Abstract
Diacylglycerol acyltransferase (DGAT) catalyses the last step in acyl-CoA-dependent triacylglycerol (TAG) biosynthesis and is an important determinant of cellular oil content and quality. In this study, a gene, designated TaDGAT2, encoding a type 2 DGAT (DGAT2)-related enzyme was identified from the oleaginous marine protist Thraustochytrium aureum. The deduced TaDGAT2 sequence contains a ~460 amino acid domain most closely related to DGAT2s from Dictyostelium sp. (45–50% identity). Recombinant TaDGAT2 restored TAG biosynthesis to the Saccharomyces cerevisiae H1246 TAG-deficient mutant, and microsomes from the complemented mutant displayed DGAT activity with C16 and C18 saturated and unsaturated fatty acyl-CoA and diacylglycerol substrates. To examine its biotechnological potential, TaDGAT2 was expressed under control of a strong seed-specific promoter in wild-type Arabidopsis thaliana and the high linoleic acid fad3fae1 mutant. In both backgrounds, little change was detected in seed oil content, but a striking increase in oleic acid content of seeds was observed. This increase was greatest in fad3fae1 seeds, where relative amounts of oleic acid increased nearly 2-fold to >50% of total fatty acids. In addition, >2-fold increase in oleic acid levels was detected in the triacylglycerol sn-2 position and in the major seed phospholipid phosphatidylcholine. These results suggest that increased seed oleic acid content mediated by TaDGAT2 is influenced in part by the fatty acid composition of host cells and occurs not by enhancing oleic acid content at the TAG sn-3 position directly but by increasing total oleic acid levels in seeds, presumably by limiting flux through phosphatidylcholine-based desaturation reactions.
Key words: Arabidopsis, diacylglycerol acyltransferase, fatty acid, oilseed, oleic acid, Thraustochytrium aureum, triacylglycerol
Introduction
Diacylglycerol acyltransferase (DGAT, EC 2.3.1.20) is a key enzyme of acyl CoA-dependent TAG biosynthesis. This enzyme catalyses esterification of the acyl group of acyl-CoAs at the sn-3 position of sn-1,2-diacylglycerol (DAG) molecules, the final step in the Kennedy pathway leading to TAG production. In plants, the DAG substrate of DGAT can also be used for the formation of the major endoplasmic reticulum (ER) phospholipid phosphatidylcholine (PC) by the enzymes CDP choline:DAG cholinephosphotransferase (CPT) (Dewey et al., 1994) and PC:DAG cholinephosphotransferase (PDCT), a recently identified enzyme that catalyses head group exchange between DAG and PC (Lu et al., 2009). Oleic acid linked to PC serves as the substrate for the ER-localized Δ12 oleic acid desaturase (FAD2), and the PC-linked linoleic acid product of this reaction is the substrate for linolenic acid synthesis via the Δ15 linoleic acid desaturase (FAD3). As a result, the balance of DGAT, CPT, and PDCT activities not only mediates cellular fatty acid storage and membrane formation, but also influences fatty acid polyunsaturation.
Two major families of DGAT enzymes have been identified, DGAT1 and DGAT2, having similar function but little sequence homology (Shockey et al., 2006; Cao, 2011). DGAT1 belongs to the membrane-bound O-acyltransferase (MBOAT) family (Cases et al., 1998), while DGAT2 belongs to the monoacylgycerol acyltransferase (MGAT1) family (Yen et al., 2002). A less studied, soluble (cytosolic) DGAT belonging to the DGAT3 family has been identified in peanut and Arabidopsis (Saha et al., 2006; Hernandez et al., 2012). Comparison of the deduced amino acid sequences of 117 DGATs from 70 organisms show them falling into two groups, presumably due to distinct structural differences (Shockey et al., 2006; Cao, 2011). The deduced amino acid sequence of DGAT1 is ~500 amino acid residues while that of DGAT2s is ~350 residues resulting in predicted 20-kDa difference in molecular mass (Cao, 2011). For example, the human DGAT1 is 488 residues (Hiramine and Tanabe, 2011) while the human DGAT2 is 335 residues (Cases et al., 2001). The N and C termini in both DGAT classes from plants extend towards the cytosol while in a mammalian DGAT1 C terminal was reported to be embedded in the ER lumen (Shockey et al., 2006; Stone et al., 2006; McFie et al., 2010). Both types of DGATs contain conserved motifs and residues in their carboxyl-terminal halves that are believed to contribute to catalytic activity (Cao, 2011). Some studies suggested that the highly divergent N-termini of DGAT1s from different species may play a role in regulation of enzyme activity and conformation as well as substrate specificity (Weselake et al., 2000; McFie et al., 2010).
DGATs have received considerable attention as biotechnological targets for enhancement of the TAG content of seeds of crops such as soybean, maize, and rapeseed to meet the growing demand for vegetable oils for food, feed, and biofuel uses (Lung and Weselake, 2006). A number of reports have described the ability to increase total oil content of seeds by transgenic expression of DGAT1 and DGAT2 enzymes of plant and fungal origin (Jako et al., 2001; Lardizabal et al., 2008; Oakes et al., 2011). Seed-specific overexpression of Arabidopsis DGAT1, for example, increased oil content by up to 15 and 46% in seeds of transgenic Brassica napus and Arabidopsis, respectively (Jako et al., 2001; Sharma et al., 2008; Taylor et al., 2009). Small increases in oil content of maize and soybean seeds were conferred by expression of the fungus Umbelopsis ramanniana DGAT2 (Lardizabal et al., 2008; Oakes et al., 2011). Moreover a relative increase of 19–26% in kernel oil was observed in transgenic maize lines expressing DGAT2 from the fungus Neurospora crassa (Oakes et al., 2011). DGAT2s from plant species such as Ricinus communis (castor bean) and Vernonia galamensis have also been reported to function as specialized enzymes for sequestration of unusual fatty acids (e.g. ricinoleic acid, vernolic acids) into TAGs (Burgal et al., 2008; Li et al., 2010, 2012).
This study isolated, characterized and evaluated a DGAT2-type enzyme from the thraustochytrid Thraustochytrium aureum, a marine protist that accumulates high levels of oil that are enriched in the nutritionally important, very-long-chain fatty acid docosahexaenoic acid (DHA) (Qiu et al., 2001; Jeh et al., 2008). T. aureum and other thraustochytrids have received increasing attention as platforms for commercial production of DHA-rich oils and as a source of genes for DHA production in recombinant hosts (Qiu et al., 2001; Lee Chang et al., 2012). Given the high oil content of T. aureum, which has been reported to be 25–37% of the cell dry weight with DHA accounting for approximately 40% of the oil (Iwao Iida, 1996; Hur et al., 2002; Taoka et al., 2011), this organism is a potentially desirable source of DGAT genes for biotechnological use.
Materials and methods
Growth conditions and generation of T. aureum ATCC 34304 cDNA clones
T. aureum ATCC 34304 cells were grown in BY+ medium (Difco, Franklin Lakes, NJ, USA) at 25 °C for 4 days under constant light and agitation at 250rpm. Biomass was harvested and rinsed in ice-cold RNase-free water followed by lysis in a French press at 69MPa and transferred into phenol (TE-buffered, saturated pH 6.7–8.0). Total RNA from the aqueous phase was precipitated at –70 °C for 30 minutes in 0.3M sodium acetate (pH 5.6) and one volume of isopropanol, followed by centrifugation at 15,000 g for 30 minutes at 4 °C and treated with DNase. Total RNA was further purified using a RNeasy Maxi kit (Qiagen, Valencia, CA, USA). A T. aureum cDNA library was generated by cloning the cDNA into EcoRI/XhoI-restricted pBluescript II SK(+) vector, pBluescript II XR library construction kit (Stratagene, La Jolla, CA, USA). For generating a genomic DNA library, genomic DNA was extracted using a genomic DNA extraction kit (Qiagen).
Cloning a putative TaDGAT2 sequence from T. aureum ATCC 34304
Random sequencing of ~5000 cDNA clones was carried out using the T7 promoter primer to generate sequences containing the 5ʹ-end of each cDNA clone. Assembled sequences were then annotated using blast2 against the appropriate GenBank divisions and using fastx against GenPept. Details on the isolation of the 5ʹ- and 3ʹ-ends of the full-length cDNA are described in Supplementary Methods (available at JXB online). The open reading frame designated TaDGAT2 of 1782bp, encoding 594 amino acids, was then amplified from T. aureum ATCC 34304 cDNA using the gene-specific primers 1424 AT EcoRI_FP (5ʹ-GTAGAATTCATGGAGCCCATAGCGTACAAG-3ʹ) and 1424 AT NotI_RP (5ʹ-ACGGCGGCCGCCTAACCCTCGGTGTACA G-3ʹ).
