Abstract
The Arabidopsis thaliana heterotrimeric G protein β subunit, AGB1, is involved in both abscisic acid (ABA) signalling and brassinosteroid (BR) signalling, but it is unclear how AGB1 regulates these signalling pathways. A key transcription factor downstream of BR, BZR1, and its gain-of-function mutant, bzr1-1, were overexpressed in an AGB1-null mutant, agb1-1, to examine their effects on the BR hyposensitivity and the ABA hypersensitivity of agb1-1, and to examine whether AGB1 regulates the functions of BZR1. Because the amino acid sequence of AGB1 contains 17 putative modification motifs of glycogen synthase kinase 3/SHAGGY-like protein kinases (GSKs), which are known components of BR signalling, the interaction between AGB1 and one of the Arabidopsis GSKs, BIN2, was examined. Expression of bzr1-1 alleviated the effects of a BR biosynthesis inhibitor, brassinazole, in both the wild type and agb1-1, and overexpression of BZR1 alleviated the effects of ABA in both the wild type and agb1-1. AGB1 did not affect the phosphorylation state of BZR1 in vivo. AGB1 interacted with BIN2 in vitro, but did not affect the phosphorylation state of BIN2. The results suggest that AGB1 interacts with BIN2, but regulates the BR signalling in a BZR1-independent manner.
Key words: Abscisic acid, Arabidopsis thaliana, brassinosteroid, glycogen synthase kinase 3/SHAGGY-like protein kinase, heterotrimeric G protein β subunit, protein–protein interaction.
Introduction
Heterotrimeric GTP-binding proteins (G proteins, consisting of subunits Gα, Gβ, and Gγ) are signalling molecules found in a variety of eukaryotic organisms. They mediate ligand-binding signals from G protein-coupled receptors (GPCRs) to downstream pathways, and thus are involved in diverse cellular processes. In contrast to humans, which have 21 Gα genes, five Gβ genes, and 12 Gγ genes, Arabidopsis thaliana has only one Gα gene (GPA1), one Gβ gene (AGB1), and three Gγ genes (AGG1–AGG3) (for a review, see Jones and Assmann, 2004; Chakravorty et al., 2011). The physiological functions of plant G proteins can be evaluated by using G protein-deficient mutants. Such studies have suggested that plant G proteins have roles in signal transduction of various stimuli such as a stress-related phytohormone, abscisic acid (ABA) (for a review, see Perfus-Barbeoch et al., 2004). GPCRs on the plasma membrane in A. thaliana were shown to bind ABA (Liu et al., 2007; Pandey et al., 2009), supporting the importance of the G proteins in ABA signal transduction, although the validity of these studies is still in dispute (Gao et al., 2007; Jaffé et al., 2012).
G proteins are thought to mediate signal transduction via interacting with effector proteins and regulating their activities (for a review, see Pierce et al., 2002). Many G protein effectors have been identified in animals, but only some of them exist in Arabidopsis or other plant species (for a review, see Jones and Assmann, 2004), suggesting that plants have plant-specific mechanisms for G protein signalling. In fact, some putative plant-specific G protein effectors have been identified. For example, THYLAKOID FORMATION 1 (THF1) physically interacts with GPA1 and is involved in GPA1-mediated sugar signalling and chloroplast development (Huang et al., 2006; Zhang et al., 2009). A cupin domain-containing protein, AtPirin1, also physically interacts with GPA1 and mediates ABA signalling (Lapik and Kaufman, 2003). NDL1 (N-MYC DOWNREGULATED-LIKE1) physically interacts with AGB1 and regulates auxin distribution in plants (Mudgil et al., 2009). An acireductone dioxygenase-like protein, ARD1, also physically interacts with AGB1, and its enzyme activity is enhanced by Gβγ dimer (Friedman et al., 2011). Overexpression of a Golgi-localized hexose transporter, SGB1, partially suppresses the phenotype of the AGB1-null mutant, agb1-2 (H.X. Wang et al., 2006). A comprehensive analysis of the G-protein interactome suggested that G proteins interact with cell wall-related proteins and thereby regulate the cell wall composition (Klopffleisch et al., 2011). However, the molecular mechanisms underlying G protein-mediated signalling remain to be elucidated.
G proteins are suggested to play roles in signal transduction of a phytohormone, brassinosteroid (BR). For example, Gα deficiency causes BR hyposensitivities in both Arabidopsis (Ullah et al., 2001, 2002) and rice (L. Wang et al., 2006), and Gα deficiency in Arabidopsis enhances the dwarf phenotypes of bri1-5 and det2-1, which have defects in BR biosynthesis and BR perception, respectively (Gao et al., 2008). An AGB1-null mutant, agb1-2, is hyposensitive to BR in seed germination (Chen et al., 2004). In addition, agb1-2 has rounder leaves, more highly branched root systems, shorter siliques (Ullah et al., 2003), and higher sensitivities to ABA (Pandey et al., 2006). All these phenotypes of agb1-2 are similar to the phenotypes of mutants that have defects in BR biosynthesis or BR signalling (for a review, see Clouse, 2011).
On the other hand, an established model of the BR signalling consists of specific types of protein kinases, protein phosphatases, and transcription factors. When a receptor kinase, BRI1 (BR INSENSITIVE1), binds BR, it phosphorylates BSK (BR signalling kinase) family protein kinases, which activate Kelch repeat-containing protein phosphatases, such as BSU1 (BRI1 SUPPRESSOR1) and BSL1 (BSU1-LIKE1). When activated, these protein phosphatases dephosphorylate and deactivate GSKs (glycogen synthase kinase 3/SHAGGY-like protein kinases). As a result, two GSK-substrate transcription factors, BZR1 (BRASSINAZOLE RESISTANT1, also known as BES2: BRI1 EMS SUPPRESSOR2) and BZR2 (also known as BES1) are dephosphorylated, and regulate their target gene expression, which leads to BR responses (Kim and Wang, 2010; for a review, see Sun et al., 2010). Type 2A protein phosphatases (PP2As) are involved in dephosphorylating BZR1 and BZR2 (Tang et al., 2011). BZR1 and BZR2 share 88% amino acid sequence identities, and redundantly function in BR signalling (Kim and Wang, 2010; for a review, see Sun et al., 2010). The phosphorylation states of BZR1 and/or BZR2 and their target gene expression have often been examined to characterize mutant and transgenic plants that have different BR responses (He et al., 2002; Yin et al., 2002; Vert and Chory, 2006; Peng et al., 2008; Kim et al., 2009; Yan et al., 2009; Rozhon et al., 2010; Tang et al., 2011).
