Abstract
The parallel (recombination) ‘R-triplex’ can accommodate any nucleotide sequence with the two identical DNA strands in parallel orientation. We have studied oligonucleotides able to fold back into such a recombination-like structure. We show that the fluorescent base analogs 2-aminopurine (2AP) and 6-methylisoxanthopterin (6MI) can be used as structural probes for monitoring the integrity of the triple-stranded conformation and for deriving the thermodynamic characteristics of these structures. A single adenine or guanine base in the third strand of the triplex-forming and the control oligonucleotides, as well as in the double-stranded (ds) and single-stranded (ss) reference molecules, was substituted with 2AP or 6MI. The 2AP*(T·A) and 6MI*(C·G) triplets were monitored by their fluorescence emission and the thermal denaturation curves were analyzed with a quasi-two-state model. The fluorescence of 2AP introduced into an oligonucleotide sequence unable to form a triplex served as a negative control. We observed a remarkable similarity between the thermodynamic parameters derived from melting of the secondary structures monitored through absorption of all bases at 260 nm or from fluorescence of the single base analog. The similarity suggests that fluorescence of the 2AP and 6MI base analogs may be used to monitor the structural disposition of the third strand. We consider the data in the light of alternative ‘branch migration’ and ‘strand exchange’ structures and discuss why these are less likely than the R-type triplex.
INTRODUCTION
Three-stranded nucleic acid alignments occur in the course of various biological processes, such as recombination or t-loop formation at telomere ends (1) The parallel (recombination) triplex or R-form DNA was predicted theoretically (2) and studied experimentally using protein-free intramolecular (3) and intermolecular (4) oligonucleotide models. Such a triplet hydrogen bond scheme is seen for the triplets G*(C·G) and C*(G·C) in the RNA mini-triple helix formed in the ribosome (5). In principle, the same kind of bonding could accommodate an arbitrary nucleotide sequence with the two identical DNA strands in parallel orientation (2).
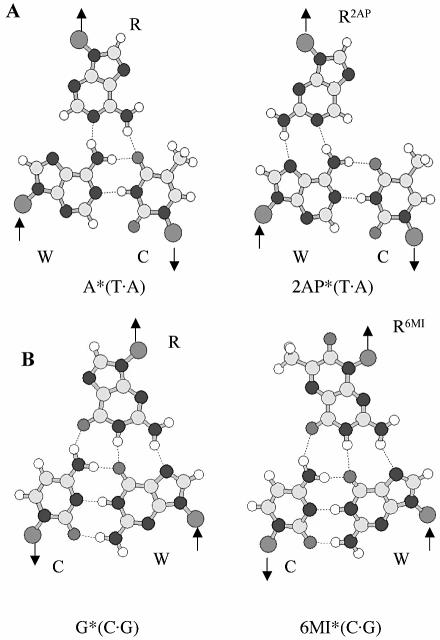
The fluorescent base analog 2-aminopurine (2AP) has been used as a site-specific probe of nucleic acid structure and dynamics because it pairs with thymine in a Watson–Crick geometry or makes a mismatch base pair with cytosine (6–12). Upon incorporation of 2AP into double-stranded (ds) B-DNA, its fluorescence is quenched in comparison to the free base (7,13–16). The local conformation and dynamics of mismatches and abasic sites in DNA have been examined using 2AP (9,10,13,14) and it has also been used to study the Mg2+-dependent conformation changes in hammerhead ribozymes (8). Different fluorescence properties of 2AP in the trans A·T base pairs of parallel-stranded DNA and in the cis A·T base pairs of antiparallel-stranded DNA make this base analog a convenient probe for strand orientation in DNA duplexes as well (A.K.Shchyolkina, D.N.Kaluzhny, O.F.Borisova, M.A.Livshits, J.J.Harvey, A.H.A.Clayton, Q.S.Hanley, D.J.Arndt-Jovin and T.M.Jovin, in preparation). 2AP can potentially substitute for adenine in the A*(T·A) triplet, forming a 2AP*(T·A) triplet consistent with the R-form (2,17) (Fig. 1A).
Figure 1.
(A) A*(T·A), 2AP*(T·A) and (B) G*(C·G) and 6MI*(C·G) base triplet schemes.
Likewise, the fluorescent guanine analog 6-methylisoxanthopterin (6MI) (18) may mimic guanine in the third strand by forming a 6MI*(C·G) triplet (Fig. 1B) (2,19). Pteridine nucleoside analog probes are highly fluorescent and have been shown to offer another approach to monitor structural perturbation of DNA molecules and subtle DNA interactions with other molecules (20). Here, we demonstrate that the fluorescent base 2AP can be used as a structural probe for monitoring formation of an AT-containing triplex and the fluorescence of 6MI pteridine can be used to probe the structure of a 5′-terminal GC-containing triplet.
MATERIALS AND METHODS
Oligonucleotide design
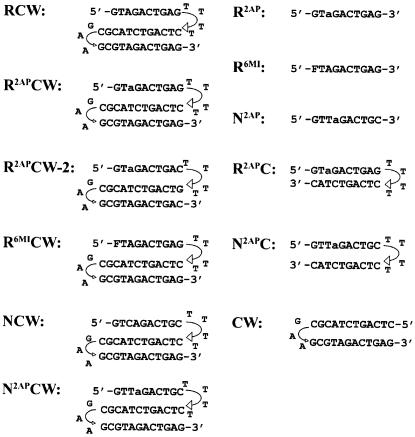
Deoxyribooligonucleotides potentially able to form the intramolecular R-triplex (RCW, R2APCW, R2APCW-2 and R6MICW) or the duplex (CW and R2APC) were studied (Fig. 2). The GAA linker together with two adjacent GC pairs serve to stabilize the duplex part of RCW, R2APCW, R2APCW-2 and R6MICW, whereas a flexible TTTT loop connects the third strand to the duplex. This design aimed (i) at minimizing displacement of the W strand from the WC duplex by the identical third strand and (ii) at resolving unstacking and dissociation of the third strand from the CW duplex. Deoxyribooligonucleotides NCW and N2APCW served as negative controls for triplex formation, as the nucleotide sequence of the N strand prevents formation of the perfect R-form or of an H and H′ type of triplex. In order to test a possible influence of the loop connecting the third strand to the CW duplex part, we studied oligonucleotides R2APCW and R2APCW-2, having -GT4C- and -CT4G- loops, respectively. R2APC and N2APC hairpins and single strands R2AP, R6MI and N2AP were used as controls in the fluorescence experiments for detection of fluorescence signals of 2AP involved in base pairing with thymine and the fluorescent base analogs incorporated into single strands.
