Abstract
The interaction of the hepatitis C virus (HCV) RNA-dependent RNA polymerase with RNA substrate is incompletely defined. We have characterized the activities of the HCV NS5B polymerase, modified by different deletions and affinity tags, with a routinely used homopolymeric substrate, and established apparent affinities of the various NS5B constructs both for the NTP and the template/primer substrates. We identified a uniquely tagged HCV NS5B RNA polymerase construct with a lower affinity (higher Km) than mature HCV NS5B for template/ primer substrate and highlighted the use of such a polymerase for the identification of inhibitors of NS5B activity, particularly inhibitors of productive RNA binding. The characterization of specific benzimidazole-5-carboxamide-based inhibitors, identified in a screening campaign, revealed that this class of compounds was non-competitive with regard to NTP incorporation and had no effect on processive elongation, but inhibited an initiation phase of the HCV polymerase activity. The potency of these compounds versus a panel of different NS5B polymerase constructs was inversely proportional to the enzymes’ affinities for template/primer substrate. The benzimidazole-5-carboxamide compounds also inhibited the full-length, untagged NS5B de novo initiation reaction using HCV 3′-UTR substrate RNA and expand the diversifying pool of potential HCV replication inhibitors.
INTRODUCTION
Discovered more than a decade ago, hepatitis C virus (HCV) prevalence is now estimated at approximately 170 million people worldwide (1,2). The plus-strand HCV RNA genome is ∼9600 nt in length and encodes at least one open-reading frame with approximately 3010 amino acids. In infected cells, this polyprotein is cleaved at multiple sites by cellular and viral proteases to produce structural and non-structural (NS) proteins (3,4). One of the NS proteins, NS5B, is an RNA-dependent RNA polymerase (RdRp) that catalyzes the replication of HCV (5). The enzyme is a prime target in the search for inhibitors of HCV replication and a variety of in vitro assays for HCV NS5B polymerase activity have been developed. Though primer-independent de novo initiation of complementary strand RNA polymerization can be reconstituted in vitro with templates that represent the 3′ end of either the plus- or minus-strand genome (6–12), many screening assays utilize synthetic homopolymeric templates/primers (5,7,13–22). Specific inhibitors of the HCV polymerase recently identified from such screening campaigns can be broadly classified as either non-nucleoside compounds that may affect an initiation step (23–26) or nucleoside analogs that inhibit polymerase elongation (27,28).
The recombinant HCV NS5B polymerase enzymes commonly used in in vitro assays are produced and isolated from either Escherichia coli or baculovirus-infected insect cells. Expression of the full-length HCV NS5B, either untagged or tagged (such as a hexa-histidine tag or GST tag), results in insoluble protein requiring extraction with detergents, salt and glycerol (5,7,14,15,18–20,22,29). The HCV NS5B protein has a highly conserved C-terminal hydrophobic segment and truncation of this portion in recombinant clones results in the expression of a soluble form of the enzyme that retains in vitro activity (16,17,19). In addition to their use in screening campaigns, these soluble forms of the enzyme have been particularly effective in crystallizing the NS5B to reveal an X-ray derived structure similar to other polymerases, but with an encircled active site (30–32). Recent X-ray derived structures of compounds bound to NS5B reveal a variety of potentially distinct inhibitor pockets, many of which localize to the thumb domain (26,33,34).
The activity of NS5B in in vitro polymerase reactions with homopolymeric RNA requires interaction with multiple substrates that include a template/primer and ribonucleotide triphosphate. Steady-state kinetic parameters, such as Km, can be determined for both the ribonucleotide triphosphate (6,14,17,19,20) and the template/primer (17) substrates. The C-terminally truncated, soluble forms of recombinant HCV polymerases have high affinity (a low Km value in the nanomolar range) for homopolymeric template/primer (17). In order to identify compounds that interfere with template/primer binding, compound concentration and/or affinity must surpass the Km of the substrate in a competitive inhibition assay. In this report we describe a modified HCV polymerase enzyme, distinct from the commonly used truncated forms, that has lower affinity for primer/template substrate. The characteristics of this modified enzyme in an in vitro assay facilitated the identification of a class of benzimidazole-5-carboxamide-based compounds that specifically inhibit HCV NS5B productive RNA binding. The compounds’ mode of inhibition is confirmed by steady state kinetics and order of addition experiments wherein we demonstrate that they interfere with the initiation process of RNA replication, rather than processive elongation. This distinct class of inhibitors would not only complement inhibitors of other HCV targets, but may also complement nucleoside analogs and other non-nucleoside NS5B inhibitors to expand the repository of potential HCV therapeutics.
MATERIALS AND METHODS
Production and purification of the different polymerase constructs
HT-NS5B. Briefly, the entire HCV NS5B region was amplified by PCR from a full-length HCV 1b genotype clone (HCV1b-40) and cloned into a pFastBacHTa vector (Invitrogen). The resulting vector, encoding the NS5B sequence with a hexa-histidine N-terminal fusion under the control of the polyhedrin promoter, was used as a donor to introduce the NS5B into a recombinant baculovirus. Sf21 insect cells were grown to a density of 1 × 106 cells/ml and infected with BacHTaNS5B16-3 recombinant baculovirus expressing the HCV polymerase. Cells expressing the HCV polymerase were harvested by centrifugation, washed with PBS and resuspended in lysis buffer (25 mM Tris–HCl pH 7.5, 2 mM 2-mercaptoethanol, 5 mM MgCl2, 1 mM EDTA, 500 mM NaCl, 50% glycerol, 2% Triton X-100, protease inhibitor cocktail). Cells were homogenized and treated with bovine pancreas DNaseI. The homogenate was sonicated and centrifuged at 105 000 g for 45 min. Supernatants were pooled and subjected to metal affinity chromatography using a Qiagen Ni-NTA column. The polymerase was eluted with an 85–400 mM imidazole linear gradient. The material was then applied onto a DEAE–Sepharose column. The flow-through and washes were pooled for subsequent purification using heparin–Sepharose chromatography. Bound protein was eluted using a linear gradient of 200–1000 mM NaCl. In order to maintain solubility of the HT-NS5B, all of the chromatography buffers contained 0.05% Triton X-100 and 0.1% NP-40. Fractions enriched (>90% purity) in NS5B (according to Coomassie-stained SDS–PAGE) were pooled. The protein concentration of this pool was determined by the micro-Bradford method (Bio-Rad) using BSA as standard. This pool was aliquoted and stored at –80°C (in 20 mM Tris–HCl pH 7.5, 1 mM EDTA, 800 mM NaCl, 20% glycerol, 0.05% Triton X-100 and 0.1% NP-40) without any significant loss of activity during 3 years of storage. The yield of purified protein was 1 mg/l of cultured Sf21 cells and it was used in the standard HCV RdRp assay at a final concentration of 10 nM.
