Abstract
Tumorigenesis is the process by which normal cells evolve the capacity to evade and overcome the constraints usually placed upon their growth and survival. To ensure the integrity of organs and tissues, the balance of cell proliferation and cell death is tightly maintained. The proteins controlling this balance are either considered oncogenes, which promote tumorigenesis, or tumor suppressors, which prevent tumorigenesis. Phosphoinositide 3-kinase enhancer (PIKE) is a family of GTP-binding proteins that possess anti-apoptotic functions and play an important role in the central nervous system. Notably, accumulating evidence suggests that PIKE is a proto-oncogene involved in tumor progression. The PIKE gene (CENTG1) is amplified in a variety of human cancers, leading to the resistance against apoptosis and the enhancement of invasion. In this review, we will summarize the functions of PIKE proteins in tumorigenesis and discuss their potential implications in cancer therapy.
Keywords: PIKE, tumorigenesis, cancer, Akt, phosphoinositide 3-kinase
Introduction
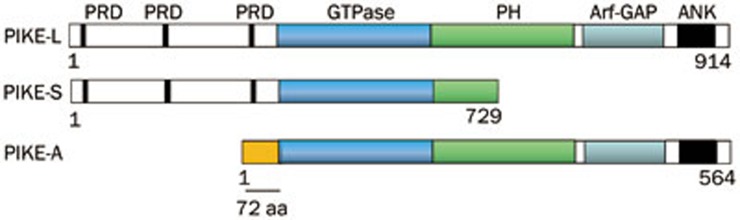
Phosphoinositide 3-kinase enhancer (PIKE) is a group of GTP-binding proteins that belong to the α1 subgroup of centaurin GTPase family1. The PIKE family includes three members, PIKE-L, PIKE-S and PIKE-A, which are originated from a single gene (CENTG1) through alternative splicing or differential transcription initiation2,3. PIKE-L is the longest isoform among the three family members, containing three proline-rich domains (PRD) at the N-terminus, a central GTP-binding domain, a pleckstrin homology (PH) domain, an Arf-GAP-like sequence, and two ankyrin repeats (ANK) in the C-terminus3. Compared to PIKE-L, PIKE-S lacks the Arf-GAP-like domain and ankyrin repeats4. PIKE-A differs from PIKE-L in that the N-terminal proline-rich domain is replaced with a unique short peptide of 72 amino acids2,5 (Figure 1). Both PIKE-S and PIKE-L bind to PI3K and enhance its activity. However, PIKE-A does not interplay with PI3K. Instead, it interacts with the downstream effector Akt and promotes its activity. These actions are mediated by their GTPase activity. Both PIKE-S and PIKE-L are prominently distributed in brain3,4, while PIKE-A shows a distinct tissue distribution pattern from other PIKE isoforms. Northern blot analysis reveals a high level of PIKE-A mRNA is expressed in brain and a small amount is also detected in liver, lung, skeletal muscle, spleen thymus, small intestine and periphery blood leukocytes6,7,8. For the cellular localization, PIKE-L and PIKE-A are present in both the nucleus and the cytoplasm, while PIKE-S resides exclusively in the nucleus9.
Figure 1.
Schematic representation of PIKE proteins. PRD, Proline-rich domain; GTPase, GTP-binding motif; PH, Pleckstrin homology domain; Arf-GAP, a domain showing sequence homology to ARF/GAP protein; ANK, ankyrin repeat.
In the last decade, our research group endeavored to delineate the physiological roles of PIKE proteins. With the availability of the whole body PIKE knockout (PIKE−/−) mice, the roles of PIKE in neuronal survival3,10,11,12, brain development13, mammary gland development14, obesity development15, insulin resistance16, and cell transformation17 have been characterized. Remarkably, PIKE proteins are involved in multiple signaling pathways in addition to the PI3K/Akt cascade. PIKEs are implicated in regulating the activity of signal transducer and activator of transcription 5A (STAT5) via prolactin (PRL)-stimulated JAK2 (Janus kinase 2) phosphorylation14. In addition, PIKE-L strongly binds SET, a DNase inhibitor, and prevented its degradation by asparaginyl endopeptidase (AEP), leading to resistance of neuronal cell death provoked by neuroexcitotoxic insults or ischemia10.
