1242/dev.125.6.1067. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Irie F, Mackem S, Yamaguchi Y. A mouse model of chondrocyte-specific somatic mutation reveals a role for Ext1 loss of heterozygosity in multiple hereditary exostoses. Proc. Natl. Acad. Sci. USA. 2010a;107:10932–10937. doi: 10.1073/pnas.0914642107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Matsumoto K, Irie F, Fukushi J-I, Stallcup WB, Yamaguchi Y. Conditional ablation of the heparan sulfate-synthesizing enzyme Ext1 leads to dysregulation of bone morphogenetic protein signaling and severe skeletal defects. J. Biol. Chem. 2010b;285:19227–19234. doi: 10.1074/jbc.M110.105338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick C, Leduc Y, Martindale D, Mattison K, Esford L, Dyer A, Tufaro F. The putative tumor suppressor EXT1 alters the expression of cell-surface heparan sulfate. Nat. Genet. 1998;19:158–161. doi: 10.1038/514. [DOI] [PubMed] [Google Scholar]
- Mundy C, Yasuda T, Kinumatsu T, Yamaguchi Y, Iwamoto M, Enomoto-Iwamoto M, Koyama E, Pacifici M. Synovial jint formation requires local Ext1 expression and heparan sulftae production in developing mous eembryo limbs and spine. Dev. Biol. 2011;351:70–81. doi: 10.1016/j.ydbio.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota KG, Kuratani S. Evolutionary origin of bone and cartilage in vertebrates. In: Pourquie O, editor. The Skeletal System. Cold Spring Harbor laboratory Press; Cold Spring Harbor, New York: 2009. pp. 1–18. [Google Scholar]
- Pacifici M, Cossu G, Molinaro M, Tato’ F. Vitamin A inhibits chondrogenesis but not myogenesis. Exp. Cell Res. 1980;129:469–474. doi: 10.1016/0014-4827(80)90517-0. [DOI] [PubMed] [Google Scholar]
- Pacifici M, Koyama E, Iwamoto M. Mechanisms of synovial joint and articular cartilage formation: recent advances, but many lingering mysteries. Birth Defects Res. 2005;75(Pt. C):237–248. doi: 10.1002/bdrc.20050. [DOI] [PubMed] [Google Scholar]
- Paine-Saunders S, Viviano B, Economides AN, Saunders S. Heparan sulfate proteoglycans retain Noggin at the cell surface. A potential mechanism for shaping bone morphogenetic protein gradients. J. Biol. Chem. 2002;277:2089–2096. doi: 10.1074/jbc.M109151200. [DOI] [PubMed] [Google Scholar]
- Pathi S, Rutenberg JB, Johnson RL, Vortkamp A. Interaction of Ihh and BMP/Noggin signaling during cartilage differentiation. Dev. Biol. 1999;209:239–253. doi: 10.1006/dbio.1998.9181. [DOI] [PubMed] [Google Scholar]
- Porter DE, Simpson AHRW. The neoplastic pathogenesis of solitary and multiple osteochondromas. J. Pathol. 1999;188:119–125. doi: 10.1002/(SICI)1096-9896(199906)188:2<119::AID-PATH321>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Rountree RB, Schoor M, Chen H, Marks ME, Harley V, Mishina Y, Kingsley DM. BMP receptor signaling is required for postnatal maintenance of articular cartilage. PLoS Biol. 2004;2:1815–1827. doi: 10.1371/journal.pbio.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuksz M, Fuster MM, Brown JR, Crawford BE, Ditto DP, Lawrence R, Glass CA, Wang LC, Tor Y, Esko JD. Surfen, a small molecule antagonist of heparan sulfate. Proc. Natl. Acad. Sci. USA. 2008;105:13075–13080. doi: 10.1073/pnas.0805862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Savage P, Hall BK. Differentiative ability of the tibial periosteum from the embryonic chick. Acta Anat. 1980;106:129–140. doi: 10.1159/000145174. [DOI] [PubMed] [Google Scholar]
- Solomon L. Diametric growth of the epiphyseal plate. J. Bone Joint Surg. 1966;48B:170–177. [PubMed] [Google Scholar]
- Solursh M, Jensen KL, Singley CT, Linsenmayer TF, Reiter RS. Two distinct regulatory steps in cartilage differentiation. Dev. Biol. 1982;94:311–325. doi: 10.1016/0012-1606(82)90350-5. [DOI] [PubMed] [Google Scholar]
- Stickens D, Zak BM, Rougier N, Esko JD, Werb Z. Mice deficient in Ext2 lack heparan sulfate and develop exostoses. Development. 2005;132:5055–5068. [Google Scholar]
- Umulis D, O’Connor MB, Blair SS. The extracellular regulation of bone morphogenetic protein signaling. Development. 2009;136:3715–3728. doi: 10.1242/dev.031534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viviano BL, Silverstein L, Pflederer C, Paine-Saunders S, Mills K, Saunders S. Altered hematopoiesis in glypican-3-deficient mice results in decreased osteoclast differentiation and a delay in endochondral ossification. Dev. Biol. 2005;282:152–162. doi: 10.1016/j.ydbio.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- Weston AD, Rosen V, Chandraratna RAS, Underhill TM. Regulation of skeletal progenitor differentiation by the BMP and retinoid signaling pathways. J. Cell Biol. 2000;148:679–690. doi: 10.1083/jcb.148.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda T, Mundy C, Kinumatsu T, Shibukawa Y, Shibutani T, Grobe K, Minugh-Purvis N, Pacifici M, Koyama E. Sulfotransferase Ndst1 is needed for mandibular and TMJ development. J. Dent. Res. 2010;89:1111–1116. doi: 10.1177/0022034510373766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak BM, Schuksz M, Koyama E, Mundy C, Wells DE, Yamaguchi Y, Pacifici M, Esko JD. Compound heterozygous loss of Ext1 and Ext2 is sufficient for formation of multiple exostoses in mouse ribs and long bones. Bone. 2011;48:979–987. doi: 10.1016/j.bone.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Wieser R, Massague J, Niswander L. Distinct roles of type I bone morphogenetic protein receptors in the formation and differentiation of cartilage. Genes Dev. 1997;11:2191–2203. doi: 10.1101/gad.11.17.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.