Abstract
Plants have evolved different strategies to resist drought, of which the best understood is the abscisic acid (ABA)-induced closure of stomatal pores to reduce water loss by transpiration. The availability of useful promoters that allow for precise spatial and temporal control of gene expression in stomata is essential both for investigating stomatal regulation in model systems and for biotechnological applications in field crops. Previous work indicated that the regulatory region of the transcription factor AtMYB60 specifically drives gene expression in guard cells of Arabidopsis, although its activity is rapidly down-regulated by ABA. Here, the activity of the full-length and minimal AtMYB60 promoters is reported in rice (Oryza sativa), tobacco (Nicotiana tabacum), and tomato (Solanum lycopersicum), using a reporter gene approach. In rice, the activity of both promoters was completely abolished, whereas it was spatially restricted to guard cells in tobacco and tomato. To overcome the negative effect of ABA on the AtMYB60 promoter, a chimeric inducible system was developed, which combined the cellular specificity of the AtMYB60 minimal promoter with the positive responsiveness to dehydration and ABA of the rd29A promoter. Remarkably, the synthetic module specifically up-regulated gene expression in guard cells of Arabidopsis, tobacco, and tomato in response to dehydration or ABA. The comparative analysis of different native and synthetic regulatory modules derived from the AtMYB60 promoter offers new insights into the functional conservation of the cis-mechanisms that mediate gene expression in guard cells in distantly related dicotyledonous species and provides novel tools for modulating stomatal activity in plants.
Key words: ABA, dehydration, guard cell-specific promoters, inducible promoters, stomata, synthetic regulatory modules.
Introduction
Drought represents a major threat to agriculture and food production. Even in the most productive cropping environment, short periods of water scarcity are responsible for considerable reductions in seed and biomass yields each year (Ciais et al., 2005). Increasing temperature and changes in rainfall are expected to exacerbate the negative effects of water deficiency in agriculture (Lobell et al., 2008). In this changing environment, yield stability will depend highly upon the ability to develop novel crop varieties with a more sustainable use of water and enhanced tolerance to water shortages.
Plants have evolved different adaptive strategies to withstand drought, including the rapid closure of the stomatal pores distributed on the surface of leaves and stems. During drought, plants accumulate the stress hormone abscisic acid (ABA), which triggers in guard cells a signalling cascade that rapidly leads to stomatal closure to minimize water loss by transpiration (Kim et al., 2010). Modelling studies predict that earlier and tighter stomatal closure would reduce desiccation and support yield stability under water stress (Sinclair and Muchov, 2001). Most importantly, data from multiple years of a field trial indicate that enhancement of ABA responses in guard cells can efficiently reduce water loss by transpiration and increase crop resilience to climate change (Wang et al., 2005, 2009).
Genetic screens and gene profiling studies have greatly improved our understanding of the molecular networks that control guard cell activity in response to internal signals and environmental cues, and have identified several candidate genes for downstream biotechnological applications (Leonhardt et al., 2004; Galbiati et al., 2008; Yang et al., 2008; Gardner et al., 2009). Evidence indicates that stomatal closure can be effectively enhanced by disrupting negative regulators of ABA responses, or by overexpressing positive regulators of the ABA signalling pathway (Pei et al., 1998; Gosti et al., 1999; Klein et al., 2003). Guard cell-related transcription factors have also proven to be valuable targets for modulating stomatal activity in plants (Cominelli et al., 2010).
Most genes involved in the regulation of guard cell responses are also expressed in other tissues and control several yield-associated traits (Schroeder et al., 2001). Consequently, genetic engineering strategies which incorporate the use of strong constitutive promoters [e.g. the Cauliflower mosaic virus (CaMV) 35S promoter] for conferring transgene expression will result in undesirable side effects on plant growth and productivity. Proper genetic manipulation of stomatal responses involves the use of effective expression systems, including guard cell-specific promoters, to confer precise spatial regulation of transgenes. Above all, regulatory modules which combine cellular specificity with responsiveness to environmental (e.g. dehydration) and/or internal (e.g. ABA) stimuli will prove invaluable in genetically engineering novel adaptive traits in crops.
In a previous work, the 1.3kb genomic region upstream of the Arabidopsis AtMYB60 gene (At1g08810) was identified as a guard cell-specific promoter (Cominelli et al., 2005). More recently, it was shown that the activity of the AtMYB60 promoter in stomata is rapidly down-regulated by exogenous application of ABA (Cominelli et al., 2011). Serial deletion analysis of the 1.3kb region allowed the discovery of the module responsible for the negative ABA-dependent regulation and identified a 246bp sequence (from –262bp to –16bp upstream of the start codon) as the minimal regulatory module sufficient to confer guard cell-specific activity (Cominelli et al., 2011).
In this work, the pattern of spatial and temporal activity of the AtMYB60 promoter in rice, tobacco, and tomato is reported. Analysis of stable transgenic lines expressing the β-glucuronidase (GUS) reporter gene under the control of either the 1.3kb or the 246bp AtMYB60 promoter revealed that these regulatory elements, although inactive in rice tissues, were specifically activated in guard cells of tobacco and tomato. Most importantly, a synthetic system which incorporates the guard cell-specific AtMYB60 module and the ABA- and drought-inducible module from the stress-regulated rd29A promoter was developed. Tobacco and tomato transgenic lines expressing GUS under the control of the chimeric promoter revealed strong activation of reporter gene expression upon ABA application or dehydration treatment exclusively in guard cells. Taken together, these results highlight the usefulness of the AtMYB60 promoter for designing modular expression systems suitable for the spatial and temporal control of gene expression in stomata, both for studying stomatal function in model systems and for engineering guard cell responses in crops.
Materials and methods
Plasmid construct
The previously described AtMYB60 pro 1.3:GUS and AtMYB60 pro 246:GUS constructs (Cominelli et al., 2011) were used to generate stable transgenic tobacco and tomato lines. For rice transformation, the AtMYB60 pro 1.3:GUS and AtMYB60 pro 246:GUS cassettes were cloned into the HindIII–EcoRI sites in the pCAMBIA 1300 binary vector (CAMBIA, Canberra, Australia), carrying resistance to hygromycin. To generate the rd29A-MYB60 pro 246 construct, the genomic region from –254bp to –40bp, located upstream of the rd29A gene, was amplified using the primers pDREABF1 (5ʹ-AAGCTTACATTTTAGGATGGAATAAATAT-3ʹ) and pDREABR1 (5ʹ-TCCCTTTATCTCTCTCAGTAAGCTT-3ʹ), both containing a HindIII site (italics). The PCR fragment was inserted in the HindIII site upstream of the 246bp promoter in the AtMYB60 pro 246:GUS construct.