It is still unclear how G proteins participate in the BR signalling, or whether the well-studied components of BR signalling interact with G proteins. Here, to gain further insights into the interaction between G proteins and BR signalling, the functions of BZR1 in agb1-1 were examined. In addition, because AGB1 has many putative GSK modification sites, the interaction between AGB1 and one of the GSKs, BIN2, was examined.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used throughout the experiments. Seeds of gpa1-4 (CS6534), agb1-1 (CS3976), and agb1-2 (CS6536) were obtained from the Arabidopsis Biological Resource Center (ABRC, http://www.arabidopsis.org). Surface-sterilized seeds were sown on 0.5× MS medium (0.8% w/v agar, 0.5× MS salts, 1% w/v sucrose, 0.5g l–1 MES, pH 5.8) with or without ABA (Wako, Japan), the brassinosteroid brassinolide (BR) (Brassino Co., Ltd., Japan), brassinazole (BRZ) (Tokyo Kasei, Japan), or bikinin (Calbiochem) (concentrations are shown in the figures), and then incubated for 3 d at 4 °C (stratification). After stratification, plants were grown at 22 °C under a 16h light/8h dark photoperiod or in the dark. To generate transgenic plants expressing BZR1–green fluorescent protein (GFP), the genomic region corresponding to the open reading frame (ORF) of BZR1 (AT1G75080) was amplified by genomic PCR as previously described (Tsugama et al., 2011) using the following primer pair: 5ʹ-GGGTCTAGAATGACTTCGGATGGAGCTACG-3ʹ and 5ʹ-TCCTCTAGAACCACGAGCCTTCCCATTTCC-3ʹ (XbaI sites are underlined). The PCR products were digested by XbaI, and inserted into the SpeI site of pBI121-35SMCS-GFP (Tsugama et al., 2012a ), generating pBI121-35S-BZR1-GFP. To generate transgenic plants expressing bzr1-1–GFP, PCR was performed using pBI121-35S-BZR1-GFP as template and either of the following two primer pairs: (i) 5ʹ-GTTTCATACCCTGGCTACTATACCTGAATGTGATG-3ʹ and 5ʹ-TCCTCTAGAACCACGAGCCTTCCCATTTCC-3ʹ; or (ii) 5ʹ-GGGTCTAGAATGACTTCGGATGGAGCTACG-3ʹ and 5ʹ-GGTATAGTAGCCAGGGTATGAAACTGGTGGCGATG-3ʹ. The two kinds of PCR products obtained with (i) and (ii) were mixed and used as template for PCR using the following primer pair: 5ʹ-GGGTCTAGAATGACTTCGGATGGAGCTACG-3ʹ and 5ʹ-TCCTCTAGAACCACGAGCCTTCCCATTTCC-3ʹ (XbaI sites are underlined). The resultant PCR products, which correspond to the bzr1-1 DNA fragment, were digested by XbaI, and inserted into the SpeI site of pBI121-35SMCS-GFP, generating pBI121-35S-bzr1-1-GFP. The wild type (WT) and agb1-1 were transformed with either pBI121-35S-BZR1-GFP or pBI121-35S-bzr1-1-GFP by the Agrobacterium-mediated floral dip method (Clough and Bent, 1998). GFP expression in T2 plants was checked by fluorescence microscopy as previously described (Zhang et al., 2008), and only GFP-positive plants were used for measuring hypocotyl lengths, scoring green cotyledons, and western blotting.
Western blot analysis of BZR1 phosphorylation states
Transgenic plants expressing BZR1–GFP were grown in the presence or absence of BR, BRZ, or bikinin as described above, and sampled at the time points indicated in the figures. Cell extracts were prepared as previously described (Tsugama et al., 2011), and used for western blotting using anti-GFP antibody (MBL, Japan). Signals were detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) and an LAS-1000 plus image analyzer (Fuji Film, Japan). Images were processed with Canvas X software (ACD Systems).
Reverse transcription–PCR (RT–PCR)
Plants were grown with 0.5 μM ABA for 20 d or without ABA for 10 d, and sampled. Total RNA was prepared as previously described (Chomczynski and Sacchi, 1987), and cDNA was synthesized from 2 μg of the total RNA with PrimeScript Reverse Transcriptase (TakaraBio, Japan) using an oligo(dT) primer. The reaction mixtures were diluted 25 times with distilled water and used as templates for PCR. GoTaq Green Master Mix (Promega) was used for semi-quantitative RT–PCR, and GoTaq qPCR Master Mix (Promega) for quantitative RT–PCR. Primers used for RT–PCR are given in Supplementary Table S1 available at JXB online. In quantitative RT–PCR, the PCR was run using a StepOne Real-Time PCR System (Applied Biosystems), and relative expression levels were calculated by the comparative CT method using UBQ5 as an internal control gene.
Prediction of three-dimensional structure and GSK modification sites of AGB1
The three-dimensional structure of AGB1 (AT4G34460.1) was predicted using SWISS-MODEL (http://swissmodel.expasy.org). The output pdb file was read by RasMol (http://www.openrasmol.org) to visualize the structure. GSK modification sites in AGB1 were predicted by the Eukaryotic Linear Motif database (ELM, http://elm.eu.org). The predicted 17 GSK modification sites in AGB1 (amino acid positions 46–53, 67–74, 93–100, 109–116, 132–139, 142–149, 146–153, 173–180, 185–192, 206–213, 208–215, 221–228, 231–238, 264–271, 296–303, 347–354, and 351–358) were highlighted using RasMol.
In vitro GST pull-down assay
Hexahistidine-tagged AGB1 (His-AGB1) was expressed in Escherichia coli and purified as previously described (Tsugama et al., 2012b ). Expression of His-AGB1 in E. coli was confirmed by western blotting using HisProbe-HRP (Thermo Fisher Scientific). To express glutathionine S-transferase (GST)-fused BIN2 (GST–BIN2), the ORF of BIN2 (AT4G18710) was amplified by RT–PCR using the following primer pair: 5ʹ-GAGGATCCATGGCTGATGATAAGGAGATGCC-3ʹ and 5ʹ-CCCACTAGTTCCAGATTGATTGATTCAAGAAGC-3ʹ (BamHI site is underlined). The PCR products were digested by BamHI and inserted into the BamHI–SmaI site of pGEX-6P-3 (GE Healthcare). This construct was transformed into the E. coli strain, BL21 (DE3). Transformed cells were cultured at 37 °C in LB medium until the OD600 reached 0.5, and was then incubated at 28 °C for 2h after addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.2mM to express GST–BIN2. Expression of GST–BIN2 was confirmed by western blotting using an anti-GST antibody (GE Healthcare). Crude E. coli extracts were prepared as previously described (Tsugama et al., 2012b ). GST–BIN2 in the E. coli extracts was bound to glutathione–Sepharose 4 Fast Flow (GE Healthcare) following the manufacturer’s instructions, and washed four times with 1× Tris-buffered saline (TBS). A solution containing purified His-AGB1 was added to the GST–BIN2-bound resin, and the mixture was incubated at room temperature for 30min with gentle shaking. The resin was then washed four times by TBS, resuspended in 20mM reduced glutathione in 50mM TRIS-HCl, pH 8.0, and incubated at room temperature for 10min to elute GST–BIN2. His-AGB1 in the elutant was analysed by western blotting using HisProbe-HRP. For negative controls, 250mM imidazole was used instead of the solution containing His-AGB1, and GST alone instead of GST–BIN2. After detecting His-AGB1, the blot was washed three times by Tween–phosphate-buffered saline (PBS)–EDTA, which was made by adding 0.5M EDTA, pH 8.0, to Tween–PBS (0.1% v/v Tween-20 in PBS) to 10mM final concentration of EDTA, for deprobing the HisProbe-HRP. The blot was then washed twice by Tween–TBS and used for a western blot analysis of phosphoproteins using Phos-tag Biotin BTL-104 (Wako, Japan) (Kinoshita et al., 2006). Signal detection and image processing were performed as described above.