Phylogenetic analysis
An unrooted phylogenetic tree of TaDGAT2 deduced amino acid sequences along with other amino acid sequences homologous to DGAT1 or DGAT2, including several functionally characterized ones, was constructed. The functional and phylogenetic relationships were identified by the neighbour-joining program in mega4 (Tamura et al., 2007). The bootstrapping was performed with 1000 replicates.
Yeast and plant vector constructs
The open reading frame encoding for TaDGAT2 was subcloned into EcoRI/NotI sites of pESC-URA (Invitrogen) vector generating pESC-TaDGAT2. For expression of TaDGAT2 in Arabidopsis seeds, the yeast expression vector pESC-TaDGAT2 was digested with EcoRI/SpeI to release the TaDGAT2 gene. The TaDGAT2 EcoRI/SpeI fragment was subsequently cloned into the EcoRI/XbaI sites of the plant binary expression vector pBinGlyRed3 to generate pBinGlyRed3-TaDGAT2. In this vector, TaDGAT2 is flanked on its 5ʹ-end by the strong seed-specific promoter for the soybean glycicin-1 gene and on its 3ʹ-end by the glycinin-1 3ʹ-untranscribed region. The backbone of this vector is derived from pCAMBIA0380 and was engineered with the DsRed marker gene under the control of the constitutively expressed cassava mosaic virus promoter for selection of transgenic seeds by fluorescence (Lu and Kang, 2008).
Yeast and Arabidopsis transformation and selection
The constructs pESCTaDGAT2, pYAtDGAT1, and pYCrDGAT2 were transformed into S. cerevisiae H1246 (W303; MATα are1-Δ::HIS3 are2-Δ::LEU2 dga1::KanMX4 lro1-Δ::TRP1 ADE2 met ura3) (Sandager et al., 2002) using the PEG/lithium acetate method (Gietz and Woods, 1998). Yeast cells harbouring the empty pESC-URA or pYes2 vector were used as negative control. Transformants were selected by uracil prototrophy on yeast synthetic medium (YSM) containing 2% (w/v) glucose and lacking uracil (Invitrogen, Carlsbad, CA, USA). For functional expression, YSM containing 2% (w/v) raffinose was inoculated with the yeast transformants and grown at 28 °C for 24h in a shaker at 350rpm. For induction, YSM containing 2% (w/v) galactose was inoculated with raffinose-grown cultures to obtain an OD of 0.2 at 600nm and grown at 28 °C for 48h. Cells were harvested by centrifugation and used for TAG analysis and microsome isolation.
The binary vector containing the cassette for seed-specific expression of TaDGAT2 was introduced into Agrobacterium tumefaciens by electroporation. Transgenic plants were generated by floral dip (Clough and Bent, 1998) of Arabidopsis Col-0 or the fad3fae1 mutant (Smith et al., 2003). DsRed marker was used for selection of transformant seeds which were also PCR confirmed. Expression of transgenes in developing seeds was confirmed by reverse-transcription PCR (Supplementary Methods).
Microscopic imaging of yeast TaDGAT2 transformants
To examine lipid bodies, pellets of transformed yeast cells were washed twice in water and stained with Nile Red (9-diethylamino-5H-benzo[α]phenoxazine-5-one) (Sigma, Saint Louis, MO, USA) as described by Kimura et al. (2004). Fluorescence microscopic images of the stained cells were recorded as described by Kimura et al. (2004) using excitation wavelength at 480–520nm and emission wavelength at 510nm.
Preparation of yeast microsomes
Yeast microsomes were prepared from cells from 150ml stationary phase culture and resuspended in 10ml lysis buffer A (50mM HEPES pH 7.5, 50mM NaCl, 1mM EDTA, 2mM DTT, 20% sucrose) containing 100 μl proteinase inhibitor cocktail (Sigma) and 5ml of 400 μm glass beads. The mixture was vortexed vigorously for 30 seconds followed by 30 seconds of incubation on ice between intervals for 10 cycles and centrifuged at 10,000 g at 4 °C for 15min. The supernatant was further centrifuged at 4 °C for 2h at 100,000 g. The pellet was resuspended in buffer B (50mM HEPES pH7.5, 50mM NaCl and 20% glycerol) and stored at –80 °C in 40 μl aliquots. The concentration of microsomal protein was determined using bovine serum albumin as standard with Bradford reagent (Fermentas, Glen Burnie, MD, USA).
Acyl-CoA synthesis
Synthesis of radiolabelled acyl-CoAs was conducted as previously described (Rajasekharan et al., 1993) (Supplementary Methods). The yield of acyl CoA was 1–2 μCi (~50% conversion). The acyl-CoA eluate was determined to be radiopure by analysis on TLC developed in butanol:acetic acid:water (50/20/30, v/v/v).
DGAT assay
The DGAT activity assay was performed using DAG and radiolabelled acyl-CoA substrates in reaction buffer containing 0.2M Tris HCl (pH 7.4), 60mM MgCl2, 40mM DTT, 120mM sucrose, 0.02mM [1-14C]acyl-CoA, 0.4mM DAG and 20 μg of microsomal protein, in a total volume of 200 μl. DGAT reactions were initiated by adding [1-14C]acyl CoA. DAG substrates (16:0/18:1, 18:1/18:1, and 18:0/18:2) were purchased from Nu-Chek Prep (Elysian, MN, USA).
The reaction mixture was incubated for 10min at 30 °C in a water bath. The reaction was stopped by adding 1ml of heptane/isopropanol (3:2, v/v) and centrifuged for 2min at 1000 g. The heptane phase (500 μl) was transferred into a new microcentrifuge tube. The extraction was repeated a second time with 500 μl heptane, mixed and centrifuged, followed by transfer of 500 μl heptane phase into the first extract. Each DAG–acyl-CoA combination was assayed three times. The heptane extract was dried under N2, resuspended in 40 μl chloroform and spotted on TLC plates (Sigma). The TAG product was resolved by TLC using a solvent system of heptane:diethyl ether:acetic acid (70/30/1, v/v/v). Lipid bands were revealed by staining with iodine vapor. The TAG band was scraped off the plate and measured by liquid scintillation counting. Similar methods were used for measurement of wax synthase activity (Supplementary Methods).
Analysis of the fatty acid composition of lipid classes from Arabidopsis lines expressing TaDGAT2
For measurement of Arabidopsis seed oil composition and content, ~10mg of seeds were weighed in a 13×100mm glass screw-cap test tube. To each tube, 1.5ml of 2% sulphuric acid in methanol, 400 μl toluene, and 100 μl of 10mg ml–1 triheptadecanoin in toluene (Nu-Chek Prep, Elysian, MN, USA) as an internal standard were added. The tubes were purged with nitrogen, capped, and heated at 90 °C for 1h. Fatty acid methyl esters (FAMEs) generated by the transesterification reaction were extracted by addition of 1ml H2O and 1.5ml heptane to each tube. The heptane layer was recovered and transferred to autosampler vials, following thorough mixing and centrifugation. The extracted FAMEs were analysed by gas chromatography using an Agilent 7890 gas chromatograph with flame ionization detection (FID). Oil content was calculated by FID response of sample components relative to 17:0 methyl ester from the internal standard. Stereospecific analyses of TAG and PC were conducted as previously described (Cahoon et al., 2006).
Electrospray ionization MS/MS analysis of PC and TAG molecular species
Mass spectrometry analysis was conducted using an 4000 QTRAP linear ion trap quadrupole mass spectrometer (Applied Biosystems, Foster City, CA, USA) to characterize PC and TAG molecular species. Seed lipid extraction protocol and detailed instrument analysis conditions are provided in Supplementary Methods.