Plant material, plant transformation, and growth conditions
Arabidopsis transgenic lines (Col-0) were generated by Agrobacterium-mediated transformation as described (A. tumefaciens strain GV3101) (Clough and Bent, 1998). Transformed seeds were sterilized overnight in a sealed chamber using 100ml of commercial bleach and 3ml of 37% HCl and selected on Murashige and Skoog (MS) medium, 1% (w/v) sucrose, 0.8% (w/v), agar, and 50 μg ml–1 kanamycin. Plants were grown in a growth chamber under long-day conditions (16h light; 8h dark at 100 μmol m–2 s–1) at 22 °C. Rice transgenics were produced in the japonica cv. Nipponbare, as described (Hiei et al., 1994), using A. tumefaciens strain EHA105. Transgenic T1 lines were selected by germinating seeds on MS medium, 1% (w/v) sucrose, 0.8% (w/v) agar and 50 μg ml–1 hygromycin. Resistant plants were transferred to pots containing a blend of loam sandy soil and peat (4:1, v/v) (VIGORPLANT, Fombio, Italy), fertilized with Guano (COMPO, Cesano Maderno, Italy) after repotting and before flowering, and grown in a greenhouse under a 12h light/12h dark cycle at 280 μmol m–2 s–1, at 26 °C. Tobacco experiments were performed in Nicotiana tabacum cv Samsun. Transgenic lines were generated as described (Horsch et al., 1985), using A. tumefaciens strain GV3101. T1 seeds were sterilized with absolute ethanol for 2min and 50% commercial bleach for 5min, rinsed with sterile distilled water, and germinated on MS medium, 1.5% (w/v) sucrose, 0.7% plant agar, and 50 μg ml–1 kanamycin. Kanamycin-resistant plants were transferred to pots and grown as described for rice. Tomato transgenic lines were produced in both the Microtom and Moneymaker backgrounds as described (McCormick, 1991; Davuluri et al., 2005) (A. tumefaciens strain AGL1). Rooted plants were transferred to pots and grown in a greenhouse as described for rice and tobacco. T1 kanamycin-resistant plants were selected as described for tobacco.
GUS assays
For detection of GUS activity, tissues were vacuum-infiltrated and incubated for 24–48h at 37 °C, in 0.5mg ml–1 X-glucuronic acid, 0.1% Triton X-100, and 0.5mM ferrocyanidine in 100mM phosphate buffer (pH 7). Tissues were cleared with a chloral hydrate:glycerol:water solution (8:1:2, v/v/v). Samples were examined using a Leica M205 FA stereomicroscope and Leica DM2500 optical microscope (Leica Microsystems GmbH, Wetzlar, Germany).
ABA and dehydration experiments
ABA treatments were performed in vivo. Plants grown in soil were sprayed with 100 μM ABA (±-cis, trans ABA; SIGMA, Milano, Italy), dissolved in 100% ethanol, or with an equal amount of ethanol (mock solution). For dehydration experiments, leaves were detached from soil-grown plants and air dried for up to 6h in a growth cabinet under continuous light, at 26 °C, with 30% relative humidity.
Quantification of mRNA expression
RNA isolation, reverse transcription, and quantitative PCRs (qPCRs) were performed as previously described (Galbiati et al., 2011). GUS expression was analysed using primers qPCR_GUSF1 and qPCR_GUSR1, and normalized using the AtACTIN2 gene (At3g18780) in Arabidopsis (Nishimura et al., 2003), the Elongation Factor 1α gene (NtEF1α) in tobacco (Liu et al., 2012), or the LeEF1α gene in tomato (Bartley et al., 2003). In rice, the presence of the transgene and GUS expression were assessed by PCR and reverse transcription–PCR (RT–PCR), respectively, using the primers GUSRTF1 and GUSRTR1. The OsActin gene was used as a control (Zhao et al., 2009). The sequences of all the primers used in expression studies are listed in Supplementary Table S1 available at JXB online.
Results
Activation pattern of the Arabidopsis AtMYB60 promoter in rice, tobacco, and tomato
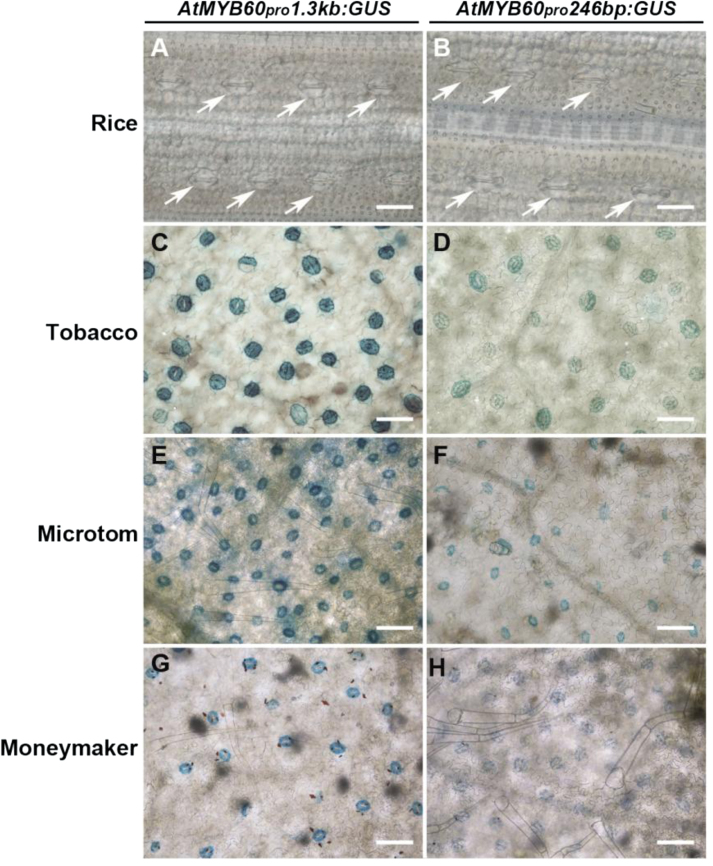
To investigate the activity of the guard cell-specific AtMYB60 promoter in cereals, stable transgenic lines of rice (spp. japonica cv. Nipponbare), expressing the reporter GUS under the control of the full-length (1.3kb) or the minimal (246bp) regulatory region of AtMYB60, were generated (AtMYB60 pro 1.3:GUS and AtMYB60 pro 246:GUS lines, respectively). Thirty hygromycin-resistant primary transformants (T0) were recovered for each construct, and the presence of the transgene was investigated by PCR (Supplementary Fig. S1 at JXB online). Staining of developing T0 AtMYB60 pro 1.3:GUS or AtMYB60 pro 246:GUS leaves did not reveal GUS expression in guard cells or in any other cell type (Fig. 1A, B). Consistently, RT–PCR analysis of the T1 seedlings did not detect expression of the reporter gene (Supplementary Fig. S1). These findings suggest the absence of a MYB60-related regulatory network in the guard cell of rice, which might reflect the functional divergence in stomata between monocots and dicots (Serna, 2011).
Fig. 1.
Histochemical localization of GUS activity in transgenic rice, tobacco, and tomato T0 plants. (A and B) Leaves from rice AtMYB60 pro 1.3:GUS and AtMYB60 pro 246:GUS plants, respectively. Arrows indicate stomata. (C and D) Leaves from tobacco AtMYB60 pro 1.3:GUS and AtMYB60 pro 246:GUS plants, respectively. (E and F) Leaves from Microtom AtMYB60 pro 1.3:GUS and AtMYB60 pro 246:GUS plants, respectively. (G and H) Leaves from Moneymaker AtMYB60 pro 1.3:GUS and AtMYB60 pro 246:GUS tomato plants, respectively. Leaves from the AtMYB60 pro 1.3:GUS lines were incubated in the staining solution for 24h (A, C, E, G), whereas leaves from the AtMYB60 pro 246:GUS lines were incubated for 48h (B, D, F, H). Bar=50 μm.
