Abstract
At seed maturity, quinoa (Chenopodium quinoa Willd.) perisperm consists of uniform, non-living, thin-walled cells full of starch grains. The objective of the present study was to study quinoa perisperm development and describe the programme of cell death that affects the entire tissue. A number of parameters typically measured during programmed cell death (PCD), such as cellular morphological changes in nuclei and cytoplasm, endoreduplication, DNA fragmentation, and the participation of nucleases and caspase-like proteases in nucleus dismantling, were evaluated; morphological changes in cytoplasm included subcellular aspects related to starch accumulation. This study proved that, following fertilization, the perisperm of quinoa simultaneously accumulates storage reserves and degenerates, both processes mediated by a programme of developmentally controlled cell death. The novel findings regarding perisperm development provide a starting point for further research in the Amaranthaceae genera, such as comparing seeds with and without perisperm, and specifying phylogeny and evolution within this taxon. Wherever possible and appropriate, differences between quinoa perisperm and grass starchy endosperm—a morphologically and functionally similar, although genetically different tissue—were highlighted and discussed.
Key words: Amaranthaceae, basal body, caspase-like protease, DNA fragmentation, endoreduplication, nucleases, quinoa, perisperm, programmed cell death (PCD), starch accumulation, TUNEL.
Introduction
The nucellus is the first structure to develop when an ovule is initiated, and, in most species, reaches its maximal development during anthesis (Werker, 1997). The nucellus provides the cellular initial for the differentiation of the megasporocyte, which undergoes meiosis to produce megaspores, one of which originates the seven-celled megagametophyte or embryo sac via mitosis (Webb and Gunning, 1990; Mansfield et al., 1991). In most angiosperms, the nucellus is partially or completely consumed during megagametophyte development but, in some Zingiberales (Monocotyledons) and Caryophyllales (Dicotyledons) genera, the nucellus persists after fertilization, becoming the main nutritive tissue of the seed, namely the perisperm. In Caryophyllales, the presence of a perisperm has been reported in genera of Amaranthaceae, Saururaceae, Caryophyllaceae, Aizoaceae, Nyctaginaceae, Phytolaccaceae, and Portulacaceae (Werker, 1997).
In Amaranthaceae sensu lato, the embryo sac acquires a horseshoe shape during development, absorbing the bordering nucellar cells (Pal et al., 1990). The ovule is amphitropous, according to Boesewinkel and Bouman (1984), because most of the nucellar tissue remains intact during seed development, forming a basal body (cf. Bouman, 1984) or perisperm. At the end of development, the perisperm accumulates storage reserves (Joshi and Kajale, 1937; Coimbra and Salerna, 1994; Prego et al., 1998).
Five subfamilies of Amaranthaceae have been identified: Amaranthoideae, Gomphrenoideae, Chenopodioideae, Salicornioideae, and Salsoloideae (APG III, 2009), although several do not seem to constitute natural groups. In this regard, Meyer (1829) distinguishes between species with spirally coiled embryos, which usually lack perisperms, and those with peripheral embryos and perisperms, which he names Spirolobeae and Cyclolobeae, respectively. Although the exact status of Spirolobeae and Cyclolobeae differs depending on the author, Spirolobeae is virtually equivalent to Salsoloideae (Salsoloideae+Suaeoidea) while Cyclolobeae encompasses Amaranthoideae, Chenopodioideae, and Gomphrenoideae (sensu APG III, 2009). To date, the destination of the nucellus in Spirolobeae remains unknown, a topic worth studying as it would help explain the phylogenetic relationship between Cyclolobeae and Spirolobeae.
In Chenopodium quinoa (Prego et al., 1998) and Ama ranthus hypocondriacus (Coimbra and Salerna, 1994), storage reserves in seeds are highly compartmentalized: protein, mineral nutrient, and lipid reserves are mainly located in the reduced micropylar endosperm and in the embryo, while carbohydrate reserves are found in the perisperm. The perisperm consists of notably large, uniform, non-living, thin-walled cells completely full of starch grains. Intriguingly, the perisperm tissue morphologically and functionally resembles grass starchy endosperm which is also dead at maturity. In view of the similarities between both tissues, it is tempting to speculate whether similar mechanisms have been used during evolution, accounting for part of the diversity found among flowering plant seed storage tissues.
There are no comprehensive studies on the subcellular processes associated with the simultaneous occurrence of cellular death and starch accumulation in any storage tissue. However, cellular death in maize, wheat, and barley endosperm has been studied, and several common programmed cell death (PCD) parameters measured, such as cellular morphological changes in nuclei and cytoplasm, endoreduplication, DNA fragmentation, and the participation of caspase-like proteases and nucleases in nucleus dismantling (Kowles et al., 1990; Domínguez and Cejudo, 1999, 2006; Chang and Gallie, 1997; Young et al., 1997; Young and Gallie, 1999, 2000; Gunawardena et al., 2001; Giuliani et al., 2002; Borén et al., 2006; Ho, 2006; Spanò et al., 2007; Radchuk et al., 2011; Domínguez et al., 2012; Sabelli and Larkins, 2012, among others).
Two simultaneous programmes occur in grass starchy endosperm and quinoa perisperm, namely cellular death and starch synthesis. The questions are whether development is comparable in these tissues, and if PCD events are involved in the development of quinoa perisperm, as they are in grass endosperm. To answer these questions, both morphological and biochemical parameters normally measured during PCD were here evaluated and compared with the same parameters previously studied in grass starchy endosperm. This study is also part of a comprehensive treatment on seeds of the Amaranthaceae family.
Materials and methods
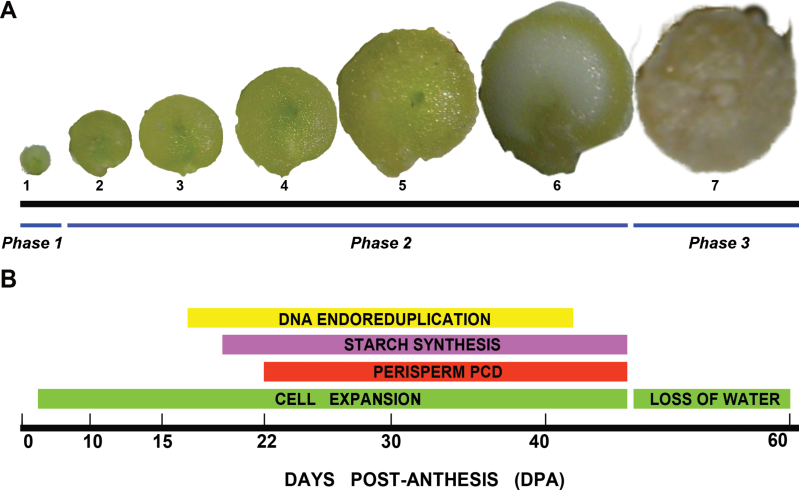
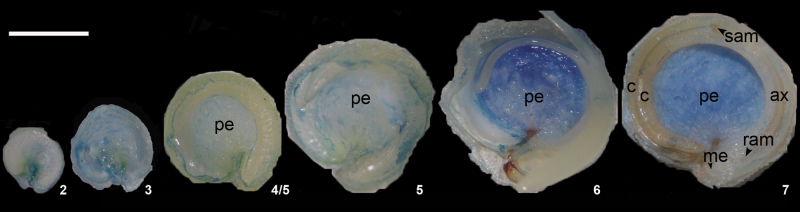
Quinoa (Chenopodium quinoa Willd.) cv. UDC plants were grown in a chamber under controlled conditions 16h light/8h dark cycles at 25 °C. Ovaries at anthesis (stage 1, Fig. 1) and grains at six different seed developmental stages (stages 2–7, Fig. 1) were collected. All materials not immediately used were stored at –80 °C. During the different procedures, materials were kept in an ice bath.
Fig. 1.
Quinoa grain development. (A) Photographs of grains at seven different developmental stages, from an ovary at anthesis to a mature grain. (B) Timeline of the main events of seed development, from anthesis to maturity. (This figure is available in colour at JXB online.)
Experiments reported here were repeated at least three times; the results were comparable across experiments, unless otherwise stated.