Figure 2.
Oligonucleotide sequences. a, 2-aminopurine, (2AP); F, 6MI pteridine. Triplex-forming oligonucleotides: RCW, positive control for the triplex with no fluorophore; R2APCW is the 2AP-containing triplex with a -GT4C- loop connecting the third strand to the CW duplex; R2APCW-2 is the 2AP-containing triplex with a -CT4G- loop connecting the third strand to the CW duplex; R6MICW, 6MI-containig triplex; NCW, non-triplex-forming control with no fluorophore; N2APCW, 2AP-containing non-triplex-forming control. Hairpin-forming oligonucleotides: R2APC, hairpin designed for detecting the fluorescence signal of the 2AP base analog involved in the 2AP*T base pair; CW, the hairpin representing the duplex part of the triplex; N2APC, non-duplex-forming control. Single-stranded control oligonucleotides: R2AP, R6MI and N2AP.
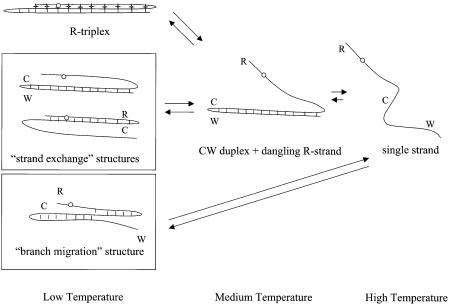
In principle, oligonucleotides RCW, R2AP CW, R2APCW-2 and R6MICW may form not only an R-triplex structure, but also a strand exchange or ‘branch migration’ structure (Fig. 3). These alternatives are considered in Results and Discussion.
Figure 3.
Schemes of temperature-dependent structural transitions for the R-triplex and alternative ‘strand exchange’ and ‘branch migration’ structures (shown in rectangles). The alternatives can involve multiple configurations, including the CW and the RC ‘strand exchange’ hairpins as well as the ‘branched’ structure (with various positions of the branch point). The open circles indicate positions of 2AP in various conformations.
Oligonucleotides containing 2AP were synthesized and purified by HPLC by the Midland Certified Reagent Co. Inc. (TX). Samples contained 0.7–1 µM oligonucleotide, 0.5 M LiCl, 10 mM Tris–HCl buffer, pH 7.6 ± 0.4 in the temperature range of the melting experiments. 6MI-containing oligonucleotides were synthesized as described (18). Samples were preheated to 90°C and fast cooled on ice. The solution conditions and the method of sample preparation were specifically chosen to ensure the predominant formation of intramolecular structures over intermolecular ones (21).
Fluorescence measurements
Measurement of the fluorescence emission and excitation spectra was carried out using a Photon Technologies International C-60SE spectrofluorometer with automatic correction of the excitation spectra, equipped with a thermostatted cuvette holder.
Thermodynamic analysis of the intramolecular triplex formation
We performed a thermodynamic analysis for the intramolecular binding of the dangling third strand of the triplex-forming oligonucleotide to its double helical CW part. We define such a process as triple helix formation. The transition curve for triplex formation was detected by two experimental techniques. One is the conventional melting curve acquired by measuring the hyperchromicity of the UV absorbance at 260 nm. All nucleotide bases of the third strand absorbing light at 260 nm contribute to this curve, whereas the transition curve measured by 2AP fluorescence emission reflects formation of a single triplet 2AP*(T·A).
The transition curve corresponds to the relationship
A(t) = B(t) + [C(t) – B(t)]·σ(t) 1
where t is the temperature (°C), B(t) and C(t) represent the temperature dependencies of the measured optical signal of the third strand in the bound and the dangling (‘single-stranded’) states, respectively, and σ(t) is shown in equation 2:
σ(t) = 1/{1 + exp[ΔH/R(1/T – 1/Tm)]} 2
where ΔH is the transition enthalpy, T is the absolute temperature (t + 273.15) and Tm is the melting temperature (K).
For UV melting curves, B(t) and C(t) are defined by a(1 – bt), the linear dependencies of extinction coefficients of the bases in the bound third strand and in the dangling third strand, respectively.
In the case of the fluorescence melting curve B(t) = a·c·(1 – bt), where c is a fraction of fluorescing 2AP in the bound third strand at 0°C. C(t) = d·(e–nt + m)/(1 + m) describes an exponential quenching of 2AP in a dangling third ‘single strand’ due to collision events with solvent. Parameters d, n and m were experimentally determined by ‘melting’ the R2AP single strand, and these values were used during fitting the theoretical curve (equation 1) to the experimental fluorescence melting curve.
Steady-state fluorescence anisotropy measurements and evaluation of relative hydrodynamic volumes of oligonucleotides using EtBr as a probe
To distinguish between intra- and intermolecular three-stranded structures, fluorescence anisotropy measurements were carried out on an SPF-1000 spectrofluorometer (AMINCO), equipped with a thermostatted cuvette holder. The steady-state anisotropy of the fluorescence emission of intercalated EtBr (r) excited by vertically polarized light was determined from the equation
r = (I|| – I⊥)/(I|| + 2I⊥) 3
where I|| and I⊥ are the parallel and perpendicular emission components, respectively. The relative dye concentration did not exceed 1 per 100 nucleotides, thereby avoiding depolarization due to energy transfer. The temperature-dependent contribution of the free EtBr fluorescence to the emission components (equation 3) was taken into account.
The hydrodynamic volume (V) of molecules with low asymmetry can be estimated by the equation:
V = kTτ/η(1/r – 1/r0) 4
where τ is the measured fluorescence lifetime of bound EtBr, r is the measured fluorescence anisotropy and r0 is the limiting anisotropy at infinite viscosity (T/η → 0), where η is the viscosity of the solution, T is the absolute temperature and k is the Boltzmann constant (22). The mean fluorescence lifetimes τ of EtBr intercalated in the parallel (recombinant) triplex, conventional antiparallel triplex and calf thymus DNA were found to be practically the same (23,24). This permitted the calculation of a relative hydrodynamic volume (V) of the oligonucleotides RCW, R2APCW and NCW with respect to the hydrodynamic volume (VCW) of the CW hairpin (see sequence in Fig. 2):
V/VCW = (1/rCW – 1/r0)/(1/r – 1/r0) 5
for the fluorescence anisotropy values determined under the same solution conditions and temperature (3,25). Under our experimental conditions, one EtBr molecule on average was intercalated in the studied oligonucleotides, causing similar induced structural perturbations. The extension of the double or triple helix on binding one intercalator molecule was assumed to be 3.4 Å (26,27) and constitutes a uniform systematic error of the estimation of the oligonucleotide hydrodynamic volume using EtBr as probe. An evaluation of the ratio V/VCW offsets the effect of the EtBr-induced extension of the oligonucleotides.