PIKE is a proto-oncogene
The accumulation of genetic damage in the forms of activated proto-oncogenes and inactivated tumor-suppressor genes is the driving force in the evolution of a normal cell to a malignant cell. Proto-oncogenes are a group of genes that cause normal cells to become cancerous when they are mutated or highly expressed18,19. Examples of proto-oncogenes include RAS, WNT, MYC, ERK, and TRK20, and they are considered as potential targets for molecular cancer therapy21,22.
The most crucial discovery of PIKE's activity in disease onset is its role in neuro-oncology. During our investigation on the mechanism of glioblastoma multiform formation, we found that the loci of GGAP2 (PIKE-A) are highly amplified in glioblastoma5. After sequence alignment analysis, we clarified that the resultant protein of GGAP2 is an isoform of PIKE, PIKE-A, which was originally identified in the human genome sequencing effort as KIAA01677. Indeed, PIKE-A displays an elevated expression in glioblastoma and astrocytoma, which result from gene amplification2. In addition, it is highly expressed in various cancers that originate from breast, prostate, skin, uterus, colon, ovary, liver, stomach, lung, cervix, rectum, testis, and kidney5,17,23,24. It is also demonstrated that PIKE-A expression is increased in 93% of brain tumors without CENTG1 amplification. Most recently, Xie et al found that PIKE-S is highly expressed in malignant human keratinocytes (SCC4 and SCC12B2) but had low expression in normal human keratinocytes25. EGF-induced squamous cell carcinoma (SCC) proliferation requires SH3 domain of Phospholipase C-γ1 (PLC-γ1), which is a guanine nucleotide exchange factor (GEF) for PIKE. Knockdown of PLC-γ1 or PIKE blocks EGF-induced SCC cell proliferation. These findings support the notion that PIKE plays a critical role in EGF-induced SCC cell proliferation and functions as a proto-oncogene in SCC.
Amplification of chromosome 12q13-q15, where CENTG1 is located, is frequently observed in numerous human cancers26,27,28,29. In 1994, Reifenberger et al revealed that CENTG1 is frequently co-amplified with cyclin-dependent kinase 4 (CDK4), which is a well-known proliferation activator that promotes E2F- and CDK2-dependent cell cycle progression in tumors28, it would be logical to surmise that PIKE-A amplification or overexpression coordinately acts with CDK4 amplification or overexpression to drive tumorigenesis. Cancer cells with this amplicon are more resistant to apoptotic stimuli compared with cells that express a normal CENTG1 copy number5. Indeed, from an automated network analysis on the core pathway of glioma formation, PIKE-A has recently been confirmed as a driver gene of glioblastoma30. These data suggest a strong correlation between PIKE-A expression and tumor formation. As a matter of act, PIKE-A overexpression is sufficient to transform NIH3T3 cells and enhance the proliferation and invasion of U87MG, a glioblastoma cell line without CDK4 amplicon and with modest PIKE-A expression17. Therefore, PIKE satisfies the criterion of a proto-oncogene, which implies its potential role in tumorigenesis.
Functions of PIKE in tumorigenesis
Three members (PIKE-L, PIKE-S, and PIKE-A) have been identified in the PIKE family so far, and accumulating evidence indicates that functions of PIKE are characterized by different isoforms at different subcellular compartments. PIKE-L and PIKE-A reside in multiple intracellular compartments, while PIKE-S localizes exclusively in the nucleus9. To understand the functions of PIKE in tumorigenesis, we will discuss the role of PIKE based on its cellular localization.