Histological analysis
Quinoa ovaries and grains were fixed in a mixture of 2% paraformaldehyde (PFA) and 0.5% glutaraldehyde in 0.1M phosphate-buffered saline (PBS), pH 7.2, for 4h (2h under vacuum in fixation solution and 2h at 4 °C). Samples were embedded with historesin according to the manufacter′s instructions (Leica Microsystems, Wetzlar, Germany). Sections of 3 μm were cut with a tungsten knife using a Leica 2155 microtome, mounted on glass slides, and stained with 0.5% toluidine blue O (Sigma-Aldrich) in aqueous solution.
Subcellular analysis
Samples were fixed for 3h at 4 °C using a mixture of 2% PFA and 0.5% glutaraldehyde in 0.1M PBS, pH 7.2. Later they were post-fixed in 1% OsO4 in the same buffer for 90min, dehydrated in a graded ethanol series followed by an ethanol–acetone series, and embedded in Spurr’s resin (Sigma-Aldrich, St Louis, MO, USA). Ultra-thin sections were mounted on grids coated with Formvar (Polyscience, Inc., Warrington, PA, USA), stained in uranyl acetate followed by lead citrate from EMS (Hatfield, PA, USA), and examined in a Zeiss M109 turbo (Zeiss, Wiesbaden, Germany) transmission electron microscope operating at an accelerating voltage of 90kV.
DNA isolation and electrophoresis
Genomic DNA was isolated from grains (stages 3–7) using the DNeasy plant mini kit (Qiagen, Germany). DNA was quantified in a NanoDrop spectrophotometer (Thermo Scientific NanoDrop 2000c). A 2 μg aliquot of DNA from each sample was separated, as described previously by Radchuk et al. (2011) with minor modifications, on a 0.8% (w/v) agarose gel at 40V for 7h and stained with SYBR® Safe DNA Gel Stain (Invitrogen, Carlsbad, CA, USA).
TUNEL assay
DNA strand breaks were detected by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labelling (TUNEL) using the in situ Cell Death Detection Kit, TMR red (Roche, Basel, Switzerland). Samples were fixed in 4% PFA in 0.1M PBS (pH 7.2) (2h under vacuum in fixation solution and 2h at 4 °C), dehydrated in an acetone series, and embedded in Technovit 8100 (Kulzerand Co., Germany). The resin was polymerized at 4 °C. Semi-thin sections (1–2 μm thickness) were obtained. TUNEL labelling was performed according to the manufacter’s instructions. Briefly, sections were permeabilized with 20 μg ml–1 proteinase K for 20min at room temperature and washed four times with PBS. The labelling reaction was performed at 37 °C in a dark, humid chamber for 1h. A negative control was included in each experiment by omitting TdT from the reaction mixture. As a positive control, permeabilized sections were incubated with DNase I (3U ml–1) for 15min before the TUNEL assay. Counterstaining was done with 0.02mg ml–1 4ʹ,6-diamidino-2-phenylindole (DAPI) staining. The sections were mounted using the Citifluor™ mounting medium (EMS).
Microscopic settings
Images for histological analysis and TUNEL were obtained by light microscopy and epifluorescence with an Axioskope 2 microscope (Carl Zeiss, Jena, Germany). The following filters were used to examine the fluorescent samples: DAPI filter (excitation 340–390nm, emission 420–470nm) and rhodamine filter (excitation 540–552nm, emission 575–640nm). Images were captured with a Cannon EOS 1000 D camera (Tokyo, Japan) and analysed using the AxioVision 4.8.2 software package (Carl Zeiss, Jena, Germany).
Images of Fig. 8B were acquired in an Olympus FV300 confocal microscope with FluoView 3.3 software (Olympus, Tokyo, Japan) and differential interference contrast (DIC). For excitation, 543 lasers for rhodamine were used, and the emission filters 585–615nm.
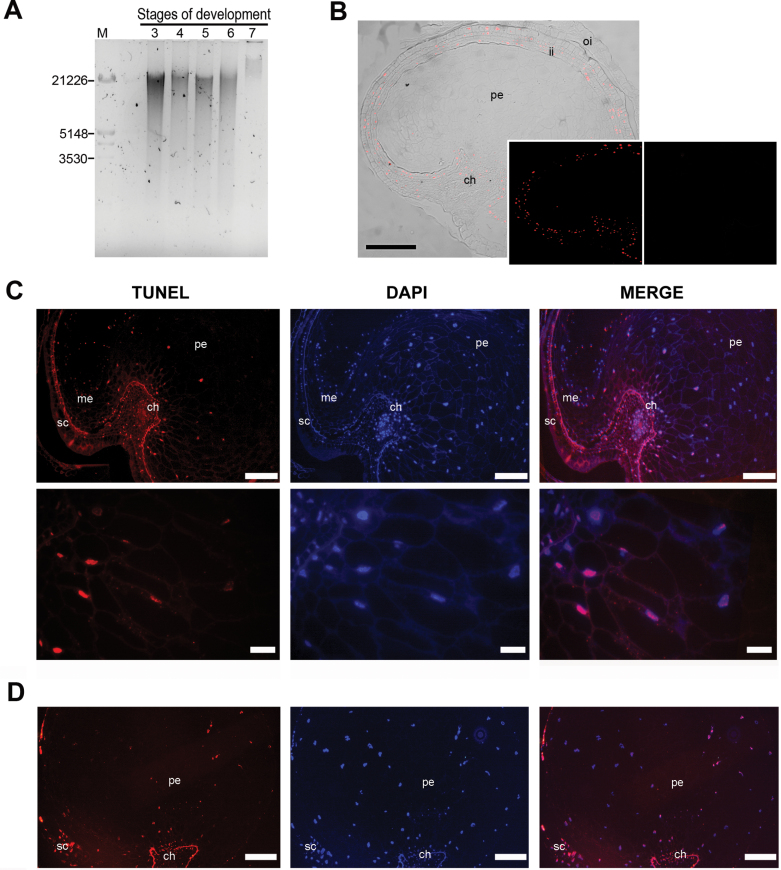
Fig. 8.
(A) DNA fragmentation in the seed during development; the DNA was isolated from the developing seeds harvested at stages 3–7. (B–D) Localization of nuclei with fragmented DNA in quinoa seeds is represented by red fluorescence: (B) TUNEL-positive nuclei in cells of the integuments (stage 2) as seen by DIC and fluorescence. Insets: left panel, fluorescence alone; right panel, negative control. (C, D) Stages 4 and 6, respectively, as revealed by DAPI TUNEL-positive nuclei were detected in some perisperm cells. Note that some nuclei of the endosperm were also TUNEL positive. In each case, the figure is a representative result of observation of at least 15 whole-mounts of quinoa seeds at each stage. Abbreviations: ch, chalaza; ii, inner integument; oi, outer integument; me, micropylar endosperm; pe, perisperm. Scale bars 100 μm; (B) 200 μm; detail 40 μm.
Evans blue staining
Evans blue is an acidic dye which has the inverse staining properties of a vital stain when determining the survival of plant cells. Cells with intact semi-permeable membranes exclude the dye, whereas the dye penetrates and stains dead cells. Therefore, Evans blue is considered a mortal stain rather than a vital stain (Crippen and Perrier, 1974).
Evans blue staining was used to reveal quinoa perisperm cells that had lost their cell membrane integrity during development. Longitudinal medial sections of grains at each stage were placed in a solution of Evans blue (0.1% in H2O) for 2min (Young and Gallie, 1999). Stained sections were washed with water for 30min, mounted, and analysed by a Zeiss Stemi 2000C stereo microscope equipped with a camera EOS 1000 D camera (Tokyo, Japan).
