Abstract
STUDY QUESTION
Can plasma microRNAs be used as a non-invasive diagnostic test for the detection of endometriosis?
SUMMARY ANSWER
Plasma miR-17-5p, miR-20a and miR-22 are down-regulated in women with endometriosis compared with those without endometriosis in mainland China.
WHAT IS KNOWN ALREADY
There is currently a pressing need to develop a non-invasive diagnostic test for endometriosis. Altered circulating microRNA profiles have already been linked to various disease states.
STUDY DESIGN, SIZE, AND DURATION
This was a prospective laboratory study in a tertiary-referral university hospital in Beijing, PR China, between January 2012 and May 2012. Twenty-three women with histologically proven endometriosis and 23 endometriosis-free controls were enrolled in this study.
PARTICIPANTS/MATERIALS, SETTING, AND METHODS
Laparoscopic inspection of the abdominopelvic cavity was performed for each patient, and peripheral blood samples were collected before laparoscopy. Microarray-based microRNA expression profiling was used to identify differentially expressed microRNAs in plasma samples between women with and without endometriosis, and quantification of selected microRNAs was performed using quantitative RT–PCR.
MAIN RESULTS AND THE ROLE OF CHANCE
Twenty-seven microRNAs were differentially expressed between women with and without endometriosis, of which six microRNAs (miR-15b-5p, miR-17-5p, miR-20a, miR-21, miR-22 and miR-26a) were selected for validation. MiR-17-5p, miR-20a and miR-22 were significantly down-regulated in women with endometriosis compared with controls (P = 0.011, 0.0020 and 0.0002, respectively), yielding an area under the receiver operator characteristics curve of 0.74 [95% confidence interval (CI): 0.58–0.90], 0.79 (95% CI: 0.65–0.93) and 0.85 (95% CI: 0.71–0.98) in discriminating endometriosis from controls, respectively.
LIMITATIONS AND REASONS FOR CAUTION
Our sample size was small and all cases were rAFS stage III–IV, which may limit generalization of plasma microRNAs for early diagnosis of endometriosis. Moreover, only six microRNAs were selected for validation.
WIDER IMPLICATIONS OF THE FINDINGS
Plasma microRNAs provide a promising opportunity for detection of endometriosis.
STUDY FUNDING/COMPETING INTEREST(S)
This research was supported by the National Natural Science Foundation of China (81170548) and Key Project for Clinical Faculty Foundation, Ministry of Health, China (2010). All authors declare no conflict of interest.
Keywords: microRNA, plasma, biomarkers, endometriosis, non-invasive diagnosis
Introduction
Endometriosis, an enigmatic disease affecting up to 10% of women of reproductive age, is a major contributor to pelvic pain and subfertility, resulting in a considerable burden in terms of healthcare costs and quality of life (Giudice, 2010; Nnoaham et al., 2011; Simoens et al., 2012). However, diagnosis of endometriosis is often difficult due to the fact that there is no definitive diagnostic biomarker yet available (May et al., 2010). Laparoscopic inspection of the pelvis is currently the gold standard for diagnosis of endometriosis (Kennedy et al., 2005), but the invasive nature, associated morbidity and cost have hampered its wide application (El-Kasti et al., 2011). Furthermore, only 70–75% of visually diagnostic lesions are confirmed histologically (Spaczynski and Duleba, 2003). On the other hand, pelvic imaging, blood markers or endometrial alterations have not yielded sufficient power for clinical use (May et al., 2010, 2011). Thus, there is a pressing need to develop a non-invasive diagnostic test for endometriosis.
The recent discovery of microRNAs, which are small non-coding RNAs of 19–25 nucleotides that inhibit mRNA translation or induce its degradation, has provided new insight into the regulation of gene expression (Ambros, 2004; Ebert and Sharp, 2012). Aberrant microRNA expression has been implicated in a number of human diseases, including endometriosis (Teague et al., 2010). Several investigators have identified the different microRNA expression profiles in the eutopic endometrium of women with and without endometriosis (Pan et al., 2007; Toloubeydokhti et al., 2008; Burney et al., 2009; Ramon et al., 2011; Dai et al., 2012) and between ectopic and eutopic endometrial samples from the same women (Pan et al., 2007; Toloubeydokhti et al., 2008; Ohlsson et al., 2009; Filigheddu et al., 2010; Ramon et al., 2011; Dai et al., 2012). They also have demonstrated that microRNAs may regulate cellular events that are critical to the pathogenesis of endometriosis, its suppression of fertility and its potential as a semi-invasive test for endometriosis (Teague et al., 2010; Hawkins et al., 2011; Dai et al., 2012).
Circulating microRNAs, first described in serum from patients with diffuse large B cell lymphoma in 2008 (Lawrie et al., 2008), have emerged as an important mine of non-invasive biomarkers (Cortez et al., 2011). Altered circulating microRNA profiles have already been linked to various disease states, including tumor burden and malignant progression (Cortez et al., 2011; Schwarzenbach et al., 2011), cardiovascular diseases (Vickers et al., 2011), neurodegenerative diseases (Salta et al., 2012), metabolic disorders (Rottiers and Naar, 2012) and pregnancy-related complications (Zhao et al., 2012). Furthermore, a strong correlation was demonstrated between the microRNA profiles of serum/plasma and cancer tissues (Resnick et al., 2009; Wang et al., 2010), suggesting that microRNAs may be released from tissues and shed into the circulation. This hypothesis was also proven in a mouse model of human prostate cancer, where human-specific microRNAs originating from human prostate cancer xenografts were readily detected in the plasma of xenografted mice, but not in controls (Mitchell et al., 2008). Considering the important roles of microRNAs in the development of endometriosis (Burney et al., 2009; Teague et al., 2010; Hawkins et al., 2011), it was hypothesized that plasma microRNAs might be differentially expressed in women with endometriosis (Teague et al., 2010). Therefore, the objective of the present study was to evaluate the feasibility of using plasma microRNAs as a non-invasive diagnostic test for the detection of endometriosis.
Materials and Methods
This study was approved by the human ethics committees of the Peking Union Medical College Hospital, Beijing, PR China, and written informed consent was obtained from all subjects before blood collection.