Yeast three-hybrid (Y3H) assays
pGBK-AGB1 (Tsugama et al., 2012b ) was digested by HpaI and SalI, and the resultant DNA fragment containing the full-length ORF of AGB1 and a partial coding sequence (CDS) of the GAL4 DNA-binding domain (GAL4BD) was inserted into pBridge (Clontech), generating pBridge-AGB1. The ORF fragment of AGG1 (AT3G63420) was obtained by PCR using pGAD-AGG1 (Tsugama et al., 2012b ) as template and the following primer pair: 5ʹ-GAGAGATCTATGCGAGAGGAAACTGTGGT-3ʹ and 5ʹ-CCTAGATCTAAGTATTAAGCATCTGCAGCC-3ʹ (BglII sites are underlined). The PCR products were digested by BglII, and inserted into the BglII site of pBridge-AGB1, generating pBridge-AGB1-AGG1. The ORF of AGG1 was obtained by digesting pGAD-AGG1 by NdeI and XhoI, and inserted into the NdeI–SalI site of pGBKT7 (Clontech), generating pGBK-AGG1. The ORF of AGG1 was obtained by PCR using pGAD-AGG1 as template and the following primer pair: 5ʹ-GAGGAATTCATGCGAGAGGAAACTGTGGT-3ʹ and 5ʹ-CC TAGATCTAAGTATTAAGCATCTGCAGCC-3ʹ (EcoRI and BglII sites are underlined). The PCR products were digested by EcoRI and BglII, and inserted into the EcoRI–BamHI site of pBridge, generating pBridge-AGG1. The ORF of AGB1 was obtained by PCR using pGBK-AGB1 as template and the following primer pair: 5ʹ-GAGGGATCCATGTCTGTCTCCGAGCTCAAAG-3ʹ and 5ʹ-C CCGGATCCTCAAATCACTCTCCTGGTCC-3ʹ (BamHI sites are underlined). The PCR products were digested by BamHI, and inserted into the BglII site of either pBridge or pBridge-AGG1, generating pBridge-2-AGB1 or pBridge-AGG1-AGB1, respectively. The ORFs of BIN2 were obtained by digesting pGEX-6P-BIN2 by NcoI and SpeI, and were inserted into the NcoI–XbaI site of pGADT7-Rec (Clontech), generating pGAD-BIN2. The ORF fragments of BIN2 were obtained again by digesting pGAD-BIN2 by NcoI and BamHI, and were inserted into the NcoI–BamHI site of pGBKT7 (Clontech), generating pGBK-BIN2. pGBK-BIN2 was digested by HpaI and PstI, and the resultant fragment containing the full-length ORF of BIN2 and a partial CDS of GAL4BD was inserted into the HpaI–PstI site of pBridge-2-AGB1, generating pBridge-BIN2-AGB1. The ORF of BZR1 was amplified by PCR using a cDNA clone of BZR1 (RAFL04-20-E20), which was obtained from RIKEN BRC Experimental Plant Division (Seki et al., 2002), as template and the following primer pair: 5ʹ-GAGGAATTCATGACTTCGGATGGAGCTACG-3ʹ and 5ʹ-TCCTCTAGAACCACGAGCCTTCCCATTTCC-3ʹ (EcoRI and XbaI sites are underlined). The PCR products were digested by EcoRI and XbaI, and inserted into the EcoRI–XbaI site of pGADT7-Rec, generating pGAD-BZR1. The Saccharomyces cerevisiae strain AH109 was transformed with combinations of pGAD and pBridge constructs. After transformation, at least four colonies grown on synthetic dextrose (SD) medium lacking leucine and tryptophan (SD/–Leu/–Trp), were streaked on SD/–Leu/–Trp and SD/–Leu/–Trp lacking histidine or lacking both histidine and adenine. Reporter gene activation was quantified by a β-galactosidase assay as described in the Yeast Protocols Handbook (Clontech).
Bimolecular fluorescence complementation (BiFC)
The ORF of BIN2 was amplified using pGBK-BIN2 as template and the following primer pair: 5ʹ-GAGTCTAGAATGGC TGATGATAAGGAGATGCC-3ʹ and 5ʹ-CCCACTAGTTCCAGA TTGATTGATTCAAGAAGC-3ʹ (XbaI and SpeI sites are underlined). The PCR products were digested by XbaI and SpeI, and inserted into the SpeI site of pBS-35SMCS-cYFP (Tsugama et al., 2012b ), generating pBS-35S-BIN2-cYFP. The ORF of AGG1 was amplified by PCR using pGBK-AGG1 as template and the following primer pair: 5ʹ-GGGACTAGTATGCGAGAGGAAACTGTGG-3ʹ and 5ʹ-CCACTAGTAAGTATTAAGCATCTGCAGCC-3ʹ (SpeI sites are underlined). The PCR products were digested by SpeI and inserted into the SpeI site of pBS-35SMCS-cYFP, generating pBS-35S-AGG1-cYFP. These cYFP (C-terminal yellow fluorescent protein) constructs and pBS-35S-nYFP-AGB1 (Tsugama et al., 2012b ) was used to co-express cYFP-fused protein and nYFP-fused AGB1 in Arabidopsis mesophyll protoplasts. Arabidopsis mesophyll protoplasts were prepared and transformed as previously described (Yoo et al., 2007; Wu et al., 2009). Recovered YFP fluorescence was observed by fluorescence microscopy 12h after transformation.
Results
ABA enhances the BR hyposensitive phenotype of agb1
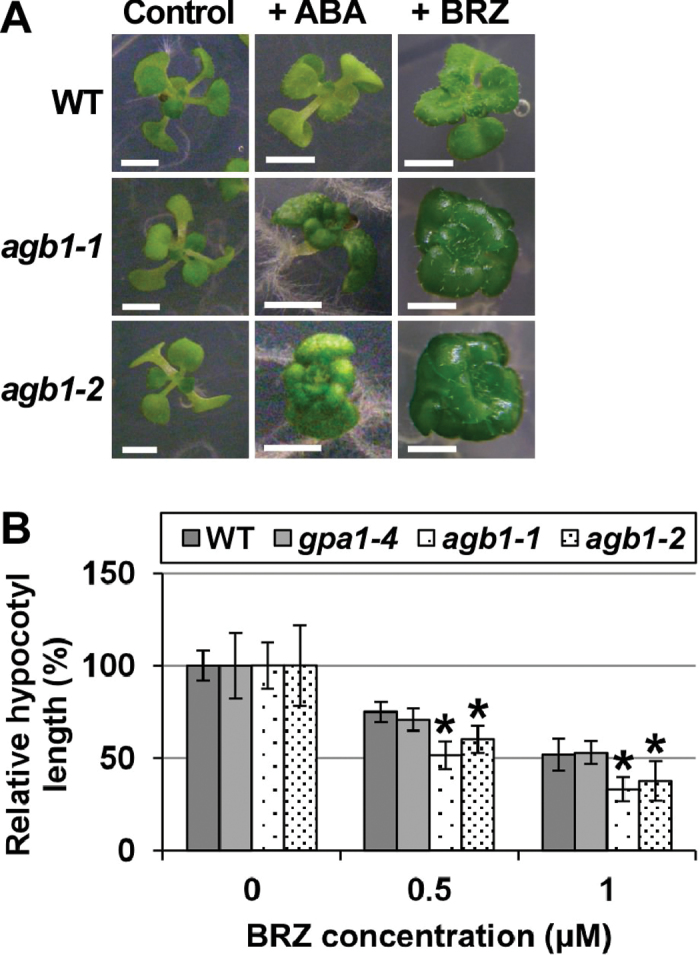
The leaves of agb1-1 and agb1-2 are rounder and their petioles are shorter than those of the WT. It was discovered that in the presence of ABA, leaves of agb1-1 and agb1-2 become even rounder and their petioles become even shorter. These ABA-induced phenotypes of agb1-1 and agb1-2 were similar to the phenotypes of mutants which have severe defects in BR biosynthesis or BR signalling (for a review, see Clouse, 2011). To examine whether the agb1 phenotypes are due to impaired BR signalling, a BR biosynthesis inhibitor, BRZ, was tested for its effects on the phenotypes of agb1-1 and agb1-2. BRZ made leaves rounder and petioles shorter in all the genotypes including the WT and gpa1-4. However, the leaf morphologies of agb1-1 and agb1-2 appeared to be more strongly affected by BRZ than those of the WT and gpa1-4 (Fig. 1A; Supplementary Fig. S1 at JXB online). BRZ also inhibited hypocotyl elongation in the dark more severely in agb1-1 and agb1-2 than in the WT and gpa1-4 (Fig. 1B; Supplementary Fig. S2), suggesting that agb1 is hypersensitive to BRZ. Exogenously added BR induced hypocotyl elongation in agb1-1 and agb1-2, but the elongation was less than that in the WT (Supplementary Fig. S3). Together, these results suggest that agb1-1 and agb1-2 are hyposensitive to BR and that their BR-hyposensitive phenotypes are enhanced by ABA. The BR hyposensitivities of agb1 in seed germination were previously reported (Chen et al., 2004), but AGB1 was not well characterized as a regulator of BR signalling. In previous studies, BR signalling mutants showed ABA hypersensitivities (for a review, see Clouse, 2011), supporting the idea that the ABA hypersensitivities of agb1 are at least partly dependent on its BR hyposensitivity. Based on these results, it was hypothesized that AGB1 regulates BR signalling, and the interactions between AGB1 and BR signalling were further examined.
Fig. 1.
BRZ hypersensitivity of agb1. (A) ABA- and BRZ-hypersensitive phenotypes of agb1. Plants were grown under a 16h light/8h dark photoperiod in the presence of either 0.5 μM ABA or 5 μM BRZ for 25 d (+ ABA or + BRZ, respectively), or in the absence of ABA or BRZ for 10 d (Control). A representative plant is shown for each genotype. Scale bars=3mm. (B) BRZ hypersensitivity of agb1 in hypocotyl elongation in the dark. Plants were grown for 5 d in the dark in the presence of 0, 0.5, or 1 μM BRZ. Relative hypocotyl lengths are shown (for absolute lengths, see Supplementary Fig. S2 at JXB online). Values are means ±SE (n=11–18). *P < 0.05 vs. the WT by Student’s t-test.