The functions of PIKE in the cell membrane
Cells transmit extracellular signals via membrane receptors. PIKE-L has been identified as a component of the netrin-1 signaling pathway, which protects neurons from apoptosis11. Traditionally, netrin-1 is a chemotropic cue for axon migration and arborization during neural development31. The most important receptors of netrin-1 are deleted in colorectal cancer (DCC) and the UNC5 family32. Recently, the roles of netrin-1 and its receptors in tumorigenesis have been broadly studied33 and DCC and UNC5 proteins are considered dependence receptors that regulate apoptosis depending on the interaction with their ligands, netrins34. They are also considered to be tumor suppressors, since they suppress tumor progression in the absence of netrin-135,36. PIKE-L/UNC5B association enhance cell survival via PI3K signaling11, which is controlled by a protein kinase Fyn. Fyn phosphorylation on both the receptor and PIKE-L is necessary for their interaction11,37. As Fyn is constitutively associated with DCC, presumably, PIKE-L may not interact with UNC5B but it may also associate with DCC38. Indeed, PIKE-L and DCC have been co-immunoprecipitated from rat brain lysates, which further supports this hypothesis11.
It has also demonstrated that PIKE-A associates with UNC5B in glioblastoma cell lines39. The PIKE-A/UNC5B binding is tightly regulated by Akt, in which Akt-induced phosphorylation of PIKE-A on Ser-472 promotes its interaction with UNC5B. PIKE-A suppresses UNC5B transcription by down-regulating p53, which is a transcriptive regulator of UNC5B40,41. As such, netrin-1 might stimulate Akt activation, which subsequently phosphorylates PIKE-A, escalating its binding to UNC5B, and thereby inhibiting the receptor cleavage and apoptosis-inducing activity. In addition, PIKE-A (S472) phosphorylation feed backs positively to further elevate the Akt activity and leads to p53 degradation through the Akt-MDM2 pathway, resulting in down-regulation of UNC5B39. Since PIKE-L associates with DCC in the brain, conceivably, other PIKE isoforms may interact with DCC and demonstrate a potential function in tumorigenesis.
Recently, we have found that PIKE-A regulates insulin receptor tyrosine kinase (IRTK) activity, leading to suppression of AMP-activated protein kinase (AMPK) activation. The association of PIKE-A with the insulin receptor is important for insulin to fully initiate the hepatic IRTK16. As IRTK also plays an important role in cancer progression42,43, it is reasonable to infer the IRTK-PIKE-A crosstalk may play a role in the regulation of PIKE-A in tumorigenesis.
The functions of PIKE in the cytoplasm
Cytoplasmic PI3-kinase regulates the membrane translocation and activation of many signaling molecules. As a PI 3-kinase enhancer, PIKE also functions through cytoplasmic molecular interactions. Our previous works demonstrate that PIKE-L reduces the anti-proliferation activity induced by merlin (schwannomin, a tumor suppressor), which is encoded by the NF2 gene. It is now recognized that mutation of the NF2 gene is not restricted to schwannomas but also extends to thyroid carcinomas, hepatocellular carcinomas, and perineurial tumors44,45,46,47. The N-terminus of PIKE-L specifically binds to the N-terminal domain of merlin48, which assists merlin's inhibition of PI3-kinase activity48. Notably, PIKEs are also implicated in regulating the activity of transcription factors such as STAT5A, which can be activated by PRL14, one important ligand of JAK249. As the JAK2/STAT5 cascade plays an important role in cancer initiation and progression50,51, it is logical to infer the roles PIKE in tumorigenesis via JAK2/STAT5 signaling regulation. PIKE-A is amplified in human cancer cells and Akt activity correlates with PIKE-A amplification5. It has been demonstrated that PIKE-A directly binds to activated Akt but not PI3-kinase in a guanine nucleotide-dependent fashion and stimulates the kinase activity of Akt, promoting cell survival, migration, and invasion52. Overexpression of PIKE-A enhances Akt activity and promotes cancer cell invasion, whereas dominant-negative PIKE-A and PIKE-A knockdown markedly inhibit these processes2. Further studies indicate that PIKA-A binds active Akt and enhances its activity through maintaining Akt active conformation or initiating its activation2. Most recently, the interaction of PIKE-A and Akt has been validated using fluorescence-based protein complementation assay in live cells. Disrupting the PIKE-A/Akt association, using a small peptide derived from the binding fragment on PIKE-A or Akt, inhibits glioblastoma cell proliferation, migration, and invasion, re-confirming the essential role of PIKE-A in Akt activation53.