Next, the conservation of the cellular specificity of the AtMYB60 promoter was investigated in dicot systems, with the two Solanaceae crop species tobacco (cv. Samsun) and tomato (cv. Microtom and cv. Moneymaker). Histochemical analysis of 10–15 independent T0 AtMYB60 pro 1.3:GUS or AtMYB60 pro 246:GUS lines for each genotype revealed specific GUS staining in guard cells distributed on developing leaves (Fig. 1C–H). In agreement with data from Arabidopsis, which demonstrate that the full-length 1.3kb AtMYB60 promoter possesses stronger activity in guard cells (Cominelli et al., 2011), stomata from AtMYB60 pro 1.3:GUS plants showed more intense GUS signals, compared with AtMYB60 pro 246:GUS individuals, in both tobacco and tomato.
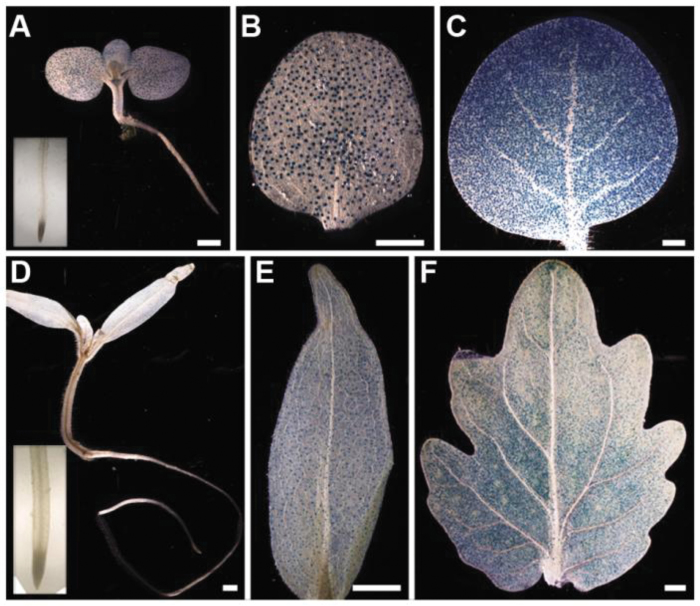
Independent T2 AtMYB60 pro 1.3:GUS and AtMYB60 pro 246:GUS tobacco and tomato lines (n=10) were selected to investigate further the cell and tissue specificity of GUS expression during plant development. Ten-day-old AtMYB60 pro 1.3:GUS seedlings displayed expression of the reporter exclusively in guard cells, distributed on cotyledons, hypocotyls, and leaf primordia, in all the tobacco (Fig. 2A, B) and tomato (Fig. 2D, E) lines analysed. No GUS signals were detected in roots (insets in Fig. 2A, D). Similarly, AtMYB60 pro 246:GUS tomato and tobacco seedlings showed GUS expression exclusively in guard cells, even though the intensity of the staining was reduced compared with seedlings harbouring the AtMYB60 pro 1.3:GUS construct (data not shown). Analysis of developing and mature leaves confirmed the guard cell-specific expression of the reporter in both species (Fig. 2C, F). In flowers, consistent GUS expression in guard cells located on sepals was detected (Supplementary Fig. S2A, E at JXB online). Occasionally, diffuse and intense staining of anthers was observed in individual flowers from tomato, whereas weak localized signals were found in the inner part of tobacco anthers (Supplementary Fig. S2). These findings are in contrast to the lack of GUS expression in reproductive tissues reported for both the AtMYB60 pro 1.3:GUS and AtMYB60 pro 246:GUS constructs in Arabidopsis (Cominelli et al., 2011). However, GUS activity was also observed in anthers of flowers from untransformed Moneymaker, Microtom, and tobacco plants (Supplementary Fig. S2). It is thus likely that GUS expression in male reproductive organs originated from an anther-specific endogenous GUS activity, which has been previously described for different members of the Solanaceae family, including tomato (Plegt and Bino, 1989).
Fig. 2.
Developmental GUS expression patterns in homozygous T2 AtMYB60 pro 1.3:GUS tobacco and tomato lines. (A) A 15-day-old tobacco seedling. The inset represents a magnified view of the primary root, (B) Detail of a cotyledon. (C) Developing tobacco leaf. (D) A 15-day-old Microtom seedling. The inset represents a magnified view of the primary root. (E) Detail of a cotyledon. (F) Developing Microtom leaflet. All tissues were incubated in the staining solution for 24h. Bar=1mm.
The activity of the AtMYB60 promoter is negatively regulated by ABA and dehydration in tobacco and tomato guard cells.
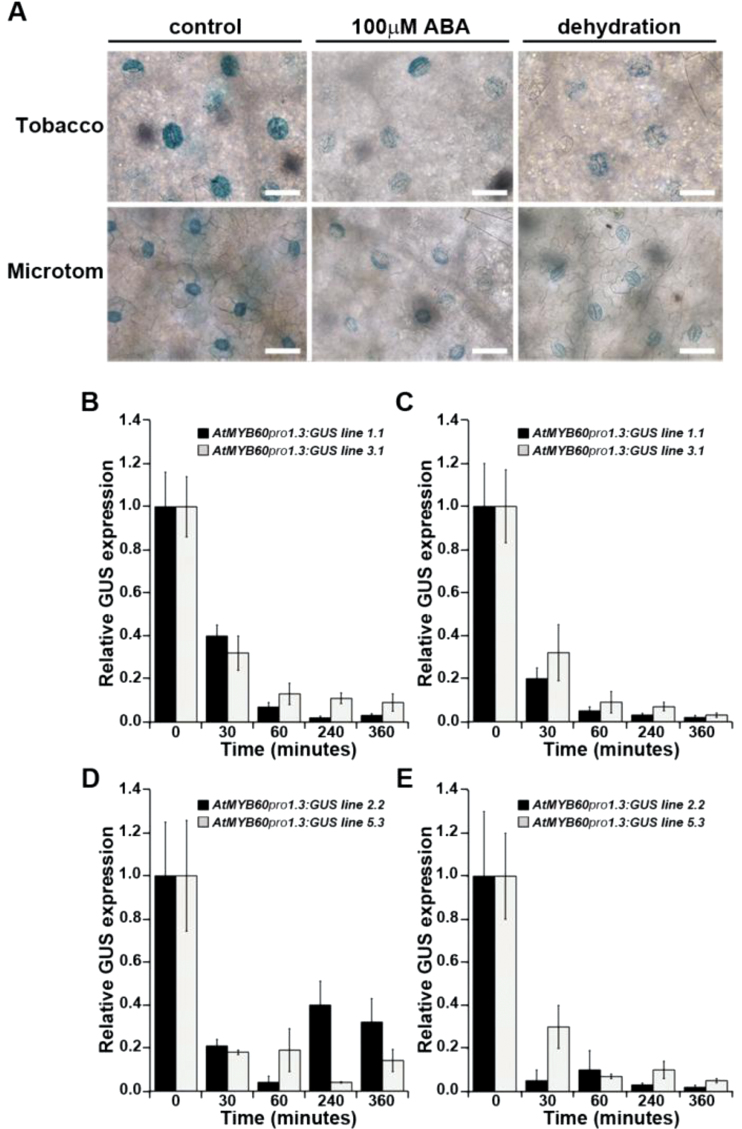
In Arabidopsis, the activity of the full-length AtMYB60 promoter is rapidly down-regulated following exogenous applications of ABA, whereas the 246bp minimal promoter is not affected by the hormone (Cominelli et al., 2011). ABA treatment of AtMYB60 pro 1.3:GUS tobacco and Microtom plants caused a marked reduction in histochemical detection of GUS signals in guard cells compared with mock-treated plants (Fig. 3A). These results were substantiated by qPCR analyses of two randomly selected lines, which showed significant down-regulation of the level of GUS transcripts upon ABA application (P < 0.001 for all time points, paired Student’s t-test) (Fig. 3B, D). A comparable down-regulation of GUS staining and GUS transcript abundance was also evident when leaves were subject to 6h of dehydration (Fig. 3A, C, E). The same results were obtained when Moneymaker AtMYB60 pro 1.3:GUS tomato plants were exposed to ABA or dehydration (data not shown).