HT-NS5BΔ21 and NS5BΔ21-HT. The recombinant HCV NS5B polymerase can be produced in soluble form by expression of a variant that lacks the C-terminal 21 amino acids (16,17,19). We expressed this NS5BΔ21 with an N-terminal hexa-histidine (termed HT-NS5BΔ21) and with a C-terminal hexa-histidine tag (termed NS5BΔ21-HT). Expression of these genes from pET vectors in E.coli strain JM109 (DE3) was induced with 0.4 mM IPTG for 3 h at 24°C. Cells were harvested and lysed in a microfluidizer. The lysate, after centrifugation, was purified according to the HT-NS5B protocol: Ni-NTA, DEAE–Sepharose and heparin–Sepharose chromatography, in buffers lacking detergent. The proteins were thereafter concentrated on a Resource S column, and applied to a Superdex 200 column where peak fractions containing highly pure (>98%) and monomeric histidine tag NS5BΔ21 were pooled and stored at –80°C in (20 mM Tris pH 7.5, 10% glycerol, 5 mM DTT and 300 mM NaCl) for more than 3 years without significant loss in activity. The yield of both enzymes was ∼2 mg/l of E.coli culture and they were used in the standard HCV RdRp assay at final concentration of 2–3 nM.
NS5BΔ57-HT. This polymerase variant that lacks the C-terminal 57 amino acids was expressed with a C-terminal hexa-histidine tag (termed NS5BΔ57-HT). Expression of this NS5B gene construct from a pET vector in E.coli strain JM109 (DE3) was induced with 0.4 mM IPTG for 3 h at 24°C. Cells were harvested and lysed in a microfluidizer. The lysate, after centrifugation, was first purified according to the HT-NS5BΔ21 protocol: Ni-NTA, DEAE–Sepharose and heparin–Sepharose chromatography and then loaded onto a Q-Sepharose column. The flow-through from this last step was collected and concentrated with centrifugal concentrators. Analytical gel filtration revealed highly pure (>95%), monomeric protein with an overall yield of 1 mg/l of culture. This enzyme was used at a concentration of 2 nM in the HCV RdRp assay.
NS5B. The mature, full-length NS5B polymerase was produced as a tagged precursor (HTaA5B) in Sf21 insect cells infected with a recombinant baculovirus encoding the HCV NS5B, with an N-terminal hexa-histidine tag linked by the C-terminal portion of NS5A (BacHTaA5B) to the NS5B segment. The N-terminal histidine tag was removed by processing with the HCV NS3 protease and a NS4A peptide cofactor, resulting in a mature, untagged form of the HCV RdRp (12). Briefly, the precursor histidine-tagged polymerase was purified according to the standard protocol in buffers containing 0.15% n-dodecyl-β-d-maltoside and then cleaved with the NS3/4A protease using a 1:50:1.25 molar ratio of NS3 protease:NS4A cofactor:HTaA5B. The reaction was performed at room temperature for 45 min followed by incubation at 4°C for 5 h. The mature NS5B RdRp was then isolated by removing the cleaved histidine-tag leader and any uncleaved HTaA5B protein with metal affinity resin. Unbound mature NS5B was subjected to heparin chromatography as described above in order to separate the NS3 protease from NS5B RdRp. The NS5B fraction obtained from heparin chromatography underwent a final step of gel filtration from which mature monomeric NS5B enzyme was recovered. The enzyme (final yield ∼0.1mg/l of Sf21 cells) was stored at –80°C for more than 3 years without loss in activity, and was used at 1–2 nM in the standard HCV RdRp assay.
Poliovirus RNA polymerase. The enzyme was produced in E.coli BL21(DE3) harboring a T7 expression plasmid encoding the poliovirus RNA polymerase (a kind gift from K. Kirkegaard, Stanford University). The protein was purified by precipitation with ammonium sulfate at 40% of saturation, followed by S-Sepharose and Q-Sepharose chromatographies. After a final gel-filtration step, the preparation was homogeneous as revealed by protein gel-electrophoresis. The enzyme was stored at –20°C in storage buffer [25 mM Tris–HCl pH 7.5, 300 mM NaCl, 5 mM DTT, 1 mM EDTA, 0.1% (v/v) Igepal CA-630 and 30% (v/v)glycerol].
Calf thymus RNA polymerase II. RNA polymerase II was purified from fresh calf thymus tissue extracts by polyethyleneimine and ammonium sulfate precipitation followed by standard chromatography techniques according to the method of Kim and Dahmus (35). The enzyme stock solution was stored in 50 mM Tris–HCl pH 7.9, 10 mM EDTA, 10 mM EGTA, 400 mM ammonium sulfate, 0.25 mM DTT and 20% glycerol (v/v).
HCV RNA-dependent RNA polymerase assay
The standard HCV RdRp assay was performed in 96-well plates using 2–10 nM of enzyme (as indicated), 0.5 µCi of [3H]UTP, 1 µM UTP, 250 nM 5′-biotinylated oligo(rU12), 10 µg/ml poly(rA) in 20 mM Tris–HCl pH 7.5, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 0.2 U/µl of RNasin, 5% DMSO, 3% glycerol, 30 mM NaCl, 0.33% dodecyl-β-d-maltoside, 0.01% IGEPAL. The 60 µl reaction was terminated after 90 min at 22°C by the addition of 20 µl of stop solution (150 µg/ml of tRNA in 0.5 M EDTA) and 30 µl of streptavidin-coated beads [8 mg/ml in 20 mM Tris–HCl pH 7.5, 25 mM KCl, 0.025% (w/v) sodium azide] for a Scintillation Proximity Assay (Perkin Elmer-Amersham). After 30 min at room temperature, 75 µl of 5 M cesium chloride were added to the wells and the plate was left at room temperature for 1 h before quantifying the radioactive UMP incorporated onto the biotinylated primer by counting for 60 s on a TopCount (Packard).
Poliovirus RNA-dependent RNA polymerase assay
The protocol was identical to the standard HCV RdRp assay, except that the poliovirus RNA polymerase enzyme was used at a final concentration of 20 nM.
Calf thymus RNA polymerase II assay
This assay measures the incorporation of 33P-UTP during the transcription of the calf thymus DNA by calf thymus RNA polymerase II (35). The assay was performed in 96-well plates using 4 µg/ml of calf thymus polymerase enzyme, 100 µg/ml of calf thymus DNA, 250 µM each of GTP, CTP and ATP, 1 µM UTP, 2.5 µCi/ml of 33P-UTP in 80 mM Tris–HCl pH 7.9, 80 mM (NH4)2SO4, 1 mM MnSO4, 2.25 mM MgCl2, 0.035 mM EDTA, 1 mM DTT, 0.1 U/µl of RNasin, 5% DMSO, 6% glycerol. The 60 µl reaction was terminated after 120 min at 37°C by TCA precipitation and filtration of the RNA transcripts. The retained RNA products were quantified using a TopCount instrument (Packard).
