Abstract
The stamen produces pollen grains for pollination in higher plants. Coordinated development of four microsporangia in the stamen is essential for normal fertility. The roles of miR165/166-directed pathways in the establishment of adaxial–abaxial polarity have been well defined in leaves. However, the molecular mechanism underlying the adaxial–abaxial polarity of the stamen is elusive. Here it is reported that HYPONASTIC LEAVES1 (HYL1), a general regulator of microRNA (miRNA) biogenesis, plays an essential role in establishing the stamen architecture of the four microsporangia in Arabidopsis thaliana. In stamens, HYL1 and miR165/6 expression are progressively restricted to the lateral region, microsporangia, microspore mother cells, and microspores, whereas HD-ZIP III genes are preferentially expressed in the middle region, vascular bundle, and stomium. Loss of HYL1 leads to the formation of two rather than four microsporangia in each stamen. In the stamen of the hyl1 mutant, miR165/6 accumulation is reduced, whereas miR165/6-targeted HD-ZIP III genes are up-regulated and FILAMENTOUS FLOWER (FIL) is down-regulated; and, specifically, REVOLUTA (REV) is overexpressed in the adaxial region and FIL is underexpressed in the abaxial regions, concomitant with the aberrance of the two inner microsporangia and partial adaxialization of the connectives. Genetic analysis reveals that FIL works downstream of HYL1, and the defects in hyl1 stamens are partially rescued by rev-9 or phv-5 phb-6 alleles. These results suggest that HYL1 modulates inner microsporangia and stamen architecture by repression of HD-ZIP III genes and promotion of the FIL gene through miR165/6. Thus, the role of HYL1 in establishment of stamen architecture provides insight into the molecular mechanism of male fertility.
Key words: Anther, Arabidopsis thaliana, FIL, HD-ZIP III, HYL1, miR165/6, polarity, stamen.
Introduction
Stamens are important reproductive lateral organs, and consist of a four-locular apical anther and a basal filament in many flowering plants, including Arabidopsis thaliana (Goldberg et al., 1993; Sanders et al., 1999). The typical anther architecture consists of an adaxial region that forms microsporangia and an abaxial region that gives rise to the connective (Dinneny et al., 2006). There are four microsporangia in the adaxial region of an anther, each of which encloses numerous pollen grains in chambers known as locules. In the abaxial area, a small lump of tissue termed the connective joins the two sets of microsporangia to the filament. During anther dehiscence, the pollen grains are released from the stomium region, which develops between the two locules in each pair (Sanders et al., 1999).
Traditionally, anther development in A. thaliana is divided into 14 stages (Sanders et al., 1999). The precise molecular developmental mechanisms at the different stages have been elucidated by previous studies. APETALA3 (AP3), PISTILLATA (PI), and AGAMOUS (AG), parts of the ‘ABCE’ model, determine stamen identity (Bowman et al., 1989; Ma, 2005). SPOROCYTELESS (SPL)/NOZZLE (NZZ) and MALE STERILE1 (MS1) play essential roles in the division and differentiation of anther cells at early anther stages (Schiefthaler et al., 1999; Yang et al., 1999). EXCESS MALE SPOROCYTES1 (EMS1) is required for tapetum differentiation (Canales et al., 2002; Zhao et al., 2002). In addition, ARF6 and ARF8, which are targeted by miR167, have important functions in filament elongation and anther dehiscence at late stages of stamen development (Ru et al., 2006; Wu et al., 2006). Changes in the expression of these key genes usually lead to severe structure abortion, reduced male fertility, and even male sterility (Sanders et al., 1999; Ma, 2005; Feng and Dickinson, 2007). Additionally, JAGGED (JAG) and NUBBIN (NUB), two transcription factors, function redundantly in adaxial cell proliferation and differentiation (Dinneny et al., 2006). A model of adaxial–abaxial polarity establishment in the stamen of rice (Oryza sativa) has been proposed. In this model, expression of PHB3 (an orthologue of Arabidopsis PHB) and ETTIN1 (an orthologue of Arabidopsis ETT, also called AUXIN RESPONSE FACTOR3, ARF3) is rearranged during anther development (Toriba et al., 2010). However, it is unclear whether this model works in Arabidopsis and other dicotyledons. Importantly, FILAMENTOUS FLOWER (FIL) directly interacts with SPL/NZZ (Sieber et al., 2004).
Along with germline determination, anther polarity establishment is a critical issue that has been widely explored in recent years (Feng and Dickinson, 2007). In some lateral organs, adaxial–abaxial polarity identity is precisely regulated by two classes of antagonistic genes. The adaxial identity genes include the HD-ZIP III family genes REVOLUTA (REV), PHABULOSA (PHB), PHAVOLUTA (PHV), CORONA (CNA), and ATHB8 (HB8), and these are repressed by miR165/166 (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003; Zhong and Ye, 2004; Kelley et al., 2009). FIL (also called YAB1) and YAB3 act redundantly to promote abaxial cell fate in lateral organs (Siegfried et al., 1999; Bowman, 2000).
HD-ZIP III family genes are the targets of the microRNA (miRNA) miR165/6. miRNAs are RNA molecules ~22 nucleotides in length and are important regulators of various processes in eukaryotes (Hake, 2003; Willams et al., 2005; Wu et al., 2006; Chuck et al., 2008; Pontes and Pikaard, 2008; Bartel, 2009). In plants, hairpin precursor RNAs are processed by DICER LIKE 1 (DCL1), an RNase III endonuclease, to generate mature miRNAs (Park et al., 2002; Reinhart et al., 2002). Mature miRNAs repress target mRNAs primarily through miRNA-directed cleavage (Llave et al., 2002; Tang et al., 2003; Schwab et al., 2005). During miRNA processing, the double-stranded RNA-binding protein HYPONASTIC LEAVES1 (HYL1) and the C2H2 Zn-finger protein SERRATE (SE) are found to promote accurate processing of pre-miRNAs by DCL1 (Dong et al., 2008). HYL1 contains two double-stranded RNA-binding domains (dsRBDs) at its N-terminal end, a putative protein–protein interaction domain at its C-terminal end, and a nuclear localization signal (NLS) in the middle (Lu and Fedoroff, 2000; Wu et al., 2007). The two N-terminal dsRBDs are sufficient for the accumulation of miRNAs and completely rescue the leaf phenotype of hyl1 mutants (Yu et al., 2005). The loss-of-function mutant hyl1 exhibits pleiotropic phenotypes (Lu and Fedoroff, 2000) such as leaf hyponasty, reduced fertility, altered root gravitropic responses, and altered responses to several hormones (Lu and Fedorrof, 2000). In the leaves of hyl1 mutants, REV expression is up-regulated while FIL expression is down-regulated (Yu et al., 2005). HYL1 coordinates the expression of REV and FIL for adaxial–abaxial polarity of leaves (Liu et al., 2010, 2011). Although reduced fertility is reported in hyl1 mutants, the developmental reasons for this defect remain unclear.
To find out the reasons for the reduced fertility of hyl1 mutants, the floral organs that govern plant fertility were investigated. It was found that hyl1 mutants have severe defects in anther polarity: two inner microsporangia were lost; and the connectives are adaxialized. The study suggests that HYL1 modulates the anther structure by coordinating the expression of REV and FIL in stamens through miR165/166.
Materials and methods
Plant materials and growth conditions
The Arabidopsis thaliana mutants hyl1 (Nossen), hyl1-2 (Columbia), rev-9 (Landsberg), phv-5 phb-6 (Landsberg), and fil-1 (Landsberg) were used in this study. hyl1 is a mutant with a Dissociation (Ds) insertion site in the second exon, corresponding to the first dsRBD (Lu and Fedoroff, 2000). hyl1-2 (SALK_064863) is a mutant with the T-DNA insertion site in the first intron. The N-terminal fragment containing only the first dsRBD does not have any function in miRNA biogenesis and fails to show any mutant phenotype (Wu et al., 2007). Therefore, both hyl1 and hyl1-2 are null mutants.
The seeds of wild-type and mutant A. thaliana were surface-sterilized in 70% ethanol for 1min followed by 1% NaOCl for 10min. Then, seeds were washed four times in sterile distilled water, mixed in molten 0.1% water agar (Biowest), and plated on top of solid Murashige and Skoog medium with 1% sucrose. The Petri dishes were sealed with Parafilm, incubated at 4 °C in darkness for 2–3 d, and then moved to a growth room and incubated at 22 °C under 12h of light and 8h of darkness per day. Two weeks later, the seedlings were transplanted to peat soil in plastic pots and moved from a growth room to a growth chamber in the SIPPE phytotron. In this growth chamber, the plants were grown at 22 °C with 16h of light per day under a light source of warm white fluorescent tubes (colour code 990), an irradiance of 150 μmol m–2 s–1, and a light intensity on the plant canopy of 75 μmol m–2 s–1. The relative humidity was 65–70% and the air velocity was ~0.9 m s–1. All of the seedlings were grouped randomly and grown under identical conditions for 6 weeks. More than 20 individual plants for each mutant were prepared, and samples were taken for various measurements.