Focal adhesion kinase (FAK) is a key player in tumor formation and progression54,55,56. Recently, Zhu et al showed that PIKE-A is a novel binding partner of FAK, and PIKE-A enhances its kinase activity in response to growth factor stimulation57. It is also revealed that PIKE-A functions as a FAK enhancer through two mechanisms: activating FAK to disassemble focal adhesions and control the trafficking of regulators to or from the focal adhesion, when the focal adhesion is being remodeled, which are key processes for cell motility and migration58,59. Moreover, phosphorylation of FAK by cyclin-dependent kinase 5 (CDK5) is critical for FAK-mediated microtubule fork formation and cell migration60 and phosphorylation of FAK by CDK5 regulates FAK's function in mediating cell migration61. Interestingly, PIKE-A has also been demonstrated as a physiological substrate of CDK5. In glioblastoma cells, CDK5 directly phosphorylates PIKE-A at Ser-279 in its GTPase domain, which stimulates PIKE-A GTPase activity and the activity of its downstream effector Akt52. Moreover, phosphorylation of PIKE-A by CDK5 mediates growth factor-induced migration and invasion of human glioblastoma cells52. Collectively, these data indicate that PIKE-A, FAK, and CDK5 are involved in cross-talk to regulate cell migration and invasion in cancer cells.
The functions of PIKE in the nucleus
While most PI3K signaling studies have focused on its activities at the plasma membrane, it is now clear that the nucleus has a distinct set of PI3K signaling machinery and effectors with different regulatory mechanisms62,63. Nuclear phosphoinositide lipids appear to regulate cell proliferation and differentiation as well64,65. PLC-γ1 is a tyrosine kinase substrate for many RTKs and non-RTKs and is essential for cell proliferation and differentiation66. It has been detected in the nucleus and is localized to the nucleus in highly transformed and proliferating cell lines67,68. It has been demonstrated that PLC-γ1 functions as a GEF (guanine nucleotide exchange factor) for PIKE-S and thereby activates PI3-kinase activity69. Moreover, Nerve growth factor(NGF) treatment elicits the membrane-associated protein, 4.1N, to translocate to the nucleus and bind PIKE, which prevents the interaction between protein 4.1N and nuclear PI3-kinase, leading to diminish the activation of PI3-kinase4.
A recent report reveals that hsa-miR26a, CDK4, and PIKE-A comprise a functional integrated oncomir/oncogene DNA cluster, which promotes the aggressiveness in glioblastoma70. Cai et al demonstrated that PIKE-A promotes tumor progression by increasing the transcriptional activity of nuclear factor κB by directly interacting with its p50 subunit23. Most recently, using a SCC system, PIKE was found to mediate epidermal growth factor receptor (EGFR)-dependent SCC cell proliferation25. In the SCC cells, PIKE is abundantly expressed in vitro and in vivo. Previous studies have indicated that PLC-γ1 is required for EGF-induced SCC cell proliferation71, and the proliferative role of PLC-γ1 relies on its SH3 domain but is independent of its lipase activity72. Conceivably, PIKE plays a role in SCC cell proliferation because of the interaction between PIKE and PLC-γ1 in the nucleus73. These data provide evidence of PIKE-EGFR signaling crosstalk; further validate the function of PIKE in tumorigenesis.
Targeting PIKE in cancer therapy
The role of a proto-oncogene has been clearly defined and targeting proto-oncogene is a strategy for cancer therapy20,21. PIKE, as a new member of the proto-oncogene family, is a potential target for cancer therapy. Similar with many known proto-oncogenes, the functions of PIKE rely on different kinds of signaling pathways associated with many binding partners (Table 1). From these molecules, we can clearly sort out the potential mechanisms for targeting PIKE in cancer therapy. Although how to manipulate PIKE activities to contribute cancer treatment is still not well clarified yet, we can speculate the regulation of PIKE in three directions.