Enzymatic characterization of the different NS5B polymerase constructs
In order to determine the Km for the template/primer [poly(rA)/oligo(U12)], a saturating amount of UTP (25–50 µM; 0.08–0.2 µCi/µl of 33P-UTP) was used in the assay in the presence of increasing concentrations of poly(rA)/oligo(U12) (with a template/primer ratio maintained at 10). Similarly, for the determination of the Km for UTP, a saturating amount of template/primer was used (250–1000 nM, depending on the enzyme) in the presence of increasing concentrations of UTP (0.1–200 µM; 0.03–0.07 µCi/µl). Conditions of other reagents in the reaction were identical to the biochemical RdRp assay. Depending on the specific activity of the different enzyme constructs, their concentration used in enzymatic characterization varied between 0.5 and 10 nM. Aliquots of 8 µl were removed at specific times, spotted onto DE81 filter paper discs, washed in phosphate buffer to remove unincorporated UTP, rinsed and counted to quantify the amount of UMP incorporated into bound product RNA. Velocities of the reactions with each concentration of poly(rA)/oligo(U12) (or UTP) were determined. The data were processed and analyzed with kinetics software (GraFit Erithacus Software) to obtain the Km for both template/primer and UTP.
Determination of the Ki and mode of compound 1 inhibition
Two series of reactions were performed to determine the mode of inhibition of compound 1. In the first series of experiments, velocities of reactions were determined at different template/primer and inhibitor concentrations. The concentration of template/primer ranged from 25 to 1000 nM with a fixed concentration of UTP at 25 µM (reactions contained up to 0.2 µCi/µl of 33P-UTP). The concentration of enzyme used in the assay was 5 nM and the concentration of inhibitor ranged from 0.25- to 4-fold the IC50 value. For each of these reactions, velocities were determined by withdrawing aliquots at defined times, transferring them on DE81 filter discs and measuring bound radioactivity on a DE81 filter as described above.
In the second series of experiments, velocities were monitored at different UTP and inhibitor concentrations. The template/primer concentration was fixed (at 250 nM), the UTP concentration ranged from 0.25 to 50 or 100 µM (with 0.02–0.2 µCi/µl of 33P-UTP), and the concentration of the inhibitor ranged from 0.25- to 4-fold the IC50 value. The NS5B polymerase used in this series of experiments ranged in concentration from 10 to 25 nM. Kinetic results were then plotted according to the method of Cornish-Bowden (36), allowing for determination of the mode of inhibition as well as the constant(s) of inhibition (Ki as the competitive part of inhibition and Ki′ as the uncompetitive part of inhibition).
Determination of IC50 values with different NS5B polymerase constructs
The IC50 values for a series of compounds were determined with the five different constructs of the NS5B polymerase. All of the RdRp assays were performed in SPA format precisely as described in the standard protocol with the indicated enzyme construct. From serial dilution of the test compound, the percentage inhibition was plotted against the compound concentration and a non-linear curve was fitted (Hill model) to the percentage inhibition–concentration data. The calculated percentage inhibition values were then used to determine the median inhibitory concentration IC50, slope factor (n) and maximum inhibition (Imax) by the non-linear regression procedure of SAS (Statistical Software System, SAS Institute Inc., Cary, NC, USA) using the following equation:
![]()
The specificity of compounds as inhibitors of HCV polymerase was evaluated through a poliovirus RNA polymerase counter screen in an SPA assay format that utilizes the identical substrates and conditions previously optimized for the standard HCV polymerase assay. The compounds were further profiled for inhibition of DNA-dependent RNA polymerase II (purified from calf thymus).
Modified order of addition single cycle HCV NS5B RdRp assay
A single-cycle of HCV RdRp initiation and elongation was assayed by modifying the order of addition, followed by the addition of heparin. The template/primer (250 nM) was first pre-incubated with the enzyme (10 nM) for 20 min at room temperature, after which heparin (final concentration of 1.2 µg/ml), the inhibitor and 0.5 µM [3H]UTP (0.013 µCi/µl) were successively added to the reaction. Following a period of 90 min incubation at room temperature, stop solution and streptavidin beads were added as in the standard HCV RdRp assay. Radioactivity in the wells was quantified on a TopCount and the inhibitory potential of a series of compounds was evaluated.
HCV NS5B de novo initiation reaction
The primer-independent de novo initiation activity of the mature full-length NS5B construct was reconstituted with substrates and conditions as previously described (12). Products were analyzed on denaturing PAGE gels or quantified on DE81 filters after washing and removal of unincorporated nucleotides.
Compound synthesis
Compounds 1, 1a–1e inclusively, and compound 2 were prepared using published procedures (37–39). All compounds had 1H-NMR and mass spectral data consistent with their assigned structures. Compound formulas are as indicated: compound 1, C37H47N5O5; compound 1a, C28H29N3O6; compound 1b, C27H27N3O5; compound 1c, C29H29N3O7; compound 1d, C29H28N4O5; compound 1e, C31H30N4O7; and compound 2, C16H10Cl3NO6S.
RESULTS
Enzymatic characterization of the different NS5B polymerase constructs
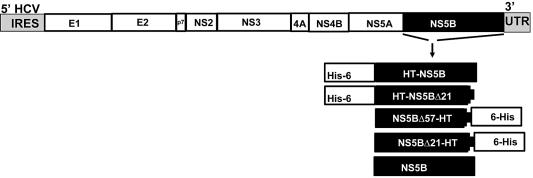
We examined a panel of purified HCV NS5B polymerase constructs that had different terminal modifications and placements of affinity tags, as depicted in Figure 1. Kinetic parameters, such as the Km, were determined for both the template/primer and the UTP. Determination of a Km for template/primer RNA with the HT-NS5B construct is exemplified in Figure 2, where the reaction velocity, as measured by UMP incorporation onto the extending RNA primer, is plotted against a range of template/primer concentrations and performed with saturating (50 µM) amounts of UTP. Similarly, the Km for UTP was determined for this and other NS5B constructs by varying the UTP concentration in the presence of saturating amounts of template/primer RNA. The Km for UTP among the different NS5B constructs ranged from 1.8 to 12 µM (Table 1). The Km for the template/primer varied <3-fold among four of the five NS5B constructs (HT-NS5BΔ21, NS5BΔ57-HT, NS5BΔ21-HT and NS5B) and ranged from 0.025 to 0.058 µM (Table 1); however, the HT-NS5B (full-length NS5B with an N-terminal hexa-histidine tag) displayed a significantly lower affinity (Km = 0.21 ± 0.02 µM).
Figure 1.
Schematic representation of the five NS5B constructs cloned and produced from an HCV 1b genome. The full-length N-terminal hexa-histidine HT-NS5B fusion and the full-length untagged NS5B were produced and purified from recombinant baculovirus-infected insect cells. Three NS5B clones were expressed in E.coli: the HT-NS5BΔ21, harboring an N-terminal hexa-histidine tag and C-terminal 21 amino acid deletion; the NS5BΔ57-HT encoding a C-terminal 57 amino acid deletion fused to hexa-histidine tag; and the NS5BΔ21-HT encoding a C-terminal 21 amino acid deletion fused to a hexa-histidine tag.
Figure 2.