Results
Isolation of a T. aureum DGAT2-related gene
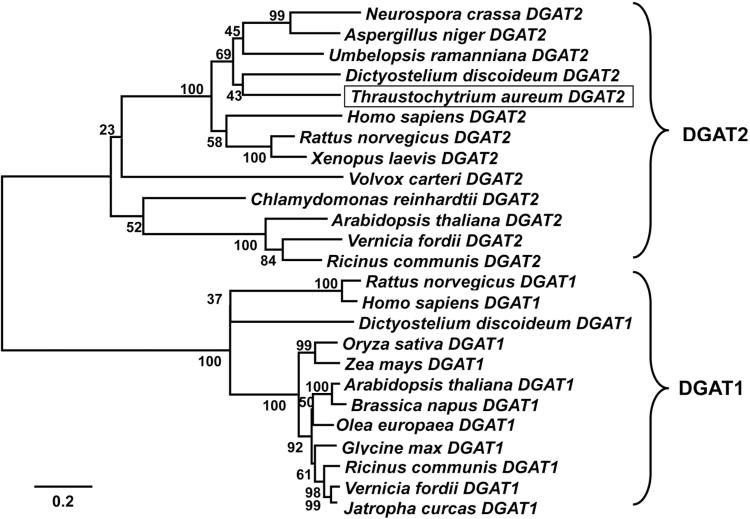
A cDNA library of T. aureum ATCC 34304 was generated that contained approximately 2.5×106 clones with an average insert size of 700bp. Random sequencing of 5ʹ-ends of cDNAs from this library resulted in the identification of a 795-bp partial clone, TA1424, with homology to known DGAT2-type cDNAs. TA1424 was subsequently used for isolation of a full-length cDNA designated TaDGAT2, encoding a 594 amino acid polypeptide (GenBank accession number JX185322). Homology searches with the coding sequence for amino acids 198–532 revealed closest identity to predicted DGAT2s from Dictyostelium species (~48% identity) and slightly lesser identity to fungal DGAT2s from species such as Umbelopsis ramanniana and Ustilago maydis (40–45% identity) (Fig. 1). The 334 amino acid domain encoded by TaDGAT2 was also related to mammalian DGAT2s (~40% identity) and more distantly related to DGAT2s from green algae and higher plants (~30% identity) (Fig. 1). The 197 amino acid N-terminus and 61 amino acid C-terminus of the TaDGAT2 have no homology to any known polypeptides. Using the SOSUI secondary structure prediction program, the entire TaDGAT2 polypeptide contains two predicted transmembrane domains encompassing amino acids 153–175 and 219–241. This is similar to plant DGAT2s, which also have two predicted transmembrane domains in the vicinity of their N-termini (Shockey et al., 2006; Liu et al., 2011). The secondary structure prediction for TaDGAT2, like that for plant and Saccharomyces cerevisiae DGAT2s, would potentially orient the N- and C-termini of this polypeptide on the cytosolic side of the ER (Shockey et al., 2006; Liu et al., 2011).
Fig. 1.
Unrooted phylogram of Thraustochytrium aureum diacylglycerol acyltransferase (DGAT) and other hypothetical or functionally characterized DGATs. The alignment was generated using clustal w and the unrooted phylogram was constructed by the neighbour-joining method in mega4 software (Tamura et al., 2007). Accession numbers of the sequences are: A. niger DGAT2, XP_001396146; A. thaliana DGAT1, CAB44774; A. thaliana DGAT2, NP_566952; B. napus DGAT1, AF164434; C. reinhardtii DGAT2, XP_001693189; D. discoideum DGAT1, XP_645633; D. discoideum DGAT2, XP_635762; G. max DGAT1, AAS78662; H. sapiens DGAT1, NP_036211.; H. sapiens DGAT2, NP_477513; J. curcas DGAT1, ABB84383; N. crassa DGAT2, XP_965438; O. europaea DGAT1, AAS01606; O. sativa DGAT1, BAD53762; R. communis DGAT1, XP_002514132; R. communis DGAT2, XP_002528531; R. norvegicus DGAT1, NP_445889; R. norvegicus DGAT2, NP_001012345; T. aureum DGAT2, JX185322; U. ramanniana DGAT2, AAK84179; V. carteri DGAT2, XP_002949151; V. fordii DGAT1, ABC94471; V. fordii DGAT2, ABC94473; X. laevis DGAT2, NP_001083204; Z. mays DGAT1_2, ABV91586.
TaDGAT2 is a functional DGAT
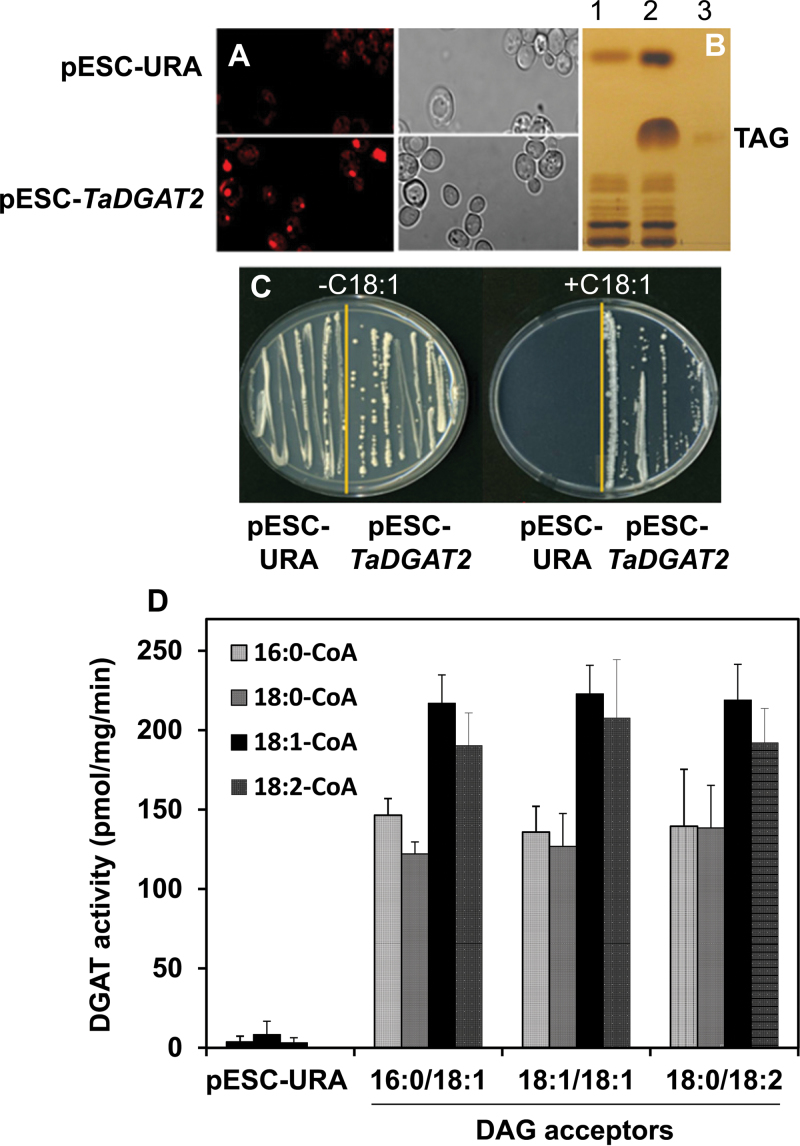
To establish the function of the TaDGAT2 polypeptide, its complete coding sequence was expressed in the TAG-deficient S. cerevisiae strain H1246, which contains disruptions of the four acyltransferase genes that contribute to TAG synthesis (Sandager et al., 2002). For these experiments, TaDGAT2 was assembled under control of the galactose-inducible GAL10 promoter in pESC-URA to generate the plasmid pESC-TaDGAT2. Large and distinct oil bodies were observed in the Nile red-stained cells harbouring pESC-TaDGAT2 after 48h of galactose induction (Fig. 2A). Little or no fluorescence was detected in the cells transformed with pESC-URA vector (Fig. 2A). In addition, TLC analysis of extracted lipids from these cells revealed a strong accumulation of TAG that was absent from cells with only the expression vector (Fig. 2B). As a further test of DGAT function, TaDGAT2-expression was found to rescue the known toxicity of exogenous oleic acid to the yeast H1246 strain (Siloto et al., 2009a, b). As shown in Fig. 2C, H1246 cells transformed with the empty pESC-URA vector were unable to grow, but these cells were fully viable in cells expressing TaDGAT2. These results indicate that TaDGAT2 has DGAT activity sufficient to restore TAG synthesis to S. cerevisiae H1246.
Fig. 2.
Thraustochytrium aureum diacylglycerol acyltransferase (DGAT) functionally complements the triacylglycerol (TAG)-deficient Saccharomyces cerevisiae H1246 mutant and displays broad substrate specificity. (A) Nile red staining (left) and interference contrast imaging (right) of transformants with an empty vector (pESC-URA) and the expression vector harbouring TaDGAT2 (pESC-TaDGAT2) at stationary phase. (B) TLC analysis of neutral lipids from yeast transformants: 1, pESC-URA; 2, pESC-TaDGAT2; 3, TAG standard. (C) Rescue of oleic acid (C18:1) toxicity of triacylglycerol-deficient S. cerevisiae H1246 cells by expression of TaDGAT2 (pESC-TaDGAT2), but not by the pESC-URA control. (D) DGAT activity in microsomal fractions from yeast cells transformed with pESC-URA (control) or pESC-TaDGAT2 vectors. Values are mean ± SD (n = 3). Shown for the pESC-URA reactions are results for assays using [1-14C]18:1-CoA, and diacylglycerol (DAG) species: 16:0/18:1, 18:1/18:1, and 18:0/18:2.