The hyl1 rev-9 and hyl1 fil-1 double mutants and hyl1 phv-5 phb-6 triple mutants were generated by crossing rev-9, fil-1, and phv-5 phb-6, respectively, to hyl1 and were identified by corresponding antibiotics and PCR tests.
Light microscopy and imaging
Samples and sections were observed using a BX 51 wide-field microscope equipped with aUPlanSAPO series objectives and a cooled DP71 camera (Olympus, Tokyo, Japan), and with a Stemi 2000 stereo microscope (Zeiss, Oberkochen, Germany). To observe the hybridization signal after in situ hybridization, slides were mounted in water and differential interference contrast (DIC) was applied. For flower and stamen imaging, Image-Pro Express version 5.1 (Media Cybernetics, Bethesda, MD, USA) software was applied to extend the depth of field. Image J (National Institutes of Health, Bethesda, MD, USA) was used to measure length and area.
Pollen viability
Alexander’s stain (Alexander, 1969) was used to examine hyl1 pollen viability as described by Guan et al. (2008).
Scanning electron microscopy (SEM)
Flowers and inflorescences were fixed in FAA [50% (v/v) ethanol, 5% (v/v) acetic acid, and 3.7% (v/v) formaldehyde], dried to critical point, and then dissected under a stereo microscope and mounted on SEM stubs. Mounted anthers were coated with palladium–gold and then examined using a JSM-6360LV SEM microscope (JEOL, Tokyo, Japan) with an acceleration voltage of 7–15kV. For pollen, fresh samples were directly mounted on SEM stubs and coated.
Histology
Flowers (inflorescences) of 5- to 6-week-old wild-type and mutant plants were fixed in FAA and embedded in paraffin (Sigma) as described (Liu et al., 2011), and 7 μm sections were stained in 0.05% (w/v) toluidine blue (Sigma) at 37 °C for 15min and then washed in water. Then, a non-toxic histological clearing agent (Histo-Clear) (National Diagnostics, Atlanta, GA, USA) was used instead of xylene to remove paraffin. For analysis of semi-thin sections, samples fixed in FAA were embedded in epoxy resin. Sections of 2 μm thickness were cut with glass knives in a ultramicrotome, fixed to glass slides, and stained in 0.05% (w/v) toluidine blue.
β-Glucuronidase (GUS) staining
The pHYL1::GUS transgenic plants have been described previously (Yu et al., 2005). Inflorescences and flower buds of the wild-type and the transgenic plants were placed in staining solution [50mM Na3PO4, pH 7.0, 0.5mM X-gluc (5-bromo-4-chloro-3-indolyl glucuronide), 20% (v/v) methanol], vacuum infiltrated, and incubated at 37 °C overnight as described previously (Yu et al., 2005). After staining, tissues were fixed in FAA for further analysis.
In situ hybridization
Flower sections (7 μm thick) from both the wild-type and mutant plants were prepared following pre-treatment and hybridization methods described previously (Brewer et al., 2006). Hybridization probes corresponding to coding sequences were defined as follows: a HYL1-specific probe located at 500–1260bp downstream of the start codon (Supplementary Table S1 available at JXB online); a REV-specific probe located at 7–2529bp; and a FIL-specific probe located at 145–567bp. Digoxigenin (DIG)-labelled probes were prepared by in vitro transcription (Roche) according to the manufacturer’s protocol. Locked nucleic acid (LNA)-modified probes of miR166 were synthesized and labelled with DIG at the 3ʹ end by TaKaRa were used for in situ hybridization of miR166.
miRNA isolation and northern blot analysis
Total RNA was extracted from inflorescences of 6-week-old wild-type and hyl1 plants. Antisense sequences of miR166 were synthesized and end-labelled as probes with biotin (TaKaRa). A 15 μg aliquot of total RNA was fractionated on a 15% polyacrylamide gel containing 8M urea and transferred to a Nitran Plus membrane (Schleicher and Schuell). Hybridization was performed at 41 °C using hybridization buffer (ULTRAhyb® Ultrasensitive Hybridizaton buffer, Ambion). Autoradiography of the membrane was performed using the LightShift Chemiluminescent EMSA Kit (Pierce). Northern blot analysis was performed according to the methods of previous studies (Liu et al., 2011).
Quantitative real-time reverse transcription–PCR (RT–PCR)
Total RNA was isolated from inflorescences (with opened flowers removed) of wild-type (Nossen) and hyl1 mutants (6 weeks) and then reverse-transcribed using oligo(dT) primers (Supplementary Table S1 at JXB online). Quantitative real-time PCR analysis was performed using the Rotor-Gene 3000 system (Corbett Research, Mortlake, NSW, Australia) using SYBR Premix Ex Taq (TaKaRa). ACTIN mRNA was used as an internal control, and relative amounts of mRNA were calculated using the comparative threshold cycle method. Primers specific for ACTIN, REV, PHB, PHV, CNA, HB8, and FIL (Supplementary Table S1) were employed to detect the expression levels in hyl1 mutants.
Results
The defects in male fertility of hyl1 mutants
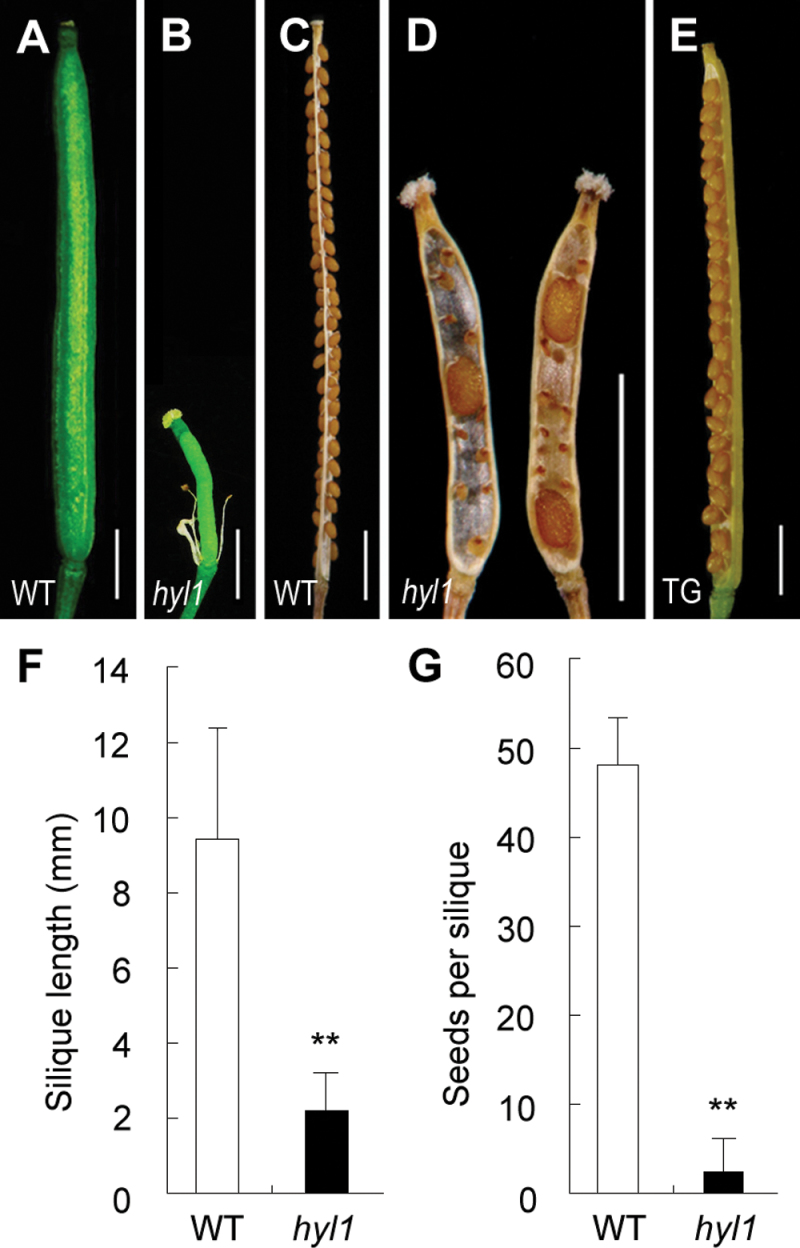
The null mutant hyl1 exhibits hyponastic leaves at the vegetative stage and poor fertility at the reproductive stage (Lu et al., 2000; Yu et al., 2005). It was observed that the fully elongated siliques of hyl1 plants were much shorter than those of the wild type (about one-fifth as long), and the number of seeds per silique was much lower in hyl1 than in wild-type plants (Fig. 1A–D). As indicated in Fig. 1F–G, the difference in seed number was significant between hyl1 and the wild type.