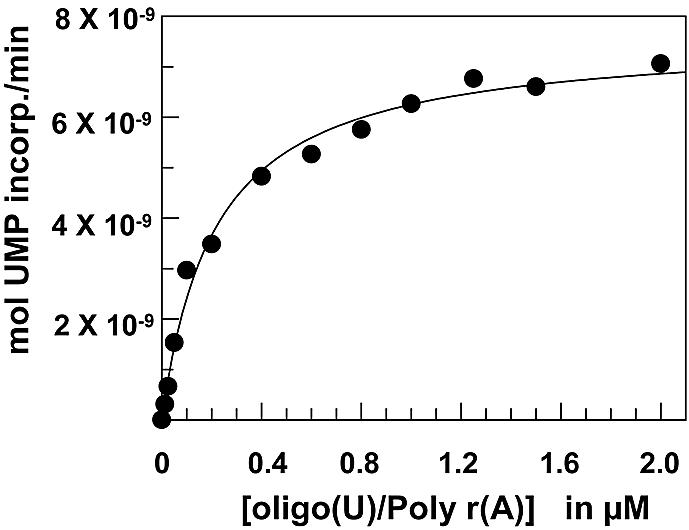
Determination of the Km for the template/primer poly(rA)/oligo(U) with the HT-NS5B enzyme. Reaction velocity, measured as 33P-UMP incorporated into product, was plotted versus increasing concentrations (ranging from 25 to 2000 nM) of poly(rA)/oligo(U) substrate. The curve was fit with Grafit Software (GraFit Erithacus Software) to obtain a Km of 214 nM.
Table 1. Kinetic analyses of different constructs of NS5B polymerases.
| HT-NS5B | HT-NS5BΔ21 | NS5BΔ57-HT | NS5BΔ21-HT | NS5B | |
|---|---|---|---|---|---|
|
Km (P/T)a (µM) |
0.21 ± 0.02 |
0.058 ± 0.01 |
0.034 ± 0.01 |
0.025 ± 0.005 |
0.025 ± 0.004 |
| Km (UTP)b (µM) | 6.2 ± 0.63 | 12 ± 2.3 | 1.8 ± 0.3 | 5.2 ± 1.3 | 3.3 ± 1.3 |
aValues are the mean of two (n = 2) determinations except for HT-NS5B (n = 3).
bValues are the mean of two (n = 2) determinations except for NS5BΔ21-HT (n = 3).
Identification and characterization of a specific HT-NS5B inhibitor
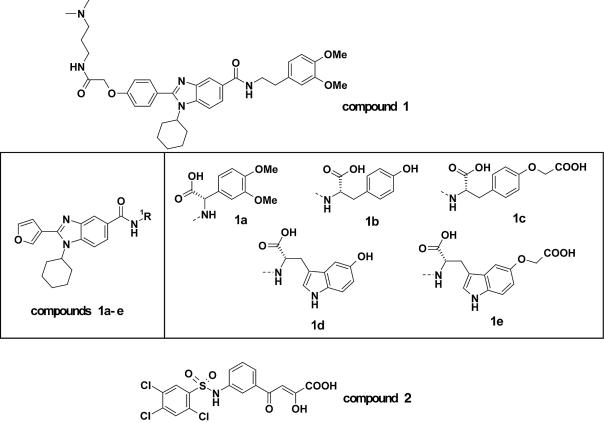
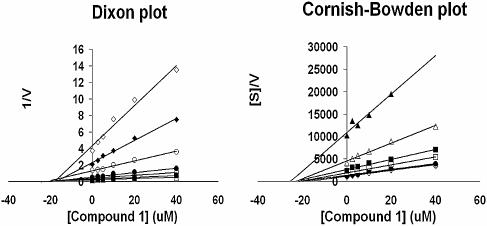
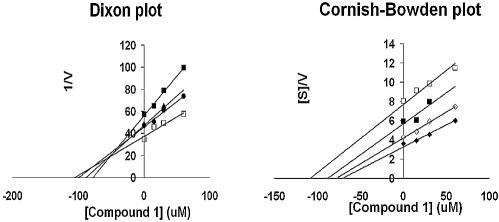
Screening of broad chemical compound collections for specific inhibitors of HCV polymerase activity was performed with the HT-NS5B construct. We postulated that the lower affinity of this construct for template/primer RNA may render it more susceptible to competitive inhibitors of template/primer binding. As a result, compound 1 (Fig. 3) a member from a benzimidazole-5-carboxamide library, was identified as a specific inhibitor of the HT-NS5B polymerase with an IC50 of 13.6 µM (Table 2). In order to determine its mode of inhibition, two series of kinetic experiments quantifying polymerase velocity were performed with HT-NS5B and compound 1. First, the concentrations of UTP and inhibitor were varied in the presence of a constant amount of template/primer (Fig. 4), and secondly, the concentrations of template/primer and inhibitor were varied in the presence of a constant amount of UTP (Fig. 5). Compound 1 demonstrated non-competitive inhibition of UTP incorporation (Ki competitive = Ki uncompetitive = 20 µM), as both Dixon and Cornish-Bowden plots intercept on the x-axis (Fig. 4). In contrast, compound 1 displayed a mixed mode of inhibition towards the template/primer, with a significant competitive component (Ki competitive = 26 µM and Ki uncompetitive = 105 µM); the mode of inhibition is characterized by the Dixon and Cornish-Bowden plots of the data in Figure 5, where the location of the intercepts on the two plots are above and below the x-axis, respectively.
Figure 3.
Structure of the compounds used in the study. Compound 1 and its analogs (1a–1e, inclusive) are a series of benzimidazole-5-carboxamide derivatives identified as specific inhibitors of HCV polymerase. Compound 2 is an unrelated di-ketoacid inhibitor of the HCV polymerase (39,49).
Table 2. IC50 valuesa (µM) for a series of compounds with different polymerase constructs.
| Compound | HT-NS5B | HT-NS5BΔ21 | NS5BΔ57-HT | NS5BΔ21-HT | NS5B | Poliovirus polymerase | CT RNA Pol II |
|---|---|---|---|---|---|---|---|
| 1 |
13.6 |
29 |
>500 |
>500 |
>500 |
>500 |
>500 |
| 1a |
0.59 |
2.7 |
13.2 |
20 |
20 |
>120 |
>200 |
| 1b |
0.49 |
3.0 |
23 |
19 |
28 |
>100 |
>200 |
| 1c |
0.14 |
1.2 |
4.5 |
7.3 |
11.2 |
100 |
>200 |
| 1d |
0.054 |
0.44 |
2.2 |
3.0 |
5.7 |
79 |
>200 |
| 1e |
0.019 |
0.16 |
0.73 |
1.1 |
1.6 |
100 |
>200 |
| 2 | 0.95 | 0.63 | 0.68 | 0.3 | 0.25 | n.d. | n.d. |
aValues are the mean of three or four determinations for each compound with coefficient of variation below 25%.
Figure 4.