Patients
Forty-six women aged 22–45 years were enrolled for this preliminary prospective study between January 2012 and May 2012. All women included in the study underwent laparoscopy for various indications, including pelvic masses, pelvic pain, infertility and uterine leiomyoma. During laparoscopy, a thorough inspection of the abdominopelvic cavity was performed to detect any typical or atypical endometriotic lesion, and all possible endometriotic lesions were excised and sent for pathological examination for confirmation of diagnosis. Women were assigned to an endometriosis group or endometriosis-free control group based on pathological reports of the excised tissues. The extent of endometriosis was determined according to the American Society of Reproductive Medicine (ASRM) revised system (Revised American Society for Reproductive Medicine, 1997). The menstrual cycle phases were recorded as proliferative phase (cycle days 6–15) or secretory phase (cycle days 16–28) after adjustment to a 28-day cycle.
Twenty-three women (mean age: 34.1 years; range: 25–44 years) had histologically confirmed endometriosis (stage III and IV). Twenty-three women (mean age: 32.1 years; range 22–45 years) served as controls, and the diagnoses of the control group were as follows: uterine leiomyoma (n = 14), mature teratoma (n = 4), simple cysts (n = 3) and unexplained infertility (n = 2). Women with post-menopausal status, previous hormonal use within 3 months, adenomyosis or malignancy were excluded from this study.
Plasma collection and RNA extraction
Peripheral blood samples (2 ml) were collected into EDTA-containing tubes immediately before administration of anesthesia for laparoscopy. Whole blood was centrifuged at 1200g for 15 min at room temperature within 30 min after blood collection, and the supernatant was transferred into microcentrifuge tubes, followed by a second centrifugation at 12 000g for 10 min at 4°C to remove cellular debris. Plasma was then aliquoted and stored at −80°C until use.
Total RNA was extracted from 400 μl of plasma using the mirVana™ RNA Isolation Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's specifications, and eluted with 100 μl of nuclease-free water. Subsequently, we concentrated the RNA in a final volume of 20 μl. The yield of RNA was determined using a NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA).
MicroRNA microarray expression profiling
Total RNAs from three endometriosis women (age: 31, 30 and 28 years, respectively; proliferative phase) and three uterine leiomyoma controls (age: 26, 36 and 27 years, respectively; proliferative phase) were used for microRNA microarray profiling. Total RNA (100 ng) was labeled with the microRNA Complete Labeling and Hyb Kit (Agilent, USA) and hybridized on the Human microRNA Microarray Kit (Release 16.0, Agilent), which contains 60 000 probes for 1205 human and 144 human viral microRNAs. Hybridization signals were detected with the Agilent Microarray Scanner (Agilent) and the scanned images were analyzed using Agilent Feature Extraction Software (Agilent).
Real-time quantitative RT–PCR
Plasma samples from 40 women (20 endometriosis and 20 control women), which were distinct from those subjected to microarray profiling, were used to validate the findings from microRNA profiling. Five microRNAs (miR-15b-5p, miR-17-5p, miR-20a, miR-21 and miR-26a) were selected because their abnormal expressions in ectopic or eutopic endometrium in endometriosis had been reported in previous studies: miR-15b-5p (Ramon et al., 2011), miR-17-5p (Toloubeydokhti et al., 2008; Ramon et al., 2011), miR-20a (Pan et al., 2007; Ramon et al., 2011), miR-21 (Pan et al., 2007; Ramon et al., 2011) and miR-26a (Pan et al., 2007). We selected miR-22 because its downstream target is hypoxia-inducible factor-1 α (HIF-1 α) (Yamakuchi et al., 2011). We selected miR-16 as an internal normalization control in plasma, since a number of studies have shown that miR-16 is expressed at similar levels in most tissues, and it has expression levels of higher stability and less variability in the circulation (Ng et al., 2009; Reid et al., 2011).
Quantification using the SYBR green quantitative RT–PCR assay (Qiagen, Germany) was performed with a two-step reaction process: reverse transcription (RT) and PCR. Each RT reaction consisted of 10 μl of plasma RNA, 4 μl of miScript RT Buffer (Qiagen, Germany) and 1 μl of miScript Reverse Transcriptase Mix (Qiagen, Germany), in a total volume of 20 μl. Reactions were performed in a Mastercycler Thermocycler (Eppendorf, Hamburg, Germany) for 60 min at 37°C, followed by heat-inactivation of RT for 5 min at 95°C. The 20 μl RT reaction mix was then diluted × 4 in nuclease-free water and held at −20°C.
Real-time PCR was performed using ABI PRISM 9700 system (Applied Biosystems) with 20 μl PCR reaction mixture that included 2 μl of the cDNA, 10 μl of 2 × QuantiTect SYBR Green PCR Master Mix (Qiagen, Germany), 2 μl of 10 × miScript Universal Primer (Qiagen, Germany), 2 μl of 10 × miScript Primer Assay and 4 μl of nuclease-free water. Reactions were incubated in a 96-well optical plate at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 55°C for 30 s and 70°C for 30 s. Each sample was run in triplicate for analysis. At the end of the PCR cycles, melting curve analyses were performed to validate the specific generation of the expected PCR product. The microRNA-specific primer sequences were designed in the laboratory and synthesized by Invitrogen Life Technologies (Invitrogen, CA, USA) based on the microRNA sequences obtained from the miRBase database (Kozomara and Griffiths-Jones, 2011) as follows:
5′-TAGCAGCACATCATGGTTTACA-3′ for miR-15b-5p;
5′-TAGCAGCACGTAAATATTGGCG-3′ for miR-16;
5′-CAAAGTGCTTACAGTGCAGGTAG-3′ for miR-17-5p;
5′-TAAAGTGCTTATAGTGCAGGTAG-3′ for miR-20a;
5′-TAGCTTATCAGACTGATGTTGA-3′ for miR-21;
5′-AAGCTGCCAGTTGAAGAACTGT-3′ for miR-22;
5′-TTCAAGTAATCCAGGATAGGCT-3′ for miR-26a.
The expression levels of microRNAs were normalized to miR-16 and were calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Statistical analyses
Data were expressed as mean ± standard deviation (SD) or proportions where appropriate. Student's t-test and Fisher's exact test were used to determine the difference of clinical characteristics between two groups where appropriate. Expression levels of plasma microRNAs were compared using the Mann–Whitney U-test. MicroRNA data are presented as fold change relative to the women without endometriosis group (control plasma = 1). Receiver operating characteristics (ROC) curves and the area under the ROC curve (AUC) were established to evaluate the diagnostic value of plasma microRNAs for differentiating between endometriosis and controls. An AUC of 0.5 indicates classifications assigned by chance. Based on ROC analysis, the best statistical cut-off values of plasma microRNAs were calculated, and the sensitivity and specificity for selected cut-off points were then assessed. All statistical analysis was performed using Graphpad Prism 5.01 (Graphpad Software Inc., San Diego, CA, USA). P-values of <0.05 (two-tailed) were considered statistically significant.