Fig. 3.
ABA- and dehydration-induced down-regulation of GUS expression in the AtMYB60 pro 1.3:GUS lines. (A) Histochemical detection of GUS activity in guard cells from tobacco and Microtom AtMYB60 pro 1.3:GUS lines following 6h of exposure to 100 μM ABA or dehydration. Control and treated tissues were incubated in the staining solution for 24h. Bar=50 μm. (B–E) qPCR analysis of GUS expression in response to 100 μM ABA (B and D) or dehydration (C and E) in two independent tobacco (B and C) or Microtom lines (D and E). Total RNA samples were extracted at the indicated time points (minutes). Relative GUS transcript levels were determined using GUS-specific primers and normalized to the expression of the tobacco or tomato EF1α genes.
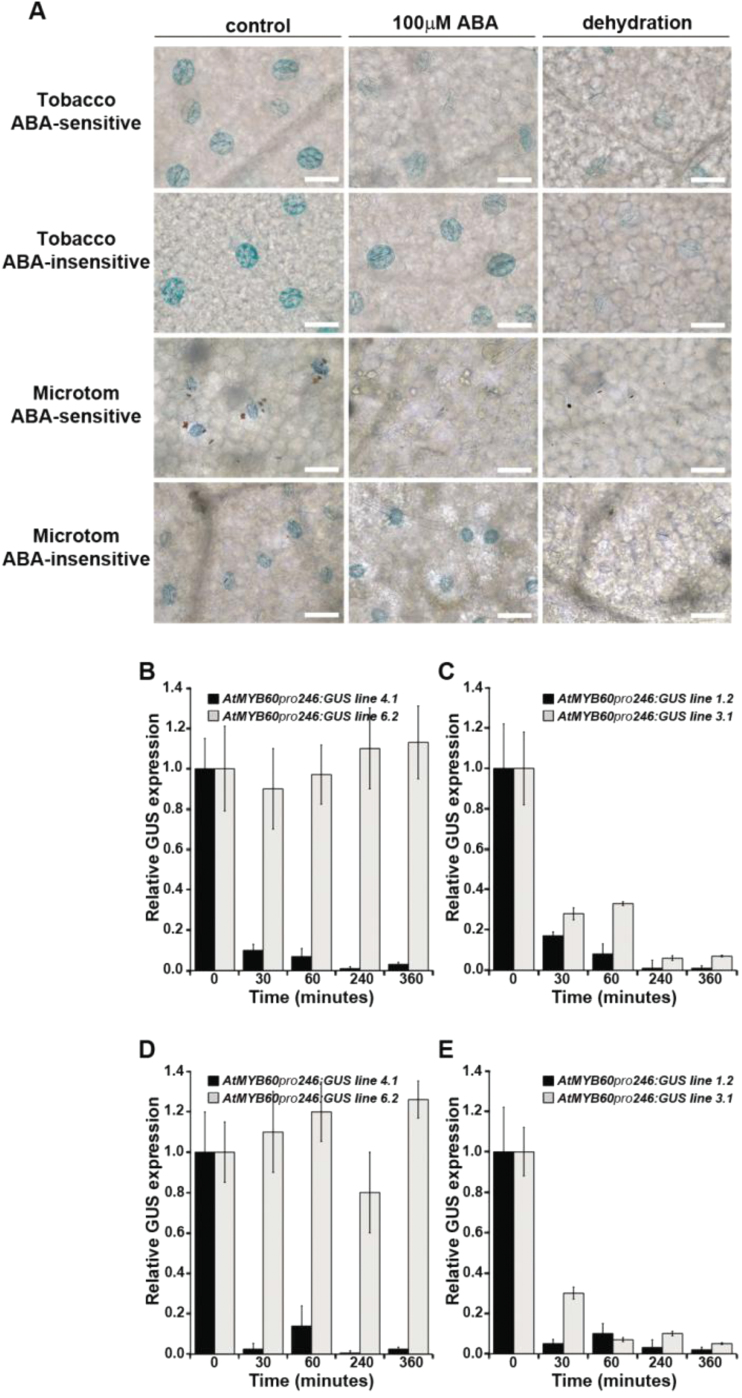
ABA treatment of the AtMYB60 pro 246:GUS lines yielded conflicting results. Nearly 80% of the Microtom (n=10), Moneymaker (n=10), or tobacco (n=15) lines analysed did not show obvious changes in the intensity of GUS staining in response to ABA (Fig. 4A). Surprisingly, the remaining 20% of the lines displayed marked down-regulation of GUS activity in guard cells, following exposure to ABA (Fig. 4A). These results were further confirmed by qPCR analysis of GUS expression in randomly selected lines (Fig. 4B, C). Nonetheless, dehydration treatments resulted in a drastic decrease of GUS expression in both ABA-insensitive and ABA-sensitive lines, suggesting a possible ABA-independent regulation of the 246bp minimal promoter in response to stress (Fig. 4A, C, D).
Fig. 4.
Analysis of GUS expression in the AtMYB60 pro 246:GUS lines in response to ABA and dehydration treatments. (A) Histochemical detection of GUS activity in guard cells from tobacco and Microtom AtMYB60 pro 246:GUS lines following 6h of exposure to 100 μM ABA or dehydration. Control and treated tissues were incubated in the staining solution for 48h. Bar=50 μm. (B and C) qPCR analysis of GUS expression in response to 100 μM ABA (B) or dehydration (C) in ABA-sensitive (black bars) and ABA-insensitive (grey bars) tobacco lines. (D and E) qPCR analysis of GUS expression in response to 100 μM ABA (D) or dehydration (E) in ABA-sensitive (black bars) and ABA-insensitive (grey bars) Microtom lines. Total RNA samples were extracted at the indicated time points (minutes). Relative GUS transcript levels were determined using GUS-specific primers and normalized to the expression of the tobacco or tomato EF1α genes.
Construction of an ABA- and dehydration-inducible guard cell-specific synthetic promoter.