Ki determination of compound 1 and mode of inhibition with regard to UTP substrate. Reaction velocities were measured at different UTP concentrations (0.25, 0.50, 1.0, 2.5, 5.0, 10, 25 and 50 µM, plotted as open diamonds, closed diamonds, open circles, closed circles, open squares, closed squares, open triangles and closed triangles, respectively) with the template/primer fixed at 0.25 µM. Dixon and Cornish-Bowden plots of the reciprocal velocity with compound 1 at concentrations of 0, 2.5, 5, 10, 20 and 40 µM. Both plots display intersects on the x-axis and reflect non-competitive inhibition with regard to UTP (Ki competitive = Ki uncompetitive = 20 µM).
Figure 5.
Ki determination of compound 1 and mode of inhibition with regard to template/primer substrate. Reaction velocities were measured at different template/primer concentrations (0.05, 0.075, 0.15 and 0.2 µM plotted as closed diamonds, open diamonds, closed squares and open squares, respectively) with the UTP fixed at 25 µM. Dixon and Cornish-Bowden plots of the reciprocal velocity with compound 1 at concentrations of 0, 15, 30 and 60 µM. Compound 1 displayed a mixed mode of inhibition towards template/primer, with a major competitive component (Ki competitive = 26 µM; intercept on the Dixon plot above the x-axis) and a minor uncompetitive component (Ki uncompetitive = 105 µM; intercept extrapolated from the Cornish-Bowden plot below the x-axis).
Compound IC50s with different HCV polymerase constructs
The IC50 values for a series of five compounds related to compound 1 (Fig. 3) were determined with the five different constructs of NS5B polymerase as illustrated in Table 2. HCV RdRp assays that were performed with the HT-NS5B enzyme used in our screening campaign and which displayed the lowest affinity for template/primer (Table 1), provided the lowest relative IC50 values for this class of compounds. Compounds (38) with modified amino acid derived R1 groups (1a–1e, Fig. 3) had improved potencies, such that compound 1e with a 5-indole-oxyacetic acid displayed an IC50 of 19 nM. The IC50 values determined under the same conditions with the other NS5B constructs were significantly higher for all six of the benzimidazole-5-carboxamide derivatives (Table 2), yet the ranking of the compound potencies among all of the constructs was identical; i.e. IC50 for compound 1e < compound 1d < compound 1c< compound 1b ∼ compound 1a < compound 1. Hence, the structure–activity relationship for this class of HCV polymerase inhibitor could be followed with any one of these enzyme constructs.
Notably, the fold increase in IC50 values of the benzimidazole-5-carboxamide class of compounds in assays performed with the different HCV polymerase constructs was inversely proportional to the Km of the respective NS5B constructs. For example, assays using the construct with the highest Km for template/primer (HT-NS5B) provided the lowest relative IC50 values, whereas constructs with a low Km (NS5B) provided the highest relative IC50 values. This shift in IC50 values for a common inhibitor among the different HCV polymerase constructs was unique to the benzimidazole-5-carboxamide series, as compound 2, an unrelated pyrophosphate product-mimic inhibitor of HCV polymerase (39) displayed less than a 4-fold difference in potency in polymerase assays with the different NS5B enzyme constructs.
Benzimidazole-5-carboxamide compounds do not inhibit HCV polymerase initiation or elongation from pre-bound RNA
The standard HCV RdRp assay is a continuous polymerization reaction with multiple cycles of initiation and elongation events. The assay can be modified by pre-incubating the enzyme with the RNA substrate and delaying the addition of NTP and test compound after polyanionic heparin is added to quench any dissociated polymerase; such a modification quantifies only a single cycle of initiation and elongation from pre-bound template/primer substrate (19,23). The potential of compound 1 and its analogs to inhibit initiation or subsequent elongation from pre-formed HCV polymerase–RNA complexes was evaluated in this assay format. Sequential, ordered addition of heparin, inhibitor and UTP to the pre-incubated NS5B–RNA complex indicated that the benzimidazole-5-carboxamide class of compounds did not inhibit HCV polymerase initiation or elongation from pre-bound RNA complexes (Table 3). Furthermore, simply pre-incubating the NS5B with RNA substrate followed by the addition of compound and UTP (without heparin) also alleviated inhibition by compound 1 (data not shown). In contrast, compound 2, the di-ketoacid derivative that specifically interferes with NTP incorporation by HCV polymerase (39), displayed similar potencies in both the standard HCV RdRp and single cycle initiation and elongation assays. The mechanism of action of the benzimidazole-5-carboxamide compounds inferred from these order of addition experiments, is consistent with the kinetic experiments that this class of compounds interferes with an early step in the polymerase reaction such as productive RNA binding.
Table 3. IC50 valuesa (µM) for a series of compounds in a single-cycle HT-NS5B elongation assay.
| Compound | HT-NS5B (elongation) |
|---|---|
| 1 |
>250 |
| 1a |
>100 |
| 1b |
>100 |
| 1c |
>100 |
| 1d |
>100 |
| 1e |
>100 |
| 2 | 1.52 ± 0.37 |
aValues are the mean of two or three determinations.
Inhibition of HCV polymerase de novo initiation
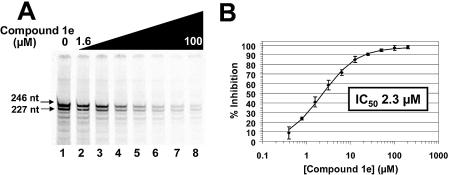
Though these compounds inhibited reactions with primer-dependent homopolymeric substrates, the HCV polymerase is proposed to commence complementary strand RNA synthesis on genomic template RNA in a primer independent de novo initiation reaction. Using a previously characterized HCV plus-strand 3′-UTR template RNA substrate (12), we examined the effect of compound 1e on product formation in a de novo initiation reaction catalyzed by the full-length, untagged NS5B construct. The major products of de novo initiation in this reaction are the 246 and 227 nt products (12,40,41) depicted in Figure 6A. Reactions reconstituted with increasing amounts of compound 1e inhibited formation of the product of de novo RNA synthesis with an IC50 of 2.3 µM (Fig. 6B). The IC50s of compound 1e in this assay and the standard HCV polymerase assay run with the mature NS5B (IC50 = 1.6 µM, Table 2) are similar and reflect the similar Km of the NS5B construct both for the homopolymeric substrate RNA (Table 1) and the 3′-UTR template (data not shown).
Figure 6.
Compound 1e inhibits de novo initiation by HCV NS5B polymerase. (A) Lane 1 shows the major products of NS5B de novo initiation using a short 3′-UTR plus strand template as 246 and 227 nt RNAs (12). Lanes 2–8 display the products of NS5B de novo initiation in the presence of 1.6, 3.13, 6.25, 12.5, 25, 50 and 100 µM of compound 1e. (B) Dose–response curve displaying the IC50 of compound 1e in a NS5B de novo initiation assay. The points on the curve represent an average (with error bars) of three independent determinations.