Expression of bzr1-1 alleviates effects of BRZ in both the wild type and agb1
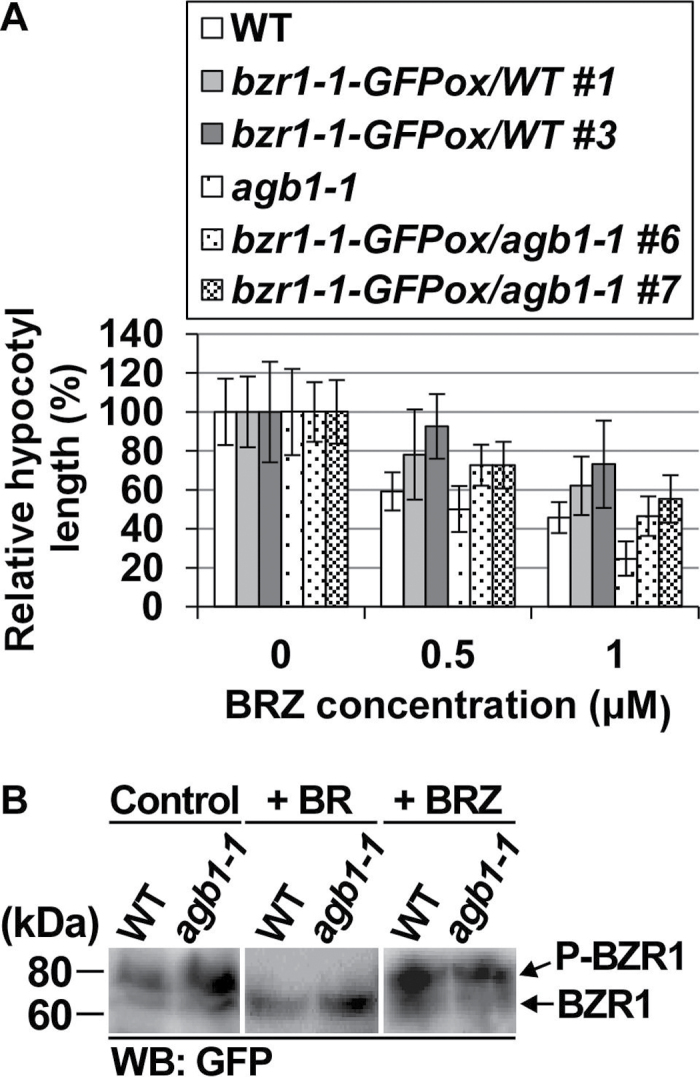
BZR1 is a transcription factor downstream of BR signalling (for a review, see Kim and Wang, 2010). To examine BZR1 functions in agb1, BZR1 and its point-mutated (P→L at amino acid position 234) version, bzr1-1, were expressed as GFP-fused proteins (BZR1–GFP and bzr1-1–GFP) in agb1-1 as well as in the WT (Table 1). Expression of BZR1-GFP and bzr1-1-GFP in transgenic plants was confirmed by RT–PCR (Supplementary Fig. S4 at JXB online). Expression of bzr1-1 is known to alleviate BRZ-induced inhibition of hypocotyl elongation in the dark (Wang et al., 2002; Ryu et al., 2007). In the presence of BRZ, the hypocotyl length of bzr1-1/a was greater than that of agb1-1 but smaller than the hypocotyl lengths of the WT and bzr1-1/WT. In the absence of BRZ, the hypocotyl length of bzr1-1/a was comparable with that of agb1-1 and slightly smaller than the hypocotyl lengths of the WT and bzr1-1/WT (Fig. 2A; Supplementary Fig. S5). These results suggest that expression of bzr1-1 partially suppresses the BRZ hypersensitivity of agb1 but does not fully suppress the agb1 phenotypes.
Table 1.
Transgenic plants used in this study
| Abbreviation | Genotype | Definition |
|---|---|---|
| BZR1/WT | BZR1-GFPox/WT | BZR1–GFP-expressing WT |
| bzr1-1/WT | bzr1-1-GFPox/WT | bzr1-1–GFP-expressing WT |
| BZR1/a | BZR1-GFPox/agb1-1 | BZR1–GFP-expressing agb1-1 |
| bzr1-1/a | bzr1-1-GFPox/agb1-1 | bzr1-1–GFP-expressing agb1-1 |
Fig. 2.
Functions of BZR1 in agb1. (A) Expression of bzr1-1 partially suppresses BRZ hypersensitivity of agb1-1. Plants were grown in the dark in the presence of 0, 0.5, or 1 μM BRZ for 4 d, and their hypocotyl lengths were measured. Relative hypocotyl lengths are shown (for absolute lengths, see Supplementary Fig. S5 at JXB online). Values are means ±SE (n=15–24). *P < 0.05 vs. non-transgenic lines by Student’s t-test. (B) AGB1 is not involved in BR-dependent regulation of BZR1 phosphorylation states. BZR1-GFPox/WT #9 and BZR1-GFPox/agb1-1 #9 (only genetic backgrounds, WT and agb1-1, are shown) were grown in the presence of 20nM BR for 15 d (+ BR), in the presence of 2.5 μM BRZ for 25 d (+ BRZ), or in the absence of BR and BRZ (Control), and used for western blotting using an anti-GFP antibody (WB: GFP). Experiments were performed in triplicate, and representative results are shown.
BR is known to induce dephosphorylation and activation of BZR1 (He et al., 2002). However, neither BR nor BRZ caused differences in BZR1–GFP phosphorylation states between BZR1/WT and BZR1/a (Fig. 2B), suggesting that AGB1 is not involved in BR-dependent changes of BZR1 phosphorylation states.
Overexpression of BZR1 suppresses effects of ABA in both the wild type and agb1
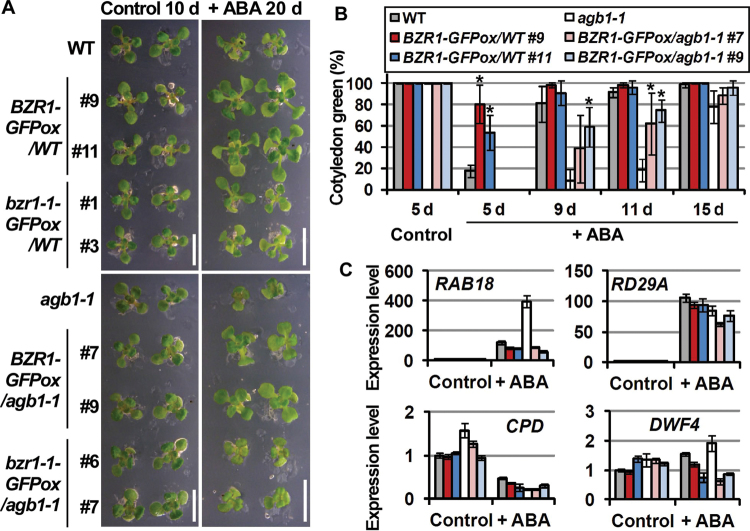
BZR1/WT, bzr1-1/WT, BZR1/a, and bzr1-1/a were grown in the presence of ABA. Interestingly, BZR1/WT but not bzr1-1/WT was larger in size than the WT. Similarly, BZR1/a but not bzr1-1/a was larger in size than agb1-1. The leaf morphology of BZR1/a was similar to that of agb1-1 rather than that of the WT (Fig. 3A). The expresson level of bzr1-1-GFP in bzr1-1/a #6 was higher than the expression level of BZR1-GFP in BZR1/a #9 (Supplementary Fig. S6 at JXB online). These results suggest that BZR1 rather than bzr1-1 alleviates the ABA responses. ABA responses of BZR1/WT and BZR1/a were further evaluated by scoring their green cotyledons in the presence of ABA. The cotyledon greening rate in the presence of ABA was in the order of BZR1/WT > WT > BZR1/a > agb1-1 (Fig. 3B), suggesting that overexpression of BZR1 alleviates ABA-induced growth retardation in both the WT and agb1, and that overexpression of BZR1 cannot fully suppress the ABA hypersensitivity of agb1. Among BZR1–GFP-overexpressing lines, BZR1 expression levels and cotyledon greening rates in the presence of ABA were not correlated (Supplementary Fig. S7), which suggests that BZR1-dependent responses are saturated in the transgenic lines studied.