TaDGAT2 is active with structurally diverse substrates
To examine the substrate properties of the TaDGAT2 enzyme, microsomal fraction from yeast H1246 cells expressing this enzyme was assayed for acyl-CoA dependent DGAT activity by measuring the incorporation of [14C]acyl-CoA into DAG acceptors. For these studies, radiolabelled acyl-CoA esters of palmitic acid (16:0), stearic acid (18:0), oleic acid (18:1), and linoleic acid (18:2) and the 1,2-DAG acceptors 16:0/18:1,18:1/18:1, and 18:0/18:2 were used as substrates. Microsomes from cells expressing TaDGAT2 were able to catalyse TAG formation from all three DAG acceptors with each of the four [14C]acyl-CoA substrates. By comparison, only trace levels of DGAT activity were detected in cells expressing the vector control (Fig. 2D). The endogenous fatty acid profile of H1246 consists mainly of 16:0, 16:1, 18:0, and 18:1. DGAT activity with [14C]16:0-CoA or [14C]18:0-CoA was similar with all three DAG substrates tested (130±10 pmol mg–1 min–1) (Fig. 2D). Higher activities were detected with [14C]18:1-CoA (220±5 pmol TAG mg–1 min–1) and [14C]-18:2-CoA (190–208 pmol TAG mg–1 min–1) with the three DAG acceptors (Fig. 2D). These results indicate that TaDGAT2 is active with a broad range of saturated, monounsaturated, and polyunsaturated acyl-CoA and DAG substrates. We also assayed microsomes from the recombinant yeast for their ability to use palmitoyl- and oleoyl-fatty alcohols as acceptors for acyl-CoA substrates in a wax synthase-type reaction. Activity was detectable with both fatty alcohol substrates (Supplementary Fig. S1), suggesting that TaDGAT2 can use substrates other than DAG as acyl-CoA substrates.
T. aureum cells naturally accumulate the long-chain polyunsaturated fatty acid DHA. Although not a direct measure of DGAT activity, this study tested the ability of yeast H1246 cells expressing TaDGAT2 to incorporate exogenously provided DHA as well as the polyunsaturated fatty acids eicosapentaenoic acid (EPA) and arachidonic acid (ARA) into TAG (Supplementary Methods, Supplementary Fig. S2 and Supplementary Table S1). Cells provided with EPA and ARA accumulated these fatty acids in TAG to amounts of 10–15% of the TAG fatty acids, whereas only low levels of DHA were detected in TAG of cells provided with this fatty acid, likely because of limited uptake of DHA by the H1246 cells. More detailed studies were conducted with TaDGAT2-expressing cells provided EPA. For comparison, the studies were also conducted with exogenously provided oleic acid as well as with yeast cells expressing Arabidopsis DGAT1 (AtDGAT1) and Chlamydomonas reinhardtii DGAT2 (CrDGAT2). EPA and oleic acid were readily incorporated into TAG by cells expressing each of these DGATs as determined by GC analysis of isolated TAG. The production of TAG was similar for TaDGAT2, AtDGAT1, and CrDGAT2 with oleic acid and EPA (Supplementary Fig. S2). Exogenous oleic acid and EPA were provided in combination to cells expressing each DGAT to gauge selectivity of the expressed DGATs and DAG-generating acyltransferases (e.g. GPAT and LPAT). No preferential incorporation of oleic acid or EPA was detected in TAG of cells expressing TaDGAT2, AtDGAT1, or CrDGAT2 (Supplementary Fig. S2), suggesting that TaDGAT2 (as well as AtDGAT1 and CrDGAT2) has broad substrate specificity and no detectable selectivity for oleic acid and EPA, which is consistent with results from the in vitro assay of TaDGAT2 with C16 and C18 substrates.
Seed-specific expression of TaDGAT2 in Arabidopsis thaliana seeds enhances oleic acid content
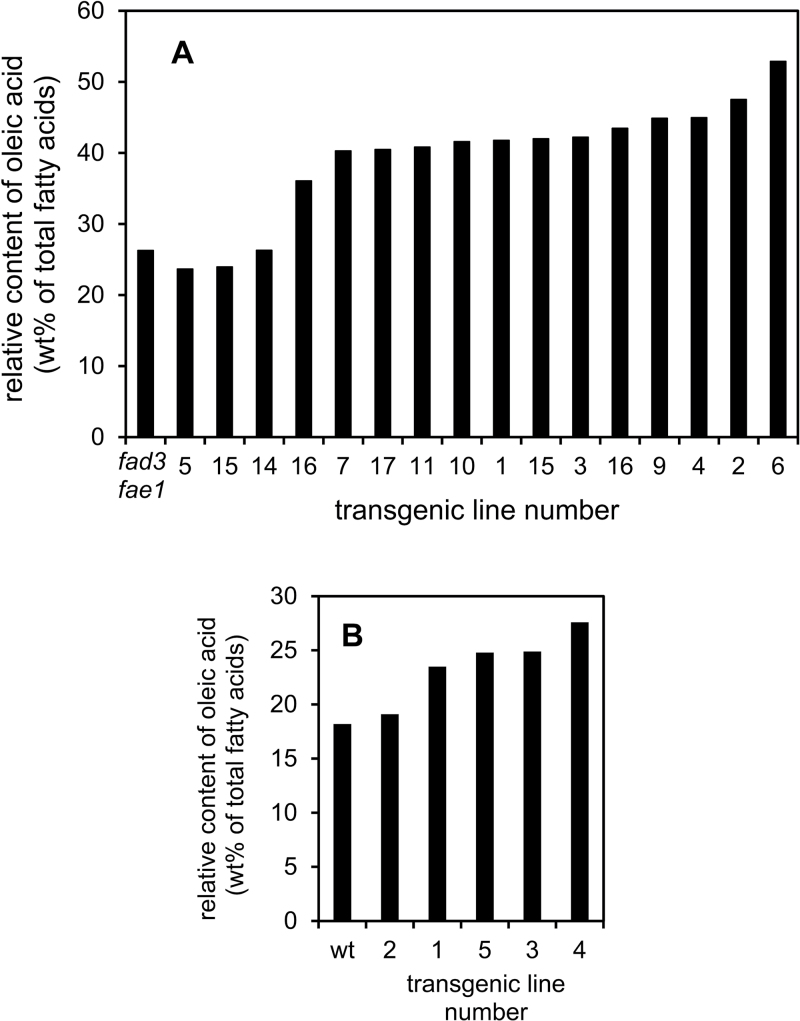
TaDGAT2 was expressed under control of the strong seed-specific glycinin-1 promoter in A. thaliana Col-0 to examine its ability to enhance seed oil content. The Arabidopsis fad3fae1 mutant was initially used for these experiments because the fatty acid composition of its seed oil more closely approximates that of seeds of vegetable oil crops such as maize, low-linolenic acid soybean, safflower, and sunflower. The fad3fae1 mutant has point mutations that inactivate FAD3, encoding the Δ15 desaturase, and FAE1, encoding fatty acid elongase-1 (Cahoon et al., 2006). These mutations result in an enrichment in linoleic acid (18:2) in the seed oil due to the loss of linolenic acid (18:3) and very-long-chain fatty acids (≥C20) (Cahoon et al., 2006). Analysis of seeds from T2 plants of 15 independent lines expressing TaDGAT2, as confirmed by reverse-transcription PCR (Supplementary Fig. S3), revealed little or no increase in total oil content (data not shown). Instead, the more striking phenotype was an increase in oleic acid content of transgenic seeds, with amounts of oleic acid exceeding 50 wt% of the total oil content (Fig. 3A). By comparison, oleic acid levels typically ranged from 26–29 wt% of the total seed oil of non-transformed fad3fael plants (Fig. 3A). This near-doubling of the oleic acid content occurred primarily at the expense of the linoleic acid content of seeds in the total lipids and TAG, which decreased by nearly the same amount as the oleic acid increase in seeds of the top TaDGAT2-expression lines (Tables 1 and 2). Oil content was re-evaluated in seeds from T4 plants of lines 2 and 6, which accumulated the highest levels of oleic acid (Table 1). Seeds from both lines contained absolute amounts of oil 1.5% greater than those of seeds from non-transformed plants grown under identical condition. This oil increase, however, was within the standard error measured for seeds from four independent plants for each line.
Fig. 3.
Distribution of oleic acid content in the total lipids of seeds from transgenic Arabidopsis fad3fae1 mutant (A) and wild-type (B) backgrounds expressing TaDGAT2. The values shown are from seeds of independent transgenic events, with the event number indicated, or from non-transgenic fad3fae1 or wild-type (wt) controls.
Table 1.