Fig. 1.
Siliques and seed set of hyl1 mutants. (A) A wild-type silique. (B) A hyl1 silique. (C) Seed set in a wild-type silique. (D) Seed set in a hyl1 silique. (E) A silique of a transgenic plant (TG) with HYL1 in the hyl1 background. (F) Quantitative analysis of silique length. The number of siliques measured is 152 for the wild type and 200 for hyl1 mutants. (G) Quantitative analysis of seed set. The number of siliques measured is 23 for both the wild type and the hyl1 mutants. Error bars indicate the SD. Two asterisks indicate a significant difference (t-test, **P < 0.01). Scale bars: 1mm in A–E. (This figure is available in colour at JXB online.)
To determine whether the male and/or female organs of hyl1 mutants were impaired by mutation of HYL1, the floral organs were carefully observed. In the wild-type flowers, each anther was composed of four microsporangia, two adjacent to the carpel (inner microsporangia) and the other two positioned away from the carpel (outer microsporangia). From these microsporangia, numerous viable pollen grains were released at anthesis (Fig. 2A–E; Supplementary Fig. S1A at JXB online). Compared with the wild-type flowers, hyl1 flowers produced small anthers, which were deficient in number, size, and function of microsporangia, and shortened filaments (both short and long) (Fig. 2F–L; Supplementary Table S2). The majority of hyl1 anthers contained two rather than four microsporangia, and were able to release a small number of viable pollen grains (Fig. 2I, L; Supplementary Fig. S1B). This type of anthers were designated as An1 anthers (anthers with only two functional microsporangia). Some of the hyl1 anthers had only one or two non-functional microsporangia and were designated as An2 anthers (anthers with non-functional mcrosporangium) (Fig. 2J; Supplementary Fig. S1C). An3 anthers exhibited a rod-shaped structure (anthers without any microsporangium) (Fig. 2K; Supplementary Fig. S1D). Functional microsporangia produced viable pollens while non-functional ones did not. The percentages of An1, An2, and An3 anthers were 61, 37, and 2% (Fig. 2Q). Under the same condition, all of the wild-type anthers contained four functional microsporangia.
Fig. 2.
Stamen phenotype of hyl1 mutants. (A–E) The wild-type. (A) Side view of the opened flower. (B) Side view of the same flower as in (A) with all sepals and petals cut off to show the stamens and pistil. (C) Overview of the opened flower. (D) An anther at late stage 12 showing two inner and two outer microsporangia in the adaxial area. (E) An anther with released pollen at stage 13. (F–L) hyl1 mutants. (F) An opened flower with one sepal and two petals cut off to show the stamen defects. (G) A flower with all sepals and petals cut off to show the stamens and pistil. (H) Overview of the opened flower. (I–K) Anthers of An1 (I) at stage 12, An2 (J), and An3 (K). (L) An1 anther at stage 13. (M–P) Flowers and stamens of a hyl1 plant transgenic for HYL1. (Q) Percentages of the three types of hyl1 anthers. The number of anthers examined is 139 for the wild type and 491 for hyl1. PG, pollen grains. Scale bars: 500 μm in A–C, F–H, and M–O, and 100 μm in D, E, I–L, and P. (This figure is available in colour at JXB online.)
To confirm that the microsporangium-deficient phenotype was caused by mutation of HYL1, HYL1 was transferred into hyl1 plants and the anthers of the transgenic plants were observed. The flowers, anthers, and microsporangia in the transgenic plants were normal (Fig. 2M–P), and the seeds in siliques were plentiful (Fig. 1E). The hyl1 phenotype was completely rescued by the exogenous HYL1 gene. This indicated that the deficiency in number and size of the microsporangia was indeed caused by the hyl1 allele.
To examine whether hyl1 pollen and/or pistils were functional, hyl1 plants were pollinated with wild-type pollen and the wild-type plants were pollinated with hyl1 pollen by hand pollination. With an excess amount of hyl1 pollen, both hyl1 and wild-type plants generated normal siliques and numerous seeds (Supplementary Fig. S2 at JXB online). With an excess amount of the wild-type pollen, hyl1 plants generated a number of seeds. This suggested that both the pollen and pistils of hyl1 were fertilized when the amount of pollen was sufficient.
Probably, the small anthers of hyl1 mutants with non-functional inner microsporangia released an insufficient amount of pollen grains for natural pollination. Meanwhile, the short filaments of hyl1 anthers should also affect natural pollination since they restricted the arrival of the pollen on the stigma. It was considered that the main reasons for the poor seed set of hyl1 plants was aberrant microsporangia and short filaments.
Impaired adaxial–abaxial polarity of hyl1 anthers
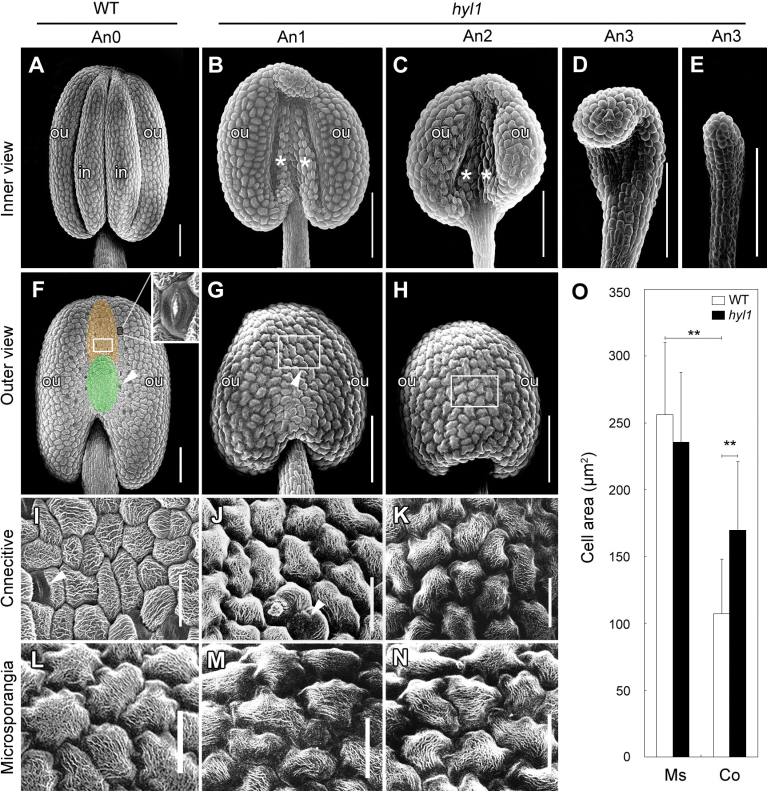
Anther locules (microsporangia) are adaxialized structures (Bowman, 1994), and the connectives between the locules have abaxial surfaces (Siegfried et al., 1999). To verify the defects in hyl1 anthers, anther formation was observed by SEM. Normal Arabidopsis anthers contained four microsporangia (Fig. 3A). As seen from the adaxial side of the anther, the inner microsporangia were smaller than the outer microsporangia. As viewed from the abaxial side, the connective base–filament boundary was precisely demarcated by the epidermal cells (Fig. 3F). The connectives formed grooves in the upper part and bulges in the lower part, with many stomata (Fig. 3F), and the epidermal cells on the smooth surface varied in shape and size. Most of the epidermal cells on the connectives were small and oblong, mixed with large rectangular cells (Fig. 3I). These cells were distinguishable from the jigsaw puzzle-shaped adaxial epidermal cells surrounding the microsporangia (Fig. 3L).
Fig. 3.