Results
Clinical characteristics
The clinical characteristics of participants are shown in Table I. The mean age (mean ± SD) of patients with and without endometriosis was 34.1 ± 5.03 and 32.1 ± 6.95 years, respectively. There were no significant differences in distribution of menstrual phase or infertility between the two groups. Women with endometriosis were more likely to report moderate-to-severe pelvic pain (VAS ≥ 5) than controls (52.17 versus 17.39%, respectively; P < 0.029). All women with endometriosis were staged III (n = 11/23) and IV (n = 12/23), and deep infiltrating endometriosis (DIE) was histologically confirmed in approximately half of the women (n = 11/23).
Table I.
Clinical characteristics of women with endometriosis and without endometriosis (controls) recruited for the study of plasma microRNA.
| Parameters | Endometriosis (n = 23) | Controls (n = 23) | P-value |
|---|---|---|---|
| Age (years) | 34.1 ± 5.03 | 32.1 ± 6.95 | 0.27 |
| Cycle phase | |||
| Proliferative | 19 (82.61) | 18 (78.26) | 1.00 |
| Secretory | 4 (17.39) | 5 (21.74) | |
| Pain intensity (VAS) | |||
| ≤4 | 11 (47.83) | 19 (82.61) | 0.01 |
| ≥5 | 12 (52.17) | 4 (17.39) | |
| Infertility | |||
| No | 18 (78.26) | 20 (86.96) | 0.70 |
| Yes | 5 (21.74) | 3 (13.04) | |
| ARSM stage | |||
| III | 10 (43.48) | — | |
| IV | 13 (56.52) | — | |
| DIE status | |||
| No | 12 (52.17) | — | |
| Yes | 11 (47.83) | — | |
Data are expressed as mean ± standard deviation or number (%).
VAS, visual analog scale; ASRM, American Society for Reproductive Medicine; DIE, deep infiltrating endometriosis.
MicroRNA microarray expression profiling
Plasma microRNA profiling detected an average of 132 microRNAs from six samples. We identified 27 plasma microRNAs with significantly different expression levels in women with endometriosis compared with those with leiomyoma (more than a 2-fold change; P < 0.05), all of which were significantly down-regulated in women with endometriosis. The list of the differentially expressed microRNAs identified by microarray analysis is shown in Table II.
Table II.
Down-regulated microRNAs (>2-fold change) identified by microarray analysis in the plasma of three women with endometriosis compared with controls free of endometriosis.
| MicroRNAs | Fold decreased | P-value | Mirbase no. |
|---|---|---|---|
| hsa-miR-122 | 8.06 | 6.2E-04 | MIMAT0000421 |
| hsa-miR-17-5p | 5.39 | 0.049 | MIMAT0000070 |
| hsa-miR-19a | 4.58 | 0.046 | MIMAT0000073 |
| hsa-miR-20a | 4.53 | 0.037 | MIMAT0000075 |
| hsa-miR-19b | 4.36 | 0.046 | MIMAT0000074 |
| hsa-miR-15b-5p | 3.63 | 0.033 | MIMAT0000417 |
| hsa-miR-451 | 3.59 | 0.045 | MIMAT0001631 |
| hsa-miR-30e | 3.48 | 0.047 | MIMAT0000692 |
| hsa-miR-27a | 3.45 | 0.010 | MIMAT0000084 |
| hsa-miR-92a | 3.21 | 0.035 | MIMAT0000092 |
| hsa-miR-26a | 3.19 | 0.025 | MIMAT0000082 |
| hsa-miR-320e | 2.99 | 0.023 | MIMAT0015072 |
| hsa-miR-320d | 2.93 | 0.027 | MIMAT0006764 |
| hsa-miR-762 | 2.83 | 0.050 | MIMAT0010313 |
| hsa-miR-1274b | 2.78 | 0.029 | MIMAT0005938 |
| hsa-miR-630 | 2.75 | 0.037 | MIMAT0003299 |
| hsa-miR-29c | 2.74 | 0.047 | MIMAT0000681 |
| hsa-miR-223 | 2.68 | 0.020 | MIMAT0000280 |
| hsa-miR-22 | 2.64 | 0.025 | MIMAT0000077 |
| hsa-miR-1268 | 2.51 | 0.040 | MIMAT0005922 |
| hsa-miR-23a | 2.49 | 0.035 | MIMAT0000078 |
| hsv2-miR-H10 | 2.40 | 0.045 | MIMAT0014353 |
| hsa-miR-21 | 2.36 | 0.034 | MIMAT0000076 |
| hsa-miR-3679-5p | 2.26 | 0.027 | MIMAT0018104 |
| hsa-miR-30d | 2.21 | 0.035 | MIMAT0000245 |
| hsa-miR-572 | 2.10 | 0.035 | MIMAT0003237 |
| hsa-miR-3665 | 2.07 | 0.029 | MIMAT0018087 |
Fold change was presented as microRNA expression in women without versus with endometriosis.
Bold font indicates microRNAs selected for validation.
In unsupervised hierarchical clustering analysis, normalized microarray expression data for the 27 microRNAs showing differential expression in the six plasma samples (three with endometriosis and three with uterine leiomyoma) were used to generate a heat map. The samples were self-segregated into the endometriosis and control clusters (Fig. 1).
Figure 1.

Heat map showing 27 differentially expressed (fold change > 2) microRNAs from plasma samples of women with endometriosis (n = 3) and controls having uterine leiomyoma but free of endometriosis (n = 3). Each row represents one microRNA, and each column represents a plasma sample. The legend on the right indicates the microRNA represented in the corresponding row. The relative microRNA expression is depicted according to the color scale. Red indicates up-regulation; green indicates down-regulation. The numbers with E indicate endometriosis; numbers with C indicate controls.
Validation of profiling data using qRT–PCR
To confirm the reliability of miR-16 as an internal normalization control in plasma, we evaluated the expression level of miR-16 in all plasma samples, and our data demonstrated that miR-16 was readily detectable in the plasma of all samples. No significant difference was observed in terms of Ct values of miR-16 (P = 0.11; Mann–Whitney U-test) between women with and without endometriosis.