Fatty acid composition and oil content of mature seeds from Arabidopsis fad3fae1 and two independent transgenic fad3fae1 lines expressing TaDGAT2 (lines 2 and 6)
Values are mean ± SE (n = 4).
| Transgenic line | Fatty acid composition (wt% of total fatty acids) | Oil content (% dry weight) | |||||
|---|---|---|---|---|---|---|---|
| 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | Others | ||
| fad3fae1 | 8.7±0.1 | 4.0±0.4 | 28.9±1.1 | 54.7±0.6 | 1.7±0.2 | 2.1±0.2 | 28.0±1.9 |
| fad3fae1 line 2 | 8.1±0.2 | 2.7±0.1 | 45.1±1.0 | 39.8±0.7 | 1.9±0.1 | 2.5±0.1 | 29.5±1.0 |
| fad3fae1 line 6 | 7.3±0.1 | 2.7±0.1 | 51.4±1.6 | 34.0±1.6 | 1.7±0.1 | 2.8±0.1 | 29.6±0.6 |
Table 2.
Fatty acid composition of TAG and PC of lipid extracts from mature seeds of A. thaliana fad3fae1 and the engineered fad3fae1 line 6. Values are mean ± SE (n = 4)
PC, phosphatidylcholine; TAG, triacylglycerol.
| Lipid class | Fatty acid composition (wt% of total fatty acids) | |||||
|---|---|---|---|---|---|---|
| 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | Others | |
| fad3fae1 | ||||||
| TAG | 9.0±0.6 | 3.4±0.0 | 30.3±1.2 | 53.7±0.7 | 1.8±0.2 | 1.8±0.3 |
| PC | 11.2±0.4 | 1.3±0.0 | 21.7±0.8 | 60.3±0.4 | 3.7±0.2 | 2.0±0.1 |
| fad3fae1 line 6 | ||||||
| TAG | 7.3±0.3 | 2.8±0.1 | 51.5±1.2 | 33.1±1.9 | 1.5±0.1 | 3.8±0.7 |
| PC | 6.9±0.5 | 0.5±0.1 | 51.3±3.0 | 37.7±2.3 | 2.5±0.1 | 1.1±0.1 |
The effect of TaDGAT2 expression was also examined in seeds of wild-type A. thaliana. In seeds of five independent lines, increased amounts of oleic acid were detected relative to the seeds from non-transformed plants (Fig. 3B). The highest amounts of oleic acid detected in seeds of these lines was 27.6 wt% of the total fatty acids, an approximately 50% increase relative to the 18 wt% oleic acid in seeds of control plants (Fig. 3B and Table 3). In contrast to the fad3fae1 background, no significant decrease was detected in linoleic acid content of the transgenic seeds. Instead, the increased levels of oleic acid were accounted for almost entirely in linolenic acid content (Table 3). As with fad3fae1 plants, no significant change in oil content was detected in seeds of the transgenic wild-type Arabidopsis (Table 3).
Table 3.
Fatty acid composition and oil content of mature seeds from Arabidopsis Col-0 and two independent transgenic Col-0 lines expressing TaDGAT2 (lines 3 and 4)
Values are mean ± SE (n = 4).
| Transgenic lines | Fatty acid composition (wt% of total fatty acids) | Oil content (% dry weight) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 20:0 | 20:1 | 20:2 | Others | ||
| Col-0 | 7.3±0.1 | 3.2±0.1 | 18.2±0.3 | 30.1±0.4 | 14.8±0.2 | 2.5±0.1 | 19.5±0.4 | 1.7±0.1 | 2.6±0.1 | 28.2±2.0 |
| Col-0 line 3 | 6.8±0.2 | 2.9±0.1 | 27.6±1.1 | 29.7±0.6 | 7.8±0.2 | 2.3±0.1 | 19.1±0.2 | 1.1±0.1 | 2.7±0.1 | 28.7±3.3 |
| Col-0 line 4 | 6.9±0.1 | 3.1±0.1 | 24.4±0.6 | 31.4±0.3 | 9.0±0.3 | 2.4±0.1 | 18.7±0.6 | 1.2±0.1 | 2.7±0.1 | 27.5±0.5 |
Strong enhancement of oleic acid content is detected in the sn-2 position of TAG and in PC of TaDGAT2-expressing fad3fae1 seeds
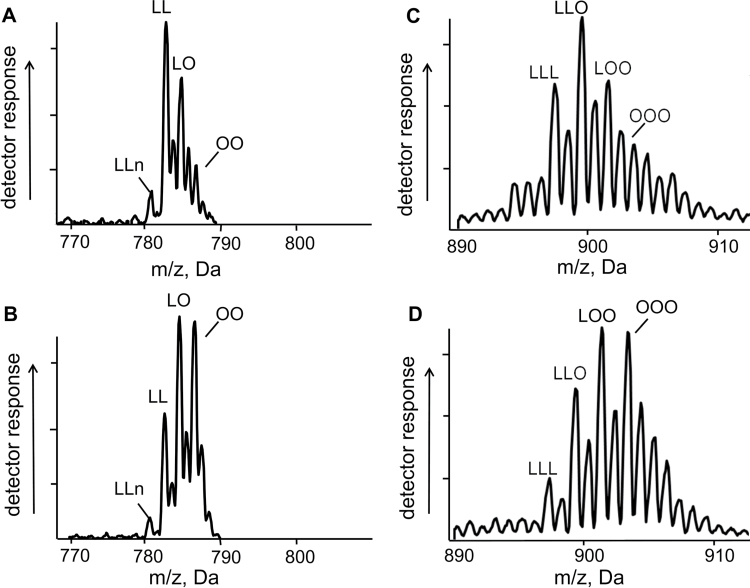
To assess the possibility that the increase in oleic acid is restricted to the sn-3 position of TAG in fad3fae1 seeds expressing TaDGAT2, stereospecific analysis of the sn-2 position was conducted following digestion of isolated TAG with a lipase specific for the sn-1 and sn-3 positions. Fatty acid analysis of the sn-2 monoacylglycerol resulting from this digestion revealed a 2.5-fold increase in the relative content of oleic acid in the sn-2 position of TAG from line 6 relative to TAG from seeds of control plants (Table 4). In TAG from the engineered seeds, nearly half of the sn-2 fatty acids were present as oleic acid. Based on this finding, it is concluded that the oleic acid increase in TaDGAT2-expressing seeds is not due to the exclusive incorporation of oleic acid at the sn-3 position via direct catalysis by TaDGAT2. Similarly, analysis of the major seed phospholipid PC revealed a 2.4-fold increase in the total oleic acid content of PC in seeds from line 6 relative to PC isolated from non-transformed fad3fae1 seeds (Table 2). The relative oleic acid content of PC was, in fact, nearly identical to that of TAG in the TaDGAT2-expressing seeds. Consistent with this, LC electrospray ionization MS/MS analysis of intact TAG and PC from seeds uncovered an increase in molecular species containing oleic acid in TaDGAT2-engineered plants relative to fad3fae1 controls (Fig. 4). These alterations included significant increases in relative amounts of triolein and di-oleoyl-PC (Fig. 4B, D). Overall, these findings indicate that expression of TaDGAT2 enhances the overall oleic acid content of seeds, while yielding corresponding reductions in polyunsaturated C18 fatty acids.
Table 4.
Fatty acid species at the sn-2 position of TAG isolated from mature seeds of A. thaliana fad3fae1 and the engineered fad3fae1 line 6
Values are mean ± SE (n = 4). ND, not detected.
| Fatty acid | Fatty acid composition (wt% of total fatty acids in sn-2 position) | |
|---|---|---|
| fad3fae1 | fad3fae1 line 6 | |
| 16:0 | ND | ND |
| 18:0 | ND | ND |
| 18:1 | 19.5±0.5 | 48.5±1.9 |
| 18:2 | 78.2±0.4 | 49.7±1.9 |
| 18:3 | 2.3±0.2 | 1.8±0.1 |
Fig. 4.
Electrospray ionization MS/MS scans showing the fatty acid components and relative abundances of intact phosphatidylcholine (PC) and triacylglycerol (TAG) species from fad3fae1 and TaDGAT2- engineered Arabidopsis seeds: (A, B) C36 species of PC in Arabidopsis fad3fae1 background (high linoleic acid) (A) and fad3/fae1 + TaDGAT2 (B). (C, D) C54 species of TAG in Arabidopsis fad3fae1 background (C) and fad3fae1 + TaDGAT2 (D). Fatty acid components of intact lipid species are indicated by abbreviations Ln (18:3), L (18:2), O (18:1); however, the labels are not intended to indicate the sn-1, 2, or 3 positional distribution of the fatty acids.