Architecture of hyl1 anthers. (A–N) Scanning electron microscope (SEM) images of anthers at stages 11/12. (A–E) Adaxial (inner) view of anthers. (A) Four microsporangia in a wild-type anther. (B–E) An1, An2, and An3 anthers. (F–H) Abaxial (outer) view of anthers. (F) The connectives of the wild-type; the groove and bulge are shown by the upper and lower ovals respectively. (G, H) The location of An1 (G) and An2 (H) connectives. (I–K) Abaxial epidermal cells of the wild-type connectives (I), An1 (J), and An2 (K) anthers from the same anthers as shown in F–H, respectively. (L–N) Adaxial (microsporangium) epidermal cells of the wild-type (L), An1 (M), and An2 (N) anthers from the same anthers shown in F–H, respectively. (O) Sizes of epidermal cells on the adaxial and abaxial surfaces of the wild-type and An1 anthers. The number of cells measured is 500 for both wild-type and hyl1 anthers. Error bars indicate the SD. Two asterisks indicate a significant difference (t-test, **P < 0.01). White asterisks in (B, C) indicate the remnants of inner microsporangia in hyl1 anthers; the white arrowheads in (F, G, I, J) indicate stomata. Co, abaxial epidermal cell on the connectives; Ms, adaxial epidermal cells on microsporangia; in, inner microsporangia; ou, outer microsporangia. Scale bars: 100 μm in A–H, 20 μm in I–N. (This figure is available in colour at JXB online.)
Compared with wild-type anthers, both the adaxial and the abaxial surfaces of hyl1 anthers were altered. The outer microsporangia of An1 anthers looked normal, although they were smaller than those of wild-type anthers; and the inner microsporangia were extremely shrunken and their remnants were visible (Fig. 3B). In An2 anthers, the inner microsporangia were invisible (Fig. 3C). The connectives of An1 anthers failed to form grooves and bulges and their epidermal cells became jigsawpuzzle-shaped and relatively enlarged (Fig. 3G, J). The epidermal cells of An2 connectives resembled the adaxial epidermal cells of the wild-type microsporangia (Fig. 3H, K). Meanwhile, An1 connectives retained a few tiny stomata on the epidermal surface, while An2 connectives had no stomata at all. Like epidermal cells of the adaxial surface of the wild type, those of An1 and An2 anthers were jigsaw puzzle-shaped (Fig. 3M, N). In addition, the size of the epidermal cells on An1 anthers were not different from those of the wild type on the adaxial surface, whereas the epidermal cells of their connectives were larger than those of the wild-type connectives (Fig. 3O). In contrast to An1 and An2 anthers, An3 anthers were rod-shaped with rectangular cells in the epidermis (Fig. 3D, E). Interestingly, the remnants of inner microsporangia in An1 anthers showed epidermal cells similar to those of An3 anthers (Fig. 3B). These results suggested that both the rod-shaped An3 anthers and the remnants of inner microsporangia in An1 anthers were extremely adaxialized.
In the anthers of hyl1-2, another hypomorphic allele of the HYL1 gene, the defects in inner microsporangia and connectives were the same as in hyl1 anthers (Supplementary Fig. S3 at JXB online).
Aberrance of inner microsporangia and connectives in stamens of hyl1 mutants
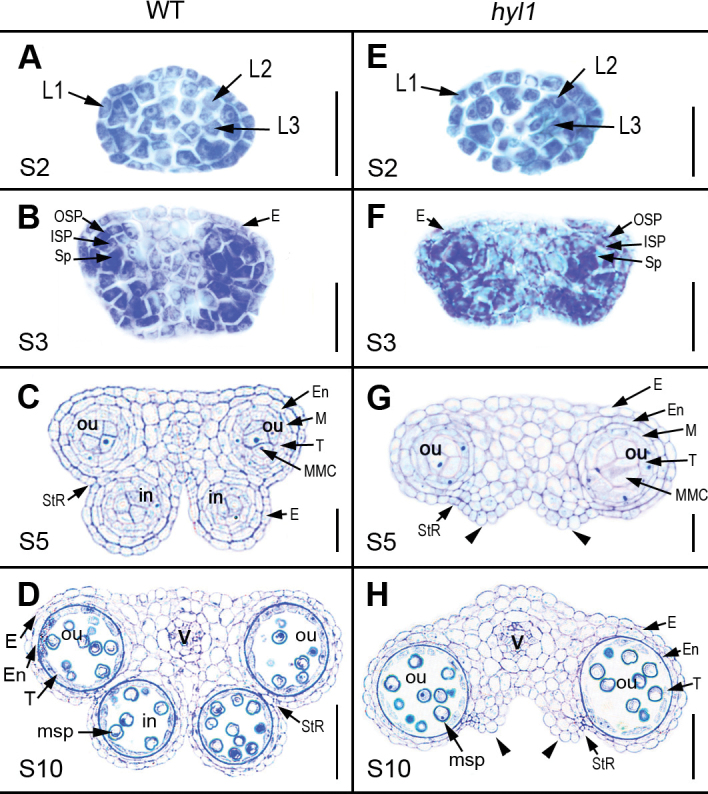
To depict the timing and position of aberrance of inner microsporangia and connectives in hyl1, anthers were traced back to the early events of stamen initiation. Anther development in Arabidopsis is divided into 14 stages (Sanders et al., 1999). According to this standard, stage 1 is characterized by divisions in the L1, L2, and L3 layers of the floral meristem, resulting in the formation of the stamen (anther) primordium. It was observed that L2 cells divided periclinally and anticlinally at the four corners of the stamen primordium at stages 2 and 3 (Fig. 4A, B), which developed into four microsporogia at later stages (Fig. 4C). The four-corner shape of hyl1 stamen primordia was not obviously different from that of the wild type (Fig. 4E, F). Owing to successive division and differentiation of archesporial cells, the four microsporangia appeared in the wild-type anthers at stage 5 (Fig. 4C), whereas the two microsporangia occurred in hyl1 anthers (Fig. 4G). At this stage, four locules were clearly defined, possessing all five specific cell layers of the microsporangium in the wild-type anthers. In contrast, only two locules were formed in hyl1 stamens. At stage 10, the microspores were generated in four locules of the wild-type anthers (Fig. 4D), whereas the others were produced in only two outer locules of hyl1 anthers (Fig. 4H).
Fig. 4.
The defects in the polarity of stamen primordia and anthers of hyl1 mutants. Cross-sections of stamen primordia and anthers at different stages showing the structures of stamen primordia and locules. (A–D) Wild-type stamens at different stages. (E–H) hyl1 stamens at different stages. Arrowheads indicate the remnants of inner microsporangia; E, epidermis; En, endothecium; in, inner locule; ISP, inner secondary parietal layer; MMC, microspore mother cell; msp, microspore; OSP, outer secondary parietal layer; ou, outer locule; PG, pollen grains; S, anther stage; Sp, sporogenous layer; StR, stomium region; V, vascular. Scale bars: 20 μm in A–C and E–G, and 50 μm in D and H. (This figure is available in colour at JXB online.)
While the microsporangia developed, the initials of the connectives in the abaxial central region of the wild-type stamens became visible at stage 3. The surface of these connectives became concave at stage 5 (Fig. 4C). In contrast, the surface of the connectives remained convex in hyl1 stamens (Fig. 4G), meaning that the connectives of hyl1 anthers were abnormal. Nevertheless, vascular bundles of hyl1 anthers were normal (Fig. 4H).
Compared with An1 anthers of hyl1 mutants, An2 anthers had one functional microsporangium and one non-functional microsporangium that did not generate viable microspores (Supplementary Fig. S4 at JXB online). An3 anthers did not produce any microsporangium. Instead, they were full of parenchymal cells.