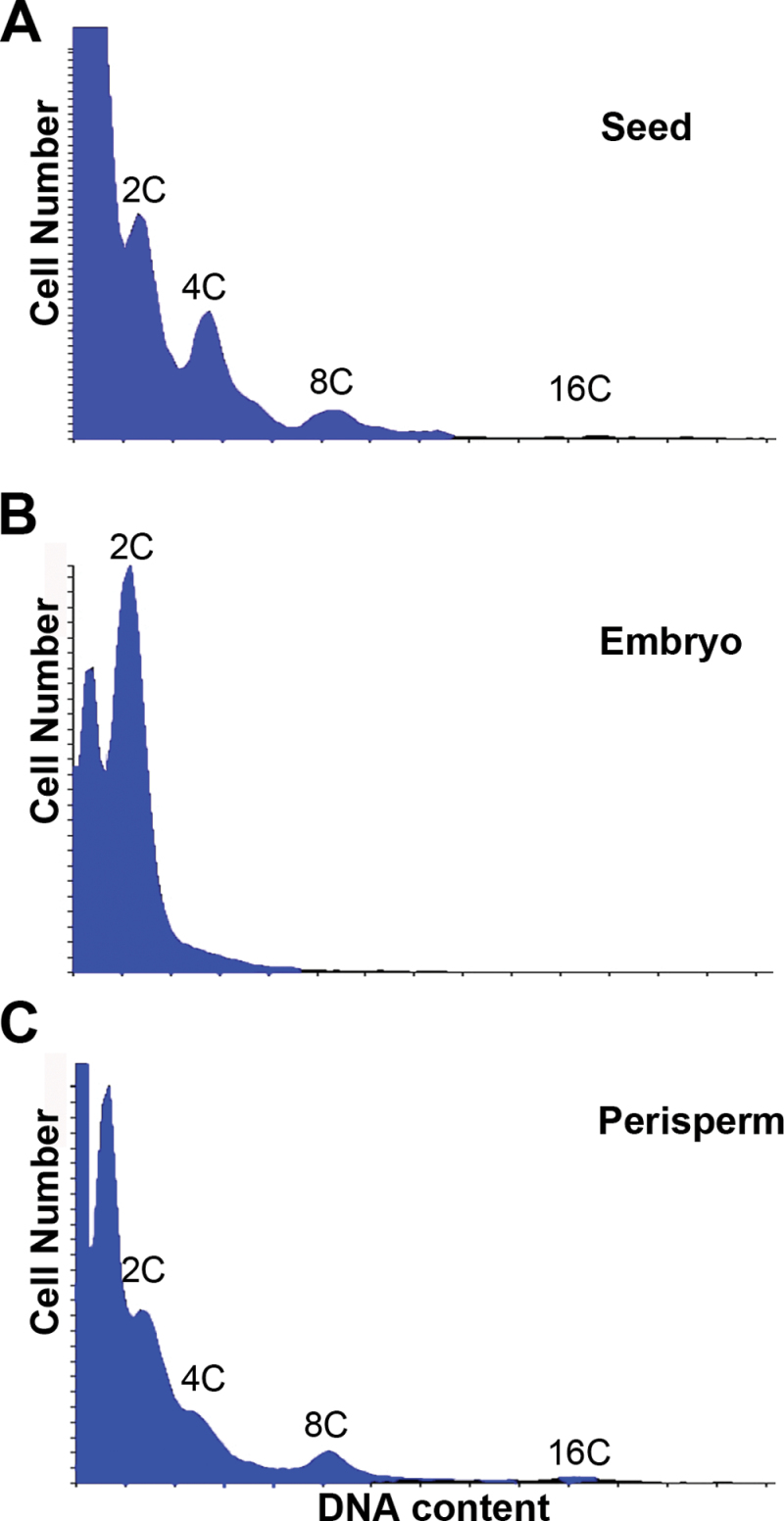
Flow cytometry analysis
Embryos, perisperms, and seeds from stage 6 were dissected and chopped with Otto I extraction buffer and the suspension containing nuclei was passed through a 50 μm filter. Then, the nuclei in the filtrate suspension were stained with 2 vols of Otto II staining solution containing DAPI (Otto, 1990). After shaking the solution gently, samples were analysed with a flow cytometer (CyFlowPloidyAnalyser, Partec). Relative DNA content was estimated according to the prominent peak in each measurement.
In-gel nuclease activity assay
DNase activity was detected according to the method defined by Thelen and Northcote (1989) with slight modifications. The samples were ground in liquid nitrogen and homogenized in 100mM HEPES (pH 7.5), 0.2% CHAPS, 10% sucrose, 0.1% Triton, and 10mM dithiothreitol (DTT). The cell extracts were centrifuged for 10min at 14 000 g, 4 °C, and the supernatant was used for the assay. The protein concentrations were determined as described by Bradford (1976) using a protein assay kit (Bio-Rad Laboratories). Protein extracts from stages 3–7 were fractionated by SDS–PAGE as previously reported by Domínguez et al. (2004), with modifications, containing 0.3mg ml–1 herring sperm DNA at 4 °C and 20 mA per plate.
Equal amounts of protein (50 μg) were incubated for 10min at 45 °C in buffer [0.125M TRIS pH 6.8, 10% (v/v) glycerol, 2% (w/v) SDS, 0.01% (w/v) bromophenol blue]. After electrophoresis, the gels were soaked in a buffer containing 25% 2-propanol and 1mM EDTA for 15min to remove SDS and residual divalent cations, and rinsed twice with water. Subsequently, the gels were washed twice for 5min and incubated overnight in 25mM sodium acetate–acetic acid buffer [pH 5.5, containing 1mM ZnSO4, 0.2mM DTT, and 1% (v/v) Triton X-100] or 10mM TRIS-HCl neutral buffer [pH 8.0, containing 10mM MgCl2, 10mM CaCl2, 0.2mM DTT, and 1% (v/v) Triton X-100] at 37 °C. After incubations, the gels were washed for 5min in cold stop buffer (10mM TRIS-HCl pH 8.0, 1mM EDTA). False nucleolytic activities associated with DNA-binding proteins were discarded by incubating the gels in 1% (w/v) SDS for 2h at room temperature and then washed in water for 10min. The gels were stained with 0.01mg ml–1 ethidium bromide to reveal the position of the nucleases and photographed using G:Box GeneSnap software from Syngene. Band intensity was analysed using Gel-Pro Analyzer Software (Media Cybernetics, Inc., Bethesda, MD, USA).
Caspase like-proteolytic (CLP) activity assay
Developing quinoa grains from stages 3 to 6 were manually dissected. Due to their small size. it was not possible to isolate the embryo and perisperm, except from stage 6. Protein extracts were obtained as described above and the reaction was performed under the following assay buffer conditions: 50mM HEPES (pH 7.5), 0.1% CHAPS, 1mM CaCl2, 1mM MgSO4, and 5mM DTT. Caspase-like proteolytic activity was measured as described by Bozhkov et al. (2004) in a FL600 fluorometer (BioTek, VI, USA; excitation 360/40; emission 460/40, sensitivity 75). The blank fluorescence readings (assay buffer and peptide alone) were subtracted. The following fluorogenic substrates were used: AcVEID-AMC, AcYVAD-AMC, and AcLEHD-AMC substrates all from Peptides International, KY, USA. All substrates were dissolved in dimethylsulphoxide (DMSO) and diluted in water to the final concentration.
For inhibitor assay, sample extracts were pre-incubated with proteinase inhibitors at 30 °C for 20min in the assay buffer before addition of AcVEID-AMC. The following concentrations of proteinase inhibitors were used: 20 μM VEID-cho, 20 μM YVAD-cho, 20 μM LEHD-cho, 20 μM lactacystin, from Peptides International; and 20 μM E-64, 20 μM calpain inhibitor 1, 10 μM pepstatin, and 250 μM phenylmethylsulphonyl fluoride (PMSF) from Roche (Mannheim, Germany). Extracts were then incubated with AcVEID-AMC.
Statistical analysis
Statistical tests were applied using GraphPad Prism version 6.00 for Windows (GraphPad Software, La Jolla, CA, USA, www.graphpad.com). For the study of caspases, data were analysed by analysis of variance (one- or two-way ANOVA), followed by post-hoc Dunnett’s or Tukey test for multiple comparisons among proteolytic activities. In all graphs, results represent the mean value of three independent experiments ±SEM. Statistical significance was defined as ***P=0.0001, **P=0.001, and *P=0.01.
Results
Perisperm development
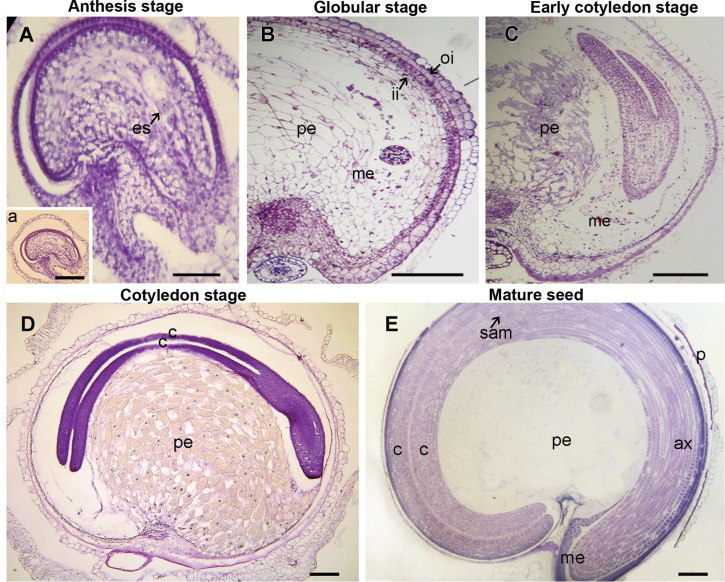
Quinoa perisperm derives from the nucellus; hence, it begins to develop as soon as the ovule does. After megasporogenesis, the growing embryo sac gradually consumes a very thin and narrow band of nucellar cells proximal to the periphery of the ovule (Fig. 2A). The remaining nucellus persists during embryo development (Fig. 2B–D), forming a basal body or perisperm, namely the main storage reserve of the seed. During development, the perisperm tissue increases around five times its size at anthesis (Fig. 2E).