Discussion
This study isolated a gene, designated TaDGAT2, from the marine protist T. aureum encoding a polypeptide with a 334 amino acid domain most related to DGAT2s from Dictyostelium species. TaDGAT2 was able to restore TAG biosynthesis to the S. cerevisiae H1246 mutant, which contains disruptions of four acyltransferase genes that contribute to TAG synthesis (Sandager et al., 2002). This study also showed that the TaDGAT2-encoded enzyme catalyses the esterification of fatty acids from acyl-CoA donors to DAG acceptors in a manner consistent with DGAT activity. In addition, TaDGAT2 displayed nearly the same activity with 14C-16:0-, 18:0-, 18:1-, and 18:2-CoAs, and DAG species containing 16:0/18:1, 18:1/18:1, and 18:0/18:1. S. cerevisiae H1246 cells expressing TaDGAT2 were also able to incorporate exogenous DHA into TAG. Similarly, broad substrate specificity has been reported for insect cell-expressed mammalian DGAT1 and DGAT2 and Umbelopsis ramanniana DGAT2A and DGAT2B. In the latter case, Umbelopsis ramanniana DGAT2A and DGAT2B were active with an 18:1-CoA substrate and DAG acceptors containing C16-C18 fatty acids (Oakes et al., 2011). Overall, the present data from yeast expression studies conclusively demonstrate that TaDGAT2 is a functional DGAT. Similar to other DGATs from both classes, TaDGAT2 does not possess the known ER retrieval motifs (–KXKXX–COOH or –KKXX–COOH) but in the vicinity of the C-terminus, the motif YKSKW is present, which is similar to a pentapeptide aromatic ER retrieval motif –ϕ–XXK/R/D/E–φ, where φ represents a large hydrophobic amino acid (McCartney et al., 2004). A notable feature of TaDGAT2 is a nearly 200 amino acid N-terminal extension that lacks significant homology to any known DGAT2 or any other known polypeptide. A 243 N-terminal extension was also recently identified in a N. crassa DGAT2 that lacks identity with any known polypeptide (Oakes et al., 2011). Like the N. crassa DGAT2, TaDGAT2 is predicted using SOSUI and TMpred databases to be largely hydrophilic but contains a transmembrane domain immediately preceding the N-terminus of the DGAT2-related domain. Although the N. crassa DGAT2 and TaDGAT2 extensions share little homology, the function, if any, of this N-terminal extension remains unclear. In the case of the N. crassa DGAT2, the complete polypeptide and a truncated form lacking the N-terminal 243-amino acids were compared for their ability to enhance DGAT activity and increase oil content upon expression in maize seeds (Oakes et al., 2011). In these experiments, the truncated form yielded transgenic maize seeds with higher DGAT activity and oil content (Oakes et al., 2011). This result does not exclude the possibility that the N-terminal extensions of TaDGAT2 and the N. crassa DGAT2 have functional significance in their native hosts.
It is also notable that wax synthase activity is detectable with TaDGAT2 in in vitro assays. Wax synthase activity has also been detected with a number of other DGATs, including mouse DGAT1 and bifunctional wax synthase/DGAT-type enzymes from organisms such as Acinetobacter calcoaceticus and Arabidopsis (Kalscheuer and Steinbuchel, 2003; Yen et al., 2005; Li et al., 2008). In addition, the human wax synthases AWAT1 and AWAT2 belong to DGAT2 family whose members are known to possess the characteristic HPHG motif which is found in AWAT1 and AWAT2 and is present in TaDGAT2 (Turkish et al., 2005)
In this study, the seed-specific expression of TaDGAT2 resulted in no detectable increase in oil content of wild-type Arabidopsis seeds. In the Arabidopsis fad3fae1 background that has seed oil enriched in linoleic acid, an increase in absolute oil content of seeds of 1.5% was detected with the expression of TaDGAT2. Although this absolute increase in oil content is similar to that reported for the Umbelopsis ramanniana DGAT2 in soybean (Lardizabal et al., 2008), it was not statistically significant in the current study. More rigorous multigenerational analysis and testing in multiple environments is needed to fully assess the efficacy of TaDGAT2 for generating modest enhancements in seed oil content, as described for the Umbelopsis ramanniana DGAT2 in soybean (Lardizabal et al., 2008).
The more striking phenotype associated with the transgenic expression of TaDGAT2 in wild-type and fad3fae1 plants was a large increase in oleic acid content. The increase was more pronounced in the fad3fae1 background where the oleic acid content was increased by ~75% from that in seeds of non-transgenic controls versus a ~50% increase in the wild-type background. The result was the generation of a ‘mid-oleic acid’ profile in the seed oil of engineered fad3fae1 background with the oil containing over 50% oleic acid. In the case of seeds of TaDGAT2 expressed in Col-0 wild-type plants, most of the oleic acid increase was at the expense of linolenic acid (18:3) content, with little or no change detected in relative amounts of linoleic acid (18:2). By contrast, the oleic acid increase in the fad3fae1 background was almost entirely at the expense of linoleic acid content. Such an increase in oleic acid was not previously reported in DGAT1 or DGAT2 overexpression studies in dicot species, including Arabidopsis, Brassica napus, and soybean (Jako et al., 2001; Lardizabal et al., 2008; Weselake et al., 2008). Oleic acid increases, however, have been observed in two studies in DGAT overexpression studies in maize (Zheng et al., 2008; Oakes et al., 2011). With overexpression of a high-activity maize DGAT1, oleic acid content of maize seed oil was doubled, resulting in ~45% oleic acid content of the oil of the engineered seeds (Zheng et al., 2008). Similarly, seed-specific expression of the Umbelopsis ramanniana and N. crassa DGAT2s resulted in as much as a 75% increase in oleic acid content of maize seed oil (Oakes et al., 2011). Interestingly, similar studies conducted with the Umbelopsis ramanniana DGAT2 in soybean yielded only small increases in oleic acid content that were within the natural variation detected for non-engineered soybean seeds (Lardizabal et al., 2008). These conflicting results for DGAT-associated oleic acid increases between plants such as maize and soybean may be related to the differences in fatty acid profiles of seeds of these species, based on the current findings in Arabidopsis. As has been shown, the oleic acid increase in TaDGAT2-engineered plants was larger in the fad3fae1 mutant, which has a similar fatty acid profile as maize seeds. Soybean seed oil, by contrast, typically contains 7–15% linolenic acid, which is similar to the linolenic acid content of wild-type Arabidopsis seeds. From these observations, one hypothesis requiring further examination is that the use of DGAT1/2 expression as a biotechnological approach for oleic acid enhancement is most effective in seeds naturally low in linolenic acid or fad3 mutants with low linolenic acid oils, such as ‘low-lin’ soybean mutants that have received recent commercial attention for improved oil-oxidative stability (Graef et al., 1988; Clemente and Cahoon, 2009).
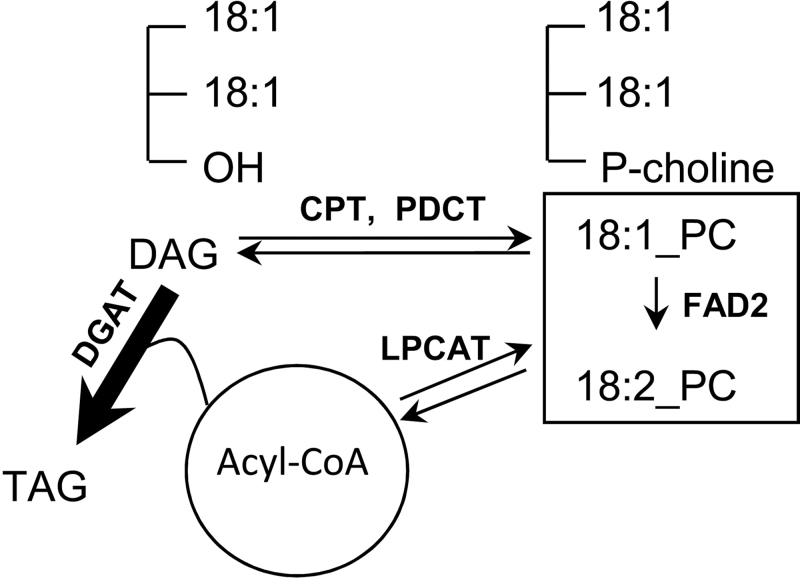
Based on this study’s findings, the enhanced oleic acid content of the TaDGAT2-engineered seeds does not result from the specific introduction of oleic acid at the sn-3 position by TaDGAT2. Although K m measurements or competition assays were not conducted with different substrates, these assays indicated that TaDGAT2 is able to use a range of acyl-CoA and DAG substrates and has no apparent strict specificity for oleoyl-CoA or oleic acid-containing DAGs. Moreover, strong enhancement of oleic acid was measured in the sn-2 position of TAG and in PC in the top fad3fae1 lines expressing TaDGAT2. These findings suggest that overall oleic acid content of seeds was increased due to the expression of TaDGAT2. As shown in Fig. 5, DAG is a common substrate for TAG and PC synthesis. Oleic acid entering PC would be available for the desaturation reactions by FAD2 and FAD3, which use PC-linked fatty acid substrates for the synthesis of linoleic and linolenic acids, respectively. It is likely that enhanced flux through DGAT, resulting from strong seed-specific TaDGAT2 expression, pulls DAG toward TAG synthesis and effectively reduces the conversion of oleic acid into polyunsaturated fatty acids via PC. The phenotype observed in the TaDGAT2-expressing seeds has similarity to the enhanced oleic acid profile of Arabidopsis PC:DAG cholinephosphotransferase (PDCT) mutant seeds, which are blocked in flux of oleic acid-DAG into oleic acid-PC for FAD2- and FAD3-catalysed desaturation (Lu et al., 2009). One possibility is that the presence of an active FAD3 may pull flux more strongly through PDCT. This may explain the differences observed between soybean versus maize and between wild-type Arabidopsis versus fad3fae1 mutant for DGAT-mediated oleic acid enhancement. It is also possible that PDCT is less active in maize seeds compared to soybean seeds, such that DGAT-overexpression more effectively shifts oleoyl-DAG flux to TAG synthesis in maize seeds. The presence of the enhanced oleic acid phenotype in PC as well as TAG suggests that TaDGAT2 may compete primarily with PDCT for DAG substrates, rather than with CPT (Fig. 5). This would effectively reduce desaturation through PC without affecting bulk PC synthesis by CPT.