DISCUSSION
The viral nucleic acid polymerase inhibitors constitute the majority of currently approved antiviral drugs (42) and portray polymerases as prime therapeutic targets for emerging viral infections. Consequently, the NS5B enzyme has become a primary target in the search for novel inhibitors of HCV replication and a variety of in vitro assays for NS5B polymerase activity have been developed for such a purpose. In this study, we have characterized steady-state kinetic parameters of both nucleotide triphosphate and template/primer RNA substrates for NS5B constructs encoding a variety of common truncations and affinity tag fusions. The Km for UTP varied from 1.8 to 12 µM, depending on the polymerase construct, which is a range consistent with previously published values using the full-length or truncated enzyme, produced in insect cells or in E.coli, with or without a histidine tag (14,19,20,43). The Km for the template/primer with our panel of NS5B constructs ranged from 25 to 214 nM. Most of the NS5B constructs have low nanomolar affinity for the homopolymeric substrate, which is in agreement with a report by Ferrari et al. (17) of a Km for a template/primer of 30 nM with the common C-terminal Δ21 truncation of the HCV polymerase using a poly(C)/oligo(G) substrate. However, the full-length N-terminally tagged polymerase, HT-NS5B, displayed a markedly lower affinity for the RNA substrate and offered a potential advantage in a screen for inhibitors.
A competitive inhibitor of enzyme activity must have affinities comparable with (or greater than) the substrate. For viral polymerases such as the NS5B, the triphosphate form of nucleoside analogs represent classic competitive inhibitors of NTP incorporation whose IC50 value is directly proportional to the substrate concentration and inversely proportional to the Km value (44). The widely established in vitro NS5B assays that use low micromolar NTP concentration are therefore sensitive to a broad range of inhibition by nucleotide mimics (27,28). Towards this end, and before the development of cell-based HCV RNA replication assays (45), nucleoside analogs (reviewed in 46) were originally identified by screening their triphosphorylated derivatives in in vitro assays with NTP concentrations at or below the Km value. The HCV RdRp assay that we used in a broad screening campaign, was optimized to detect competitive inhibitors of both NTP and template/primer RNA substrates, by using the HT-NS5B enzyme construct under conditions that reconstituted in vitro activity with the concentration of both substrates at or below their Km value. Hence, the low affinity that the HT-NS5B construct had for the poly(rA)/oligo(U) homopolymeric substrate, supported the design of a screening assay with a relatively high RNA substrate concentration [i.e. 250 nM of oligo(U) primer and a 10-fold molar-nucleotide excess of poly(rA) template] that was within the Km value. In contrast, the optimized standard assay that used the other NS5B polymerases constructs (Fig. 1 and Table 1) had template/primer concentrations that were 4–10-fold higher than the Km values, and accounted (44) for the significantly higher IC50 (Table 2) values displayed by inhibitors of RNA binding.
Using the HT-NS5B-based assay, compound 1 was identified in a broad screen of the Boehringer Ingelheim proprietary compound collection, as a non-nucleoside inhibitor (IC50 = 13.6 µM) originating from a benzimidazole-5-carboxamide combinatorial chemistry library. Compound 1 and derivatives displayed specific and reversible inhibition of the HCV polymerase and failed to inhibit poliovirus RdRp and a DNA-dependent RNA polymerase II activity (Table 2). Moreover, inhibition by compound 1 was only observed with the N-terminally tagged NS5B and not with the other NS5B constructs (Table 2). In order to determine the mode of HT-NS5B inhibition by compound 1, reaction velocities were measured over a range of template/primer, NTP and inhibitor concentrations. Reciprocal plots of velocity (36) revealed a mixed mode of inhibition for compound 1 towards the template/primer with a significant competitive component, and non-competitive inhibition with UTP. HIV-1 RT non-nucleoside inhibitors, such as nevirapine, are non-competitive inhibitors with UTP, but in contrast to compound 1, are also non-competitive with template/primer substrate (47,48).
At least three other chemically distinct series of non-nucleoside inhibitors of the HCV polymerase have been recently reported: (i) a benzothiadiazine series (23); (ii) phenylalanine derivatives (26); and (iii) a cyclopentyl dihydropyran-2-one inhibitor (34). The latter two bind to a distinct allosteric site in the thumb domain distally removed from the central active site (26,34). Representative compounds of these three non-nucleoside inhibitors are all non-competitive inhibitors of NTP incorporation, and their mode of action with respect to template/primer are incompletely characterized. Though the benzothiadiazine series binding site has not been reported, an uncompetitive mode of inhibition with respect to template/primer RNA is suggested from the unproductive enzyme–substrate–inhibitor complex formed by the NS5B, template RNA substrate and benzothiadiazine (24). The benzimidazole-5-carboxamide compounds’ mode of action clearly differed from the benzothiadiazine inhibitor. The latter series effectively inhibits NS5B initiation from pre-bound RNA (24), whereas the benzimidazole-5-carboxamides did not inhibit the activity of pre-bound NS5B–RNA complexes (Table 3). Besides the modified order of addition experiments, the steady-state kinetic data revealed that benzimidazole-5-carboxamides interfere with RNA binding. Combination experiments with a benzothiadiazine and a benzimidazole-5-carboxamide showed that the compounds were not mutually exclusive and probably bind to separate sites (McKercher,G., unpublished observations).
The distinct mode of inhibition by the benzimidazole-5-carboxamide class of compounds was also manifested by the dramatically different IC50 values obtained with the modified enzyme constructs. Though the potency ranking of the related derivatives was identical between the different constructs, increases in IC50s ranged from ∼6-fold with the HT-NS5BΔ21 enzyme up to 72-fold with the untagged full-length NS5B. Notably, the magnitude of the increases in IC50s was inversely proportional to the Km of the respective enzyme constructs for template/primer RNA [Km (P/T)] and is fully consistent with inhibitors that are predicted to interfere with RNA binding. As a result, compound screening with the higher affinity NS5B constructs would have greatly diminished the assay’s sensitivity to inhibitors of template/primer binding and failed to detect weak to moderately potent compounds of this class. Conversely, compounds that function by an alternative mechanism, such as the di-ketoacid class (compound 2) that mimic pyrophosphate product (39,49), would be unaffected by an enzyme’s Km (P/T) property and thereby displayed similar potencies against this group of modified enzymes.
The position of the hexa-histidine tag on the NS5B affected the enzyme’s affinity for RNA substrate and, in turn, the IC50 of the benzimidazole-5-carboxamides. Lower IC50 values were effectively obtained with the two polymerase constructs that had the hexa-histidine tag at the N-terminus. The tagged, full-length polymerase (HT-NS5B) had the lowest affinity for RNA substrate and we ensured that this was not an artifact of reduced enzyme solubility by extracting, purifying and maintaining the protein (without any significant loss in activity) in detergent. The truncated, soluble polymerase that can be purified without detergent also had slightly lower affinity for RNA substrate, relative to the untagged NS5B, only when the tag was positioned at the N-terminus (HT-NS5BΔ21); notably, inhibition by compound 1 was also detected with this construct.