Fig. 3.
Overexpression of BZR1 alleviates effects of ABA. (A) Faster growth of BZR1-overexpressing plants in the presence of ABA. Plants were grown in the presence of 0.5 μM ABA for 20 d (+ ABA 20 d) or in the absence of ABA for 10 d (Control 10 d). Two representative plant are shown for each genotype. Scale bars=1cm. (B) Faster cotyledon greening of BZR1-overexpressing plants in the presence of ABA. Plants were grown in the presence of 0 (Control) or 1 μM ABA (+ ABA). Green cotyledons were scored at the indicated time points. More than 30 plants were used for scoring green cotyledons in each genotype. Experiments were performed in triplicate. Values are means ±SE. *P < 0.05 vs. non-transgenic lines by Student’s t-test. (C) Quantitative RT–PCR analysis of ABA- and BR-responsive gene expression in BZR1-overexpressing plants. Plants were grown in the presence of 0.5 μM ABA for 20 d (+ ABA) or in the absence of ABA for 10 d (Control), and sampled. Relative expression levels of ABA-responsive genes (RAB18 and RD29A) and BR-responsive genes (CPD and DWF4) were calculated by the comparative CT method using UBQ5 as an internal control gene and the WT sample as a reference sample. Experiments were performed in triplicate. Values are means ±SE.
The expression levels of ABA-responsive genes, RAB18 and RD29A (Umezawa et al., 2006), were lower in BZR1/WT and BZR1/a than in the WT and agb1-1 in the presence of ABA (Fig. 3C, upper panels), which is in agreement with the finding that overexpression of BZR1 alleviates the effects of ABA on cotyledon greening and subsequent growth (Fig. 3A, B). Under control conditions, the expression of BZR1 target (thereby BR-responsive) genes, CPD and DWF4 (He et al., 2005), was higher in agb1-1 than in the WT, which is consistent with the finding that agb1 is hyposensitive to BR. The CPD expresson level was lower in BZR1/a than in agb1-1, but no significant difference was observed in the DWF4 expression levels between agb1-1 and BZR1/a (Fig. 3C, lower panels). ABA increased the DWF4 expression levels in both the WT and agb1-1, but both BZR1/WT and BZR1/a showed lower expression levels of DWF4 than either the WT or agb1-1 in the presence of ABA (Fig. 3C, lower right panel), again supporting the idea that BZR1 overexpression alleviates the effects of ABA. ABA significantly decreased the CPD expression levels and, in the presence of ABA, no difference was observed in the CPD expression levels among all the genotypes studied (Fig. 3C, lower left panel).
AGB1 interacts with BIN2
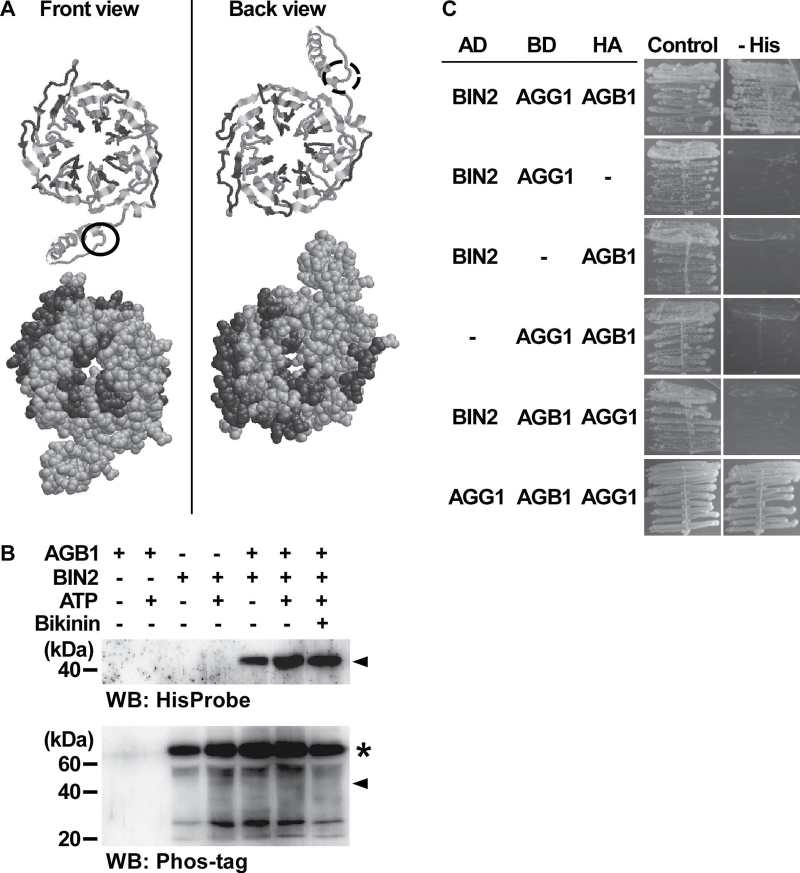
A motif-scanning program (ELM, http://elm.eu.org) identified 17 possible GSK modification sites in the amino acid sequence of AGB1. GSKs are known to regulate BR signalling negatively in Arabidopsis (He et al., 2002; Rozhon et al., 2010). Some of the putative GSK modification sites are located on the surface of the predicted three-dimensional structure of AGB1 (Fig. 4A), raising the possibility that AGB1 interacts with GSKs and is phosphorylated by them. To test this idea, the interaction between AGB1 and BIN2, the best-characterized plant GSK, was examined by an in vitro GST pull-down assay using His-AGB1 and GST–BIN2. His-AGB1 and GST–BIN2 were expressed in E. coli (Supplementary Fig. S8 at JXB online). GST–BIN2 was bound to resin and mixed with purified His-AGB1. After incubation, GST–BIN2 was eluted from the resin and His-AGB1 in the elutant was analysed by western blotting. His-AGB1 was detected only when both His-AGB1 and GST–BIN2 were present in the reaction mixture (Supplementary Fig. S9), suggesting that AGB1 and BIN2 interact in vitro. Neither ATP, which is required for phosphorylation by protein kinases, nor a GSK inhibitor, bikinin (De Rybel et al., 2009), affected the level of His-AGB1 detected in the pull-down assay (Fig. 4B, upper panel), suggesting that the kinase activity of BIN2 does not affect the interaction between AGB1 and BIN2. After the pull-down assay, phosphorylated proteins on the same blot were detected using a phosphoprotein probe, Phos-tag (Kinoshita et al., 2006). However, no clear difference was observed in the banding patterns of phosphoproteins in the presence or absence of His-AGB1 (Fig. 4B, lower panel), suggesting that AGB1 is not phosphorylated by BIN2, and that AGB1 does not affect BIN2 autophosphorylation.
Fig. 4.
AGB1 interacts with BIN2 in vitro. (A) GSK modification sites are present on the surface of the three-dimensional structure of AGB1. The three-dimensional structure of AGB1 is shown as cartoon models (upper) and space-filling models (lower). Putative GSK modification sites are shown in dark grey and the other amino acids in light grey. Circles indicate the N-termini. (B) In vitro GST pull-down assay. Hexahistidine-tagged AGB1 (His-AGB1) and GST-fused BIN2 (GST–BIN2) were expressed in E. coli and used for the analysis. For AGB1–, 250mM imidazole was used instead of a solution containing purified His-AGB1. For BIN2–, GST alone was used instead of GST–BIN2. ATP was used in 0 (ATP –) or 1mM (ATP +) final concentration. Bikinin was used in 0 (Bikinin –) or 100 μM (Bikinin +) final concentration. His-AGB1 was analysed by western blotting using a polyhistidine probe, HisProbe-HRP (WB: His). After detecting His-AGB1, the same blot was deprobed, and phosphoproteins on the blot were analysed using a Phos-tag (WB: Phos-tag). The positions of His-AGB1 (in both the upper and lower panels) are indicated by arrowheads. Putative GST–BIN2 signals are indicated by *. Experiments were performed in triplicate, and a representative result is shown. (C) Y3H interaction between BIN2, AGB1, and AGG1. The yeast reporter strain AH109 was transformed with pGADT7-Rec containing BIN2 to express the GAL4 activation domain (AD)-fused BIN2, and pBridge containing either or both of AGB1 and AGG1 to express them as GAL4 DNA-binding domain (BD)-fused and HA-tagged forms. The combinations of the expressed proteins are shown at the left. Transformed cells were grown on SD medium (Control) and SD medium lacking histidine (– His). For each transformed cell line, four individual colonies that had appeared after transformation were tested, and representative results are shown. The combination of AD-fused AGG1, BD-fused AGB1, and HA-tagged AGG1 (bottom) is shown as a positive control.