Temporal and spatial expression of HYL1 in anthers
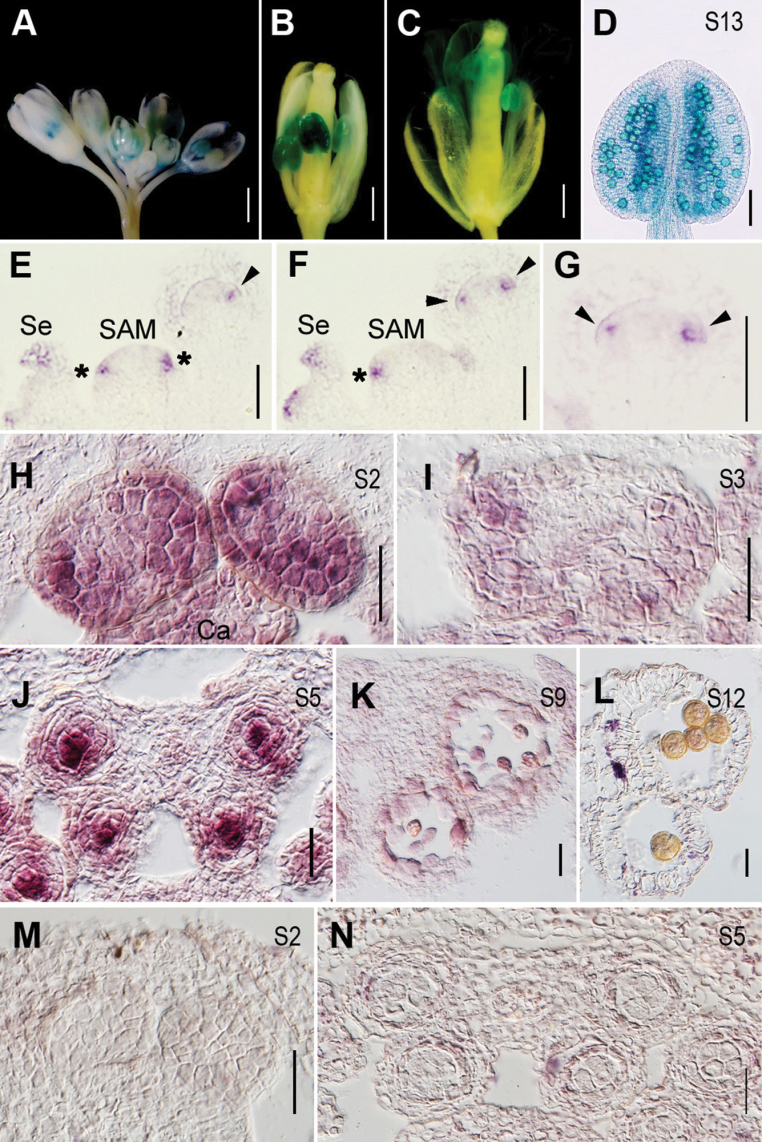
To examine the temporal and spatial expression of HYL1 in reproductive organs, transgenic plants carrying pHYL1::GUS reported previously (Yu et al., 2005) were used. In the wild-type inflorescences, HYL1 was preferentially expressed in anthers of all stages observed (Fig. 5A–C) and in pollen grains (Fig. 5D).
Fig. 5.
Temporal and spatial expression of HYL1. (A–D) GUS staining of HYL1 in pHYL1::GUS transgenic plants. (A) Flower buds showing HYL1 expression in anthers. (B, C) The flowers showing HYL1 expression in anthers at pre- (B) and post-anthesis (C). (D) An anther at stage 13 showing GUS signals in pollen grains. (E–L) In situ hybridization showing expression of HYL1 in an inflorescence and anthers of the wild type (E) Longitudinal section 1 of an inflorescence. (F) Longitudinal section 2 of an inflorescence. (G) Magnification of the flower in F. (H) Anthers at stage 2. (I) Anthers at stage 3. (J) Anthers at stage 5. (K) Anthers at stage 9. (L) Anthers at stage 12. (M and N) The sense probe as the negative control. Asterisks indicate stamen primordia. Arrowheads show flower primordium initiation sites. SAM, shoot apical meristem; ca, carpel; msp, microspores; pe, petal; PG, pollen grains; S, anther stage; se, sepal; tds, tetrads. Scale bars: 1mm in A; 500 μm in B, C; 100 μm in D; and 50 μm in E–N.
To examine the temporal and spatial patterns of HYL1 expression, in situ hybridization was performed. HYL1 was expressed preferentially in the presumed flower primordium initiation sites, sepals of older flower primordia, and stamen primordia (Fig. 5E–G). In the stamens of stages 2 and 3, HYL1 was expressed in the lateral regions where future microsporangia developed (Fig. 5H, I). At stage 5, HYL1 was expressed mainly in microsporangia (Fig. 5J), especially in microspore mother cells. HYL1 expression decreased dramatically at stage 9, but was detected at a relatively high level in developing microspores (Fig. 5K). However, HYL1 transcript was not seen in mature pollen grains at stage 12 (Fig. 5L).
Functions of miR165/166 and their target genes in stamen polarity
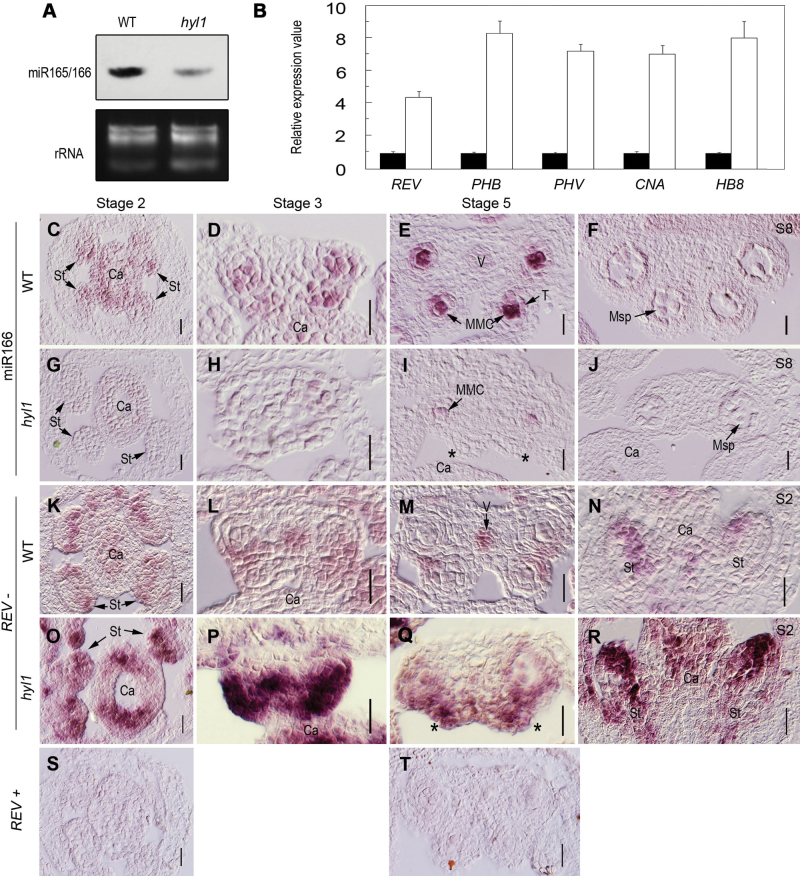
HYL1 mutation causes reduced accumulation of miR165/166 and increased expression levels of miR165/166-targeted HD-ZIP III genes in rosette leaves (Wu et al., 2007). To clarify whether HYL1 mutation alters the accumulation of miR166 and the expression levels of HD-ZIP III genes in anthers, the abundance of miRNA165/166 and the expression levels of its target genes in hyl1 inflorescences were monitored. Northern hybridization indicated that smaller quantities of miR166 were present in inflorescences of hyl1 mutants at the bolting stage than in the wild type (Fig. 6A). Real-time PCR demonstrated that all HD-ZIP III genes targeted by miR165/166 were up-regulated in anthers at stage 3 (Fig. 6B).
Fig. 6.
Expression patterns of miR166 and HD-ZIP III genes in hyl1 mutants. (A) Accumulation of miR165/166 in inflorescences of hyl1 mutants shown by northern blotting. (B) Real-time RT–PCR of HD-ZIP III genes in hyl1 anthers at stage 3. Three biological replicates were performed. Error bars indicate the SD. (C–J) In situ hybridization of miR166 with LNA probes. (C–F) miR166 signals in transverse sections of wild-type stamens at stage 2 (C), stage 3 (D), stage 5 (E), and stage 8 (F). (G–J) miR166 signals in transverse sections of hyl1 stamens at stage 2 (G), stage 3 (H), stage 5 (I), and stage 8 (J). (K–R) In situ hybridization of REV. (K–M) REV signals in cross-sections of wild-type stamens at stage 2 (K), stage3 (L), and stage 5 (M). (N) REV signals in a longitudinal section of a wild-type stamen at stage 2. (O–Q) REV signals in cross-sections of hyl1 stamens at stage 2 (O), stage 3 (P), and stage 5 (Q). (R) REV signals in a longitudinal section of a hyl1 stamen at stage 2. (S and T) The sense probe as the negative control. Asterisks indicate the remnants of the inner locule. ca, carpels; MMC, microspore mother cell; msp, microspores; S, anther stage; st, stamens; T, tapetum; V, vascular bundle. Scale bars: 20 μm in C–T.