Fig. 2.
Ovule at anthesis (A) and seeds in four different developmental stages (B–E). (A) Stage 1 (0 DPA); (B) stage 3 (around 17 DPA); (C) stage 4 (around 25 DPA); (D) stage 5 (around 32 DPA); (E) stage 7 (around 58–60 DPA). In each case, the figure is a representative result of observation of at least 30 whole-mounts of quinoa seeds at each stage. Abbreviations: ax, hypocotyl–radicle axis; c, cotyledon; es, embryo sac; ii, inner integument; me, micropylar endosperm; pe, perisperm; oi,outer integument; sam, shoot apical meristem. In A, arrow indicates embryo sac. Scales bars 50 μm (A), 200 μm (B–E and inset of A). (This figure is available in colour at JXB online.)
Three major developmental phases could be distinguished: phase 1, early development of the nucellus, including mitotic activity; the last stage of mitosis takes place before anthesis, leaving the tissue with its final cell number and configuration; phase 2, cellular differentiation which can be broken down into the more or less simultaneous processes of cellular expansion, endoreduplication, accumulation of starch reserves, and PCD (Fig. 2A–D); and phase 3, the maturation phase (Fig. 2E), which comprises the shutdown of biosynthetic processes, desiccation induction, and quiescence. To study the three phases, seven stages were selected (Fig. 1).
Endoreduplication took place between stages 3 and 6; the nucleus changed from circular to lobe-shaped and grew considerably in size; the chromatin was found to be partially condensed in the lobes of the nucleus, but scattered in the middle; and the nucleoplasm became more translucent, although nucleoli retained their original shape (Fig. 3A–C). The DNA content peaked mostly at 8C (Fig. 4) but some nuclei reached 16C and 32C. The progression of the nuclear changes included, first, the complete dispersion of the chromatin (Fig. 3C), followed by the disappearance of the nucleolus and nuclear membrane (Fig. 3D).
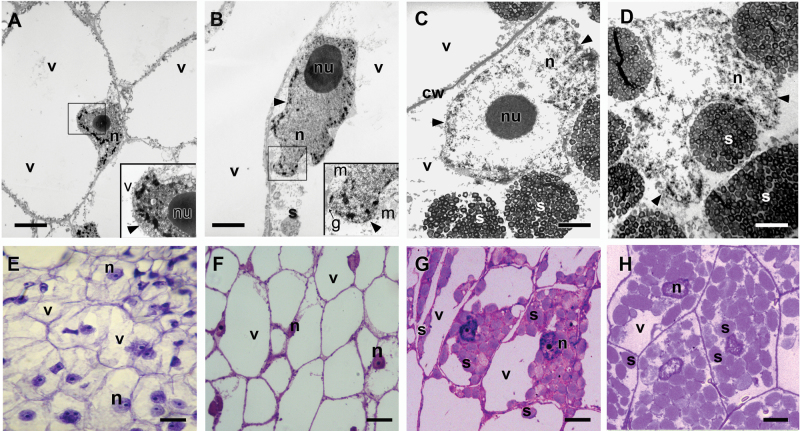
Fig. 3.
Quinoa perisperm tissue analysed by transmission electron microscopy (A–D) and light microscopy (E–H). (A–D) Changes in shape, size, and chromatin state of nuclei during seed development: (A) stage 1; (B) stage 3; (C) stage 5; (D) stage 6. (E–H) Sections of perisperm tissue corresponding to stages 1, 3, 5, and 6, respectively. Progressive changes in nuclei (disappearance of nucleolus, dispersion of chromatin) and vacuoles (disappearance), and an increase in starch accumulation (which continued until completely filling the cell lumen, occupying the vacuole space) can be observed. In each case, the figure is a representative result of observation of at least 30 whole-mounts of quinoa seeds at each stage. Abbreviations: g, Golgi apparatus; m, mitochondrion; n, nucleus; nu, nucleolus; s, compound starch grain; v, vacuole. The arrowhead indicates the nuclear membrane; Scale bars 0.5 μm (A, B); 2.5 μm (C, D); 30 μm (E–H). (This figure is available in colour at JXB online.)
Fig. 4.
Flow cytometric measurement of mean nuclear ploidy at stage 6 of quinoa seed development. Histograms show the C DNA levels in: (A) complete seeds; (B) isolated embryos; and (C) isolated perisperms. The x-axes are the log of the fluorescence intensity and the y-axes correspond to the relative frequency of a given intensity. The C value indicates the number of genome copies. Histograms revealed endopolyploidy in perisperm, but not in embryo tissue. The 2C DNA level corresponds to the diploid state of the genome and the 4C peaks could indicate the capacity of cells to enter mitosis. However, in the perisperm, cell divisions cease at anthesis, thus peaks of 4C and 8C are indicators of endoreduplication. In embryo, cell division finishes before stage 6. (This figure is available in colour at JXB online.)
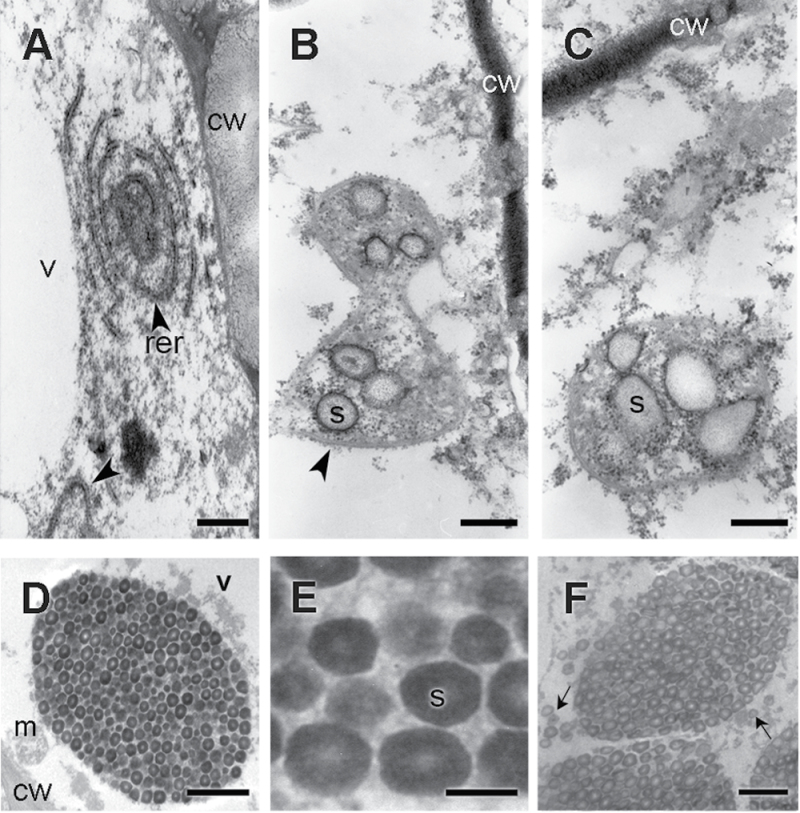
During cellular differentiation, cells expanded to form a large central vacuole, dramatically growing until reaching around 4–5 times their size at anthesis (Fig. 3E–H).The cells reached their final size at ~35–40 DPA (days post-anthesis). Cellular expansion was followed by starch accumulation, which continued until completely filling the cell lumen (Figs 3G, H, 5, 6). At early stages, the cytoplasm appeared rich in mitochondria, Golgi, and cisternae of the rough endoplasmic reticulum (RER). Frequently, cisternae formed concentric circles defining a denser central area (Fig. 5A). From the beginning, the grains of starch accumulated, forming compound structures, in the dense area bordered by RER cisternae (Fig. 5B, C). Starch deposition continued, reducing the central vacuole until it disappeared (Figs 3G, H, 5D–F, 6). The grain-filling period continued until the death of the perisperm was complete (Figs 2E, 5F). At this point, the cell lumens were completely full of starch grains. The PCD of the perisperm marked the end of its development.