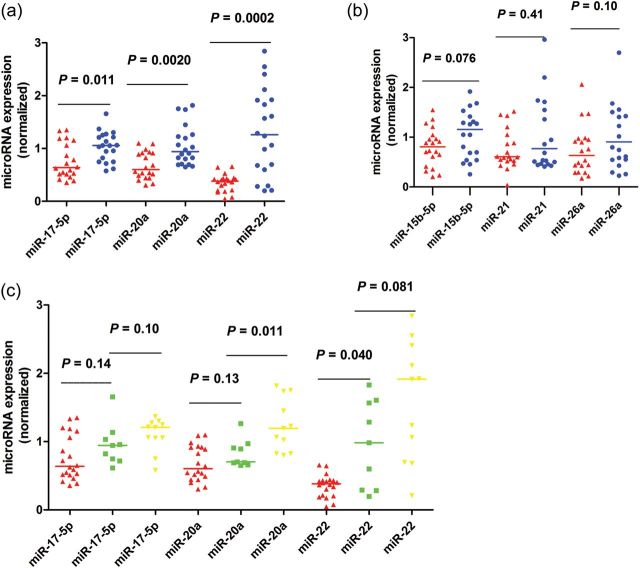
Using miR-16 as normalization control, the trends for down-regulation of microRNA expression were consistent in all six qRT–PCR measurements (Fig. 2a and b). Our data indicated that miR-17-5p, miR-20a and miR-22 were significantly down-regulated in women with endometriosis compared with those without endometriosis (P = 0.011, 0.0020 and 0.0002, respectively) (Fig. 2a). The remaining three microRNAs, miR-15b-5p, miR-21 and miR-26a, did not demonstrate a statistically significant difference in expression between plasma from women with and without endometriosis (P = 0.076, 0.41 and 0.10, respectively) (Fig. 2b).
Figure 2.
Validation of microRNA expression by qRT–PCR analysis in plasma samples from women with (n = 20) and without endometriosis (n = 20). MicroRNA expression data are presented as fold change relative to the women without endometriosis group (control plasma = 1). Data are represented by scatter plots. The line represents the median value. The P-values were calculated by the Mann–Whitney U-test. Red indicates women with endometriosis and blue indicates women that are endometriosis-free (a and b). In c, red indicates women with endometriosis, green indicates endometriosis-free women without leiomyoma and yellow indicates women with leiomyoma.
Compared with women with leiomyoma (n = 11/20), those without leiomyoma (n = 9/20) showed a significantly lower expression of plasma miR-20a (P = 0.011, Mann–Whitney U-test). No significant difference was observed for the remaining five microRNAs, including miR-15b-5p, miR-17-5p, miR-21, miR-22 and miR-26a (P = 0.38, 0.10, 0.68, 0.07 and 0.97, respectively) (Fig. 2c). We also performed a subgroup analysis between women with rAFS stage III and stage IV, women with simple endometrioma and those with complicated DIE. In general, no significant association between the six selected plasma microRNAs and rAFS stages or DIE status was found (data not shown).
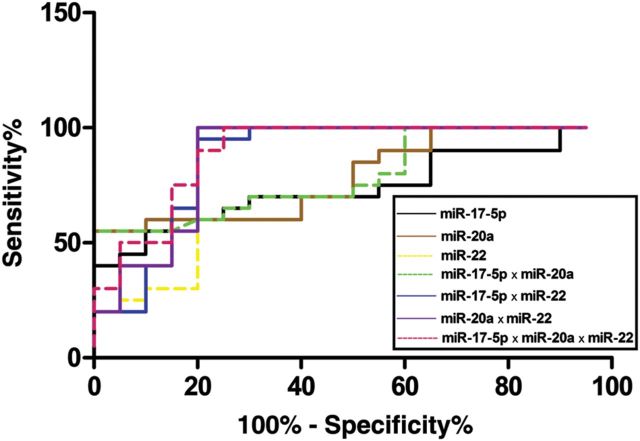
ROC curve analyses revealed that the plasma levels of both miR-17-5p, miR-20a and miR-22 were useful biomarkers for differentiating women with and without endometriosis, with AUC values of 0.74 [95% confidence interval (CI): 0.58–0.90], 0.79 (95% CI: 0.65–0.93) and 0.85 (95% CI: 0.71–0.98), respectively. At the cut-off value of 0.9057 for miR-17-5p, the sensitivity and specificity were 70.0 and 70.0%, respectively. At the cut-off value of 0.6879 for miR-20a, the sensitivity and specificity were 60.0 and 90.0%, respectively. With respect to miR-22, a sensitivity of 90.0% and a specificity of 80.0% were obtained at the cut-off value of 0.5647. We also examined if the combination of a panel of microRNAs could improve the diagnostic power. When the three microRNAs were combined by multiplication (miR-17-5p × miR-20a, miR-17-5p × miR-22, miR-20a × miR-22 and miR-17-5p × miR-20a × miR-22, respectively), the AUC values for differentiating women with and without endometriosis were 0.79 (95% CI: 0.65–0.93), 0.87 (95% CI: 0.74–0.99), 0.88 (95% CI: 0.76–1.00) and 0.90 (95% CI: 0.80–1.00), respectively (Fig. 3).
Figure 3.
ROC curve analysis using plasma microRNAs for discriminating endometriosis. ROC curve analyses revealed that the plasma levels of miR-17-5p, miR-20a and miR-22 were useful biomarkers for differentiating women with and without endometriosis, with AUC values of 0.74 (95% CI: 0.58–0.90), 0.79 (95% CI: 0.65–0.93) and 0.85 (95% CI: 0.71–0.98), respectively. When the three microRNAs were combined by multiplication, the AUC values for differentiating women with and without endometriosis were 0.79 (95% CI: 0.65–0.93), 0.87 (95% CI: 0.74–0.99), 0.88 (95% CI: 0.76–1.00) and 0.90 (95% CI: 0.80–1.00), respectively.
Discussion
Endometriosis is associated with a 6.7 year average diagnostic delay, resulting in serious progress of the disease and impairment in quality of life (Nnoaham et al., 2011; Tokushige et al., 2011). It is estimated that 10% of women in their reproductive age suffer from endometriosis (Giudice, 2010); however, the precise epidemiology is unknown due to a lack of reliable non-invasive tests. Thus, discovery of an accurate and sensitive biomarker is one of the main priorities in current endometriosis research (Rogers et al., 2009). Circulating biomarkers in blood and urine, which are inherently non-invasive, are considered paramount. Considerable effort has been invested in studying the levels of cytokines and growth factors in serum, plasma and urine (May et al., 2010; Tokushige et al., 2011; Cho et al., 2012; Reis et al., 2012); however, none of these are sufficiently sensitive or specific to be translated into clinical diagnosis for endometriosis (May et al., 2010). CA-125 is still the only serum marker of endometriosis used in clinical practice in the past 20 years, despite its low sensitivity (Mol et al., 1998).