RESULTS AND DISCUSSION
Formation of a triple-stranded structure as monitored by 2-aminopurine fluorescence
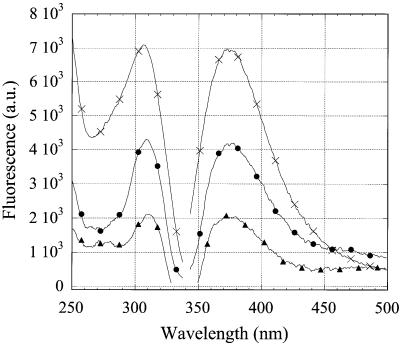
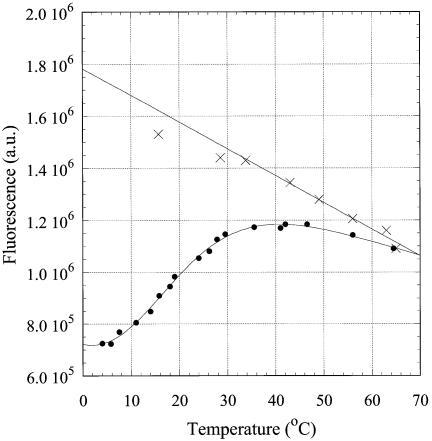
The fluorescence emission and excitation spectra of 2AP were found to be different for the triplex-forming (R2APCW), double-helical hairpin-forming (R2APC) and single-stranded (ss) (R2AP) oligonucleotides (Fig. 4). As the molar absorption of the three oligonucleotides at the excitation wavelength 310 nm was found to be the same (data not shown), the intensity of the fluorescence emission is proportional to the relative quantum yield (RQY) of 2AP incorporated in the three structures. The excitation maximum for emission at 370 nm for R2APC, R2APCW and R2AP is positioned around 311, 309 and 306 nm, respectively. A blue shift of the excitation maximum reflects an increased exposure to solvent of the 2AP base analog in R2APCW as compared to the ds R2APC (11) and this effect is still more pronounced in the case of the ss R2AP. Fluorescence of the 2AP nucleotide is well known to be considerably quenched by base stacking and hydrogen bonding interactions with other bases (compare the R2AP and R2APC temperature profiles in Fig. 5). Thus, by comparing the RQY of 2AP fluorescence as well as the blue shift of the excitation maximum in R2APCW and R2APC (at low temperatures) we conclude that 2AP is more exposed to solvent in the R2APCW structure than in the R2APC duplex. This finding is consistent with relatively weak stacking of bases in the third strand of the ‘R-triplex’ as predicted theoretically, especially for a 5′-pyrimidine-purine-3′ step (see fig. 5B in ref. 2). It should be noted that in the R2APCW construct the 2AP nucleotide is preceded by T, i.e. 2AP is expected to be more accessible to the solvent than in the duplex (Fig. 2). For brevity, we denote the triple-stranded structure stabilized at low temperature as ‘R-triplex’. Whether this is indeed a triplex or rather a branch migration structure (Fig. 3) is discussed in the following two sections, where the experimental data are presented.
Figure 4.
Fluorescence emission and excitation spectra of oligonucleotides at 8°C. R2APCW (filled circles), R2APC (filled triangles) and R2AP (crosses). The excitation wavelength for emission spectra was 310 nm, the emission wavelength for emission spectra was 370 nm. Samples contained 1 µM oligonucleotide, 0.5 M LiCl, 10 mM Tris–HCl buffer, pH 7.6.
Figure 5.
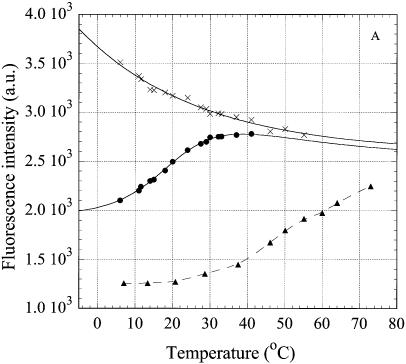
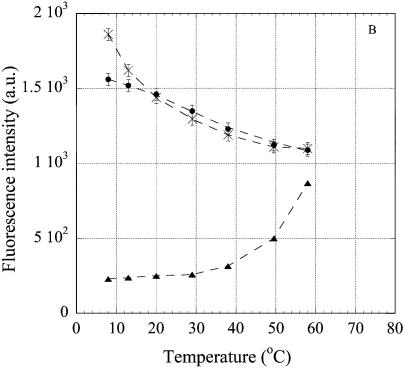
Temperature dependence of intensity of the fluorescence emission at 370 nm of 2AP, incorporated in the oligonucleotides. The excitation wavelength was 310 nm. (A) oligonucleotides R2APCW (filled circles), R2APC (filled triangles) and R2AP (crosses); (B) control N2APCW (filled circles), ds N2APC (filled triangles), ss N2AP (crosses); (C) oligonucleotides R2APCW-2 (filled circles) and N2APCW (crosses). Solid curves are the best theoretical fits (see Materials and Methods); dashed curves are the smoothed experimental data. Derived thermodynamic parameters are given in Table 1. Solution conditions as in Figure 4.