Fig. 5.
Quinoa perisperm cells analysed by transmission electron microscopy during development. (A) Cytoplasm before starch deposition (stage 1, detail of Fig. 3A); (B, C) cytoplasm at the beginning of starch deposition (late stage 3, detail of Fig. 3B); (D) stage 5 (detail of Fig. 3C); (E) detail of the starch grain shown in D; (F) cell lumen of a perisperm at stage 6, almost completely full of starch (detail of Fig. 3D). Abbreviations: cw, cell wall; m, mitochondrion; rer, rough endoplasmic reticulum; v, vacuole. In B, the arrowhead indicates the endoplasmic reticulum; in F, arrows indicate single starch grains occupying the spaces among the compound starch grains. Scale bars 400nm (A–C); 1 μm (D); 500nm (E); 2 μm (F).
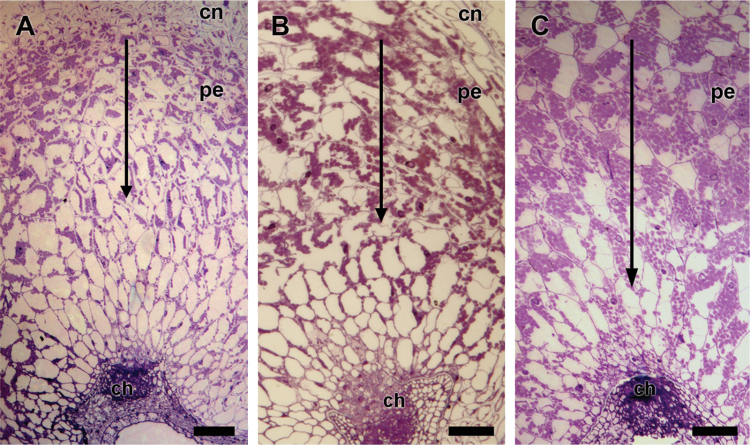
Fig. 6.
Sections of the quinoa perisperm at: (A) stage 4; (B) stage 5; and (C) stage 6, revealed an apical–basal starch deposition pattern (arrow). In each case, the figure is a representative result of observation of at least 30 whole-mounts of quinoa seeds at each stage. Abbreviations: ch, chalaza. Scale bare 100 μm. (This figure is available in colour at JXB online.)
Starch deposition began in the apical perisperm prior to stage 4, continued towards the centre, and then towards the chalazal region (Fig. 6A–C). As detected by Evans blue (Fig. 7), nucellar cell death initiates at early stages near the chalazal region and moved forward to the peripheral region (stages 2–3, Fig 7). Cell death occurred stochastically throughout the whole perisperm during the next developmental stages. Staining of the perisperm started at stage 4 (25 DPA) until stage 7, when the cells were no longer capable of excluding the dye, indicating their death.
Fig. 7.
Evans blue staining in quinoa grains revealed PCD in cells of the chalaza and nucellus (stages 2–3) and perisperm (stages 4–7). Abbreviations: ax, hypocotyl–radicle axis; c, cotyledon; me, micropylar endosperm; pe, perisperm; ram, root apical meristem; sam, shoot apical meristem. Scale bar 1mm. (This figure is available in colour at JXB online.)
Isolation of DNA and electrophoresis
DNA fragmentation is a typical outcome during cell death in many systems. Total DNA was isolated from grains from stage 3 to 7 and analysed by DNA gel electrophoresis. The results were: (i) a smear for DNA of all the stages analysed, except stage 7; and (ii) at stage 3, degradation was minor compared with the following stages 4–6. This pattern indicated DNA degradation throughout seed development.
In situ detection of DNA fragmentation by TUNEL
During seed development, cell death occurs in different seed tissues according to a controlled temporal and specific pattern. Nuclear degradation was detected in situ by TUNEL assay, accompanying the rapid expansion of the different seed tissues.
Figure 8B shows a representative TUNEL assay images of stage 2 where the first detectable events of DNA fragmentation began. At this stage, only nuclei of the integument cells and a few from the chalazal region presented TUNEL-positive labelling, whereas the rest of the seed tissues were TUNEL negative. At stage 3, only TUNEL-positive nuclei were visible in integuments and chalaza.
At later developmental stages (stages 4–6), a TUNEL labelling could be observed in a few nuclei of the micropylar endosperm and in nuclei of the perisperm (Fig. 8C, D). Likewise the integuments and the chalazal region had a weak signal, consistent with their previous initiation of PCD.
Control treatments were conducted for each set of slides. TUNEL labelling was absent when TdT enzyme was omitted (Fig.8B, inset, right panel). In positive controls previously treated with DNase, all the nuclei were labelled, rendering the procedure valid.
Detection of nuclease activities during seed development
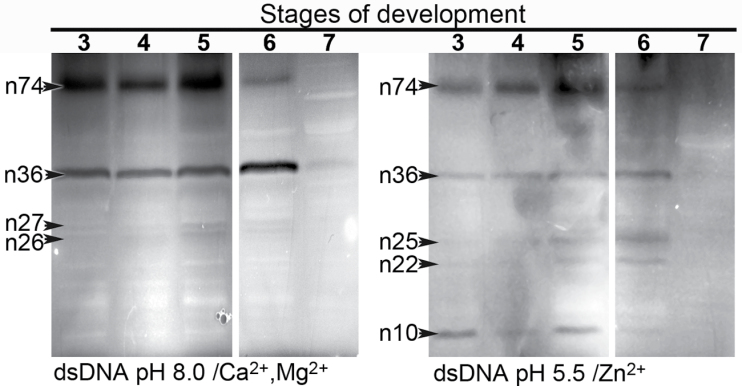
DNA-SDS–PAGE was used to identify the activities of the DNases expressed in the perisperm during development. These enzymes represent several classes based on their pH and ion dependency and are associated with nuclear DNA fragmentation. As shown in Fig. 9, different patterns of Ca2+/Mg2+- or Zn2+-dependent nucleases were specific for different stages, as follows: (i) DNase activity gel staining in the presence of Ca2+ and Mg2+ at pH 8.0 revealed four bands with masses of 74, 36, 27, and 26kDa. At stage 6, when dispersion of chromatin is followed by the disappearance of the nucleolus and nuclear membrane (Fig. 3) the activities of n74 and n36 were opposite: while n74 decreased its activity ~15 times, disappearing at stage 7, n36 duplicated its activity. The activity of n27 and n26 was slightly higher towards stages 5 and 6 (Fig. 9). (ii) Five Zn2+-specific nucleases which digested double-stranded DNA were identified (n74, n36, n25, n22, and n10). At earlier stages, n10 and n74 were clearly active while n25 and n22 increased their activity during stages 5 and 6. It is worth noting that n74 and n36 seem to be activated by both Ca2+ and Zn2+.
Fig. 9.
Identification of nucleases in quinoa perisperm tissue undergoing programmed cell death. Total proteins (50 μg of protein) were extracted from different stages (3–7) of quinoa grain development and were analysed by in-gel nuclease assay at the indicated pH. Nucleases active in the presence of Mg2+ and Ca2+ (left panel), and of Zn2+ (right panel). Double-stranded DNA was used as a substrate.