The AGB1–BIN2 interaction was further examined by a yeast three-hybrid (Y3H) assay. In the Y3H system, three kinds of protein of interest were co-expressed as GAL4 activation domain (AD)-fused, GAL4 DNA-binding domain (BD)-fused, and haemagglutinin (HA)-tagged forms, respectively, in yeast cells, and subsequent reporter gene activation was checked by culturing the transformed cells on a selection medium. Yeast cells could grow on the selection medium when transformed with AD-fused BIN2 (AD-BIN2), BD-fused AGG1 (BD-AGG1), and HA-tagged AGB1 (HA-AGB1), whereas yeast cells could not grow when one of AD-BIN2, BD-AGG1, or HA-AGB1 was not expressed (Fig. 4C, upper four panels), suggesting that these three proteins can form a complex in yeast cells. Yeast cells also did not grow when the combination AD-BIN2, BD-AGB1, and HA-AGG1 was used (i.e. AGB1 and AGG1 for BD/HA fusions were swapped) (Fig. 4C, bottom). In this case, the BD may have interrupted the interaction between BIN2, AGB1, and AGG1.
A BiFC assay was also performed. The ORF of AGB1 was fused downstream of the ORF of nYFP and the ORF of BIN2 was fused upstream of the ORF of cYFP. When nYFP-fused AGB1 (nYFP–AGB1) and cYFP-fused BIN2 (BIN2–cYFP) were expressed together in Arabidopsis mesophyll protoplasts, speckled YFP signals were detected. The speckled signals were small in number and large in size (Supplementary Fig. S10 at JXB online), and might have resulted from aggregation of nYFP–AGB1 and BIN2–cYFP. It is unclear whether the fluorescence pattern reflects the interaction between AGB1 and BIN2 under physiological conditions.
Effects of AGB1 on the interaction between BIN2 and BZR1
BZR1 is a substrate of GSKs (He et al., 2002; Rozhon et al., 2010). The Y3H system was used to evaluate the effect of HA-AGB1 on the reporter gene activations, which are dependent on co-expressed AD-BZR1 and BD-BIN2. In either the presence or absence of HA-AGB1, yeast cells transformed with AD-BZR1 and BD-BIN2 could grow on high-stringency selection medium, but the activity of β-galactosidase, one of the reporter gene products, was lower in the presence of HA-AGB1 than in its absence (Supplementary Fig. S11 at JXB online), suggesting that HA-AGB1 can interfere to some extent with the interaction between BZR1 and BIN2 in yeast.
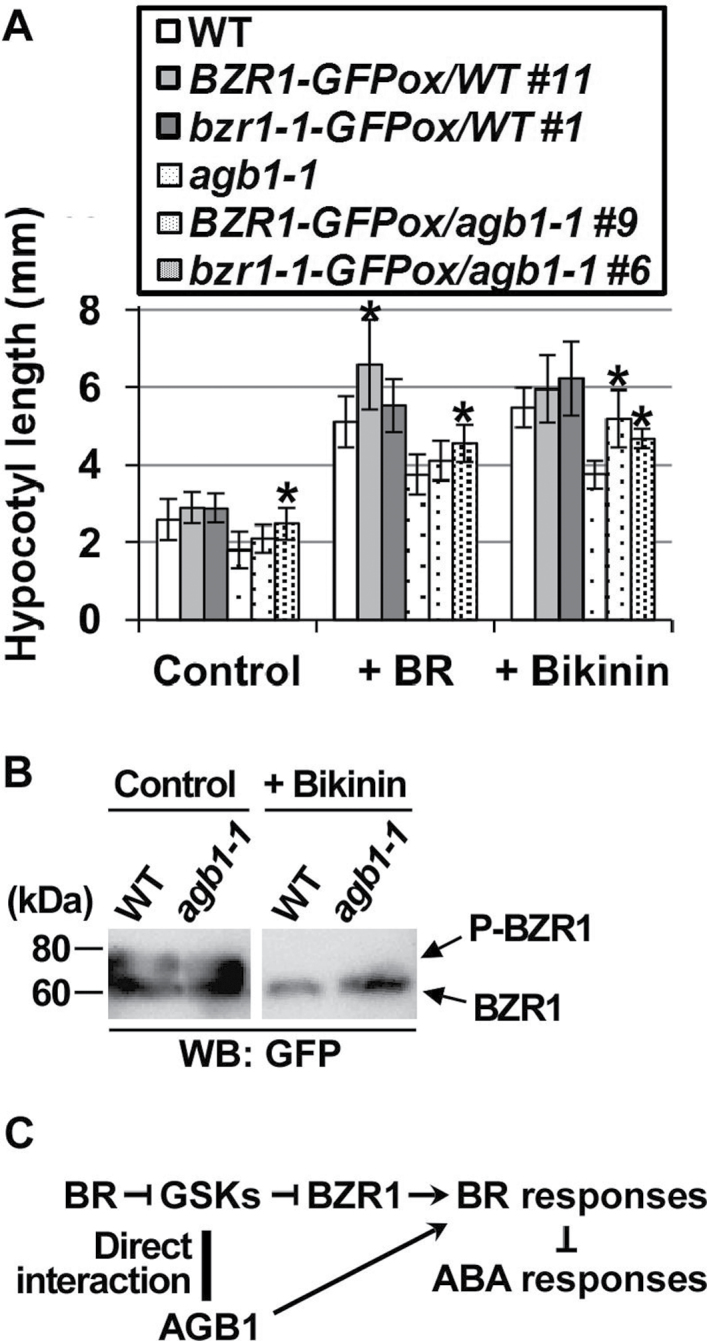
To examine the effects of AGB1 on the functions of GSKs in vivo, BZR1/WT, bzr1-1/WT, BZR1/a, and bzr1-1/a were grown in the presence of BR or bikinin, and their phenotypes were compared. BR- or bikinin-induced hypocotyl elongation was greater in BZR1/WT and bzr1-1/WT than in the WT. Similarly, BR- or bikinin-induced hypocotyl elongation was greater in BZR1/a and bzr1-1/a than in agb1-1. The effect of bzr1-1 overexpression on the BR- or bikinin-induced hypocotyl elongation was similar to the effect of BZR1 overexpression (Fig. 5A). In a previous study, bzr1-1 overexpression caused greater BR-induced hypocotyl elongation than BZR1 overexpression (Ryu et al., 2007), which conflicts with the present result. Although the expression level of bzr1-1-GFP in bzr1-1/a #6 was higher than the expression level of BZR1-GFP in BZR1/a #9 (Supplementary Fig. S6 at JXB online), the expression levels of bzr1-1-GFP still might have been insufficient to cause the greater hypocotyl elongation. In the presence of bikinin, BZR1–GFP was detected as a single, lower sized band in both BZR1/WT and BZR1/a (Fig. 5B). In the presence of BR, BRZ, or bikinin, no clear difference was observed in BZR1–GFP localization between the WT and agb1-1 (Supplementary Fig. S12). These results suggest that AGB1 is not involved in GSK-dependent BZR1 phosphorylation in vivo.
Fig. 5.