In the current study, we used array-based profiling of plasma microRNAs to identify potential biomarkers for endometriosis. Our data demonstrated the feasibility of using plasma microRNAs to discriminate endometriosis. We identified significantly reduced levels of miR-17-5p, miR-20a and miR-22 in the plasma of Han Chinese women with moderate-to-severe endometriosis, which yielded AUC values of 0.74, 0.79 and 0.85, respectively. To the best of our knowledge, this is the first study on the quantitative assessment of plasma microRNAs in women with endometriosis, which provides a promising new opportunity for detection of endometriosis.
Circulating microRNAs, which are non-invasive biomarkers for cancer detection, were first described by Lawrie et al. (2008). The authors demonstrated that serum levels of miR-21 were associated with relapse-free survival in patients with diffuse large B-cell lymphoma. Since then, an increasing number of studies have reported changes in circulating microRNAs in different diseases (Schwarzenbach et al., 2011). In addition, unique expression profiles have been identified for different cancer types, although some of the microRNAs are commonly detected in a number of different cancer types, such as miR-21, miR-92 and miR-155 (Chen et al., 2008; Cortez et al., 2011; Reid et al., 2011). Altered circulating microRNA profiles are also associated with hepatic injury (miR-122) (Wang et al., 2009), atherosclerosis (miR-223) (Vickers et al., 2011), type 2 diabetes (miR-126) (Zampetaki et al., 2010), essential hypertension (let -7e) (Li et al., 2011) and ectopic pregnancy (miR-323-3p) (Zhao et al., 2012). In comparison to the traditional protein-based biomarkers, circulating microRNAs meet many requisite features of good biomarkers, such as stability in various bodily fluids (Ng et al., 2009), tissues or biological specificity (Wang et al., 2009), relatively modest number (Kozomara and Griffiths-Jones, 2011) and easy measurement (Cortez et al., 2011; McDonald et al., 2011).
There is growing evidence that specific microRNAs are involved in the development and progression of endometriosis through the regulation of broad signaling pathways, including inflammation (Lin et al., 2012), local estrogen biosynthesis (Toloubeydokhti et al., 2008), progesterone resistance (Burney et al., 2009), cell invasion (Dai et al., 2012), extracellular matrix remodeling (Ohlsson et al., 2009), angiogenesis (Hawkins et al., 2011; Ramon et al., 2011) and epigenetic regulation (Filigheddu et al., 2010) in endometriotic tissues. In the present study, we observed significantly lower expression of miR-17-5p and miR-20a in plasma from women with endometriosis compared with controls. Both miR-17-5p and miR-20a comprise a down-regulated microRNA cluster in endometriotic lesions (Ramon et al., 2011). Low levels of these microRNAs may ease post-transcriptional suppression of their up-regulated mRNA targets, including anti-apoptotic protein B-cell CLL/lymphoma 2 (BCL2) (Inomata et al., 2009) and cell cycle repressor cyclin-dependent kinase inhibitor 1A (CDKN1A/p21) (Cloonan et al., 2008), resulting in enhanced cell survival and repressed cell proliferation in endometriosis (Flores et al., 2007). Furthermore, miR-17-5p/20a targets transforming growth factor-β (Volinia et al., 2006) and interleukin-8 (Yu et al., 2010), and the reduced expression of these microRNAs may contribute to the local inflammation environment and tissue remodeling during endometriotic lesion development (Kyama et al., 2006). In addition, down-regulation of miR-17-5p/20a is associated with overexpression of hypoxia-inducible transcription factors (HIF-1α) (Taguchi et al., 2008) and vascular endothelial growth factor (VEGF-A) (Lei et al., 2009), leading to neoangiogenesis (Tsuzuki et al., 2012). In contrast, two reports from the same group showed that miR-17 and miR-20a were up-regulated in ectopic endometrium compared with eutopic endometrium and normal endometrium (Pan et al., 2007; Toloubeydokhti et al., 2008). It is possible that the small sample size and different origins of ectopic endometrium of those studies contributed to this discrepancy (Ramon et al., 2011).
Little is currently known about the function and role of miR-22 in endometriosis. Significantly lower expression of miR-22 has been observed in women with endometriosis compared with those with leiomyoma using microarray analysis (Pan et al., 2007). The expression of miR-22 was also found to be significantly lower in isolated endometrial glandular epithelial cells than endometrial stromal cells (Pan et al., 2007). However, no confirmed and functional study has been conducted to explore this phenomenon. Down-regulation of miR-22 has been observed in various tumor types (Xiong, 2012), including those involved in the malignant transformation of endometriosis (Nagaraja et al., 2010). In vitro studies have demonstrated that knockdown of miR-22 enhances hypoxic and NF-kappa B pathways (Takata et al., 2011; Yamakuchi et al., 2011), which have been linked to the pathogenesis of endometriosis (Gonzalez-Ramos et al., 2007; Becker et al., 2008). Furthermore, miR-22 targets HIF-1α in colon cancer cells (Taguchi et al., 2008), and hypoxia-induced HIF-1α expression is associated with VEGF overexpression in human endometrial stromal cells in vitro (Tsuzuki et al., 2012). Thus, miR-22 may be associated with the pathogenesis of endometriosis, which is a potentially important topic that can be explored in future studies.
Notably, all 27 plasma microRNAs identified during the microarray process were significantly down-regulated in women with endometriosis. Little is known about the mechanisms by which microRNAs are generated in plasma and the biologic impact of these molecules in distant sites of the body (Cortez et al., 2011). Tissue studies of the normal endometrium as well as paired eutopic and ectopic endometrium samples have also shown a similar progressive decline in microRNA expression from endometrium to ectopic endometrium (Pan et al., 2007). This phenomenon was also observed in Burney et al.'s microarray profiling study (2009), which identified a total of six microRNAs differentially expressed and down-regulated (fold change ≥1.5) in women with versus without endometriosis. Furthermore, a strong association between microRNA profiles of the serum/plasma and cancer tissues has been demonstrated in studies in cancer (Resnick et al., 2009; Wang et al., 2010), and tumor resection can restore the altered microRNA expression profiles of serum/plasma in patients with liver, breast and colorectal cancer (Ng et al., 2009; Yamamoto et al., 2009; Heneghan et al., 2010). Thus, it is possible that the expression of plasma microRNAs may return to normal after surgical excision of endometriotic lesions, but this hypothesis requires further evaluation.