Detection of caspase- like proteolytic (CLP) activities during seed development
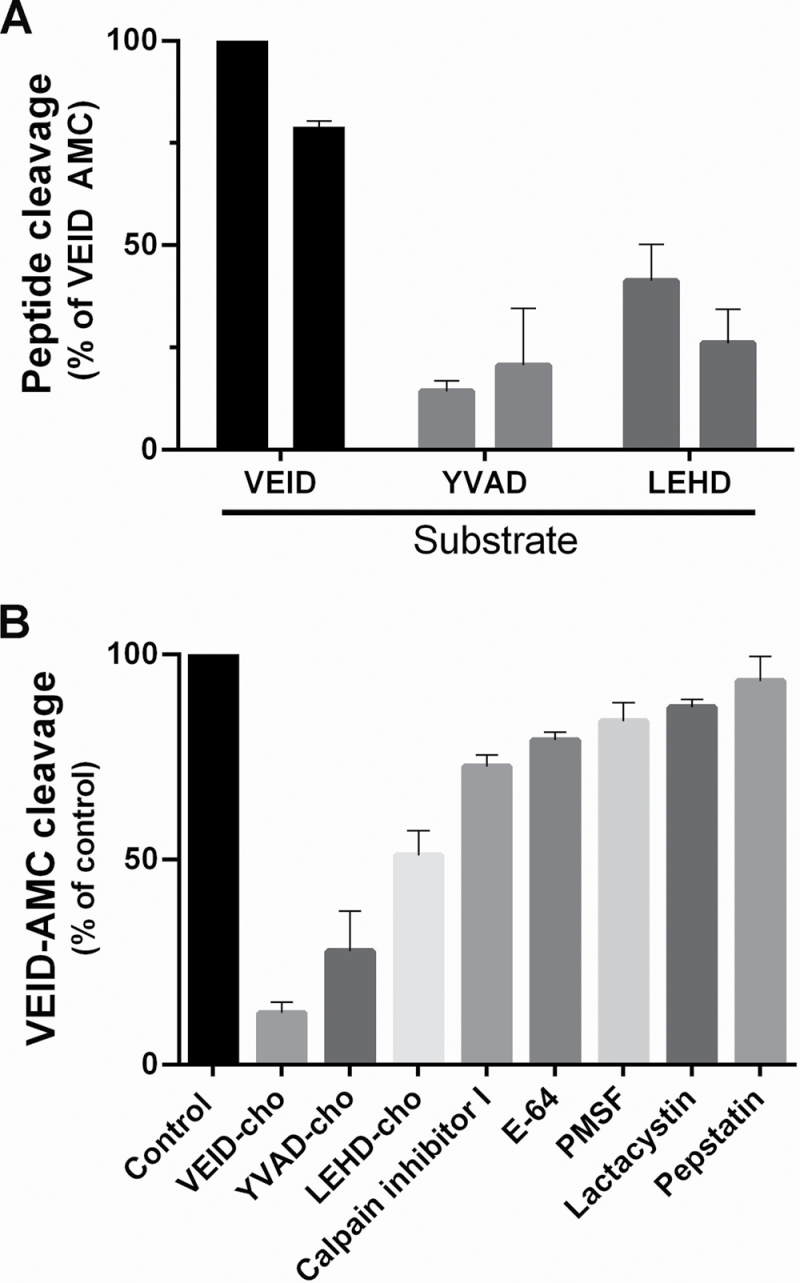
The enzymatic activity of three caspase-like proteases (1, 6, and 9) during perisperm development was analysed by measuring their cleavage of tetrapeptide-AMC substrates. As Fig. 10A illustrates, caspase 6-like proteolytic activity (Ac-VEID substrate) was significantly different from caspase 1- and 9-like proteolytic activities at stages 4 and 5 (***P < 0.0001). YVADase (caspase 1) and LEHDase (caspase 9) activity was not modified when the respective inhibitor had been added, suggesting no specific cleavage related to CLP activity. There was no CLP activity at stages 3 or 6 (data not shown).
Fig. 10.
Caspase-like protease activity during quinoa perisperm development. (A) In vitro cleavage of peptide substrates specific to different mammalian caspases by cell extracts prepared at stages 4 and 5. The graph represents mean rates (SEM) of peptide cleavage relative to the highest activity, VEID-AMC (%). The assay was performed under standard conditions (see the Materials and methods). (B) Effects of protease inhibitors on quinoa perisperm VEIDase like-protease activity in vitro. Cell extracts were incubated for 20min with a range of protease inhibitors in the presence of substrate VEID-AMC, and their ability to inhibit VEIDase activity was tested in stage 4. The data are expressed as a percentage of mean VEIDase activity relative to control (no inhibitor; SEM, n=3).
The highest caspase 6-like proteolytic activity was detected at stage 4, namely at the beginning of starch deposition, when the vacuole occupies almost all the cell lumen and the first detectable events of nuclear fragmentation were evident (Fig. 6A). To characterize further the presence of VEIdase activity, the cell extracts at this stage were exposed to different protease inhibitors; only Ac-VEID-CHO, a caspase 6 inhibitor, significantly reduced the VEIDase activity by 89% (***P < 0.0001). The statistical analysis (Fig. 10B) showed that VEID activity was also 50% and 30% inhibited (**P < 0.001) by Ac-LEHD-CHO (caspase 9 inhibitor) and by Ac-YVAD-CHO (caspase 1 inhibitor), respectively. The different proteases inhibitors, E-64, lactacystin, pepstatin, calpain inhibitor I, and PMSF, did not significantly differ from the control, suggesting that the reduction in the cleavage of the Ac-VEID substrate was due to a true caspase-like proteolytic activity during quinoa grain development.
Discussion
The species C. quinoa belongs to Meyer’s (1829) Cyclolobeae group. To date, no information is available as to the destination of the nucellus in Spirolobeae, a topic worth studying as it would help to explain the phylogenetic relationship between Cyclolobeae and Spirolobeae. This is not a short-term goal, but, when achieved, it will improve our understanding of the phylogeny of the Amaranthaceae family, as well as the evolution of nucellar cell death towards the creation of new seed forms.
A comparison was drawn between quinoa perisperm and grass starchy endosperm, two tissues similar in terms of general characteristics and function, although genetically different. In both tissues, death and dismantling are temporally separated, a separation that can take years depending on when the seeds germinate (Young et al., 1997; Young and Gallie, 1999, 2000). In both cases, cell death occurs during seed formation and is not due to a complete process of autophagy since the cell remains until germination, when the starch reserves are mobilized and the dead tissue is finally dismantled.
This type of cellular death does not seem to be autophagic in the strict sense of the term (van Doorn and Woltering, 2005). In a more recent classification, van Doorn et al. (2011) recognize two major classes of cell death occurring in plant tissues: vacuolar cell death and necrosis; however, in grass starchy endosperm, according to van Doorn et al. (2011), and in quinoa perisperm (present study) PCD does not strictly fall into those two categories. This is a topic worth pursuing in the future.
During quinoa seed development, different cell death types can be recognized depending on the seed tissue: (i) one variety of cellular death is that which occurs before anthesis in nucellar cells bordering the growing embryo sac, as well as in endosperm cells located on either side of the growing embryo. In both cases, cells collapse immediately following death; this type of death has been reported in cotton (Jensen, 1975), barley (Norstog, 1974; Chen and Foolad, 1997), Oenothera biennis (de Halac, 1980), and Ricinus communis (Greenwood et al., 2005) nucellus and is associated with proteolytic and nucleolytic activities (Domínguez and Cejudo, 1998, 2006; Domínguez et al., 2001). In all these species, cellular death removes cells that will not be necessary later, during germination. (ii) Another type of cell death detected during quinoa seed development is that occurring in the micropylar endosperm during germination, which is also characteristic of the aleurone layer of grass endosperms, and, in general, of endosperms that store lipids and proteins. In this type, cells survive until germination, their contents become mobilized, and they die by autophagy; that is, they direct their own deterioration by secreting hydrolases that are released from the vacuole (Greenwood et al., 2005; DeBono and Greenwood, 2006). (iii) A third type of cell death, observed in the lasting layers of quinoa integuments, is also characteristic of the quinoa perisperm and grass starchy endosperm: in these tissues, death and dismantling are temporally divorced, a separation that can take years, depending on when the seeds germinate. As mentioned above, cell death occurs during seed formation and is not due to a complete process of autophagy since the cell remains intact until germination, at which time the starch reserves are mobilized and the dead tissue is finally dismantled.
During differentiation in quinoa perisperm, cells expanded to form a large central vacuole, dramatically growing until reaching at least five times their size during anthesis (Fig. 2). Endopolyploidy is generally considered the most common way to increase nuclear DNA content in plants (D’Amato, 1984). Furthermore, a clear relationship has been established between endopolyploidy and cell size (Melaragno, 1993). In quinoa seeds, parallel to perisperm cellular growth by expansion, the nuclei began a process of endoreduplication accompanied by a change from circular to lobe-shaped and a considerable increase in size (Figs 3, 4). As in grass starchy endosperm, endoreduplication was strongly associated with this increase in cell size, and with the high metabolic rates associated with starch accumulation. In fact, high levels of ploidy have also been detected in maize starchy endosperm (Kowles et al., 1990) and also in the cotyledonary tissues of common beans (Bino et al., 1993).