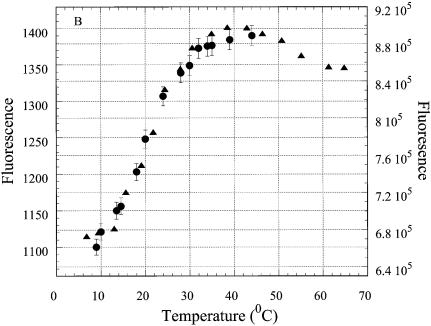
The temperature dependencies of the steady-state intensity of the fluorescence emission at 370 nm for the three structures R2APCW, R2APC and R2AP are shown in Figure 5A. The temperature-dependent quenching of the fluorescence of ss R2AP is attributable to collision events with solvent and other species in solution (Fig. 5A, crosses). The emission intensity of R2APCW displays a sigmoidal curve in the temperature range 8–40°C. Above 50°C, R2AP and R2APCW display the fluorescence expected for an unpaired 2AP. In the temperature range 8–40°C R2APC remains in the double-stranded conformation, with melting starting above 35–38°C (as revealed by the UV temperature profiles; data not shown). The changes in fluorescence of 2AP incorporated into the duplex were minor between 3 and 38°C. The sigmoidal temperature dependence of RQY in the case of oligonucleotide R2APCW apparently reflects a conformational change in the structure or formation/disruption of the single 2AP*(T·A) triplet. This conformational transition occurs in the same temperature range as unstacking of the bases in the third strand, as detected independently by UV hyperchromicity (see below), and therefore can be attributed to melting of the three-stranded structure to a duplex with a ‘dangling’ third strand and unstacked 2AP bases (Fig. 3).
To verify the specific character of this structural transition, we examined the temperature dependence of fluorescence emission of 2AP incorporated in the third strand of the control oligonucleotide N2APCW (Fig. 5B, filled circles). The third strand N2AP of the control is not able to bind specifically to the duplex-forming CW part of the oligonucleotide and does not form runs of base triplets of R- or H-form. Consequently, throughout the temperature range the curves for N2APCW (Fig. 5B, filled circles) and ss N2AP (crosses) coincide. We conclude that 2AP in the third strand of N2APCW remains in an environment similar to that of 2AP in the single strand. Below 15°C a small decline in the N2APCW curve as compared to that of ss N2AP presumably indicates the presence of non-specific random interactions of 2AP with other bases. The fluorescence of 2AP in ds N2APC indicates an involvement in base pairing (Fig. 5B) even though the two strands in this experiment are not fully complementary. The nucleotide sequence of the N2APC hairpin allows formation of an 8 bp intramolecular double helix by strand slippage thereby forming a 2AP·T base pair, which explains the temperature dependence of its RQY (Fig. 5B, triangles). Analysis of the N2APC UV melting curve (not shown) yielded thermodynamic parameters consistent with this conclusion (Table 1).
Table 1. Thermodynamic parameters of triplex and duplex formation.
| Experimental technique to monitor thermal denaturation | Triplex-forming oligonucleotide | Hairpin-forming oligonucleotide | ΔH (kJ mol–1) | Tm (°C) |
|---|---|---|---|---|
| 2AP fluorescence |
R2APCW |
|
–103 ± 16 |
22 ± 5 |
| |
R2APCW-2 |
|
–107 ± 14 |
21 ± 6 |
| 6MI fluorescence |
R6MICW |
|
–76 ± 10 |
17 ± 4 |
| A260 |
RCW |
|
–97 ± 10 |
18.0 ± 3 |
| |
R2APCW |
|
–102 ± 15 |
23.5 ± 1 |
| |
R6MICW |
|
–91 ± 10 |
19 ± 4 |
| |
|
R2APC |
–243 ± 9 |
79.5 ± 0.2 |
| |
|
N2APC (imperfect 8-bp hairpin) |
–148 ± 3 |
60.6 ± 0.1 |
| CW | NA | >85 |
For the triplex-forming oligonucleotides, the transition from a three-stranded structure to a CW duplex with a dangling third strand was studied. Parameters were determined as the best fits to the theoretical two-state model equation 1 (see Materials and Methods). Experimental conditions: 0.5 M LiCl, 10 mM Tris–HCl buffer, pH 7.6, 20°C.
Comparison of the data from the triplex-forming oligonucleotide R2APCW and the triplex-negative control N2APCW confirms the sequence-specific nature of the fluorescence melting profile of R2APCW and supports the interpretation of this process as ‘melting’ of the single 2AP*( T·A) triplet. The shape of the curve (Fig. 5A, filled circles) agrees with the assumption of a quasi-two-state mode of this transition. Fitting the fluorescence data (see Materials and Methods) yielded the thermodynamic parameters for 2AP*(T·A)·triplet formation in R2APCW tabulated in Table 1.
In general, the structural properties of the nucleotide loop (5′-NTTTTM-3′) that links the third strand to the duplex might affect the thermodynamics or kinetics of folding of the intramolecular ‘triplex’ (28). With this in mind, we compared folding of the two specific triplex-forming oligonucleotides, R2APCW and R2APCW-2, containing 5′-GTTTTC-3′ and 5′-CTTTTG-3′ linkers, respectively (Fig. 2). A comparison of the R2APCW-2 oligonucleotide to the control N2APCW was also made, as both oligonucleotides have the same 5′-CTTTTG-3′ linker that attaches the third strand to the duplex part (Fig. 5C). The thermodynamic parameters of the R2APCW-2 triplex are presented in Table 1. In spite of the different G*C·G and C*G·C 3′-end triplets which might influence TTTT loop conformation, we found no difference in either the enthalpy or Tm of the R2APCW and R2APCW-2 triplexes. The data clearly demonstrate that the nucleotide sequence specificity of the third strand and not a difference in the linkers promotes third strand binding to the duplex.