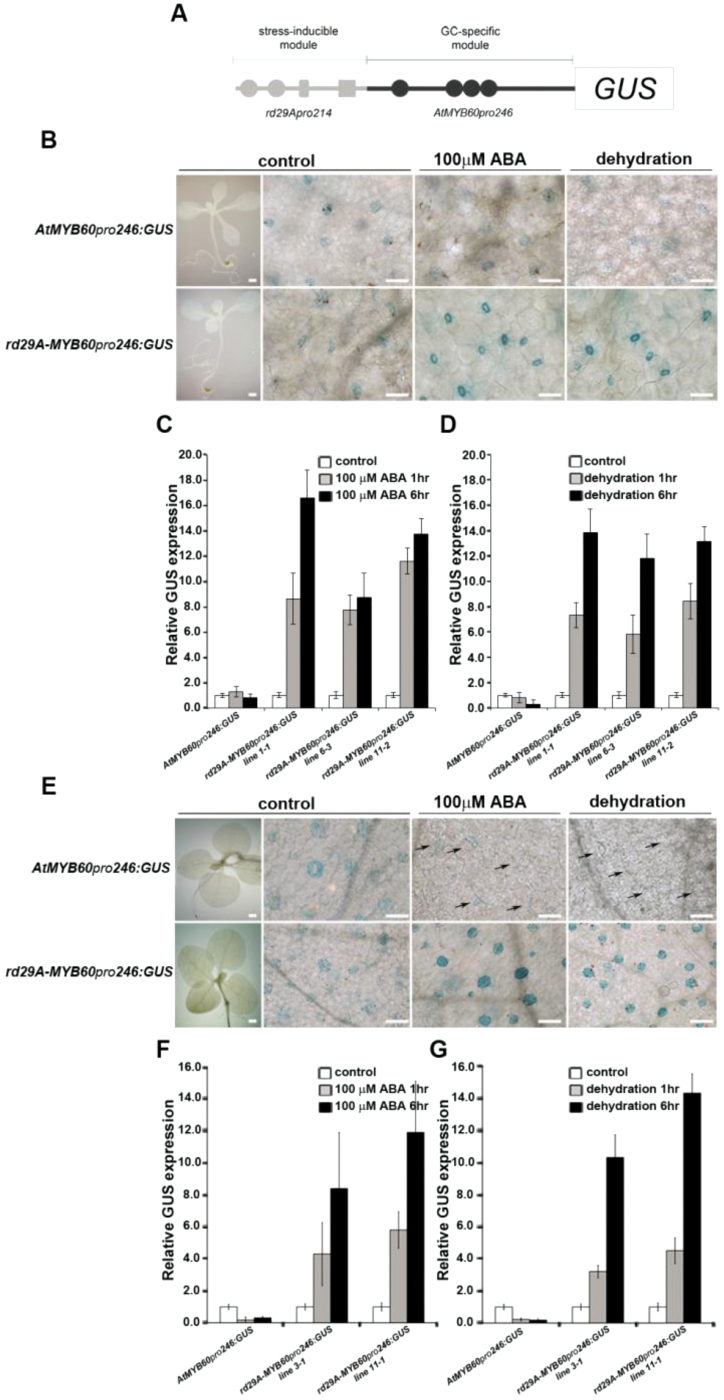
The conserved cellular specificity of the AtMYB60 promoter makes it a potentially valuable tool to manipulate guard cell activity in Solanaceae crops. However, an obvious pitfall for the general applicability of this tool resides in the strong ABA- and dehydration-induced down-regulation of its activity. In the attempt to reprogram the negative response of the AtMYB60 promoter to ABA and dehydration, a novel chimeric promoter was constructed. Such a synthetic regulatory element combined the AtMYB60 guard cell-specific module with the ABA- and stress-inducible rd29A promoter (At5g52310) (Yamaguchi-Shinozaki and Shinozaki, 1994). In more detail, the 246bp AtMYB60 minimal promoter was fused to the 214bp region of the rd29A promoter (from –254bp to –40bp), which contains two dehydration-responsive elements (DREs; TACCGACAT), a DRE-core motif (GCCGAC), one activator sequence (as1; TGACGTCA), and one ABA-responsive element (ABRE; TACGTGTC) (Yamaguchi-Shinozaki and Shinozaki, 1994; Narusaka et al., 2003). This regulatory region has been shown to activate gene expression strongly in response to osmotic stress through both ABA-dependent and ABA-independent pathways in different plant species (Kasuga et al., 1999, 2004; Wang et al., 2005). The resulting rd29A-MYB60 pro 246 chimeric promoter was fused to the reporter GUS and transformed in Arabidopsis (Fig. 5A). Fifteen independent T2 rd29A-MYB60 pro 246:GUS lines were selected for analysis of GUS expression. A previously described AtMYB60 pro 246:GUS line was used as a control for the experiments (Cominelli et al., 2011). Under standard growth conditions, all the transgenic lines expressing GUS under the control of the chimeric promoter showed a weak stomatal GUS pattern, comparable in intensity and distribution to that of the control line (Fig. 5B). In agreement with a previous report (Cominelli et al., 2011), the activity of the 246bp control promoter was largely unaffected by ABA, albeit that it was down-regulated by dehydration. Conversely, both ABA and dehydration treatments triggered a strong increase of GUS expression in the rd29A-MYB60 pro 246:GUS lines (Fig. 5B). Importantly, augmented GUS signals were only observed in guard cells and not in other cell types. qPCR analysis of three randomly selected lines confirmed the significant induction of GUS transcripts following ABA application or exposure to dehydration (up to 16-fold, P < 0.001, paired Student’s t-test) (Fig. 5C, D).
Fig. 5.
Rewiring of the activity of the AtMYB60 promoter in guard cells. (A) Schematic representation of the rd29A-MYB60 pro 246:GUS construct (not to scale). Grey circles represent DRE-core motifs (GCCGAC), the grey rectangle represents the as1 motif (TGACGTCA), the grey square represents the ABRE motif (TACGTGTC), and black circles represent DOF motifs ([A/T]AAAG). (B) Histochemical localization of GUS activity in guard cells from a 15-day-old AtMYB60 pro 246:GUS Arabidopsis control plant (upper left panel) and from a rd29A-MYB60 pro 246:GUS plant (lower left panel), grown under standard conditions (bar=1mm). Leaves from the AtMYB60 pro 246:GUS control line did not show evident changes in the intensity of the GUS staining following 6h of exposure to 100 μM ABA, whereas they showed reduced staining after 6h of dehydration treatment (upper panels). Conversely, ABA and dehydration tratments induced a drastic increase in GUS activity in the rd29A-MYB60 pro 246:GUS line (lower panels) (bar=50 μm). (C and D) qPCR analysis of GUS expression in response to 100 μM ABA (C) or dehydration (D) in three independent Arabidopsis rd29A-MYB60 pro 246:GUS lines. (E) Histochemical localization of GUS expression in guard cells of 15-day-old tobacco plants from an ABA-sensitive AtMYB60 pro 246:GUS control line (upper left panel) and from a rd29A-MYB60 pro 246:GUS line (lower left panel), grown under standard conditions (bar=1mm). After 6h of exposure to 100 μM ABA or dehydration, AtMYB60 pro 246:GUS control plants showed a severe decrease in the intensity of the GUS staining (upper panels, arrows indicate stomatal guard cells). In contrast, ABA and dehydration treatments resulted in enhanced GUS activity in the rd29A-MYB60 pro 246:GUS lines (lower panels) (bar=50 μm). (F and G) qPCR analysis of GUS expression in response to 100 μM ABA (C) or dehydration (D) in two independent tobacco rd29A-MYB60 pro 246:GUS lines. qPCR experiments in Arabidopsis (C and D) and tobacco (F and G) included an AtMYB60 pro 246:GUS line as a control. Relative GUS transcript levels were determined using gene-specific primers and normalized to the expression of the AtACTIN2 gene (At3g18780) in (C) and (D), or using the NtEF1α gene in (F) and (G).