Cells of quinoa perisperm and grass starchy endosperm undergo a process of PCD while storing starch reserves. In quinoa perisperm, PCD continued until all cytoplasm, nuclei, and vacuoles had disappeared and all cell lumens had been filled with starch. The RER participated in the synthesis of compound starch grains through cisternae that in the cytoplasm partially bordered circular spaces of denser content where the single grains were deposited (Fig. 5A–C). During this stage, mitochondria and RER cisternae were widespread. To the authors’ knowledge, this is the first report on subcellular aspects associated with the synthesis of compound starch grains in storage seed tissues. This is a topic worth investigating in the future.
In quinoa perisperm, starch deposition followed an established pattern, initiating in the periphery near the developing embryo (i.e. distally to the chalaza) and continuing through the centre towards the chalaza (Fig. 6). A pattern of starch deposition is recognized in maize starchy endosperm (Young and Gallie, 2000) but not in wheat starchy endosperm (Young and Gallie, 1999).
In contrast to starch deposition, the onset of PCD in quinoa perisperm was not confined to a specific region. Likewise, PCD progression during subsequent development appeared to be a random process that continued until the entire endosperm was affected, by stage 7 (Fig. 7). Therefore, the PCD pattern was similar to the wheat pattern and different from that reported in maize (Young and Gallie, 1999, 2000).
According to Rogers (2005), a significant decline in DNA content often occurs during plant PCD without forming the characteristic DNA ladder. In cereals, a typical DNA ladder has been reported for maize (Young et al., 1997; Young and Gallie, 2000) and wheat (Young and Gallie, 1999) starchy endosperm during development. Here, no clear internucleosomal DNA fragmentation was observed, but rather a smear, indicating DNA degradation, which increased throughout development. The lack of detection of a clear laddering could be interpreted as a result of the DNA analysed being from the heterogeneous tissues constituting the quinoa seed, namely the embryo, endosperm, perisperm, and integuments, with the embryo tissue not undergoing PCD and the latter three with different timing of PCD.
The occurrence of PCD is more precisely determined when structural analyses are associated with the TUNEL assay (Zakeri et al., 1995). In this study, both analyses were used to validate and interpret TUNEL data and finally to conclude that cell death in quinoa perisperm occurs by means of a programme displaying the typical features of PCD (Fig. 8B–D).
Domínguez et al. (2012) report that the proteases and nucleases, of which little is known to date, are directly involved in the execution of nucleus dismantling in plants, namely chromatin condensation, internucleosomal DNA fragmentation, and nuclear envelope disorganization. Plant nucleases are usually classified according to metal ion cofactors and the optimum pH required for their activation. Accordingly, nucleases belong to the Zn2+- or Ca2+-dependent class. Among the Ca2+-dependent nucleases, some can also be activated by Mg2+ (Domínguez and Cejudo, 2006), Mn2+ (He and Kermode, 2003), or Co2+ (Langston et al., 2005).
According to Lesniewics et al. (2010), practically nothing is known about the processes of DNA degradation in storage organs. To date, studies with maize kernel indicate that the activities of Ca2+-dependent endonucleases are elevated in association with internucleosomal DNA fragmentation during the first stages of starchy endosperm degeneration. In maize endosperms, Young et al. (1997) studied the changes in activity of endonucleases with masses of 33.5, 36.0, and 38.5kDa, comparing the nuclease profiles of the wild type and the shrunken2 (shr2) mutant, in which internucleosomal fragmentation is accelerated. It could be inferred that the three nucleases may belong to the Ca2+ class since they are active in the presence of Ca2+ at pH 6.8 and not below pH 5.0. In this study, seven DNases with different electrophoretic mobility were detected. These enzymes represent several classes based on pH and ion dependency. In fact, three Zn2+- and two Ca2+/Mg2+-dependent enzymes and two other nucleases whose activity seems to be independent of any divalent cation were identified.
Proteases govern several important processes at the cell and tissue level during the growth and development of the organism and during initiation and execution of their death. In regard to caspases, homologues were not found in plants, but sequencing of the Arabidopsis genome has revealed the presence of several metacaspase genes (Uren et al., 2000), and true caspase-like activities have been reported during plant PCD (Rotari et al., 2005, and references within). Caspase 1-, 6-, and/or 9-like proteolytic activities have been correlated with both developmental and chemical-induced plant PCD, and the latter can be abolished or delayed by the use of caspase inhibitors (Woltering et al., 2002; Sanmartín et al., 2005). The VEIDase is the protease that cleaves the substrate Val-Glu-Ile-Asp; its activity resembles that of mammalian caspase 6 and it is involved as a principal caspase-like protease during PCD in varied eukaryotic systems (Bozhkov et al., 2004; Borén et al., 2006). Here, VEIDase activity was detected during quinoa perisperm development, although caspase 1- and caspase 9-like activities were not significantly induced, suggesting that only VEIDase activity is involved in executing the cell death programme in quinoa perisperm.
Similar to barley starchy endosperm (Borén et al., 2006), VEIDase activity is positively correlated with the occurrence of PCD in quinoa perisperm (present study). However, while in barley starchy endosperm VEIDase activity peaks ~1 week before nuclear DNA fragmentation and loss of plasma membrane integrity (Borén et al., 2006), VEIDase activity in quinoa perisperm is strongly induced and almost repressed by their specific inhibitors throughout nearly all PCD, not only at the beginning of the process. In fact, greater VEIDase actvity was detected at stages 4 and 5; that is, at the beginning of starch deposition, when the vacuole occupies almost all the cell lumen and the first detectable events of nuclear fragmentation are evident.
Conclusion
A number of parameters typically measured during PCD, such as cellular morphological changes in nuclei and cytoplasm, endoreduplication, DNA fragmentation, and the participation of caspases-like proteases and nucleases in nucleus dismantling were evaluated. This, together with the analysis of morphological changes in the cytoplasm including subcellular aspects related to starch compound grain accumulation demonstrates that two programmes simultaneously occur during quinoa seed development, i.e. PCD and starch accumulation. In this sense, quinoa perisperm development resembles that of grass starchy endosperm. On the other hand, it is proposed that the data generated here are invaluable as they provide novel insight into the Amaranthaceae, as illustrated by the results discussed above, as well a starting point for phylogenetic studies.
Acknowledgements
Funding for this work was provided by the Universidad de Buenos Aires (X125 to SM). For her fellowships, MPL-F thanks the Universidad de Buenos Aires and Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET).