Fig. 5.
A model of triacylglycerol (TAG) biosynthesis from diacylglycerol (DAG) in seeds of Arabidopsis fad3fae1 + TaDGAT2 transgenic lines. As shown, enhanced DGAT flux mediated by TaDGAT2 pulls oleoyl-DAG moieties toward TAG synthesis and away from phosphatidylcholine (PC)-linked desaturation reactions catalysed by Δ12 oleic acid desaturase (FAD2) and initiated by PC:DAG cholinephosphotransferase (PDCT), which catalyses head group exchange between DAG and PC. DAG is also converted to PC by the enzyme CDP choline:DAG cholinephosphotransferase (CPT). Lysophosphatidylcholine acyltransferase (LPCAT) catalyses the reversible exchange of fatty acids between the sn-2 position of PC and the acyl-CoA pool and contributes along with PDCT to PC-linked desaturation of oleic acid.
Oleic acid enhancement of vegetable oils has been a major biotechnological target. Oils enriched in oleic acid have improved oxidative stability for food processing as well as biofuel and bio-based products. These oils have also been in increasing demand for use in foods with hydrogenated oils that are low in trans-fats (Damude and Kinney, 2008a, b). High oleic acid phenotypes have been achieved primarily by seed-specific suppression of FAD2 (Buhr et al., 2002). The current findings combined with those from previous reports suggest that an alternative route to high oleic oils would involve a strategy of seed-specific enhancement of DGAT activity (Zheng et al., 2008; Lu et al., 2009; Oakes et al., 2011), possibly using TaDGAT2 or a fungal DGAT2, and seed-specific suppression of PDCT in a low linolenic acid background, such as maize or low-lin soybean seeds, to shift oleoyl-DAG strongly to the synthesis of TAG and away from PC-based desaturation pathways. Seed-specific suppression of additional enzyme activities such as lysophosphatidylcholine acyltransferase (LPCAT) (Xu et al., 2012) that are associated with oleic acid flux through PC may also contribute to further enhancement of high oleic acid phenotypes in seeds.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Table S1. Arachidonic acid and docosahexaenoic acid content of TAG from TaDGAT2- or AtDGAT1-expressing yeast H1246 cells.
Supplementary Fig. S1. Wax ester synthase activity in microsomal fractions from yeast cells transformed with pESC-URA or pESC-TaDGAT2 vectors.
Supplementary Fig. S2. Eicosapentaenoic acid and oleic acid content of TAG from TaDGAT2-, AtDGAT1-, or CrDGAT2-expressing yeast H1246 cells.
Supplementary Fig. S3. TaDGAT2 expression in transgenic Arabidopsis fad3/fae1 plants.
Supplementary Methods. Supplementary experimental procedures.
Acknowledgements
The authors thank the Center for Advanced Biofuel Systems (CABS), an Energy Frontier Research Center funded by the US Department of Energy, Office of Basic Energy Sciences for funding under award number DE-SC0001295 for biochemical characterization of TaDGAT2 and Abbott Nutrition for support of the Arabidopsis engineering studies. The authors also thank the National Natural Science Foundation of China (grant no. 31071453) and the 111 Project (grant no. B07041).
References
- Buhr T, Sato S, Ebrahim F, Xing A, Zhou Y, Mathiesen M, Schweiger B, Kinney A, Staswick P. 2002. Ribozyme termination of RNA transcripts down-regulate seed fatty acid genes in transgenic soybean. The Plant Journal 30, 155–163 [DOI] [PubMed] [Google Scholar]
- Burgal J, Shockey J, Lu C, Dyer J, Larson T, Graham I, Browse J. 2008. Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechology Journal 6, 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon EB, Dietrich CR, Meyer K, Damude HG, Dyer JM, Kinney AJ. 2006. Conjugated fatty acids accumulate to high levels in phospholipids of metabolically engineered soybean and Arabidopsis seeds. Phytochemistry 67, 1166–1176 [DOI] [PubMed] [Google Scholar]
- Cao H. 2011. Structure-function analysis of diacylglycerol acyltransferase sequences from 70 organisms. BMC Research Notes 4, 249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases S, Smith SJ, Zheng YW, et al. 1998. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proceedings of the National Academy of Sciences, USA 95, 13018–13023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases S, Stone SJ, Zhou P, Yen E, Tow B, Lardizabal KD, Voelker T, Farese RV., Jr 2001. Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. Journal of Biological Chemistry 276, 38870–38876 [DOI] [PubMed] [Google Scholar]
- Clemente TE, Cahoon EB. 2009. Soybean oil: genetic approaches for modification of functionality and total content. Plant Physiology 151, 1030–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Camelina sativa-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743 [DOI] [PubMed] [Google Scholar]
- Damude HG, Kinney AJ. 2008a. Engineering oilseeds to produce nutritional fatty acids. Physiologia Plantarum 132, 1–10 [DOI] [PubMed] [Google Scholar]
- Damude HG, Kinney AJ. 2008b. Enhancing plant seed oils for human nutrition. Plant Physiology 147, 962–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey RE, Wilson RF, Novitzky WP, Goode JH. 1994. The AAPT1 gene of soybean complements a cholinephosphotransferase-deficient mutant of yeast. The Plant Cell 6, 1495–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. 1998. 4 Transformation of yeast by the lithium acetate/single-stranded carrier DNA/PEG method. Methods in Enzymology 26, 53–66 [DOI] [PubMed] [Google Scholar]
- Graef GL, Fehr WR, Miller LA, Hammond EG, Cianzo SR. 1988. Inheritance of fatty acid composition in a soybean mutant with low linolenic acid. Crop Science 28, 55–58 [Google Scholar]
- Hernandez ML, Whitehead L, He Z, Gazda V, Gilday A, Kozhevnikova E, Vaistij FE, Larson TR, Graham IA. 2012. A cytosolic acyltransferase contributes to triacylglycerol synthesis in sucrose-rescued Arabidopsis seed oil catabolism mutants. Plant Physiology 160, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramine Y, Tanabe T. 2011. Characterization of acyl-coenzyme A:diacylglycerol acyltransferase (DGAT) enzyme of human small intestine. Journal of Biological Chemistry 67, 259–264 [DOI] [PubMed] [Google Scholar]
- Hur BK, Cho DW, Kim HJ, Park CI, Suh HJ. 2002. Effect of culture conditions on growth and production of docosahexaenoic acid (DHA) using Thraustochytrium aureum ATCC 34304. Biotechnology and Bioprocess Engineering 7, 10–15 [Google Scholar]
- Iwao Iida TN, Yokochi T, Kamisaka Y, Yagi H, Yamaoka M, Suzuki O. 1996. Improvement of docosahexaenoic acid production in a culture of Thraustochytrium aureum by medium optimisation. Journal of Fermentation and Bioengineering 81, 76–78 [Google Scholar]
- Jako C, Kumar A, Wei Y, Zou J, Barton DL, Giblin EM, Covello PS, Taylor DC. 2001. Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiology 126, 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeh E-J, Kumaran RS, Hur B-K. 2008. Lipid body formation by Thraustochytrium aureum (ATCC 34304) in response to cell age. Korean Journal of Chemical Engineering 25, 1103–1109 [Google Scholar]
- Kalscheuer R, Steinbuchel A. 2003. A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. Journal of Biological Chemistry 278, 8075–8082 [DOI] [PubMed] [Google Scholar]
- Kimura K, Yamaoka M, Kamisaka Y. 2004. Rapid estimation of lipids in oleaginous fungi and yeasts using Nile red fluorescence. Journal of Microbiological Methods 56, 331–338 [DOI] [PubMed] [Google Scholar]
- Lardizabal K, Effertz R, Levering C, Mai J, Pedroso MC, Jury T, Aasen E, Gruys K, Bennett K. 2008. Expression of Umbelopsis ramanniana DGAT2A in seed increases oil in soybean. Plant Physiology 148, 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Chang KJ, Dunstan GA, Abell GC, Clementson LA, Blackburn SI, Nichols PD, Koutoulis A. 2012. Biodiscovery of new Australian thraustochytrids for production of biodiesel and long-chain omega-3 oils. Applied Microbiology and Biotechnology 93, 2215–2231 [DOI] [PubMed] [Google Scholar]