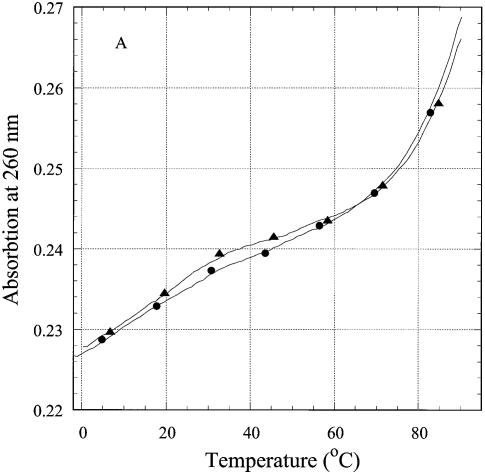
UV thermal denaturation of the R-forming oligonucleotides
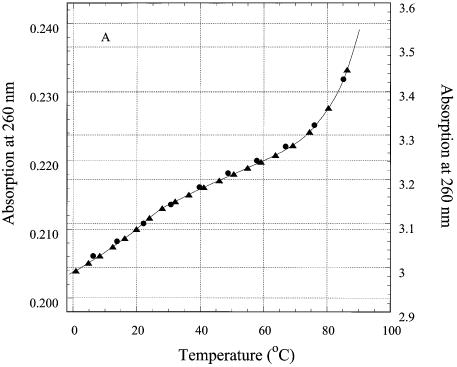
To correlate the fluorescence signal from the single triplet with the behavior of the third strand, we studied thermal denaturation of the oligonucleotides by UV absorption. Every base of the third strand should contribute to hyperchromicity at 260 nm during ‘triplex’ melting, the contribution of the 2AP analog being smallest as its extinction coefficient at 260 nm is about 0.1 that of adenine (29). Thermal denaturation curves of R2APCW, control N2APCW and their first derivatives are shown in Figure 6A and B. Denaturation of CW, the ds hairpin, does not contribute to absorption changes below 50°C (Fig. 6A, triangles). The behavior of the R2AP and N2AP strands in the melting of oligonucleotides R2APCW and N2APCW was different (Fig. 6A), differences further accentuated in the first derivatives of these melting curves (Fig. 6B). The R2APCW melting curve displayed a two-state structural transition, which can be interpreted as melting of a ‘triplex’ to a structure containing the CW hairpin with a dangling third strand, followed by melting of the CW hairpin at high temperature (Fig. 3). In contrast, the first derivative of the N2APCW melting curve was devoid of a maximum (Fig. 6B, open circles), suggesting a gradual temperature-induced unstacking of the single third strand rather than triplex melting.
Figure 6.
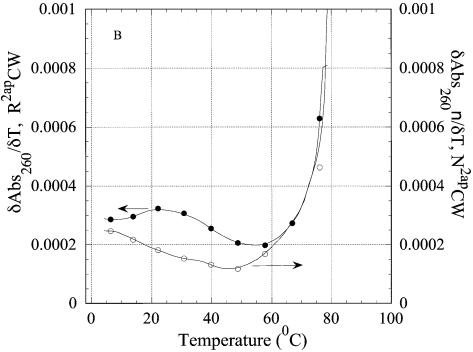
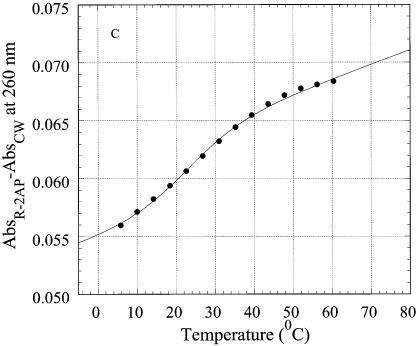
Thermal denaturation of R2APCW and control oligonucleotide N2APCW monitored by absorption at 260 nm. (A) Melting curves of R2APCW (filled circles) and of N2APCW (open circles), left ordinate; melting curve of CW hairpin (filled triangles), right ordinate; (B) first derivatives of the melting curves of R2APCW (filled circles) and N2APCW (open circles); (C) determination of the thermodynamic parameters of R2APCW triplex formation from the absorption thermal denaturation experiments. The triplex melting curve (filled circles) is a result of subtraction of the CW contribution to the melting profile of R2APCW (see filled circle and filled triangle in curves Fig. 5A). The solid curve is the best theoretical fit for a two-state model (see Materials and Methods). The derived thermodynamic parameters are given in Table 1. The concentration of each oligonucleotide was 0.77 µM.
To rule out possible intermolecular structures contributing to the observed triplex melting behavior, we measured melting of the R2APCW oligonucleotide at three different concentrations (0.3, 0.97 and 12 µM). Figure 7 compares thermal denaturation profiles of R2APCW at 0.97 and 12 µM (Fig. 7A) and at 0.3 and 12 µM (Fig. 7B) detected by UV absorption or 2AP fluorescence, respectively. We found both the UV absorption melting profiles and the 2AP base fluorescence changes to be superimposable at all three concentrations.
Figure 7.
Thermal denaturation of R2APCW at different oligonucleotide concentrations. (A) UV absorption melting profiles. R2APCW concentrations were 0.97 (filled circles) and 12 µM (filled triangles). The 12 µM sample was measured in a 1 mm cell and the plotted OD values are calculated for a 1 cm path. (B) Temperature dependence of 2AP fluorescence in R2APCW at 370 nm: oligonucleotide concentrations were 0.3 (filled circles) and 12 µM (filled triangles).
Because melting of the CW hairpin continues far beyond the experimental temperature range, treating the R2APCW melting in terms of a three-state transition from ‘triplex’ to hairpin and then to the open ss oligonucleotide appears impossible (Fig. 6A). One way to extract thermodynamic parameters from the ‘triplex’ melting curve (Fig. 6A, filled circles) is to subtract the duplex CW melting curve (Fig. 6A, triangles). Such an exercise was performed as shown in Figure 6C (filled circles) with the best fit theoretical curve (equation 1). The parameters from R2APCW and RCW ‘triplex’ melting are given in Table 1. The formation enthalpies and melting temperatures Tm of the two structures were the same within experimental error, with a tendency for a higher Tm for R2APCW. We obtained remarkably similar parameters for triplex formation derived from the absorption of bases at 260 nm and from the fluorescence of 2AP (Table 1). One may interpret this as a structural quasi-two-state transition with all bases of the third strand simultaneously participating in stabilization of the triple-stranded conformation (in our interpretation, the ‘R-triplex’). The similarity also implies that fluorescence of the 2AP base analog alone may be used to monitor ‘triplex’ integrity.
Formation of a triple-stranded structure as monitored by fluorescence of 6MI pteridine
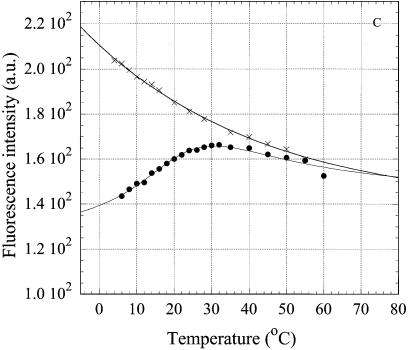
In the 5′-terminal position of the R6MICW oligonucleotide, 6MI minimally affects stability of the triple-stranded structure, as revealed by the Tm, and apparently allows formation of a 6MI*(C·G) triplet (Fig. 8). Quenching of 6MI fluorescence is known to take place upon stacking with other bases (18) and proceeds upon cooling in the case of R6MICW but not in the case of ss R6MI (Fig. 8). The formation enthalpy of the ‘triplex’ derived from the melting curve in Figure 8 (filled circles) was ∼25% less than that for RCW and R2APCW (Table 1). This may be accounted for by a terminal position of the triplet under consideration. Furthermore, the electronic properties of 6MI are markedly different from those of guanine, which may alter the energy contribution of stacking interactions of 6MI*(C·W) with the adjacent triplet. Moreover, minor structural perturbations of the triplet terminus due to substitution of 6MI for G could weaken ‘triplex’ stability. Nevertheless, formation of a terminal 6MI*(C·G) triplet as measured by fluorescence of this base analog is an additional argument in favor of the R-like triplex structure.