Although the results of our pilot study are promising, there were several limitations. First, the sample size was small and all cases were rAFS stage III–IV, and therefore further validation in larger cohorts of patients with early stages of the disease is necessary. Second, our control group had various other benign diseases, which may have impacted the levels of plasma microRNAs. However, choosing adequate control groups is a complex task in endometriosis research (Cho et al., 2012). In the current study, all of the control subjects were laparoscopically and histologically confirmed to be endometriosis-free, which may strengthen our findings. Furthermore, only miR-20a was significantly differentially expressed in the subgroup analysis between women with and without leiomyoma. Third, we selected three homogenous uterine leiomyoma (intramural leiomyoma, proliferative phase) patients for the initial microarray profiling, to eliminate the potential influence of the menstrual cycle and local hyper-estrogenic state on the expression of plasma microRNA. Women with uterine leiomyoma were involved in previous studies that compared microRNA expression in the endometrium of women with and without endometriosis (Burney et al., 2009; Hawkins et al., 2010). Furthermore, these two disorders may be associated with each other (Huang et al., 2010; Uimari et al., 2011), and a hyper-estrogenic state might have a role in the development of both fibroids and endometriosis (Uimari et al., 2011).
There are still pre-analytical and analytical challenges in the analysis of circulating microRNAs. We used plasma microRNAs in our analysis, as studies have shown that microRNA concentrations were higher in plasma than in serum (McDonald et al., 2011). Another issue that remains to be addressed is how to normalize or compare microRNA measurement results between samples. Frequently used reference genes, such as U6 small nuclear RNA and 5S ribosomal RNA, have been shown to have lower expression stability and can be degraded in serum samples (Chen et al., 2008; Cortez et al., 2011). We selected miR-16 as the normalization control because of its higher stability and less variability in circulation compared with other microRNAs (Ng et al., 2009; Cortez et al., 2011; Reid et al., 2011). Although the addition of synthetic, single-stranded microRNA may represent an interesting approach to circumventing normalization issues, more studies are necessary to determine an accurate normalization protocol (Cortez et al., 2011).
In summary, our pilot study has shown that plasma miR-17-5p, miR-20a and miR-22 are down-regulated in women with endometriosis, which raises the potential clinical utility of plasma microRNA profiling in endometriosis diagnosis. Although this approach is technically challenging, and the exact mechanisms of how microRNAs enter plasma as well as their biological functions remain unknown and will require further investigation, we believe that plasma microRNAs are a promising diagnostic tool for endometriosis.
Authors’ roles
S.-z.J. contributed substantially to conception and design analysis, interpretation of data, drafted the article and revised and approved the final version to be published. Y.Y. performed the analysis and interpretation of data, revised the article and approved the final version to be published. J.L. contributed substantially to conception and design and approved the final version to be published. P.S. contributed to the acquisition of data and approved the final version to be published. Jinhua Leng contributed substantially to conception and design, revised the article and approved the final version to be published.
Funding
This research was supported by grants to Jinhua Leng from the National Natural Science Foundation of China (81170548) and Key Project for Clinical Faculty Foundation, Ministry of Health, China (2010).
Conflict of interest
None declared.
Acknowledgements
We thank Shan Huang, PhD, at the Center of Excellence in Tissue Engineering, Chinese Academy of Medical Sciences and Peking Union Medical College for her excellent technical assistance. We thank all of the patients for agreeing to participate in our study.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. doi:10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Becker CM, Rohwer N, Funakoshi T, Cramer T, Bernhardt W, Birsner A, Folkman J, D'Amato RJ. 2-methoxyestradiol inhibits hypoxia-inducible factor-1{alpha} and suppresses growth of lesions in a mouse model of endometriosis. Am J Pathol. 2008;172:534–544. doi: 10.2353/ajpath.2008.061244. doi:10.2353/ajpath.2008.061244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burney RO, Hamilton AE, Aghajanova L, Vo KC, Nezhat CN, Lessey BA, Giudice LC. MicroRNA expression profiling of eutopic secretory endometrium in women with versus without endometriosis. Mol Hum Reprod. 2009;15:625–631. doi: 10.1093/molehr/gap068. doi:10.1093/molehr/gap068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. doi:10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Cho S, Choi YS, Yim SY, Yang HI, Jeon YE, Lee KE, Kim H, Seo SK, Lee BS. Urinary vitamin D-binding protein is elevated in patients with endometriosis. Hum Reprod. 2012;27:515–522. doi: 10.1093/humrep/der345. doi:10.1093/humrep/der345. [DOI] [PubMed] [Google Scholar]
- Cloonan N, Brown MK, Steptoe AL, Wani S, Chan WL, Forrest AR, Kolle G, Gabrielli B, Grimmond SM. The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008;9:R127. doi: 10.1186/gb-2008-9-8-r127. doi:10.1186/gb-2008-9-8-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. doi:10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Gu L, Di W. MiR-199a attenuates endometrial stromal cell invasiveness through suppression of the IKKbeta/NF-kappaB pathway and reduced interleukin-8 expression. Mol Hum Reprod. 2012;18:136–145. doi: 10.1093/molehr/gar066. doi:10.1093/molehr/gar066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for MicroRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. doi:10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kasti MM, Wright C, Fye HK, Roseman F, Kessler BM, Becker CM. Urinary peptide profiling identifies a panel of putative biomarkers for diagnosing and staging endometriosis. Fertil Steril. 2011;95:1261–1266. doi: 10.1016/j.fertnstert.2010.11.066. doi:10.1016/j.fertnstert.2010.11.066. [DOI] [PubMed] [Google Scholar]