- Li F, Wu X, Lam P, Bird D, Zheng H, Samuels L, Jetter R, Kunst L. 2008. Identification of the wax ester synthase/acyl-coenzyme A: diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiology 148, 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu K, Hatanaka T, Hildebrand DF. 2010. Vernonia DGATs increase accumulation of epoxy fatty acids in oil. Plant Biotechnology Journal 8, 184–195 [DOI] [PubMed] [Google Scholar]
- Li R, Yu K, Wu Y, Tateno M, Hatanaka T, Hildebrand DF. 2012. Vernonia DGATs can complement the disrupted oil and protein metabolism in epoxygenase-expressing soybean seeds. Metabolic Engineering 14, 29–38 [DOI] [PubMed] [Google Scholar]
- Liu Q, Siloto RM, Snyder CL, Weselake RJ. 2011. Functional and topological analysis of yeast acyl-CoA:diacylglycerol acyltransferase 2, an endoplasmic reticulum enzyme essential for triacylglycerol biosynthesis. Journal of Biological Chemistry 286, 13115–13126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Kang J. 2008. Generation of transgenic plants of a potential oilseed crop Camelina sativa by Camelina sativa-mediated transformation. Plant Cell Reports 27, 273–278 [DOI] [PubMed] [Google Scholar]
- Lu C, Xin Z, Ren Z, Miquel M, Browse J. 2009. An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of Arabidopsis . Proceedings of the National Academy of Sciences, USA 106, 18837–18842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung SC, Weselake RJ. 2006. Diacylglycerol acyltransferase: a key mediator of plant triacylglycerol synthesis. Lipids 41, 1073–1088 [DOI] [PubMed] [Google Scholar]
- McCartney AW, Dyer JM, Dhanoa PK, Kim PK, Andrews DW, McNew JA, Mullen RT. 2004. Membrane-bound fatty acid desaturases are inserted co-translationally into the ER and contain different ER retrieval motifs at their carboxy termini. The Plant Journal 37, 156–173 [DOI] [PubMed] [Google Scholar]
- McFie PJ, Stone SL, Banman SL, Stone SJ. 2010. Topological orientation of acyl-CoA:diacylglycerol acyltransferase-1 (DGAT1) and identification of a putative active site histidine and the role of the n terminus in dimer/tetramer formation. Journal of Biological Chemistry 285, 37377–37387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes J, Brackenridge D, Colletti R, et al. 2011. Expression of fungal diacylglycerol acyltransferase2 genes to increase kernel oil in maize. Plant Physiology 155, 1146–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hong H, MacKenzie SL. 2001. Identification of a Δ4 fatty acid desaturase from Thraustochytrium sp. involved in the biosynthesis of docosahexanoic acid by heterologous expression in Saccharomyces cerevisiae and Brassica juncea. Journal of Biological Chemistry 276, 31561–31566 [DOI] [PubMed] [Google Scholar]
- Rajasekharan R, Marians RC, Shockey JM, Kemp JD. 1993. Photoaffinity-labeling of acyl-CoA oxidase with 12-azidooleoyl-CoA and 12-[(4-Azidosalicyl)Amino]dodecanoyl-CoA. Biochemistry 32, 12386–12391 [DOI] [PubMed] [Google Scholar]
- Saha S, Enugutti B, Rajakumari S, Rajasekharan R. 2006. Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiology 141, 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandager L, Gustavsson MH, Stahl U, Dahlqvist A, Wiberg E, Banas A, Lenman M, Ronne H, Stymne S. 2002. Storage lipid synthesis is non-essential in yeast. Journal of Biological Chemistry 277, 6478–6482 [DOI] [PubMed] [Google Scholar]
- Sharma N, Anderson M, Kumar A, Zhang Y, Giblin EM, Abrams SR, Zaharia LI, Taylor DC, Fobert PR. 2008. Transgenic increases in seed oil content are associated with the differential expression of novel Brassica-specific transcripts. BMC Genomics 9, 619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM. 2006. Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. The Plant Cell 18, 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siloto RM, Truksa M, Brownfield D, Good AG, Weselake RJ. 2009a. Directed evolution of acyl-CoA:diacylglycerol acyltransferase: development and characterization of Brassica napus DGAT1 mutagenized libraries. Plant Physiology and Biochemistry 47, 456–461 [DOI] [PubMed] [Google Scholar]
- Siloto RM, Truksa M, He X, McKeon T, Weselake RJ. 2009b. Simple methods to detect triacylglycerol biosynthesis in a yeast-based recombinant system. Lipids 44, 963–973 [DOI] [PubMed] [Google Scholar]
- Smith MA, Moon H, Chowrira G, Kunst L. 2003. Heterologous expression of a fatty acid hydroxylase gene in developing seeds of Arabidopsis thaliana. Planta 217, 507–516 [DOI] [PubMed] [Google Scholar]
- Stone SJ, Levin MC, Farese RV., Jr 2006. Membrane topology and identification of key functional amino acid residues of murine acyl-CoA:diacylglycerol acyltransferase-2. Journal of Biological Chemistry 281, 40273–40282 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. mega4: Molecular Evolutionary Genetics Analysis (mega) software version 4.0. Molecular Biology and Evolution 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- Taoka Y, Nagano N, Okita Y, Izumida H, Sugimoto S, Hayashi M. 2011. Effect of Tween 80 on the growth, lipid accumulation and fatty acid composition of Thraustochytrium aureum ATCC 34304. Journal of Bioscience and Bioengineering 111, 420–424 [DOI] [PubMed] [Google Scholar]
- Taylor DC, Francis T, Guo Y, Brost JM, Katavic V, Mietkiewska E, Michael Giblin E, Lozinsky S, Hoffman T. 2009. Molecular cloning and characterization of a KCS gene from Cardamine graeca and its heterologous expression in Brassica oilseeds to engineer high nervonic acid oils for potential medical and industrial use. Plant Biotechnology Journal 7, 925–938 [DOI] [PubMed] [Google Scholar]
- Turkish AR, Henneberry AL, Cromley D, Padamsee M, Oelkers P, Bazzi H, Christiano AM, Billheimer JT, Sturley SL. 2005. Identification of two novel human acyl-CoA wax alcohol acyltransferases: members of the diacylglycerol acyltransferase 2 (DGAT2) gene superfamily. Journal of Biological Chemistry 280, 14755–14764 [DOI] [PubMed] [Google Scholar]
- Weselake RJ, Nykiforuk CL, Laroche A, Patterson NA, Wiehler WB, Szarka SJ, Moloney MM, Tari LW, Derekh U. 2000. Expression and properties of diacylglycerol acyltransferase from cell-suspension cultures of oilseed rape. Biochemical Society Transactions 28, 684–686 [PubMed] [Google Scholar]
- Weselake RJ, Shah S, Tang M, Quant PA, Snyder CL, Furukawa-Stoffer TL, Zhu W, Taylor DC, Zou J, Kumar A. 2008. Metabolic control analysis is helpful for informed genetic manipulation of oilseed rape (Brassica napus) to increase seed oil content. Journal of Experimental Botany 59, 3543–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Carlsson AS, Francis T, Zhang M, Hoffman T, Giblin ME, Taylor DC. 2012. Triacylglycerol synthesis by PDAT1 in the absence of DGAT1 activity is dependent on re-acylation of LPC by LPCAT2. BMC Plant Biology 12, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CL, Monetti M, Burri BJ, Farese RV., Jr 2005. The triacylglycerol synthesis enzyme DGAT1 also catalyzes the synthesis of diacylglycerols, waxes, and retinyl esters. Journal of Lipid Research 46, 1502–1511 [DOI] [PubMed] [Google Scholar]
- Yen CL, Stone SJ, Cases S, Zhou P, Farese RV., Jr 2002. Identification of a gene encoding MGAT1, a monoacylglycerol acyltransferase. Proceedings of the National Academy of Sciences, USA 99, 8512–8517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Allen WB, Roesler K, et al. 2008. A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nature Genetics 40, 367–372 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.