To define the temporal and spatial patterns of miR166 and its target genes, in situ hybridization was performed. Dynamic patterns of miR165/6 signal in the wild-type stamens were similar to that of HYL1. In the stamens of stages 2 and 3, miR165/6 preferentially accumulated in the lateral regions (Fig. 6C, D). At stage 5, the miR166 signal was strong in microsporangia, especially in microspore mother cells (Fig. 6E). At stage 8 and thereafter, miRNA accumulation became much weaker (Fig. 6F). Compared with the wild-type plants, hyl1 mutants displayed a sharp decrease of miR165/6 accumulation in the stamens (Fig. 6G–J) where miR165/6 was present in outer microsporangia albeit to a much lesser extent (Fig. 6I). This revealed that HYL1 controlled the accumulation level rather than the location of miR165/6.
To determine whether increased accumulation of miR166 in the adaxial regions of hyl1 stamens causes any change in the temporal and spatial expression of its target genes, in situ hybridization of REV was performed. REV in the wild-type stamen at stage 2 was expressed in the middle regions where the future vascular bundle and stomium tissue developed (Fig. 6K). From stage 3 to 5, its sites of expression were restricted to the vascular bundles and stomium tissue (Fig. 6L, M). Longitudinal sections of stamens showed that REV was expressed in the middle region (Fig. 6N). In hyl1 stamens at stage 2, the expression level of REV was much stronger and the sites of expression were much broader than in the wild type (Fig. 6O). At stage 3, REV was apparently overexpressed and even extended to the connectives (Fig. 6P). At stage 5, REV expression was stronger in the remnants of inner microsporangia than in outer microsporangia (Fig. 6Q). The longitudinal section of stamens of stage 2 showed extremely strong expression of REV (Fig. 6R). In An3 anthers, REV was apparently overexpressed in the entire anther body (Fig. 6O). In combination with the remnants of inner microsporangia, it is suggested that the portions of inner microsporangia in An1 and An2 anthers were severely adaxialized.
Genetic interaction between HYL1 and FIL in anther polarity
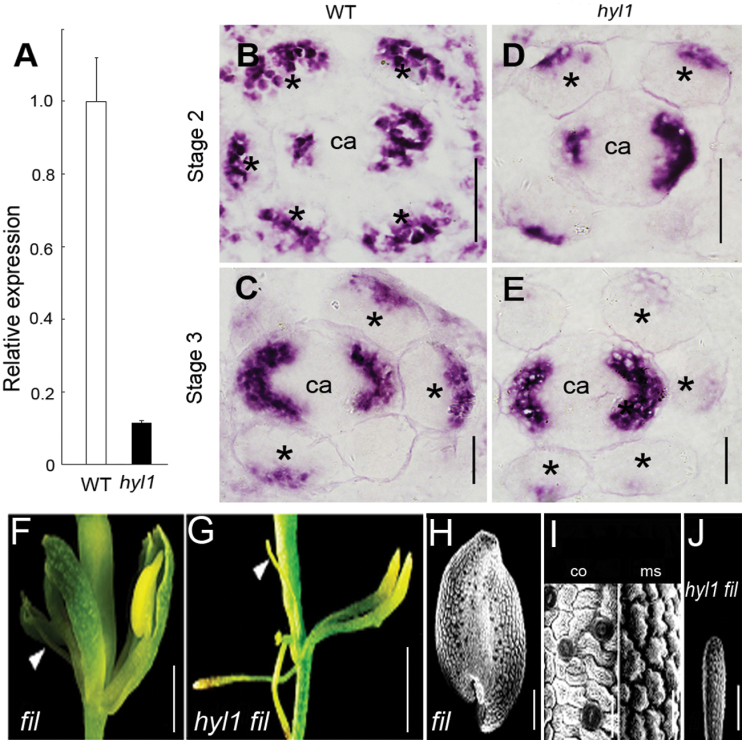
FIL is expressed abaxially in all lateral organs and can promote abaxial cell fates when ectopically expressed in adaxial positions (Sawa et al., 1999). FIL expression is localized at the abaxial surface of the connective tissue in maturing anthers (Siegfried et al., 1999), consistent with the idea that anther locules are adaxialized structures (Bowman, 1994). To examine whether the activities of abaxial identity genes are altered upon repression of miR165/166 accumulation, FIL expression patterns in hyl1 anthers were analysed. Corresponding to the increased expression of REV, the temporal and spatial expression patterns of FIL in hyl1 anthers were altered. Real-time PCR showed that the expression level of the FIL gene in the hyl1 anthers of stage 3 was 8-fold lower, compared with that of wild-type anthers, indicating a marked down-regulation of abaxial identity genes in early anthers (Fig. 7A). In situ hybridization demonstrated that FIL was preferentially expressed in abaxial regions of the wild-type stamen at stages 2. From stage 2 to stage 3, there were fewer sites of expression and the expression levels were reduced. Nevertheless, FIL expression was not restricted to the connective and might be expanded to the regions in which future microsporangia developed. Given that expression levels of FIL genes in the hyl1 carpel were indistinguishable from those observed in the wild-type carpel, the temporal and spatial expression of this gene was compared in stamen primordia. In comparison with wild-type connectives, expression levels of FIL in the abaxial regions of hyl1 anthers were remarkably reduced and there were fewer sites of FIL expression as it was restricted to a small region (Fig. 7D, E). The reduction of the FIL signal in hyl1 anthers at stage 3 was greater than that at stage 2. In combination with the adaxial character in the location of the connectives of hyl1 stamens, it was inferred that the connectives of hyl1 anthers were partially adaxialized.
Fig. 7.
Temporal and spatial expression of the FIL gene in hyl1 anthers. (A) Relative expression levels of HYL1 in anthers at stage 3, analysed by real-time PCR. (B–E) Cross sections showing in situ hybridization of the FIL gene in hyl1 anthers. (B) Wild-type anthers at stage 2. (C) An1 anthers of hyl1 at stage 2. (D) Wild-type anthers at stage 3. (E) An1 anthers of hyl1 at stage 3. (F and G) Flowers of the fil-1 single mutant (F) and hyl1 fil-1 double mutant (G). (H–J) SEM showing epidermal cells of the fil-1 anther (H), the connective and microsporangium of the fil-1 anther (I), and of the hyl1 fil-1 anther (J). Asterisks indicate stamens. ca, carpels; co, connective; ms, microsporangium. Scale bars: 20 μm in B–E, I; 500 μm in F, G; 100 μm in H, J.
To define the genetic relationship between HYL1 and FIL, hyl1 were crossed with fil-1 mutants. Anthers of fil-1 single mutants were arrested as radial filamentous stamens (Fig. 7F), but occasionally develop one or two microsporangia (Fig. 7H), which showed the epidermal cells of the connectives and microsporagia (Fig. 7I), as described previously (Sawa et al., 1999). Anthers of hyl1 fil-1 double mutants resembled those of the fil-1 single mutant in terms of radial filamentous stamens (Fig. 7G, J). These filamentous anthers were thinner than fil-1 anthers and they never developed locules. This indicated that fil-1 and hyl1 alleles were synergistic and fil-1 was epistatic to hyl1.
Partial rescue of hyl1 stamen defects by rev-9 and phv-5 phb-6 alleles
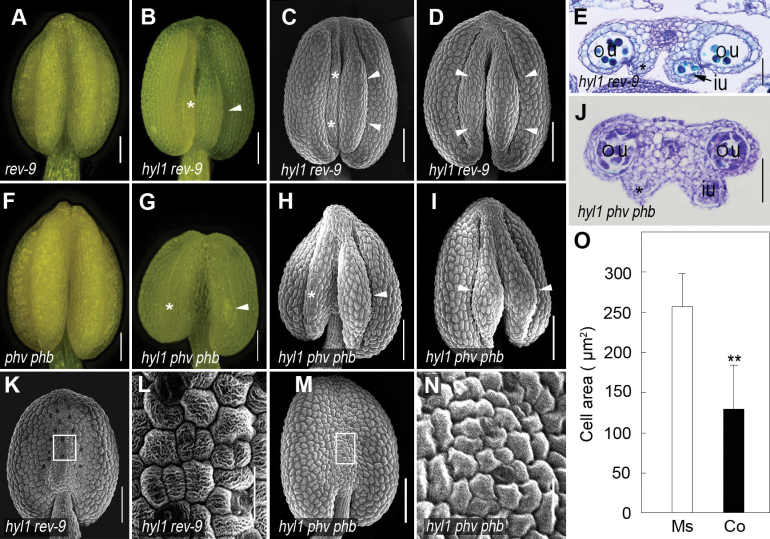
To understand the genetic relationship between HD-ZIP III genes and HYL1 in anther polarity, hyl1 plants were crossed with loss-of-function mutants of REV, PHB, and PHV, the three targets of miR165/166. Normal stamens were observed in rev-9 single mutants (Fig. 8A) and phv-5 phb-6 double mutants (Fig. 8F). Among hyl1 rev-9 double mutants, 49% of anthers produced three microsporangia (Fig. 8B, C), 2% of anthers generated four microsporangia with normal filament lengths (Fig. 8D), and the rest looked similar to An1 anthers. In three-microsporangium anthers, the two outer locules were functional while the one inner locule was partly functional (Fig. 8E). Among hyl1 phv-5 phb-6 triple mutants, 35% of anthers produced three or four microsporangia with viable pollen (Fig. 8G–I), and the rest looked similar to An1 anthers. The inner microsporangia of these anthers looked big but remained underdeveloped. While one inner microsporangium was able to generate male germline cells, another inner microsporangium did not contain any locule (Fig. 8J). The connectives of hyl1 rev-9 double mutants became nearly normal since they displayed small and oblong epidermal cells without stomata (Fig. 8K, L), as observed in the wild-type. The size of the connective epidermal cells in hyl1 rev-9 was partly recovered (Fig. 8O). Similarly, the defect in hyl1 connectives was recovered by phv-5 phb-6 (Fig. 8M, N). In summary, two-microsporangium anthers of the hyl1 phenotype are partially rescued by the rev-9 and phv-5 phb-6 alleles. All of these data indicated that the defects in inner microsporangia and the connectives were largely caused by overexpression of HD-ZIP III genes.