Next, the rd29A-MYB60 pro 246:GUS construct was introduced in tobacco and tomato to test whether the synthetic promoter retained its cellular specificity and its responsiveness in these two species. Histochemical analysis of 10 independent T2 tobacco lines revealed expression of the reporter exclusively in stomatal guard cells, with an intensity of staining similar to the AtMYB60 pro 246:GUS line, used as a control (Fig. 5E). Following application of ABA or exposure to dehydration, all the rd29A-MYB60 pro 246:GUS lines showed significant up-regulation of GUS expression, as demonstrated by both the intensity of the staining (Fig. 5E) and the level of the GUS transcripts (Fig. 5F, G, up to 14-fold, P < 0.001, paired Student’s t-test). Notably, after 6h of exposure to dehydration, plants showed severe symptoms of wilting (Supplementary Fig. S3 at JXB online). Yet, the rd29A-MYB60 pro 246:GUS lines still showed intense GUS staining in stomata distributed on the damaged tissue. Comparable results, in terms of both cellular specificity and ABA- and dehydration-induced up-regulation of GUS expression, were obtained in tomato plants transformed with the rd29A-MYB60 pro 246:GUS construct (Supplementary Fig. S4).
As a whole, these findings validate the use of the AtMYB60 minimal promoter to engineer synthetic regulatory modules to activate gene expression in guard cells in response to hormonal signals and environmental cues, in both model plant systems and crops.
Discussion
The availability of a wide repertoire of cell-specific and inducible promoters has become increasingly important for all levels of genetic engineering in plants, from primary research to development of commercial biotech crops. Previous studies indicated that the AtMYB60 transcription factor is highly expressed in stomatal guard cells (Leonhardt et al., 2004; Galbiati et al., 2008; Bates et al., 2012) and demonstrated that its promoter sequence specifically activates transgene expression in stomata of Arabidopsis (Cominelli et al., 2005, 2011; Nagy et al., 2009; Meyer et al., 2010; Bauer et al., 2013). In this work the cellular specificity of the AtMYB60 promoter was investigated in rice, tobacco, and tomato. Analysis of several independent rice lines carrying the GUS gene under the control of either the minimal or the full-length AtMYB60 promoter did not detect reporter activity in guard cells or in any other cell type (Fig. 1A, B). Stomata found in grasses and in dicots are highly divergent in terms of cell morphology and tissue patterning (Serna, 2011). The lack of activity of the AtMYB60 promoter in rice probably reflects an evolution-driven difference in the transcriptional mechanisms that mediate gene expression in guard cells from monocots and dicots. Control of gene expression is largely determined by cis-regulatory modules localized in the promoter sequence of regulated genes and their cognate transcription factors. Clusters of DNA consensus sequences for DOF proteins ([A/T]AAAG), found upstream of the ATG codon of AtMYB60, have proved essential to activate gene expression in stomata (Cominelli et al., 2011). Consistently, a DOF-type transcription factor (Stomatal Carpenter 1; SCAP1) has been shown to bind the AtMYB60 promoter directly and to regulate AtMYB60 expression in guard cells (Negi et al., 2013). One likely possibility is that rice guard cells lack the trans-acting factors which bind to the [A/T]AAAG motifs in the AtMYB60 promoter, suggesting that grasses employ cis-elements other than DOF motifs to regulate gene expression in stomata. Further evidence indicates the lack of a MYB60-related guard cell-specific regulatory network in rice. Two putative AtMYB60 orthologues have been identified in the rice genome, namely LOC_Os11g03440 and LOC_Os12g03150 (Kawahara et al., 2013). A comprehensive transcriptomic analysis of several cell types from rice revealed that both genes are widely expressed in leaves but their expression is strongly down-regulated in guard cells compared with other cell types, including blade mesophyll, bundle sheath, and vein (Jiao et al., 2009).
In contrast to rice, in tobacco and tomato, activity of the AtMYB60 promoter was regulated to the same developmental, spatial, and cell-specific stringency as in Arabidopsis. Transgenic lines harbouring the AtMYB60 pro 1.3:GUS or the AtMYB60 pro 246:GUS construct revealed GUS expression exclusively in guard cells throughout plant development (Fig. 2). Oh and colleagues reported that a 1.2kb region of the AtMYB60 promoter can drive reporter gene expression in roots of Arabidopsis (Oh et al., 2011). Nevertheless, patchy patterns of GUS activity in root tissues were only detected in seedlings upon prolonged treatment (up to 24h) with indole acetic acid (IAA), a conditon which might not reflect the physiological role of the AtMYB60 promoter. Conversely, previous analysis of nearly 100 independent Arabidopsis lines carrying serial deletions of the AtMYB60 promoter fused to the GUS gene did not reveal expression of the reporter in root tissues under standard growth conditions or following exposure to ABA (Cominelli et al., 2011). Fully consistent with these results, no GUS staining was observed in roots from the tobacco and tomato lines described in this study (Fig. 2).
As previously observed in Arabidopsis (Cominelli et al., 2011), the 246bp AtMYB60 regulatory region showed weaker activity in guard cells from tobacco and tomato, compared with the 1.3kb full-length promoter (Fig. 1C–H). Different regions of the AtMYB60 promoter can thus be exploited to produce cell-specific expression systems tailored to achieve various level of transgenes expression in stomata.
Results from tobacco and tomato point to the conservation of the cis- and possibly trans-mechanisms that modulate gene expression in the guard cell of Arabidopsis and Solanaceae. Interestingly, a preliminary survey of the closest homologue of AtMYB60 found in the tomato genome (Solyc10g081490) revealed a high degree of similarity in the number, organization, and localization of DOF target sites with the promoter region of AtMYB60 (Supplementary Fig. S5 at JXB online). As clusters of [A/T]AAAG motifs have also been identified in the promoter of the guard cell-specific potassium channel KST1 from potato (Plesch et al., 2001), as well as in the regulatory region of VvMYB60, the functional orthologue of AtMYB60 from grape (Galbiati et al., 2011), it is intriguing to speculate that the conservation of guard cell-specific cis-regulatory elements extends across a wide range of dicotyledonous plants.