Figure 8.
Temperature dependence of intensity of the fluorescence emission at 430 nm of 6MI, incorporated into the oligonucleotides. The excitation wavelength was 350 nm. The oligonucleotides were R6MICW (filled circles) and R6MI (crosses); solid curves are the best theoretical fits (see Materials and Methods). Derived thermodynamic parameters are given in the Table 1. Solution conditions as in Figure 4.
Alternative triple-stranded structures
Figure 3 shows some alternative structures possible where no triplex occurs but rather various duplex configurations are formed by ‘branch migration’ or ‘strand exchange’. Our biphasic melting data, in which the higher temperature melt is not affected by the presence of a third strand (see Table 1 and Fig. 6), is not compatible with various duplex structures whereby the R strand invasion would shorten the 12 bp CW duplex. The latter should lead to a broader and lower temperature melting curve. Furthermore, we demonstrated a direct involvement of the terminal base in stabilization of the triple-stranded structure by using 6MI substitution, which strongly undermines a model with fraying ends.
If the structure is rather an equilibrium between two ‘strand exchange’ duplexes (Fig. 3), the first step of the melting curve should be assigned to melting of the shorter 10 bp RC hairpin and the second step to melting of the 12 bp CW hairpin (as described above). As we demonstrated, an RC hairpin unfolds closer to 80°C than to the low temperature transition of ∼20°C (Fig. 6).
Neither the branch migration nor the strand exchange structures alone are consistent with the experimental data. On the other hand, we cannot exclude that an equilibrium may exist at low temperature between the R-triplex and the other structures. Nevertheless, estimation of the hydrodynamic volume of the RCW and R2APCW oligonucleotides (see below) suggests close alignment of the third strand along the CW duplex at this low temperature such that no true ‘dangling strand’ can be present.
Estimation of the hydrodynamic volume of the oligonucleotides by intercalated dye fluorescence anisotropy
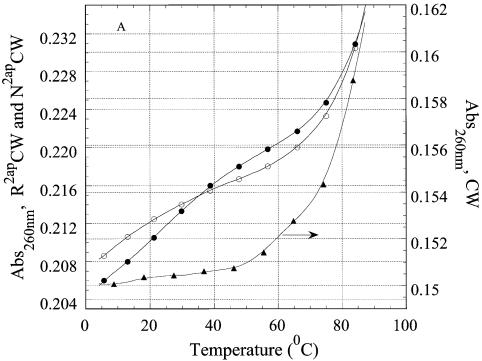
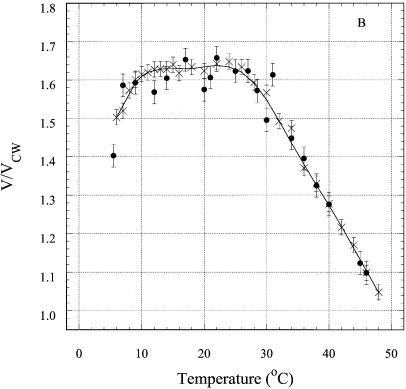
The fluorescence anisotropy of an intercalated DNA dye reflects the hydrodynamic volume of the DNA and, thus, is a probe for the intra- or intermolecular nature of the structures. The steady-state fluorescence of EtBr intercalated in short DNA duplexes (21,30), triplexes (3) and tetraplexes (31) has proved to be a convenient tool for the detection of multi-stranded structures (see Materials and Methods). EtBr does not destabilize DNA triplexes at low occupancy (23). The effect of EtBr intercalation on the melting curves of R2APCW is shown in Figure 9A. As expected, binding of one EtBr molecule (on average) to R2APCW had a minor effect on its stability and did not influence the value of the absorption hyperchromicity. The measured steady-state fluorescence anisotropy values were used to calculate ratios of the hydrodynamic volume of RCW or R2APCW to that of CW at different temperatures according to equation 5 and are plotted in Figure 9B. The resulting ratios (V/VCW) at 6°C are 1.4–1.5. These values are very close to those found in an earlier study (32) for the ‘canonical’ antiparallel purine intramolecular triplex anti-AG [5′-d(CT)5-L-d(AG)5-L-d(GA)5-3′] of V/VCW = 1.40 and for the parallel ‘recombination-like’ triplex par-AG [5′-d(GA)5-L-d(TC)5-L-d(GA)5-3′] of V/VCW = 1.45, where L stands for the linker pO(CH2CH2O)3p. The ratio V/VCW has been shown to be sensitive to formation of intermolecular structures, for example in the presence of magnesium ions (33). In such a case, dimerization of the anti-AG or par-AG oligonucleotides (mentioned above), increases the value of V/VCW up to 2.0–2.3 (33). Thus, we interpret the ratios for the data at 6°C (Fig. 9B) as reflecting an intramolecular triple-stranded structure with the third strand tightly arranged along the double-helical part of the oligonucleotide. Importantly, the similarity of the thermal denaturation curves of R2APCW at concentrations of 0.3, 0.97 and 12 µM independently supports the assumption that the structures are intramolecular in nature (see Fig. 7).
Figure 9.
Probing the RCW and R2APCW structures with EtBr. (A) Effect of EtBr binding on the thermal denaturation profiles of the oligonucleotides. RCW (triangles) and RCW in the presence of EtBr (circles). The concentration of bound EtBr calculated from binding isotherms was not more than one EtBr molecule per oligonucleotide, on average. (B) Temperature dependence of the relative hydrodynamic volume of the oligonucleotide–EtBr complexes: RCW (crosses) and R2APCW (filled circles) compared to the hydrodynamic volume of CW under the same conditions.