- Filigheddu N, Gregnanin I, Porporato PE, Surico D, Perego B, Galli L, Patrignani C, Graziani A, Surico N. Differential expression of microRNAs between eutopic and ectopic endometrium in ovarian endometriosis. J Biomed Biotechnol. 2010;2010:369549. doi: 10.1155/2010/369549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores I, Rivera E, Ruiz LA, Santiago OI, Vernon MW, Appleyard CB. Molecular profiling of experimental endometriosis identified gene expression patterns in common with human disease. Fertil Steril. 2007;87:1180–1199. doi: 10.1016/j.fertnstert.2006.07.1550. doi:10.1016/j.fertnstert.2006.07.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice LC. Clinical practice. Endometriosis. N Engl J Med. 2010;362:2389–2398. doi: 10.1056/NEJMcp1000274. doi:10.1056/NEJMcp1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ramos R, Donnez J, Defrere S, Leclercq I, Squifflet J, Lousse JC, Van Langendonckt A. Nuclear factor-kappa B is constitutively activated in peritoneal endometriosis. Mol Hum Reprod. 2007;13:503–509. doi: 10.1093/molehr/gam033. doi:10.1093/molehr/gam033. [DOI] [PubMed] [Google Scholar]
- Hawkins SM, Creighton CJ, Han DY, Zariff A, Anderson ML, Gunaratne PH, Matzuk MM. Functional microRNA involved in endometriosis. Mol Endocrinol. 2011;25:821–832. doi: 10.1210/me.2010-0371. doi:10.1210/me.2010-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. doi:10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- Huang JQ, Lathi RB, Lemyre M, Rodriguez HE, Nezhat CH, Nezhat C. Coexistence of endometriosis in women with symptomatic leiomyomas. Fertil Steril. 2010;94:720–723. doi: 10.1016/j.fertnstert.2009.03.052. [DOI] [PubMed] [Google Scholar]
- Inomata M, Tagawa H, Guo YM, Kameoka Y, Takahashi N, Sawada K. MicroRNA-17-92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood. 2009;113:396–402. doi: 10.1182/blood-2008-07-163907. doi:10.1182/blood-2008-07-163907. [DOI] [PubMed] [Google Scholar]
- Kennedy S, Bergqvist A, Chapron C, D'Hooghe T, Dunselman G, Greb R, Hummelshoj L, Prentice A, Saridogan E. ESHRE guideline for the diagnosis and treatment of endometriosis. Hum Reprod. 2005;20:2698–2704. doi: 10.1093/humrep/dei135. doi:10.1093/humrep/dei135. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. doi:10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyama CM, Overbergh L, Debrock S, Valckx D, Vander PS, Meuleman C, Mihalyi A, Mwenda JM, Mathieu C, D'Hooghe TM. Increased peritoneal and endometrial gene expression of biologically relevant cytokines and growth factors during the menstrual phase in women with endometriosis. Fertil Steril. 2006;85:1667–1675. doi: 10.1016/j.fertnstert.2005.11.060. doi:10.1016/j.fertnstert.2005.11.060. [DOI] [PubMed] [Google Scholar]
- Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. doi:10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- Lei Z, Li B, Yang Z, Fang H, Zhang GM, Feng ZH, Huang B. Regulation of HIF-1alpha and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS One. 2009;4:e7629. doi: 10.1371/journal.pone.0007629. doi:10.1371/journal.pone.0007629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhu J, Zhang W, Chen Y, Zhang K, Popescu LM, Ma X, Lau WB, Rong R, Yu X, et al. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation. 2011;124:175–184. doi: 10.1161/CIRCULATIONAHA.110.012237. doi:10.1161/CIRCULATIONAHA.110.012237. [DOI] [PubMed] [Google Scholar]
- Lin SC, Wang CC, Wu MH, Yang SH, Li YH, Tsai SJ. Hypoxia-induced MicroRNA-20a expression increases ERK phosphorylation and angiogenic gene expression in endometriotic stromal cells. J Clin Endocrinol Metab. 2012;97:E1515–E1523. doi: 10.1210/jc.2012-1450. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. doi:10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- May KE, Conduit-Hulbert SA, Villar J, Kirtley S, Kennedy SH, Becker CM. Peripheral biomarkers of endometriosis: a systematic review. Hum Reprod Update. 2010;16:651–674. doi: 10.1093/humupd/dmq009. doi:10.1093/humupd/dmq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May KE, Villar J, Kirtley S, Kennedy SH, Becker CM. Endometrial alterations in endometriosis: a systematic review of putative biomarkers. Hum Reprod Update. 2011;17:637–653. doi: 10.1093/humupd/dmr013. doi:10.1093/humupd/dmr013. [DOI] [PubMed] [Google Scholar]
- McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57:833–840. doi: 10.1373/clinchem.2010.157198. doi:10.1373/clinchem.2010.157198. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. doi:10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol BW, Bayram N, Lijmer JG, Wiegerinck MA, Bongers MY, van der Veen F, Bossuyt PM. The performance of CA-125 measurement in the detection of endometriosis: a meta-analysis. Fertil Steril. 1998;70:1101–1108. doi: 10.1016/s0015-0282(98)00355-0. doi:10.1016/S0015-0282(98)00355-0. [DOI] [PubMed] [Google Scholar]
- Nagaraja AK, Creighton CJ, Yu Z, Zhu H, Gunaratne PH, Reid JG, Olokpa E, Itamochi H, Ueno NT, Hawkins SM, et al. A link between mir-100 and FRAP1/mTOR in clear cell ovarian cancer. Mol Endocrinol. 2010;24:447–463. doi: 10.1210/me.2009-0295. doi:10.1210/me.2009-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. doi:10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- Nnoaham KE, Hummelshoj L, Webster P, D'Hooghe T, de Cicco NF, de Cicco NC, Jenkinson C, Kennedy SH, Zondervan KT. Impact of endometriosis on quality of life and work productivity: a multicenter study across ten countries. Fertil Steril. 2011;96:366–373. doi: 10.1016/j.fertnstert.2011.05.090. doi:10.1016/j.fertnstert.2011.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson TE, Van der Hoek KH, Van der Hoek MB, Perry N, Wagaarachchi P, Robertson SA, Print CG, Hull LM. MicroRNA-regulated pathways associated with endometriosis. Mol Endocrinol. 2009;23:265–275. doi: 10.1210/me.2008-0387. doi:10.1210/me.2008-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Luo X, Toloubeydokhti T, Chegini N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Reprod. 2007;13:797–806. doi: 10.1093/molehr/gam063. doi:10.1093/molehr/gam063. [DOI] [PubMed] [Google Scholar]