The optimized compound 1e inhibited the primer- independent de novo initiation activity and demonstrated efficacy and similar potencies in assays with homopolymeric or heteropolymeric RNA template using the untagged, full-length NS5B. Compound 1e was also tested for inhibition of HCV RNA replication in a cell-based replicon assay (45,50) and demonstrated marginal potency (i.e. 63% inhibition at 50 µM; data not shown). Current medicinal chemistry efforts are directed towards improving the cell permeability of these compounds (Beaulieu,P., manuscript in preparation).
A uniquely modified NS5B enzyme construct facilitated the identification of the benzimidazole-5-carboxamide series of compounds as a novel class of HCV RNA polymerase inhibitors with a mode of action distinct from any previously characterized viral polymerase inhibitors. Studies are on-going to improve their potency and assess their benefit as a potential therapy in the treatment of chronic HCV infection.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Samuel April, Dr Professor Volkhard Austel, Yves Bousquet, Michael Bös, James Gillard, Gulrez Fazal, Jean Gauthier, Sylvie Goulet and Marc-André Poupart for the medicinal chemistry contributions. We thank Carol Homon, Paul Kaplita, Gustavo Rodriguez and George Rogers for high-throughput screening. We are grateful for the discussions with Pierre Bonneau, Michael Cordingley and Peter White throughout the course of this work.
REFERENCES
- 1.Choo Q.L., Kuo,G., Weiner,A.J., Overby,L.R., Bradley,D.W. and Houghton,M. (1989) Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science, 244, 359–362. [DOI] [PubMed] [Google Scholar]
- 2.Wasley A. and Alter,M.J. (2000) Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis., 20, 1–16. [DOI] [PubMed] [Google Scholar]
- 3.Hijikata M., Kato,N., Ootsuyama,Y., Nakagawa,M. and Shimotohno,K. (1991) Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl Acad. Sci. USA, 88, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grakoui A., Wychowski,C., Lin,C., Feinstone,S.M. and Rice,C.M. (1993) Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol., 67, 1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens S.E., Tomei,L. and De Francesco,R. (1996) Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J., 15, 12–22. [PMC free article] [PubMed] [Google Scholar]
- 6.Luo G., Hamatake,R.K., Mathis,D.M., Racela,J., Rigat,K.L., Lemm,J. and Colonno,R.J. (2000) De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol., 74, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh J.W., Ito,T. and Lai,M.M. (1999) A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol., 73, 7694–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong W., Ferrari,E., Lesburg,C.A., Maag,D., Ghosh,S.K., Cameron,C.E., Lau,J.Y. and Hong,Z. (2000) Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J. Virol., 74, 9134–9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong W., Uss,A.S., Ferrari,E., Lau,J.Y. and Hong,Z. (2000) De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol., 74, 2017–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kao C.C., Yang,X., Kline,A., Wang,Q.M., Barket,D. and Heinz,B.A. (2000) Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol., 74, 11121–11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kao C.C., Singh,P. and Ecker,D.J. (2001) De novo initiation of viral RNA-dependent RNA synthesis. Virology, 287, 251–260. [DOI] [PubMed] [Google Scholar]
- 12.Pellerin C., Lefebvre,S., Little,M.J., McKercher,G., Lamarre,D. and Kukolj,G. (2002) Internal initiation sites of de novo RNA synthesis within the hepatitis C virus polypyrimidine tract. Biochem. Biophys. Res. Commun., 295, 682–688. [DOI] [PubMed] [Google Scholar]
- 13.Yuan Z.H., Kumar,U., Thomas,H.C., Wen,Y.M. and Monjardino,J. (1997) Expression, purification and partial characterization of HCV RNA polymerase. Biochem. Biophys. Res. Commun., 232, 231–235. [DOI] [PubMed] [Google Scholar]
- 14.Lohmann V., Roos,A., Korner,F., Koch,J.O. and Bartenschlager,R. (1998) Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology, 249, 108–118. [DOI] [PubMed] [Google Scholar]
- 15.Lohmann V., Korner,F., Herian,U. and Bartenschlager,R. (1997) Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol., 71, 8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamashita T., Kaneko,S., Shirota,Y., Qin,W., Nomura,T., Kobayashi,K. and Murakami,S. (1998) RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem., 273, 15479–15486. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari E., Wright-Minogue,J., Fang,J.W., Baroudy,B.M., Lau,J.Y. and Hong,Z. (1999) Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol., 73, 1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishii K., Tanaka,Y., Yap,C.C., Aizaki,H., Matsuura,Y. and Miyamura,T. (1999) Expression of hepatitis C virus NS5B protein: characterization of its RNA polymerase activity and RNA binding. Hepatology, 29, 1227–1235. [DOI] [PubMed] [Google Scholar]
- 19.Tomei L., Vitale,R.L., Incitti,I., Serafini,S., Altamura,S., Vitelli,A. and De Francesco,R. (2000) Biochemical characterization of a hepatitis C virus RNA-dependent RNA polymerase mutant lacking the C-terminal hydrophobic sequence. J. Gen. Virol., 81, 759–767. [DOI] [PubMed] [Google Scholar]
- 20.Johnson R.B., Sun,X.L., Hockman,M.A., Villarreal,E.C., Wakulchik,M. and Wang,Q.M. (2000) Specificity and mechanism analysis of hepatitis C virus RNA-dependent RNA polymerase. Arch. Biochem. Biophys., 377, 129–134. [DOI] [PubMed] [Google Scholar]
- 21.Qin W., Luo,H., Nomura,T., Hayashi,N., Yamashita,T. and Murakami,S. (2002) Oligomeric interaction of hepatitis C virus NS5B is critical for catalytic activity of RNA-dependent RNA polymerase. J. Biol. Chem., 277, 2132–2137. [DOI] [PubMed] [Google Scholar]
- 22.Qin W., Yamashita,T., Shirota,Y., Lin,Y., Wei,W. and Murakami,S. (2001) Mutational analysis of the structure and functions of hepatitis C virus RNA-dependent RNA polymerase. Hepatology, 33, 728–737. [DOI] [PubMed] [Google Scholar]
- 23.Dhanak D., Duffy,K.J., Johnston,V.K., Lin-Goerke,J., Darcy,M., Shaw,A.N., Gu,B., Silverman,C., Gates,A.T., Nonnemacher,M.R. et al. (2002) Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem., 277, 38322–38327. [DOI] [PubMed] [Google Scholar]