The V/VCW ratios for R2APCW and RCW increased with temperature to a value of 1.60–1.64 at ∼26°C, after which the value decreased to 1.05–1.1 at temperatures of 48–50°C (Fig. 8B). We interpret the data as follows. Below 26–28°C the third strand of the RCW and R2APCW oligonucleotides remains attached to the duplex, presumably due to a triplet-stabilizing effect of the EtBr, though local bulging of bases may cause the slight increase in V/VCW observed. At temperatures >30°C the triplex third strand begins to melt, as shown in Figures 5 and 6, decreasing contact with the duplex. Correspondingly, the EtBr molecules would redistribute from triplex binding sites to duplex sites. Above 48–50°C we would expect the third strand to be freely rotating relative to its duplex part, thereby contributing little to the hydrodynamic volume of the complex and thus the anisotropy value diminishes to a value similar to that for the CW duplex. These data further support the intramolecular nature of RCW and R2APCW triplex formation at low temperatures and the melting observed by spectroscopic means presented in the previous sections.
Concluding remarks
Compared to conventional antiparallel triplexes Py*(Pu·Py), the parallel three-stranded structure [or putative recombination R-triplex (2)] is less stable (3), therefore its experimental detection is more complicated. The structure involving the R strand is relatively weak, with a formation enthalpy of ∼–100 kJ (i.e. ∼2.5-fold less than the enthalpy of the RC duplex; see Table 1). In other words, the low temperature transition is principally different from the duplex melting. The low stability of the ‘R-triplex’ is probably related to less favorable base–base interactions (compared to Hoogsteen-type interactions) with heterogeneous and weak stacking in the third strand. In addition to the planar base–base interactions, the stereochemistry of R-triplets allows for formation of inter-planar interactions between the third strand bases and the Watson–Crick duplex (2). Such a dynamic behavior and relatively low stability of the base triplets, however, are major factors making the ‘R-triplex’ an ideal candidate for a transient intermediate in homologous recombination (2,34–36) or in t-loop formation at the mammalian telomere ends (1).
In this study we have used two different fluorescent probes that enabled us for the first time to follow the behavior of the individual R strand in the course of triplex melting. Importantly, the oligonucleotides RCW and R2APCW were specially designed to have exceptionally stable CW hairpins, which facilitated equilibrium formation of the R-type triplex at low temperature. Although we cannot completely exclude formation of alternative (non-R-triplex) structures, our data strongly disfavor ‘branch migration’ or ‘strand exchange’ structures under our experimental conditions. The results demonstrate the potential of 2AP and 6MI as structural probes to monitor the rearrangement of nucleic acid strands upon formation and dissociation of multi-stranded complexes.
Acknowledgments
ACKNOWLEDGEMENTS
The authors appreciate the helpful discussions of V. L. Florent’ev, M. Livshits, I. Panyutin and N. Ulyanov and the assistance of D. Cherny with the figures. The study was supported by RFBR grants nos 01-04-48561 and 02-04-49182, a Collaborative Linkage NATO grant SA (LST.CLG.978296) and the Max Planck Society.
REFERENCES
- 1.Stansel R.M., de Lange,T. and Griffith,J.D. (2001) T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J., 20, 5532–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhurkin V.B., Raghunathan,G., Ulyanov,N.B., Camerini-Otero,R.D. and Jernigan,R.L. (1994) A parallel DNA triplex as a model for the intermediate in homologous recombination. J. Mol. Biol., 239, 181–200. [DOI] [PubMed] [Google Scholar]
- 3.Shchyolkina A.K., Timofeev,E.N., Borisova,O.F., Il’icheva,I.A., Minyat,E.E., Khomyakova,E.B. and Florent’ev,V.L. (1994) R-form does exist. FEBS Lett., 339, 113–118. [DOI] [PubMed] [Google Scholar]
- 4.Shchyolkina A.K., Timofeev,E.N., Lysov,Y.P., Florentiev,V.L., Jovin,T.M. and Arndt-Jovin,D.J. (2001) Protein-free parallel triple-stranded DNA complex formation. Nucleic Acids Res., 29, 986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wimberly B.T., Guymon,R., McCutcheon,J.P., White,S.W. and Ramakrishnan,V.A. (1999) A detailed view of a ribosomal active site: the structure of the L11-RNA complex. Cell, 97, 491–502. [DOI] [PubMed] [Google Scholar]
- 6.Hochstrasser R.A., Carver,T.E., Sowers,L.C. and Millar,D.P. (1994) Melting of a DNA helix terminus within the active site of a DNA polymerase. Biochemistry, 33, 11971–11979. [DOI] [PubMed] [Google Scholar]
- 7.Law S.M., Eritja,R., Goodman,M.F. and Breslauer,K.J. (1996) Spectroscopic and calorimetric characterizations of DNA duplexes containing 2-aminopurine. Biochemistry, 35, 12329–12337. [DOI] [PubMed] [Google Scholar]
- 8.Menger M., Tuschl,T., Eckstein,F. and Pörschke,D. (1996) Mg(2+)-dependent conformational changes in the hammerhead ribozyme. Biochemistry, 35, 14710–14716. [DOI] [PubMed] [Google Scholar]
- 9.Rachofsky E.L., Seibert,E., Stivers,J.T., Osman,R. and Ross,J.B.A. (2001) Conformation and dynamics of abasic sites in DNA investigated by time-resolved fluorescence of 2-aminopurine. Biochemistry, 40, 957–967. [DOI] [PubMed] [Google Scholar]
- 10.Sowers L.C., Boulard,Y. and Fazakerley,G.V. (2000) Multiple structures for the 2-aminopurine-cytosine mispair. Biochemistry, 39, 7613–7620. [DOI] [PubMed] [Google Scholar]
- 11.Xu D., Evans,K.O. and Nordlund,T.M. (1994) Melting and premelting transitions of an oligomer measured by DNA base fluorescence and absorption. Biochemistry, 33, 9592–9599. [DOI] [PubMed] [Google Scholar]