Fig. 8.
Rescue of hyl1 stamen defects by the rev-9 and phv-5 phb-6 alleles. (A and B) Anthers of rev-9 and hyl1 rev-9 mutants. (C and D) SEM images of hyl1 rev-9 anthers at stages 11/12. (E) Cross-section of a hyl1 rev-9 anther at stage 12. (F and G) Anthers of phv-5 phb-6 (phv phb) and hyl1 phv phb mutants. (H and I) SEM images of hyl1 phv phb anthers at stages 11/12. (J) Cross-section of a hyl1 phv phb anther at stage 6. (K–N) SEM images of the connectives of hyl1 rev-9 and hyl1 phv phb anthers at stages 11/12. (K and M) The connective and its epidermal cells of hyl1 rev-9 at stage 11/12. (L and N) A magnification of the boxed regions in K and M. (O) Cell size of the epidermis of microsporangia (Ms) on the adaxial side and of the connectives (Co) on the abaxial side of the hyl1 rev-9 anthers at stage 11/12. The number of cells measured is 450 for the adaxial microsporangia (Ms) and 478 for the abaxial connective (Co). Error bars indicate the SD. Two asterisks indicate a significant difference (t-test, **P < 0.01). in, inner locule; ou, outer locule. Scale bars: 100 μm in K and M; and 20 μm in L and N. (This figure is available in colour at JXB online.)
Discussion
HYL1 is required for the establishment of stamen polarity
All floral organ primordia originate from floral meristems (Bowman et al., 1989; Smyth et al., 1990). Organ identity is established after initiation of these primordia. During primordium initiation and differentiation, stamens establish polarity (Feng and Dickinson, 2007). Unlike leaves, in which the adaxial and abaxial regions are easily characterized as two regions, anthers can be divided into three regions: two inner microsporangia, two outer microsporangia, and the connectives. These structures are disrupted in hyl1 stamen primordia, where the two inner microsporangia are underdeveloped and the connectives are irregular. The hyl1 stamens of stage 2 are indistinct from those of the wild type. This fact reflects that the early stage of stamen primordia at which differentiation and division of four clusters of archesporial cells in four corners of hyl1 stamens occur are not affected. The obvious changes in shape and size of hyl1 stamens are at stage 5, thus reminding us that the time when inner microsporangia are underdeveloped is before stage 5. Mutation of HYL1 impairs inner microsporangia. The general layout of the outer microsporangia remains largely unchanged in hyl1 anthers. This fact implies that HYL1 functions in defining the inner microsporangia. Otherwise it suggests that the microsporangia are ‘partitioned’ into outer and inner microsporangia.
HYL1 is preferentially expressed in floral meristems and stamen primordia in developing flowers. Specifically, HYL1 domains are in lateral regions, but excluded in abaxial regions at stage 2 and 3. From the lateral regions, four microsporangia are differentiated and developed. Interestingly, HYL1 expression is restricted to the microsporangia at stage 5, especially in microspore mother cells, and then further restricted to microspores at stage 8. The changes in the sites of HYL1 expression with time reveal that HYL1 serves for development of microsporangia, microsporocytes, and microspores.
miR165/166 regulates anther polarity
The sites of miR165/6 accumulation in stamens are the same as those of HYL1. That is, the sites at which miR165/6 accumulates are in microsporangia, and pre- and post-microsporangium tissues. In hyl1 anthers, accumulation of miR165/6 is remarkably reduced. Nevertheless, miR165/6 remains visible in microsporangia, microsporocytes, and microspores. This indicates that HYL1 controls dynamic expression levels of miR165/6 in stamens.
In mature embryos, miR166 accumulates in the adaxial regions of cotyledons (Williams et al., 2005) and miR165 accumulates in the abaxial regions of 7-day-old leaves (Kidner and Martienssen, 2004). The data show that miR165/6 accumulated preferentially in the lateral regions of stamen primordia at early stages, and in microsporangia, microsporocytes, and microspores at later stages. This indicates that the sites of accumulation of miR165/6 are different in the adaxial–abaxial regions of cotyledon, leaves, and stamens.
In hyl1 mutants, most anthers have only two outer microsporangia. This defect may be associated with miR165/166. The sharp reduction in miR165/166 accumulation causes stronger expression of HD-ZIP III genes in the adaxial region of hyl1 anthers. REV overexpression is especially prominent in parts of the inner microsporangia. This overexpression may cause severe adaxialization of the inner microsporangia and even partial adaxialization of the connectives. rev-10d, a gain-of-function allele of REV that causes overexpression of the REV gene, results in adaxialization of lateral organs and alterations in the radial patterning of vascular bundles in the stem (Emery et al., 2003). In phb-1d, the gain-of-function mutant of PHB (McConnell and Barton, 1998), the anthers shows only two microsporangia, which make pollen sacs develop with their lines of dehiscence oriented laterally (data not shown). Therefore, the remnants of inner microsporangia are probably because the innermost area of the adaxial region is adaxialized so severely that inner microsporangia are arrested. In this case, REV overexpression may have a similar influence on the remnants of inner microsporangium tissue as that on rod-shaped An3 anthers. Meanwhile, up-regulation and expansion of the sites of expresion of HD-ZIP III genes in hyl1 anthers are concomitant with partial adaxialization of the connectives.
Overexpression of HD-ZIP III genes in hyl1 anthers impairs inner microsporangia and the connectives. This suggests that HD-ZIP III transcription factors negatively regulate development of microsporangia. REV expression is localized in the middle regions of stamens at early stages and vascular bundles and stomium at later stages. Therefore, HD-ZIP III transcription factors may function in development of vascular bundles and stomium. It might be thought that vascular bundles and stomium are the adaxial regions according to the original function of HD-ZIP III genes in leaves. However, this is not true in stamens of Arabidopsis. REV domains are excluded from adaxial regions of stamen primordia. Therefore, HD-ZIP III genes may function in arrest of microsporangium and/or promotion of vascular tissues and stomium in stamen.
Anther polarity depends on abaxial identity gene FIL
FIL directly interacts with SPL/NZZ, the major regulator of microsporangia and microspore mother cells (Sieber et al., 2004). In the adaxial regions of hyl1 anthers, REV expression is so high that it expands to the abaxial regions, whereby FIL expression is remarkably reduced and its expression is restricted to smaller regions. As a result, FIL expression is very weak in outer microsporangia, and relatively strong in the small regions of the connectives. FIL down-regulation is associated with REV up-regulation in the stamens of hyl1 anthers. Although the FIL domains are in abaxial regions of hyl1 stamens, as in the wild type, the size and strength of them decreased.
The anther of hyl1 fil-1 mutants resembles that of fil-1, in that all of the anthers are radially filamentous, without any microsporangium. Epistasis of fil-1 to the hyl1 allele in anther polarity indicates that deregulation of FIL in hyl1 anthers may be the molecular reason for aberrant inner microsporangia and the adaxialized connectives. One reasonable explanation for the shrunken stamen in hyl1 anthers is that strong HD-ZP III genes antagonize FIL function in the abaxial regions. Adaxial identity genes and abaxial identity genes are antagonistic (Sawa et al., 1999; Siegfried et al., 1999; Eshed et al., 2001; Kerstetter et al., 2001; Emery et al., 2003). In this sense, regulation by HYL1 of stamen development is through the balance between miR165/6, HD-ZIP III genes, and FIL gene.