Previous data demonstrated that the activity of the full-length AtMYB60 promoter is negatively regulated by ABA in Arabidopsis (Cominelli et al., 2011). In agreement with this observation, exogenous application of the hormone or exposure to severe dehydration resulted in the rapid down-regulation of GUS expression in tobacco and tomato AtMYB60 pro 1.3:GUS lines (Fig. 3). The sequence comprised between –366bp and –262bp from the ATG start codon of AtMYB60 has been identified as the region responsible for the negative effect of ABA on gene expression (Cominelli et al., 2011). Consistent with this finding, Arabidopsis lines carrying the reporter gene under the control of the 246bp minimal promoter (devoid of the –366/–262bp region) did not show significant changes in GUS expression in response to ABA (Cominelli et al., 2011). Even though the majority of the tobacco and tomato AtMYB60 pro 246:GUS lines analysed in this study displayed insensitivity to ABA in terms of regulation of GUS activity, a few lines retained a negative response to the hormone (Fig. 4). This result could be the consequence of positional effects on the activity of the transgene, or could advocate the involvement of divergent cis-mechanisms that mediate ABA-induced gene repression in Arabidopsis and Solanaceae. Notably, both ABA-insensitive and ABA-sensitive lines revealed a drastic decrease of GUS activity in response to dehydration (Fig. 4A, C, D). This indicates that the AtMYB60 minimal promoter probably encompasses cis-acting elements capable of down-regulating gene expression in response to stress in an ABA-independent manner and demonstrates the functional conservation of such elements in Arabidopsis, tobacco, and tomato.
The observed ABA- and dehydration-induced down-regulation of the AtMYB60 promoter activity poses obvious limits to its applicability. Guard cell-specific regulatory modules suitable for the selective up-regulation of transgene expression upon stress imposition are highly desirable both for functional studies and for biotechnological applications in crops. Taking advantage of the well-characterized modular organization of the stress-activated rd29A promoter, a chimeric regulatory element (rd29A-MYB60 pro 246) intended for rewiring the activity of the AtMYB60 promoter was constructed. To this end, the 214bp stress-responsive module from the rd29A promoter was conjugated to the 246bp AtMYB60 guard cell-specific element (Fig. 5A). Analysis of several independent transgenic lines demonstrated that, in contrast to the native 246bp AtMYB60 promoter, the chimeric rd29A-MYB60 pro 246 system was capable of boosting gene expression in response to ABA or dehydration not only in Arabidopsis (Fig. 5B–D), but also in tobacco (Fig. 5E–G) and tomato (Supplementary Fig. S3 at JXB online). This implies that the dehydration-induced down-regulation of GUS expression mediated by the AtMYB60 minimal promoter is over-ruled by the stress-activated rd29A module. One likely possibility is that, under stress, repression of the 246bp AtMYB60 promoter is counteracted by the activity of the dehydration-induced CBF/DREB transcriptional activators, which directly bind the DRE motifs in the rd29A promoter (Shinozaki and Yamaguchi-Shinozaki, 2000).
Most importantly, the rd29A-MYB60 pro 246 regulatory module retained the tight cellular specificity of the AtMYB60 minimal promoter, as ABA- and dehydration-induced up-regulation of reporter gene expression exclusively occurred in guard cells (Fig. 5B, E; Supplementary Fig. S3 at JXB online). Interestingly, the 214bp region of the rd29A promoter, incorporated in the chimeric system, contains the root-specific activator sequence as1 (Lam et al., 1989). Lam and colleagues, demonstrated that the insertion of a single as1 motif in a green tissue-specific promoter is sufficient to confer root expression (Lam et al., 1989). Despite the presence of the as1 motif, GUS expression in roots (or in any other tissue devoid of stomata) was not observed in any of the rd29A-MYB60 pro 246 lines analysed in this study. This suggests that the control exerted by the rd29A-MYB60 pro 246 synthetic promoter over gene expression is predominantly mediated by the cell-specific module and employs trans-regulatory mechanisms that are differentially expressed in guard cells. One possible scenario is that, in its default state, the AtMYB60 promoter is inactivated by the binding of one or more transcriptional repressors. In the guard cell, the absence of such repressors (e.g. due to lack of expression or to selective protein degradation) allows for the binding of guard cell-specific trans-activators (e.g. SCAP1), to promote transcription.
A modular synthetic promoter for the spatio-temporal control of transgene expression in stomata has been reported by fusing a guard cell-specific element from the promoter of the potato phosphoenolpyruvate carboxylase (PEPC) gene with the ethanol-inducible gene switch AlcR/alcA (Xiong et al., 2009). This system resulted in reliable activation of transgene expression upon ethanol application in Arabidopsis stable transformants. Yet, expression of the transgene was not restricted to mature guard cells, as it was also observed in the guard cell lineage, including meristemoids and guard mother cells (Xiong et al., 2009). Even though the PEPC–AlcR/alcA module represents a valuable tool to investigate stomatal development and activity in model systems, its exploitation in field crops is rather difficult as it relies on exogenous application of ethanol to activate the expression of downstream genes. Conversely, the rd29A-MYB60 pro 246 system described in this study provides a more suitable tool to engineer stomatal activity in crops. It only employs plant-specific cis-elements and it is directly activated in response to stress, allowing for the spatial and temporal regulation of gene expression in a more physiological context. Most importantly, its activity in crop species, including tobacco and tomato commercial varieties, has been directly validated. Several biotechnological applications that employ the use of this regulatory module can be envisaged. For instance, recent evidence indicates that the regulation of the guard cell-autonomous ABA synthetic pathway plays a major role in modulating stomatal activity in response to stress (Bauer et al., 2013). In this respect, the rd29A-MYB60 pro 246 promoter represents a suitable tool to modulate the cell-specific and stress-regulated expression of key ABA biosynthetic genes (e.g. ABA3) in guard cells to tailor plant adaptation to the prevailing climatic conditions.
Taken together, results from this work corroborate the value of the AtMYB60 promoter as a tool to design novel flexible expression systems suitable for modulating stomatal activity in dicotyledonous plant model systems and crops. In addition, they rationalize the combinatorial engineering of hormone- and stress-responsive cis-motifs upstream of cell-specific core promoters for the accurate control of gene expression.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. PCR and RT–PCR analysis of independent T1 rice transgenic seedlings.
Figire S2. GUS expression pattern in flowers from tomato and tobacco AtMYB60 pro 1.3:GUS lines.
Figure S3. Histochemical localization of GUS expression in tobacco rd29A-MYB60 pro 246:GUS plants exposed to severe dehydration.
Figure S4. Rewiring of the activity of the AtMYB60 promoter in tomato guard cells.
Figure S5. Occurrence of [A/T]AAAG motifs in the 1.3kb regulatory region located upstream of the translational start codon of the AtMYB60 and Solyc10g081490 genes.
Table S1. Sequence of oligonucleotides used in this study for analysis of GUS expression.
Acknowledgements
This work was supported by ‘Progetto AGER, bando Viticoltura da Vino’, project SERRES 2010–2105, by Regione Lombardia, Fondo per la Promozione di Accordi Istituzionali, project BIOGESTECA 15083/RCC, and by Fondazione Umberto Veronesi per il Progresso delle Scienze, Milano, Italy, project AGRISOST.