- 12.Jiao Y., Stringfellow,S. and Yu,H. (2002) Distinguishing “looped-out” and “stacked-in” DNA bulge conformation using fluorescent 2-aminopurine replacing a purine base. J. Biomol. Struct. Dyn., 19, 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guest C.R., Hochstrasser,R.A., Sowers,L.C. and Millar,D.P. (1991) Dynamics of mismatched base pairs in DNA. Biochemistry, 30, 3271–3279. [DOI] [PubMed] [Google Scholar]
- 14.Stivers J.T. (1998) 2-Aminopurine fluorescence studies of base stacking interactions at abasic sites in DNA: metal-ion and base sequence effects. Nucleic Acids Res., 26, 3837–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jean J.M. and Hall,K.B. (2001) 2-Aminopurine fluorescence quenching and lifetimes: role of base stacking. Proc. Natl Acad. Sci. USA, 98, 37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jean J.M. and Hall,K.B. (2002) 2-Aminopurine electronic structure and fluorescence properties in DNA. Biochemistry, 41, 13152–13161. [DOI] [PubMed] [Google Scholar]
- 17.Dagneaux C., Gousset,H., Shchyolkina,A.K., Ouali,M., Letellier,R., Liquier,J., Florentiev,V.L. and Taillandier,E. (1996) Parallel and antiparallel A*A-T intramolecular triple helices. Nucleic Acids Res., 24, 4506–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawkins M.E., Pfleiderer,W., Balis,F.M., Porter,D. and Knutson,J.R. (1997) Fluorescence properties of pteridine nucleoside analogs as monomers and incorporated into oligonucleotides. Anal. Biochem., 244, 86–95. [DOI] [PubMed] [Google Scholar]
- 19.Van Meervelt L., Vlieghe,D., Dautant,A., Gallois,B., Precigoux,G. and Kennard,O. (1995) High-resolution structure of a DNA helix forming (C·G)*G base triplets. Nature, 374, 742–744. [DOI] [PubMed] [Google Scholar]
- 20.Hawkins M.E. (2001) Fluorescent pteridine nucleoside analogs: a window on DNA interactions. Cell. Biochem. Biophys., 34, 257–281. [DOI] [PubMed] [Google Scholar]
- 21.Shchyolkina A.K., Borisova,O.F., Livshits,M.A., Pozmogova,G.E., Chernov,B.K., Klement,R. and Jovin,T.M. (2000) Parallel-stranded DNA with mixed AT/GC composition: role of trans G·C base pairs in sequence dependent helical stability. Biochemistry, 39, 10034–10044. [DOI] [PubMed] [Google Scholar]
- 22.Weber G. and Anderson,S. (1969) The effects of energy transfer and rotational diffusion upon the fluorescence polarization of macromolecules. Biochemistry, 8, 361–371 [DOI] [PubMed] [Google Scholar]
- 23.Shchyolkina A.K. and Borisova,O.F. (1997) Stabilizing and destabilizing effects of intercalators on DNA triplexes. FEBS Lett., 419, 27–31. [DOI] [PubMed] [Google Scholar]
- 24.Mergny J.L., Collier,D., Rougee,M., Montenay-Garestier,T. and Helene,C. (1991) Intercalation of ethidium bromide into a triple-stranded oligonucleotide. Nucleic Acids Res., 19, 1521–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duhamel J., Kanyo,J., Dinter-Gottlieb,G. and Lu,P. (1996) Fluorescence emission of ethidium bromide intercalated in defined DNA duplexes: evaluation of hydrodynamics components. Biochemistry, 35, 16687–16697. [DOI] [PubMed] [Google Scholar]
- 26.Jain S.C., Tsai,C.C. and Sobell,H.M. (1977) Visualization of drug-nucleic acid interactions at atomic resolution. II. Structure of an ethidium/dinucleoside monophosphate crystalline complex, ethidium:5-iodocytidylyl (3′-5′) guanosine. J. Mol. Biol., 114, 317–331. [DOI] [PubMed] [Google Scholar]
- 27.Tuite E. and Norden,B. (1995) Intercalative interactions of ethidium dyes with triplex structures. Bioorg. Med. Chem., 3, 701–711. [DOI] [PubMed] [Google Scholar]
- 28.Roberts R.W. and Crothers,D.M. (1996) Kinetic discrimination in the folding of intramolecular triple helices. J. Mol. Biol., 260, 135–146. [DOI] [PubMed] [Google Scholar]
- 29.Ward D.C., Reich,E. and Stryer,L. (1969) Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside and their derivatives. J. Biol. Chem., 244, 1228–1237. [PubMed] [Google Scholar]
- 30.Shchyolkina A.K., Borisova,O.F., Livshits,M.A., Klement,R. and Jovin,T.M. (2001) The telomeric dG(GT)4G sequence can adopt a parallel-stranded double helical conformation. J. Biomol. Struct. Dyn., 18, 493–501, 503. [DOI] [PubMed] [Google Scholar]
- 31.Borisova O.F., Shchyolkina,A.K., Timofeev,E.N. and Florentiev,V.L. (1992) Evidence for the tetraplex structure of the d(GT)n repetitive sequences in solution. FEBS Lett., 306, 140–142. [DOI] [PubMed] [Google Scholar]
- 32.Shchyolkina A.K., Borisova,O.F., Minyat,E.E., Timofeev,E.N., Il’icheva,I.A., Khomyakova,E.B. and Florentiev,V.L. (1995) Parallel purine-pyrimidine-purine triplex: experimental evidence for existence. FEBS Lett., 367, 81–84. [DOI] [PubMed] [Google Scholar]
- 33.Borisova O.F., Shchelkina,A.K., Timofeev,E.N. and Florentiev,V.L. (1995) Dimers of DNA ‘triplexes’. Mol. Biol. (Mosk.), 29, 1076–1085. [PubMed] [Google Scholar]
- 34.Malkov V.A., Panyutin,I.G., Neumann,R.D., Zhurkin,V.B. and Camerini-Otero,R.D. (2000) Radioprobing of a RecA-three-stranded DNA complex with iodine 125: evidence for recognition of homology in the major groove of the target duplex. J. Mol. Biol., 299, 629–640. [DOI] [PubMed] [Google Scholar]
- 35.Howard-Flanders P., West S.C. and Stasiak,A. (1984) Role of RecA protein spiral filaments in genetic recombination. Nature, 309, 215–219. [DOI] [PubMed] [Google Scholar]
- 36.Nishinaka T., Shinohara,A., Ito,Y., Yokoyama,S. and Shibata,T. (1998) Base pair switching by interconversion of sugar puckers in DNA extended by proteins of RecA-family: a model for homology search in homologous genetic recombination. Proc. Natl Acad. Sci. USA, 95, 11071–11076. [DOI] [PMC free article] [PubMed] [Google Scholar]