- Ramon LA, Braza-Boils A, Gilabert-Estelles J, Gilabert J, Espana F, Chirivella M, Estelles A. microRNAs expression in endometriosis and their relation to angiogenic factors. Hum Reprod. 2011;26:1082–1090. doi: 10.1093/humrep/der025. doi:10.1093/humrep/der025. [DOI] [PubMed] [Google Scholar]
- Reid G, Kirschner MB, van Zandwijk N. Circulating microRNAs: association with disease and potential use as biomarkers. Crit Rev Oncol Hematol. 2011;80:193–208. doi: 10.1016/j.critrevonc.2010.11.004. doi:10.1016/j.critrevonc.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Reis FM, Luisi S, Abrao MS, Rocha AL, Vigano P, Rezende CP, Florio P, Petraglia F. Diagnostic value of serum activin A and follistatin levels in women with peritoneal, ovarian and deep infiltrating endometriosis. Hum Reprod. 2012;27:1445–1450. doi: 10.1093/humrep/des055. doi:10.1093/humrep/des055. [DOI] [PubMed] [Google Scholar]
- Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–59. doi: 10.1016/j.ygyno.2008.08.036. doi:10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil Steril. 1997;67:817–821. doi: 10.1016/s0015-0282(97)81391-x. doi:10.1016/S0015-0282(97)81391-X. [DOI] [PubMed] [Google Scholar]
- Rogers PA, D'Hooghe TM, Fazleabas A, Gargett CE, Giudice LC, Montgomery GW, Rombauts L, Salamonsen LA, Zondervan KT. Priorities for endometriosis research: recommendations from an international consensus workshop. Reprod Sci. 2009;16:335–346. doi: 10.1177/1933719108330568. doi:10.1177/1933719108330568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. doi:10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salta E, De Strooper B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol. 2012;11:189–200. doi: 10.1016/S1474-4422(11)70286-1. doi:10.1016/S1474-4422(11)70286-1. [DOI] [PubMed] [Google Scholar]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. doi:10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- Simoens S, Dunselman G, Dirksen C, Hummelshoj L, Bokor A, Brandes I, Brodszky V, Canis M, Colombo GL, DeLeire T, et al. The burden of endometriosis: costs and quality of life of women with endometriosis and treated in referral centres. Hum Reprod. 2012;27:1292–1299. doi: 10.1093/humrep/des073. doi:10.1093/humrep/des073. [DOI] [PubMed] [Google Scholar]
- Spaczynski RZ, Duleba AJ. Diagnosis of endometriosis. Semin Reprod Med. 2003;21:193–208. doi: 10.1055/s-2003-41326. doi:10.1055/s-2003-41326. [DOI] [PubMed] [Google Scholar]
- Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, Goto H, Takahashi T. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 microRNA cluster. Cancer Res. 2008;68:5540–5545. doi: 10.1158/0008-5472.CAN-07-6460. doi:10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
- Takata A, Otsuka M, Kojima K, Yoshikawa T, Kishikawa T, Yoshida H, Koike K. MicroRNA-22 and microRNA-140 suppress NF-kappaB activity by regulating the expression of NF-kappaB coactivators. Biochem Biophys Res Commun. 2011;411:826–831. doi: 10.1016/j.bbrc.2011.07.048. doi:10.1016/j.bbrc.2011.07.048. [DOI] [PubMed] [Google Scholar]
- Teague EM, Print CG, Hull ML. The role of microRNAs in endometriosis and associated reproductive conditions. Hum Reprod Update. 2010;16:142–165. doi: 10.1093/humupd/dmp034. doi:10.1093/humupd/dmp034. [DOI] [PubMed] [Google Scholar]
- Tokushige N, Markham R, Crossett B, Ahn SB, Nelaturi VL, Khan A, Fraser IS. Discovery of a novel biomarker in the urine in women with endometriosis. Fertil Steril. 2011;95:46–49. doi: 10.1016/j.fertnstert.2010.05.016. doi:10.1016/j.fertnstert.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Toloubeydokhti T, Pan Q, Luo X, Bukulmez O, Chegini N. The expression and ovarian steroid regulation of endometrial micro-RNAs. Reprod Sci. 2008;15:993–1001. doi: 10.1177/1933719108324132. doi:10.1177/1933719108324132. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tsuzuki T, Okada H, Cho H, Tsuji S, Nishigaki A, Yasuda K, Kanzaki H. Hypoxic stress simultaneously stimulates vascular endothelial growth factor via hypoxia-inducible factor-1alpha and inhibits stromal cell-derived factor-1 in human endometrial stromal cells. Hum Reprod. 2012;27:523–530. doi: 10.1093/humrep/der405. doi:10.1093/humrep/der405. [DOI] [PubMed] [Google Scholar]
- Uimari O, Jarvela I, Ryynanen M. Do symptomatic endometriosis and uterine fibroids appear together? J Hum Reprod Sci. 2011;4:34–38. doi: 10.4103/0974-1208.82358. doi:10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. doi:10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. doi:10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. doi:10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zheng Z, Guo J, Ding X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol. 2010;119:586–593. doi: 10.1016/j.ygyno.2010.07.021. doi:10.1016/j.ygyno.2010.07.021. [DOI] [PubMed] [Google Scholar]
- Xiong J. Emerging roles of microRNA-22 in human disease and normal physiology. Curr Mol Med. 2012;12:247–258. doi: 10.2174/156652412799218886. doi:10.2174/156652412799218886. [DOI] [PubMed] [Google Scholar]
- Yamakuchi M, Yagi S, Ito T, Lowenstein CJ. MicroRNA-22 regulates hypoxia signaling in colon cancer cells. PLoS One. 2011;6:e20291. doi: 10.1371/journal.pone.0020291. doi:10.1371/journal.pone.0020291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Kosaka N, Tanaka M, Koizumi F, Kanai Y, Mizutani T, Murakami Y, Kuroda M, Miyajima A, Kato T, et al. MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers. 2009;14:529–538. doi: 10.3109/13547500903150771. doi:10.3109/13547500903150771. [DOI] [PubMed] [Google Scholar]
- Yu Z, Willmarth NE, Zhou J, Katiyar S, Wang M, Liu Y, McCue PA, Quong AA, Lisanti MP, Pestell RG. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc Natl Acad Sci USA. 2010;107:8231–8236. doi: 10.1073/pnas.1002080107. doi:10.1073/pnas.1002080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. doi:10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zhao Q, Warrick J, Lockwood CM, Woodworth A, Moley KH, Gronowski AM. Circulating MicroRNA miR-323–3p as a Biomarker of Ectopic Pregnancy. Clin Chem. 2012;58:896–905. doi: 10.1373/clinchem.2011.179283. doi:10.1373/clinchem.2011.179283. [DOI] [PMC free article] [PubMed] [Google Scholar]