References
- APG III 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society 161, 122–127 [Google Scholar]
- Bino RJ, Lanteri S, Verhoeven HA, Kraak HL. 1993. Flow cytometric determination of nuclear replication stages in seed tissues. Annals of Botany 72, 181–187 [Google Scholar]
- Boesewinkel FD, Bouman F. 1984. The seed structure. In: Johri BM, ed. Embryology of angiosperms. Berlin: Springer-Verlag, 567–610 [Google Scholar]
- Borén M, Höglund A-S, Bozhkov P, Jansson C. 2006. Developmental regulation of a VEIDase caspase-like proteolytic activity in barley caryopsis. Journal of Experimental Botany 57, 3747–3753 [DOI] [PubMed] [Google Scholar]
- Bouman F. 1984. The ovule. In: Johri BM, ed. Embryology of angiosperms. Berlin: Springer-Verlag, 123–157 [Google Scholar]
- Bozhkov P, Filonova LH, Suárez MF, Helmersson A, Smertenko AP, Zhivotovsky B, von Arnold S. 2004. VEIDase is a principal caspase-like activity involved in plant programmed cell death and essential for embryonic pattern formation. Cell Death and Differentiation 11, 175–182 [DOI] [PubMed] [Google Scholar]
- Bozhkov P, Jansson C. 2007. Autophagy and cell-death proteases in plants. Autophagy 3, 136–138 [DOI] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry 72, 248–254 [DOI] [PubMed] [Google Scholar]
- Chang SC, Gallie DR. 1997. RNase activity decreases following a heat shock in wheat leaves and correlates with its posttranslational modification. Plant Physiology 113, 1253–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Foolad MR. 1997. Molecular organization of a gene in barley which encodes a protein similar to aspartic protease and its specific expression in nucellar cells during degeneration. Plant Molecular Biology 35, 821–831 [DOI] [PubMed] [Google Scholar]
- Coimbra S, Salema R. 1994. Amaranthus hypochondriacus: seed structure and localization of seed reserves. Annals of Botany 74, 373–379 [Google Scholar]
- Crippen RW, Perrier JL. 1974. The use of neutral red and Evans blue for live–dead determinations of marine plankton. Staining Technology 49, 97–104 [DOI] [PubMed] [Google Scholar]
- D’Amato F. 1984. Role of polyploidy in reproductive organs and tissues. In: Johri BM, ed. Embryology of angiosperms. Berlin: Springer, 519–566 [Google Scholar]
- DeBono AG, Greenwood JS. 2006. Characterization of programmed cell death in the endosperm cells of tomato seed: two distinct death programs. Canadian Journal of Botany 84, 791–804 [Google Scholar]
- de Halac IN. 1980. Fine structure of nucellar cells during development of the embryo sac in Oenotherabiennis L. Annals of Botany 45, 515–521 [Google Scholar]
- Domínguez F, Cejudo FJ. 1998. Germination related genes encoding proteolytic enzymes are expressed in the nucellus of developing wheat grains. The Plant Journal 15, 569–574 [Google Scholar]
- Dominguez F, Cejudo FJ. 1999. Patterns of starchy endosperm acidification and protease gene expression in wheat grains following germination. Plant Physiology 119, 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez F, Cejudo FJ. 2006. Identification of a nuclear-localized nuclease from wheat cells undergoing programmed cell death that is able to trigger DNA fragmentation and apoptotic morphology on nuclei from human cells. Biochemical Journal 307, 529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez F, Moreno J, Cejudo FJ. 2001. The nucellus degenerates by a processs of programmed cell death during the early stages of wheat grain development. Planta 213, 352–360 [DOI] [PubMed] [Google Scholar]
- Domínguez F, Moreno J, Cejudo FJ. 2004. A gibberellin-induced nuclease is localized in the nucleus of wheat aleurone cells undergoing programmed cell death. Journal of Biological Chemistry 279, 11530–11536 [DOI] [PubMed] [Google Scholar]
- Domínguez F, Moreno J, Cejudo FJ. 2012. The scutellum of germinated wheat grains undergoes programmed cell death: identification of an acidic nuclease involved in nucleus dismantling. Journal of Experimental Botany 63, 5475–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani C, Consonni G, Gavazzi G, Colombo M, Dolfini S. 2002. Programmed cell death during embryogenesis in maize. Annals of Botany 90, 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood JS, Helm M, Gietl C. 2005. Ricinosomes and endosperm transfer cell structure in programmed cell death of the nucellus during Ricinus seed development. Proceedings of National Academy of Sciences, USA 102, 2238–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena AH, Pearce DM, Jackson MB, Hawes CR, Evans DE. 2001. Characterisation of programmed cell death during aerenchyma formation induced by ethylene or hypoxia in roots of maize (Zea mays L.). Planta 212, 205–214 [DOI] [PubMed] [Google Scholar]
- He X, Kermode AR. 2003. Nuclease activities and DNA fragmentation during programmed cell death of megagametophyte cells of white spruce (Picea glauca) seeds. Plant Molecular Biology 51, 509–521 [DOI] [PubMed] [Google Scholar]
- Ho A. 2006. Developmental regulation of a VEIDase caspase-like proteolytic activity in barley caryopsis. Access 57, 3747–3753 [DOI] [PubMed] [Google Scholar]
- Jensen WA. 1975. The composition and ultrastructure of the nucellus in cotton. Journal of Ultrastructural Research 13, 112–128 [Google Scholar]
- Joshi AC, Kajale LB. 1937. Fertilization and seed development in Amaranthaceae. Proceedings of the Indian Academy of Sciences 5, 91–100 [Google Scholar]
- Kowles RV, Srienc F, Phillips RL. 1990. Endoreduplication of nuclear DNA in the developing maize endosperm. Developmental Genetics 11, 125–132 [Google Scholar]
- Langston BJ, Bail S, Jones ML. 2005. Increase in DNA fragmentation and induction of a senescence-specific nuclease are delayed during corolla senescence in ethylene-insensitive (etr1-1) transgenic petunias. Journal of Experimental Botany 56, 15–23 [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. 1991. Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Canadian Journal of Botany 69, 461–476 [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW. 1993. Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis . The Plant Cell 5, 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer CA. 1829. Genera e Chenopodearum. In: Ledebour CF, ed. Flora Altaica, Vol. 1 Berlin: Reimer, 370–371 [Google Scholar]
- Norstog K. 1974. Nucellus during early embryogeny in barley: fine structure. Botanical Gazette 135, 97–103 [Google Scholar]
- Otto F. 1990. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. In: Crissman HA, Darzynkiewicz Z, eds. Methods in cell biology. New York: Academic Press, 105–110 [DOI] [PubMed] [Google Scholar]
- Pal A, Singh RP, Pal M. 1990. Development and structure of seeds in Amaranthus hypochaondriacus L. and its wild progenitor A. hybridus L. Phytomorphology 40, 145–150 [Google Scholar]
- Prego I, Maldonado S, Otegui M. 1998. Seed structure and localization of reserves in Chenopodium quinoa . Annals of Botany 82, 481–488 [Google Scholar]
- Radchuk V, Weier D, Radchuk R, Weschke W, Weber H. 2011. Development of maternal seed tissue in barley is mediated by regulated cell expansion and cell disintegration and coordinated with endosperm growth. Journal of Experimental Botany 62, 1217–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HJ. 2005. Cell death and organ development in plants. Current Topics in Development 71, 225–261 [DOI] [PubMed] [Google Scholar]
- Rotari VI, He R, Gallois P. 2005. Death by proteases in plants: whodunit. Physiologia Plantarum 123, 376–385 [Google Scholar]
- Sabelli PA, Larkins BA. 2012. The development of endosperm in grasses. Plant Physiology 149, 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmartín M, Jaroszewski L, Raikhel NV, Rojo E. 2005. Caspases. Regulating death since the origin of life. Plant Physiology 137, 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanò C, Buselli R, Ruffini, Castiglione M, Bottega S, Grilli I. 2007. RNases and nucleases in embryos and endosperms from naturally aged wheat seeds stored in different conditions. Journal of Plant Physiology 164, 487–495 [DOI] [PubMed] [Google Scholar]
- Thelen MP, Northcote DH. 1989. Identification and purification of a nuclease from Zinnia elegans L.: a potential molecular marker for xylogenesis. Planta 179, 181–195 [DOI] [PubMed] [Google Scholar]
- Uren AG, O’Rourke K, Aravind L, Pisabarro MT, Seshagiri S, Koonin EV, Dixit VM. 2000. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Molecular Cell 6, 961–967 [DOI] [PubMed] [Google Scholar]
- Van Doorn WG, Beers EP, Dangl JL, et al. 2011. Morphological classification of plant cell deaths. Cell Death and Differentiation 18, 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorn WG, Woltering EJ. 2005. Many ways to exit? Cell death categories in plants. Trends in Plant Science 10, 1360–1385 [DOI] [PubMed] [Google Scholar]
- Webb MC, Gunning BES. 1990. Embryo sac development in Arabidopsis thaliana . Sexual Plant Reproduction 3, 244–256 [Google Scholar]
- Werker E. 1997. Seed anatomy. Encyclopedia of plant anatomy. Bd. 10, Teil 3. SpeziellerTeil, Berlin: Gebrüder Borntraeger [Google Scholar]
- Woltering EJ, van der Bent A, Hoeberichts FA. 2002. Do plant caspases exist? Plant Physiology 130, 1764–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TE, Gallie DR. 1999. Analysis of programmed cell death in wheat endosperm reveals differences in endosperm development between cereals. Plant Molecular Biology 39, 915–926 [DOI] [PubMed] [Google Scholar]
- Young TE, Gallie DR. 2000. Programmed cell death during endosperm development. Plant Molecular Biology 44, 283–301 [DOI] [PubMed] [Google Scholar]
- Young TE, Gallie DR, DeMason DA. 1997. Ethylene mediated programmed cell death during maize endosperm development of Su and sh2 genotypes. Plant Physiology 115, 737–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakeri Z, Bursch W, Tenniswood M, Lokshin RA. 1995. Cell death: programmed, apoptosis, necrosis, or other? Cell Death and Differentiation 2, 87–96 [PubMed] [Google Scholar]