AGB1 is not involved in GSK-dependent regulation of BZR1 functions. (A) Overexpression of BZR1 and bzr1-1 partially suppresses the BR hyposensitivity of agb1-1. Plants were grown under a 16h light/8h dark photoperiod for 15 d in the presence of either 20nM BR (+ BR) or 40 μM bikinin (+ Bikinin), or in the absence of BR or bikinin (Control), and their hypocotyl lengths were measured. Values are means ±SE (n=15–22). *P < 0.05 vs. non-transgenic lines by Student’s t-test. (B) AGB1 is not involved in GSK-dependent phosphorylation of BZR1. BZR1-GFPox/WT #9 and BZR1-GFPox/agb1-1 #9 (only genetic backgrounds, WT and agb1-1, are shown) were grown for 15 d in the presence of 0 (Control) or 40 μM bikinin (+ Bikinin), and used for western blotting using an anti-GFP antibody (WB: GFP). Experiments were performed in triplicate and a representative result is shown. (C) A model proposed by this study.
Discussion
This study has shown that the ABA hypersensitivity of agb1 is partially suppressed by overexpressing BZR1 (Fig. 3), suggesting that the ABA hypersensitivity of agb1 is at least partly dependent on the BR hyposensitivity of agb1. Although AGB1 is not phosphorylated by BIN2 (Fig. 4B) and does not affect the phosphorylation states of BZR1 or BIN2 (Figs 2, 4B, 5B), AGB1 has putative GSK modification sites on its surface (Fig. 4A) and physically interacts with BIN2 (Fig. 4B, C), supporting the idea that AGB1 regulates BR responses via interaction with GSKs. A model proposed by this study is shown in Fig. 5C.
Overexpression of bzr1-1 suppressed BRZ-hypersensitive phenotypes of agb1-1, but the hypocotyl lengths of bzr1-1/a were still lower than those of the WT and bzr1-1/WT (Fig. 2A). Similarly, BR- or bikinin-induced hypocotyl elongation was not as great in BZR1/a and bzr1-1/a as it was in the WT, BZR1/WT, and bzr1-1/WT (Fig. 5A). These results indicate that BZR1 can enhance BR responses even in agb1, although AGB1 is required to activate BR responses fully. Because AGB1 is not necessary for regulating either BZR1 phosphorylation states or the subcellular localization of BZR1 (Figs 2B, 5B; Supplementary Fig. S10 at JXB online), AGB1 does not seem to be involved in regulating the functions of BZR1. BZR2 is another key transcription factor regulating the BR responses. It is also unlikely that AGB1 regulates the functions of BZR2 because BZR2 is similar to BZR1 in several respects, including amino acid sequence (88% identity), physiological function (Wang et al., 2002; Yin et al., 2002; for a review, see Kim and Wang, 2010), BR- and GSK-dependent phosphorylation states (He et al., 2002; Yin et al., 2002; Vert and Chory, 2006; Yan et al., 2009; Rozhon et al., 2010), and target genes (He et al., 2005; Vert and Chory, 2006; Sun et al., 2010).
AGB1 was originally identified as ELK4 (ERECTA-LIKE4), a gene responsible for the erecta (er) phenotype. agb1/elk4 shows several er phenotypes (e.g. rounder leaves with shorter petioles, shorter stems, more highly clustered flower buds, and shorter and wider siliques) (Lease et al., 2001). The well-studied BR-related mutants have these phenotypes as well (for a review, see Clouse, 2011), although the phenotypes of the BR-related mutants seem much more severe than those of er and agb1. ER encodes a receptor-like kinase (RLK) which has 20 extracellular leucine-rich repeats (LRRs), a single transmembrane domain, and an intracellular serine/threonine protein kinase domain (Lease et al., 2001). Interestingly, a BR receptor, BRI1, and its co-receptor, BAK1, are both LRR-RLKs. BR-mediated heterodimerization of BRI1 and BAK1 is thought to promote BR signalling (for a review, see Kim and Wang, 2010). Further studies are required to determine whether AGB1 and ER interact or whether ER is involved in BR signalling.
Overexpression of BZR1 alleviated the effects of ABA on cotyledon greening and subsequent growth in both the WT and agb1-1 (Fig. 3). This is consistent with previous studies suggesting that mutants which have impaired BR responses are hypersensitive to ABA (for a review, see Clouse, 2011). BZR1-GFPox/agb1-1 showed lower cotyledon greening rates in the presence of ABA than the WT (Fig. 3B). It is unclear how BR signalling decreases the ABA responses, but BZR1-mediated BR responses are thought to be insufficient to suppress the ABA hypersensitivity of agb1 fully. It is also interesting that BZR1 overexpression rather than bzr1-1 overexpression alleviated the ABA effects (Fig. 3A). BZR1 and bzr1-1 have been suggested to differ in their protein stabilities (He et al., 2002; Wang et al., 2002; Tang et al., 2011), phosphorylation states (He et al., 2002; Tang et al., 2011), and subcellular localizations (Ryu et al., 2007; this study, Supplementary Fig. S12 at JXB online). In addition, bzr1-1 has a higher affinity for a subunit of PP2A than does BZR1 (Tang et al., 2011). BZR1 and bzr1-1 may have different affinities for other interaction partners that affect the expression of BZR1 target genes. These differences might lead to the different effects of BZR1 and bzr1-1 on the gene expression involved in the ABA responses. Further studies are required to elucidate the molecular mechanisms underlying BZR1-mediated ABA signalling.
AGB1 was shown to interact with BIN2 in vitro (Fig. 4B). However, AGB1 did not affect the kinase activity of BIN2 (Figs 4B, 5B) and was not phoshorylated by BIN2 (Fig. 4B). Thus the physiological relevance of the interaction between AGB1 and BIN2 is unclear. One possibility is that AGB1 regulates the phosphorylation of BIN2 substrates other than BZR1 and BZR2. ARF2 and YODA are two examples of BIN2 substrates other than BZR1. ARF2 is a transcription factor regulating auxin-responsive gene expression (Vert et al., 2008). YODA is a mitogen-activated protein kinase kinase kinase (MAPKKK) regulating stomatal development, and, interestingly, YODA has been suggested to be under the control of ER signalling as well as BIN2-dependent signalling (Kim et al., 2012). AGB1 is also involved in auxin signalling in Arabidopsis (Ullah et al., 2003), and Pisum sativum Gβ interacts with a MAPK (Bhardwaj et al., 2011). Therefore, it will be interesting to examine whether AGB1 regulates the BIN2-dependent phosphorylation of ARF2 and YODA.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Phenotypes of agb1 grown in different concentrations of BRZ.
Figure S2. Absolute hypocotyl lengths of agb1 grown in the presence of BRZ.
Figure S3. BR-induced hypocotyl elongation in agb1.
Figure S4. Semi-quantitative RT–PCR analysis of BZR1-GFP and bzr1-1-GFP expression.
Figure S5. Absolute hypocotyl lengths of bzr1-1–GFP-overexpressing plants grown in the presence of BRZ.
Figure S6. Expression levels of BZR1-GFP and bzr1-1-GFP in transgenic plants.
Figure S7. BZR1 expression levels in BZR1-GFP-overexpressing lines and their responses to ABA.
Figure S8. Expression of His-AGB1 and GST–BIN2 in E. coli.
Figure S9. In vitro GST pull-down assay.
Figure S10. BiFC between AGB1 and BIN2.
Figure S11. Effects of AGB1 on the interaction between BIN2 and BZR1 in a Y3H system.
Figure S12. Subcellular localizations of BZR1–GFP in agb1.
Table S1. Primer pairs used for RT–PCR analyses.
Acknowledgements
This work was supported by a Grant-in-aid for Scientific Research (21380002) to TT and (22–2144) to DT. We are grateful to the ABRC for providing the Arabidopsis mutant seeds. An Arabidopsis full-length cDNA clone (RAFL04-20-E20) was developed by the plant genome project of RIKEN Genomic Sciences Center, and provided by RIKEN BRC which is participating in the National Bio-Resource Project of the MEXT, Japan.