- 24.Gu B., Johnston,V.K., Gutshall,L.L., Nguyen,T.T., Gontarek,R.R., Darcy,M.G., Tedesco,R., Dhanak,D., Duffy,K.J., Kao,C.C. et al. (2003) Arresting initiation of hepatitis C virus RNA synthesis using heterocyclic derivatives. J. Biol. Chem., 278, 16602–16607. [DOI] [PubMed] [Google Scholar]
- 25.Chan L., Reddy,T.J., Proulx,M., Das,S.K., Pereira,O., Wang,W., Siddiqui,A., Yannopoulos,C.G., Poisson,C., Turcotte,N. et al. (2003) Identification of N,N-disubstituted phenylalanines as a novel class of inhibitors of hepatitis C NS5B polymerase. J. Med. Chem., 46, 1283–1285. [DOI] [PubMed] [Google Scholar]
- 26.Wang M., Ng,K.K., Cherney,M.M., Chan,L., Yannopoulos,C.G., Bedard,J., Morin,N., Nguyen-Ba,N., Alaoui-Ismaili,M.H., Bethell,R.C. et al. (2003) Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J. Biol. Chem., 278, 9489–9495. [DOI] [PubMed] [Google Scholar]
- 27.Carroll S.S., Tomassini,J.E., Bosserman,M., Getty,K., Stahlhut,M.W., Eldrup,A.B., Bhat,B., Hall,D., Simcoe,A.L., LaFemina,R. et al. (2003) Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem., 278, 11979–11984. [DOI] [PubMed] [Google Scholar]
- 28.Stuyver L.J., Whitaker,T., McBrayer,T.R., Hernandez-Santiago,B.I., Lostia,S., Tharnish,P.M., Ramesh,M., Chu,C.K., Jordan,R., Shi,J. et al. (2003) Ribonucleoside analogue that blocks replication of bovine viral diarrhea and hepatitis C viruses in culture. Antimicrob. Agents Chemother., 47, 244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al R.H., Xie,Y., Wang,Y. and Hagedorn,C.H. (1998) Expression of recombinant hepatitis C virus non-structural protein 5B in Escherichia coli. Virus Res., 53, 141–149. [DOI] [PubMed] [Google Scholar]
- 30.Ago H., Adachi,T., Yoshida,A., Yamamoto,M., Habuka,N., Yatsunami,K. and Miyano,M. (1999) Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold. Des., 7, 1417–1426. [DOI] [PubMed] [Google Scholar]
- 31.Bressanelli S., Tomei,L., Roussel,A., Incitti,I., Vitale,R.L., Mathieu,M., De Francesco,R. and Rey,F.A. (1999) Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl Acad. Sci. USA, 96, 13034–13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lesburg C.A., Cable,M.B., Ferrari,E., Hong,Z., Mannarino,A.F. and Weber,P.C. (1999) Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nature Struct. Biol., 6, 937–943. [DOI] [PubMed] [Google Scholar]
- 33.Bressanelli S., Tomei,L., Rey,F.A. and De Francesco,R. (2002) Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol., 76, 3482–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love R.A., Parge,H.E., Yu,X., Hickey,M.J., Diehl,W., Gao,J., Wriggers,H., Ekker,A., Wang,L., Thomson,J.A. et al. (2003) Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol., 77, 7575–7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim W.Y. and Dahmus,M.E. (1988) Purification of RNA polymerase IIO from calf thymus. J. Biol. Chem., 263, 18880–18885. [PubMed] [Google Scholar]
- 36.Cornish-Bowden A. (1974) A simple graphical method for determining the inhibition constants of mixed, uncompetitive and non-competitive inhibitors. Biochem. J., 137, 143–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beaulieu P.L., Haché,B. and von Moos,E. (2003) A practical Oxone®-mediated, high-throughput, solution-phase synthesis of benzimidazoles from 1,2-phenylenediamines and aldehydes and its application to preparative scale synthesis. Synthesis, 2003, 1683–1692. [Google Scholar]
- 38.Beaulieu P.L., Fazal,G., Gillard,J., Kukolj,G. and Austel,V. (2002) Viral Polymerase Inhibitors. Patent US 6448281, US 6479508. PCT/WO 02/04425.
- 39.Altamura S., Tomei,L., Koch,U., Neuner,P.J.S. and Summa,V. (2000) Diketoacid-Derivatives as Inhibitors of Polymerases. Patent US 6492423. PCT/WO 00/06529.
- 40.Oh J.W., Sheu,G.T. and Lai,M.M. (2000) Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem., 275, 17710–17717. [DOI] [PubMed] [Google Scholar]
- 41.Reigadas S., Ventura,M., Sarih-Cottin,L., Castroviejo,M., Litvak,S. and Astier-Gin,T. (2001) HCV RNA-dependent RNA polymerase replicates in vitro the 3′ terminal region of the minus-strand viral RNA more efficiently than the 3′ terminal region of the plus RNA. Eur. J. Biochem., 268, 5857–5867. [DOI] [PubMed] [Google Scholar]
- 42.De Clercq E. (2001) Antiviral drugs: current state of the art. J. Clin. Virol., 22, 73–89. [DOI] [PubMed] [Google Scholar]
- 43.Carroll S.S., Sardana,V., Yang,Z., Jacobs,A.R., Mizenko,C., Hall,D., Hill,L., Zugay-Murphy,J. and Kuo,L.C. (2000) Only a small fraction of purified hepatitis C RNA-dependent RNA polymerase is catalytically competent: implications for viral replication and in vitro assays. Biochemistry, 39, 8243–8249. [DOI] [PubMed] [Google Scholar]
- 44.Cheng Y. and Prusoff,W.H. (1973) Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 per cent inhibition (IC50) of an enzymatic reaction. Biochem. Pharmacol., 22, 3099–3108. [DOI] [PubMed] [Google Scholar]
- 45.Lohmann V., Korner,F., Koch,J., Herian,U., Theilmann,L. and Bartenschlager,R. (1999) Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science, 285, 110–113. [DOI] [PubMed] [Google Scholar]
- 46.Beaulieu P.L. and Llinàs-Brunet,M. (2002) Therapies for hepatitis C infection: targeting the non-structural proteins of HCV. Curr. Med. Chem. Anti-Infect. Agents, 1, 163–176. [Google Scholar]
- 47.Merluzzi V.J., Hargrave,K.D., Labadia,M., Grozinger,K., Skoog,M., Wu,J.C., Shih,C.K., Eckner,K., Hattox,S., Adams,J. et al. (1990) Inhibition of HIV-1 replication by a nonnucleoside reverse transcriptase inhibitor. Science, 250, 1411–1413. [DOI] [PubMed] [Google Scholar]
- 48.Kopp E.B., Miglietta,J.J., Shrutkowski,A.G., Shih,C.K., Grob,P.M. and Skoog,M.T. (1991) Steady state kinetics and inhibition of HIV-1 reverse transcriptase by a non-nucleoside dipyridodiazepinone, BI-RG-587, using a heteropolymeric template. Nucleic Acids Res., 19, 3035–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Francesco R., Tomei,L., Altamura,S., Summa,V. and Migliaccio,G. (2003) Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antiviral Res., 58, 1–16. [DOI] [PubMed] [Google Scholar]
- 50.Pause A., Kukolj,G., Bailey,M., Brault,M., Do,F., Halmos,T., Lagace,L., Maurice,R., Marquis,M., McKercher,G. et al. (2003) An NS3 serine protease inhibitor abrogates replication of subgenomic hepatitis C virus RNA. J. Biol. Chem., 278, 20374–20380. [DOI] [PubMed] [Google Scholar]