References
- Bartley G, Ishida B. 2003. Developmental gene regulation during tomato fruit ripening and in-vitro sepal morphogenesis. BMC Plant Biology 3, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates GW, Rosenthal DM, Sun J, Chattopadhyay M, Peffer E, Yang J, Ort DR, Jones AM. 2012. A comparative study of the Arabidopsis thaliana guard-cell transcriptome and its modulation by sucrose. PLoS One 7, e49641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H, Ache P, Lautner S, et al. 2013. The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Current Biology 23, 1–5 [DOI] [PubMed] [Google Scholar]
- Ciais P, Reichstein M, Viovy N, et al. 2005. Europe-wide reduction in primary productivity caused by the heat and drought in 2003. Nature 437, 529–533 [DOI] [PubMed] [Google Scholar]
- Clough S, Bent A. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743 [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Albertini A, Fornara F, Conti L, Coupland G, Tonelli C. 2011. DOF-binding sites additively contribute to guard cell-specificity of AtMYB60 promoter. BMC Plant Biology 11, 162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Tonelli C. 2010. Transcription factors controlling stomatal movements and drought tolerance. Transcription 1, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C. 2005. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Current Biology 15, 1196–1200 [DOI] [PubMed] [Google Scholar]
- Davuluri G, van Tuinen A, Fraser P, et al. 2005. Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nature Biotechnology 23, 890–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati M, Matus JT, Francia P, Rusconi F, Canon P, Medina C, Conti L, Cominelli E, Tonelli C, Arce-Johnson P. 2011. The grapevine guard cell-related VvMYB60 transcription factor is involved in the regulation of stomatal activity and is differentially expressed in response to ABA and osmotic stress. BMC Plant Biology 11–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati M, Simoni L, Pavesi G, Cominelli E, Francia P, Vavasseur A, Nelson T, Bevan M, Tonelli C. 2008. Gene trap lines identify Arabidopsis genes expressed in stomatal guard cells. The Plant Journal 53, 750–762 [DOI] [PubMed] [Google Scholar]
- Gardner M, Bakerù A, Assie J, Poethig R, Haseloff J, Webb A. 2009. GAL4GFP enhancer trap lines for analysis of stomatal guard cell development and gene expression. Journal of Experimental Botany 60, 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J. 1999. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. The Plant Cell 11, 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal 6, 271–282 [DOI] [PubMed] [Google Scholar]
- Horsch R, Fry J, Hoffmann N, Eichholtz D, Rogers S, Fraley R. 1985. A simple and general method for transferring genes into plants. Science 227, 1229–1231 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Tausta SL, Gandotra N, et al. 2009. A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nature Genetics 41, 258–263 [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. 1999. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotechnology 17, 287–291 [DOI] [PubMed] [Google Scholar]
- Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K. 2004. A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant and Cell Physiology 45, 346–350 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton J, et al. 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Bohmer M, Hu H, Nishimura N, Schroeder J. 2010. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annual Review of Plant Biology 61, 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M, Perfus-Barbeoch L, Frelet A, Gaedeke N, Reinhardt D, Mueller-Roeber B, Martinoia E, Forestier C. 2003. The plant multidrug resistance ABC transporter AtMRP5 is involved in guard cell hormonal signalling and water use. The Plant Journal 33, 119–129 [DOI] [PubMed] [Google Scholar]
- Lam E, Benfey P, Gilmartin P, Fang R-X, Chua N-H. 1989. Site-specific mutations alter in vitro factor binding and change promoter expression pattern in transgenic plants. Proceedings of the National Academy of Sciences, USA 86, 7890–7894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak MJ, Robert N, Waner D, Leonhardt G, Schroeder JI. 2004. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscissic acid hyperseneitive protein phosphatase 2C mutant. The Plant Cell 16, 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Shi L, Han C, Yu J, Li D, Zhang Y. 2012. Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS One 7, e46451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell DB, Burke MB, Tebaldi C, Mastrandrea MD, Falcon WP, Naylor RL. 2008. Prioritizing climate change adaptation needs for food security in 2030. Science 319, 607–610 [DOI] [PubMed] [Google Scholar]
- McCormick S. 1991. Transformation of tomato with Agrobacterium tumefaciens. Plant Tissue Culture Manual B6, 1–9 [Google Scholar]
- Meyer S, Mumm P, Imes D, Endler A, Weder B, Al-Rasheid K, Geiger D, Marten I, Martinoia E, Hedrich R. 2010. AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. The Plant Journal 63, 1054–1062 [DOI] [PubMed] [Google Scholar]
- Nagy R, Grob H, Weder B, Green P, Klein M, Frelet-Barrand A, Schjoerring J, Brearley C, Martinoia E. 2009. The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. Journal of Biological Chemistry 284, 33614–33622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari Z, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K. 2003. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. The Plant Journal 34, 137–148 [DOI] [PubMed] [Google Scholar]
- Negi J, Moriwaki K, Konishi M, et al. 2013. A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Current Biology 23, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Yokota E, Wada E, Shimmen T, Okada K. 2003. An Arabidopsis ACT2 dominant-negative mutation, which disturbsF-actin polymerization, reveals its distinctive function in root development. Plant and Cell Physiology 44, 1131–1140 [DOI] [PubMed] [Google Scholar]
- Oh J, Kwon Y, Kim J, Noh H, Hong S-W, Lee H. 2011. A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Molecular Biology 77, 91–103 [DOI] [PubMed] [Google Scholar]
- Pei Z-M, Ghassemian M, Kwak C, McCourt P, Schroeder J. 1998. Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282, 287–290 [DOI] [PubMed] [Google Scholar]
- Plegt L, Bino R. 1989. β-Glucoronidase activity during development of the male gametophyte from transgenic and nontransgenic plants. Molecular and General Genetics 216, 321–327 [Google Scholar]
- Plesch G, Ehrhardt T, Mueller-Roeber B. 2001. Involvement of TAAAG elements suggests a role for Dof transcription factors in guard cell-specific gene expression. The Plant Journal 28, 455–464 [DOI] [PubMed] [Google Scholar]
- Schroeder J, Kwak J, Allen G. 2001. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410, 327–333 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. 2000. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Current Opinion in Plant Biology 3, 217–223 [PubMed] [Google Scholar]
- Serna L. 2011. Stomatal development in Arabidopsis and grasses: differences and commonalities. International Journal of Developmental Biology 55, 5–10 [DOI] [PubMed] [Google Scholar]
- Sinclair TR, Muchov RC. 2001. System analysis of plant traits to increase grain yield on limited water supplies. Agronomy Journal 93, 263–270 [Google Scholar]
- Wang Y, Beaith M, Chalifoux M, Ying J, Uchacz T, Sarvas C, Griffiths R, Kuzma M, Wan J, Huang Y. 2009. Shoot-specific down-regulation of protein farnesyltransferase (α-subunit) for yield protection against drought in canola. Molecular Plant 2, 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ying J, Kuzma M, et al. 2005. Molecular tailoring of farnesylation for plant drought tolerance and yield protection. The Plant Journal 43, 413–424 [DOI] [PubMed] [Google Scholar]
- Xiong T, Hann C, Chambers J, Surget M, Ng C-Y. 2009. An inducible, modular system for spatio-temporal control of gene expression in stomatal guard cell. Journal of Experimental Botany 60, 4129–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 1994. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell 6, 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel R, Schroeder JI. 2008. Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 19, 4–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hu Y, Dai M, Huang L, Zhoub D-X. 2009. The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. The Plant Cell 21, 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.