References
- Bhardwaj D, Sheikh AH, Sinha AK, Tuteja N. 2011. Stress induced β subunit of heterotrimeric G-proteins from Pisum sativum interacts with mitogen activated protein kinase. Plant Signaling and Behavior 6, 287––292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty D, Trusov Y, Zhang W, Acharya BR, Sheahan MB, McCurdy DW, Assmann SM, Botella JR. 2011. An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana . The Plant Journal 67, 840––851 [DOI] [PubMed] [Google Scholar]
- Chen J-G, Pandey S, Huang J, Alonso M, Ecker JR, Assmann SM, Jones AM. 2004. GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiology 135, 907––915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Analytical Biochemistry 162, 156––159 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735––743 [DOI] [PubMed] [Google Scholar]
- Clouse SD. 2011. Brassinosteroids. The Arabidopsis book 9, e0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Audenaert D, Vert G, et al. 2009. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chemistry and Biology 16, 594––604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman EJ, Wang HX, Jiang K, Perovic I, Deshpande A, Pochapsky TC, Temple BR, Hicks SN, Harden TK, Jones AM. 2011. Acireductone dioxygenase 1 (ARD1) is an effector of the heterotrimeric G protein β subunit in Arabidopsis. Journal of Biological Chemistry 286, 30107––30118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wang S, Asami T, Chen J-G. 2008. Loss-of-function mutations in the Arabidopsis heterotrimeric G-protein α subunit enhance the developmental defects of brassinosteroid signaling and biosynthesis mutants. Plant and Cell Physiology 49, 1013––1024 [DOI] [PubMed] [Google Scholar]
- Gao Y, Zeng Q, Guo J, Cheng J, Ellis BE, Chen J-G. 2007. Genetic characterization reveals no role for the reported ABA receptor, GCR2, in ABA control of seed germination and early seedling development in Arabidopsis. The Plant Journal 52, 1001––1013 [DOI] [PubMed] [Google Scholar]
- He J-X, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang Z-Y. 2005. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634––1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J-X, Gendron JM, Yang Y, Li J, Wang Z-Y. 2002. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proceedings of the National Academy of Sciences, USA 99, 10185––10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Taylor JP, Chen J-G, Uhrig JF, Schnell DJ, Nakagawa T, Korth KL, Jones AM. 2006. The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. The Plant Cell 18, 1226––1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffé FW, Freschet G-E, Valdes BM, Runions J, Terry MJ, Williams LE. 2012. G protein-coupled receptor-type G proteins are required for light-dependent seedling growth and fertility in Arabidopsis. The Plant Cell 24, 3649––3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Assmann SM. 2004. Plants: the latest model system for G-protein research. EMBO Reports 5, 572––578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-W, Guan S, Sun Y, Deng Z, Tang W, Shang J-X, Sun Y, Burlingame AL, Wang Z-Y. 2009. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nature Cell Biology 11, 1254––1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-W, Michniewicz M, Bergmann DC, Wang Z-Y. 2012. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419––422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-W, Wang Z-Y. 2010. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annual Review of Plant Biology 61, 681––704 [DOI] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. 2006. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Molecular and Cellular Proteomics 5, 749––757 [DOI] [PubMed] [Google Scholar]
- Klopffleisch K, Phan N, Augustin K, et al. 2011. Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Molecular Systems Biology 7, 532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapik YR, Kaufman LS. 2003. The Arabidopsis cupin domain protein AtPirin1 interacts with the G protein α-subunit GPA1 and regulates seed germination and early seedling development. The Plant Cell 15, 1578––1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease KA, Wen J, Li J, Doke JT, Liscum E, Walker JC. 2001. A mutant Arabidopsis heterotrimeric G-protein β subunit affects leaf, flower, and fruit development. The Plant Cell 13, 2631––2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yue Y, Li B, Nie Y, Li W, Wu W-H, Ma L. 2007. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science 315, 1712––1716 [DOI] [PubMed] [Google Scholar]
- Mudgil Y, Uhrig JF, Jiping Z, Temple B, Jiang K, Jones AM. 2009. Arabidopsis N-MYC DOWNREGULATED-LIKE1, a positive regulator of auxin transport in a G protein-mediated pathway. The Plant Cell 21, 3591––3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Chen J-G, Jones AM, Assmann SM. 2006. G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiology 141, 243––256 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pandey S, Nelson DC, Assmann SM. 2009. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136, 136––148 [DOI] [PubMed] [Google Scholar]
- Peng P, Yan Z, Zhu Y, Li J. 2008. Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Molecular Plant 1, 338––346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfus-Barbeoch L, Jones AM, Assmann SM. 2004. Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Current Opinion in Plant Biology 7, 719––731 [DOI] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ. 2002. Seven-transmembrane receptors. Nature Reviews Molecular Cell Biology 3, 639––650 [DOI] [PubMed] [Google Scholar]
- Rozhon W, Mayerhofer J, Petutschnig E, Fujioka S, Jonak C. 2010. ASK θ, a group-III Arabidopsis GSK3, functions in the brassinosteroid signalling pathway. The Plant Journal 62, 215––223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I. 2007. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. The Plant Cell 19, 2749––2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, et al. 2002. Functional annotation of a full-length Arabidopsis cDNA collection. Science 296, 141––145 [DOI] [PubMed] [Google Scholar]
- Sun Y, Fan X-Y, Cao D-M, et al. 2010. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Developmental Cell 19, 765––77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, et al. 2011. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nature Cell Biology 13, 124––131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugama D, Liu S, Takano T. 2011. A rapid chemical method for lysing Arabidopsis cells for protein analysis. Plant Methods 7, 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugama D, Liu S, Takano T. 2012. a A putative myristoylated 2C-type protein phosphatase, PP2C74, interacts with SnRK1 in Arabidopsis. FEBS Letters 586, 693––698 [DOI] [PubMed] [Google Scholar]
- Tsugama D, Liu S, Takano T. 2012. b Arabidopsis heterotrimeric G protein β subunit interacts with a plasma membrane 2C-type protein phosphatase, PP2C52. Biochimica et Biophysica Acta 1823, 2254––2260 [DOI] [PubMed] [Google Scholar]
- Ullah H, Chen J-G, Temple B, Boyes DC, Alonso JM, Davis KR, Ecker JR, Jones AM. 2003. The β-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. The Plant Cell 15, 393––409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen J-G, Wang S, Jones AM. 2002. Role of a heterotrimeric G protein in regulation of arabidopsis seed germination. Plant Physiology 129, 897––907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen J-G, Young JC, Im K-H, Sussman MR, Jones AM. 2001. Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292, 2066––2069 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Okamoto M, Kushiro T, Nambara E, Oono Y, Seki M, Kobayashi M, Koshiba T, Kamiya Y, Shinozaki K. 2006. CYP707A3, a major ABA 8ʹ-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana . The Plant Journal 46, 171––182 [DOI] [PubMed] [Google Scholar]
- Vert G, Chory J. 2006. Downstream nuclear events in brassinosteroid signalling. Nature 441, 96––100 [DOI] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL. 2008. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proceedings of the National Academy of Sciences, USA 105, 9829––9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HX, Weerasinghe RR, Perdue TD, Cakmakci NG, Taylor JP, Marzluff WF, Jones AM. 2006. A Golgi-localized hexose transporter is involved in heterotrimeric G protein-mediated early development in Arabidopsis. Molecular Biology of the Cell 17, 4257––4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xu Y-Y, Ma Q-B, Li D, Xu Z-H, Chong K. 2006. Heterotrimeric G protein α subunit is involved in rice brassinosteroid response. Cell Research 16, 916––922 [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Nakano T, Gendron J, et al. 2002. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Developmental Cell 2, 505––513 [DOI] [PubMed] [Google Scholar]
- Wu F-H, Shen S-C, Lee L-Y, Lee S-H, Chan M-T, Lin C-S. 2009. Tape–Arabidopsis Sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Zhao J, Peng P, Chihara RK, Li J. 2009. BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiology 150, 710––721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang Z-Y, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. 2002. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181––191 [DOI] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565––1572 [DOI] [PubMed] [Google Scholar]
- Zhang C-Q, Nishiuchi S, Liu S, Takano T. 2008. Characterization of two plasma membrane protein 3 genes (PutPMP3) from the alkali grass, Puccinellia tenuiflora, and functional comparison of the rice homologues, OsLti6a/b from rice. BMB Reports 41, 448––454 [DOI] [PubMed] [Google Scholar]
- Zhang L, Wei Q, Wu W, Cheng Y, Hu G, Hu F, Sun Y, Zhu Y, Sakamoto W, Huang J. 2009. Activation of the heterotrimeric G protein α-subunit GPA1 suppresses the ftsh-mediated inhibition of chloroplast development in Arabidopsis. The Plant Journal 58, 1041––1053 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.