As indicated above, the two-microsporangium phenotype of hyl1 mutants is because of the overexpression of HD-ZIP III genes in parts of the inner microsporangia, while adaxialization of the connective is caused by the reduced expression of the FIL gene in the abaxial regions. On the basis of the relationship between adaxial and abaxial identity genes, a model is proposed for HYL1-regulated establishment of anther architecture. For normal stamen development, the juxtaposition of HD-ZIP III genes and FIL should be properly controlled. HYL1 determines the progressive activities of miR165/6 in the lateral region, microsporangia, microspore mother cells, and microspores. Actually, miR165/6 controls the expression level of FIL in the adaxial regions and the connectives through the antagonistic function of REV. The development of inner microsporangia and the connectives is largely dependent on the activity of the FIL gene. By a balance between HD-ZIP III genes and FIL, HYL1 modulates inner microsporangia and the connectives in stamen.
Undeveloped anther mutants have stamens with filament-like structures that do not contain anthers (Sanders et al., 1999). Two receptor kinases, BAM1 and BAM2, are predicted to function during the first cell division of archesporial cells and subsequently during their periclinal divisions to produce the somatic cell layers (Hord et al., 2006). Because HYL1, SPL/NZZ, and BAM1/BAM2 are all required for early anther development, including aspects of cell division and differentiation, it should be tested whether they interact in stamen ontogeny. Exploration of the molecular mechanisms by which HYL1 coordinates with miR165/6 and SPL/NZZ will provide further insight into plant developmental networks.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Detection of pollen viability in anthers stained by Alexander’s red.
Figure S2. Siliques of the female plants after crossing by hand pollination.
Figure S3. Anther phenotype of the hyl1-2 mutant.
Figure S4. Cross-sections of An2 and An3 anthers of hyl1 mutants at stage 10.
Table S1. Sequences of probes and primers used in this study.
Table S2. Parameters of hyl1 flower organs.
Acknowledgements
We thank Dr Fedoroff (Pennsylvania State University, USA) and Dr Fletcher (Plant Gene Expression Center, USA) for providing the mutant seeds. This work was supported by grants from the Major State Basic Research Development Program of China (grant no. 30730053) and the Natural Science Foundation of China (grant no. 90508005)
References
- Alexander MP. 1969. Differential staining of aborted and nonaborted pollen. Stain Technology 44, 117–122 [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL. 1994. Flowers. In: Bowman JL, ed. Arabidopsis: an atlas of morphology and development. New York, Springer-Verlag, 156–161 [Google Scholar]
- Bowman JL. 2000. The YABBY gene family and abaxial cell fate. Current Opinion in Plant Biology 3, 17–22 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. 1989. Genes directing flower development in Arabidopsis . The Plant Cell 1, 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Heisler MG, Hejátko J, Friml J, Benková E. 2006. In situ hybridization for mRNA detection in Arabidopsis tissue sections. Nature Protocols 1, 1462–1467 [DOI] [PubMed] [Google Scholar]
- Canales C, Bhatt AM, Scott R, Dickinson H. 2002. EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis . Current Biology 12, 1718–732 [DOI] [PubMed] [Google Scholar]
- Chuck G, Candela H, Hake S. 2008. Big impacts by small RNAs in plant development. Current Opinion in Plant Biology 12, 1–6 [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Weigel D, Yanofsky MF. 2006. NUBBIN and JAGGED define stamen and carpel shape in Arabidopsis . Development 133, 1645–1655 [DOI] [PubMed] [Google Scholar]
- Dong Z, Han M, Fedoroff N. 2008. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proceedings of the National Academy of Sciences, USA 105, 9970–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. 2003. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Current Biology 13, 1768–1774 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL. 2001. Establishment of polarity in lateral organs of plants. Current Biology 11, 1251–1260 [DOI] [PubMed] [Google Scholar]
- Feng X, Dickinson HG. 2007. Packaging the male germline in plants. Trends in Genetics 23, 503–510 [DOI] [PubMed] [Google Scholar]
- Goldberg R, Beals T, Sanders P. 1993. Anther development: basic principles and practical applications. The Plant Cell 5, 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Huang X, Zhu J, Gao J, Zhang H, Yan Z. 2008. RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis . Plant Physiology 147, 852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S. 2003. MicroRNAs: a role in plant development. Current Biology 13, 851–852 [DOI] [PubMed] [Google Scholar]
- Hord CL, Chen C, Deyoung BJ, Clark SE, Ma H. 2006. The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. The Plant Cell 18, 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Skinner DJ, Gasser CS. 2009. Roles of polarity determinants in ovule development. The Plant Journal 57, 1054–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS. 2001. KANADI regulates organ polarity in Arabidopsis . Nature 411, 706–709 [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. 2004. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428, 81–84 [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau K, Carrington J. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056 [DOI] [PubMed] [Google Scholar]
- Liu, Jia L, Mao Y, He Y. 2010. Classification and quantification of leaf curvature. Journal of Experimental Botany 61, 2757–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Jia L, Wang H, He Y. 2011. HYL1 regulates the balance between adaxial and abaxial identity for leaf flattening via miRNA-mediated pathways. Journal of Experimental Botany 62, 4367–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Fedoroff N. 2000. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. The Plant Cell 12, 2351–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. 2005. Molecular genetic analyses of microsporogenesis and moicrogametogenesis in flowering plants. Annual Review of Plant Biology 56, 393–434 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. 1998. Leaf polarity and meristem formation in Arabidopsis . Development 125, 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman JH, Barton MK. 2001. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713 [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in micro-RNA metabolism in Arabidopsis thaliana . Current Biology 12, 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Pikaard C. 2008. siRNA and miRNA processing: new functions for Cajal bodies. Current Opinion in Genetics and Development 18, 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B, Weinstein E, Rhoades M, Bartel B, Bartel D. 2002. MicroRNAs in plants. Sexual Plant Reproduction 16, 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru P, Xu L, Ma H, Huang H. 2006. Plant fertility defects induced by the enhanced expression of microRNA167. Cell Research 16, 457–465 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Robert B, Goldberg RB. 1999. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sexual Plant Reproduction 11, 297–322 [Google Scholar]
- Sawa S, Ito T, Shimura Y, Okada K. 1999. FILAMENTOUS FLOWER controls the formation and development of arabidopsis inflorescences and floral meristems. The Plant Cell 11, 69–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S, Watanabe K, Goto K, Kanaya E, Morita EH, Okada K. 1999. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes and Development 13, 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K. 1999. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 96, 11664–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Palatnik J, Riester M, Schommer C, Schmid M, Weigel D. 2005. Specific effects of microRNAs on the plant transcriptome. Developmental Cell 8, 517–527 [DOI] [PubMed] [Google Scholar]
- Sieber P, Petrascheck M, Barberis A, Schneitz K. 2004. Organ polarity in Arabidopsis. NOZZLE physically interacts with members of the YABBY family. Plant Physiology 135, 2172–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL. 1999. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis . Development 126, 4117–4128 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JH, Meyerowitz EM. 1990. Early flower development in Arabdopsis . The Plant Cell 2, 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Reinhart B, Bartel D, Zamore P. 2003. A biochemical framework for RNA silencing in plants. Genes and Development 17, 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriba T, Suzaki T, Yamaguchi T, Ohmori Y, Tsukaya H, Hirano HY. 2010. Distinct regulation of adaxial–abaxial polarity in anther patterning in rice. The Plant Cell 22, 1452–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Grigg S, Xie M, Christensen S, Fletcher J. 2005. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132, 3657–3668 [DOI] [PubMed] [Google Scholar]
- Wu F, Yu L, Cao W, Mao Y, Liu Z, He Y. 2007. The N-terminal double-stranded RNA binding domains of Arabidopsis HYPONASTIC LEAVES1 are sufficient for pre-microRNA processing. The Plant Cell 19, 914–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Tian Q, Reed J. 2006. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218 [DOI] [PubMed] [Google Scholar]
- Yang W, Ye D, Xu J, Sundaresan V. 1999. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes and Development 13, 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Yu X, Shen R, He Y. 2005. HYL1 gene maintains venation and polarity of leaves. Planta 221, 231–242 [DOI] [PubMed] [Google Scholar]
- Zhao D, Wang G, Speal B, Ma H. 2002. The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes and Development 16, 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye Z. 2004. Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant and Cell Physiology 45, 369–385 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.