Table 1. PIKE interaction partners summary.
| PIKE isoforms | Interacting proteins | Functions |
|---|---|---|
| PIKE-S | PI3K | Enhancing kinase activity of nuclear PI3K |
| PLCγ1 | Enhancing PIKE-S GTPase activity | |
| 4.1N | Abolishing PIKE-S/PI3K association | |
| BRAG2 | Accumulating BRAG2 in nucleoli | |
| PIKE-L | PI3K | Enhancing kinase activity of PI3K |
| Homer 1c | Activating PI3K by quisqualate | |
| Merlin | Inhibiting merlin-induced PI3K activity | |
| UNC5B | Activating PI3K by netrin-1 | |
| SET | Preventing SET from proteolytic cleavage | |
| PIKE-A | Akt | Enhancing Akt kinase activity |
| Arf1 | Enhancing GTPase activity of Arf1 | |
| AP1 | Inhibiting GAP activity of PIKE-A; | |
| Altering the intracellular localization of AP1 | ||
| Fyn | Preventing PIKE-A degradation | |
| AMPK | Regulating AMPK activity | |
| CDK5 | Enhancing the GTPase of PIKE-A | |
| FAK | Enhancing the kinase activity of FAK | |
| UNC5B | Regulating UNC5B apoptosis-inducing effect | |
| Insulin receptor | Inhibiting AMPK phosphorylation | |
| STAT5a | Enhancing STAT5a phosphorylation |
Regulating PIKE GTPase activity
In response to stimulatory signals, GTPase activities are controlled by the GTP/GDP ratio and subcellular distribution in the cell through the joint action of multiple regulatory molecules: GEFs, which activate GTPases by promoting GDP-to-GTP exchange, and GAPs, which inactivate the GTPases by enhancing intrinsic GTP hydrolysis activity74. It has been reported that Dynamin 2 (a potent activator of metastatic migration) enhances invasive migration of pancreatic tumor cells through stabilization of the Rac1 GEF Vav175. The anticancer agent cucurbitacin I, a Jak2 inhibitor, reduces the activation of Rac1 in response to the ErbB3 ligand heregulin in breast cancer cells76. As a GTPase, PIKE can be activated by intrinsic or extrinsic cues, setting off a signaling cascade. Therefore, regulating the GEFs and GAPs of PIKE is a potential way to interfere with the functions of PIKE. So far, we have characterized that PLC-γ1 is a GEF of PIKE73, accordingly, functions of PIKE may be able to be regulated by manipulating PLC-γ1 activity. Discovering other GEFs and GAPs of PIKE in the future may also contribute to the regulation of PIKE functions.
The PH domain is a structural motif that binds phosphatidylinositol lipids to biological membranes77. The PH domain of PIKE plays a regulatory role in regard to the cellular localization and GTPase activity of the protein78. Binding of phospholipids to the PIKE PH domain increases the GTPase activity. It is suggested that the phospholipid/PIKE interaction causes a conformational change in the protein, rendering the C-terminal Arf-GAP domain accessible to its N-terminal domain, thus inducing the GTPase activity. This hypothesis is supported by the observation that the enzymatic activity of PIKE-A is low when the Arf-GAP domain is deleted8. Moreover, a recent structural study of the PIKE PH domain also suggests that the motif is responsible for binding the head groups of phosphoinositides in the cell membrane79. Together, these results indicate that PIKE cellular localization and activity are controlled by the interaction between phospholipids and the protein's PH domain.
Cytoplasm-nucleus shuttling of PIKE is known to correlate with its GTPase activity and cellular functions. Based on the structure of its PH domain, PIKE has been considered as one of the split PH domain containing proteins79. The split PH domain mediates the cytoplasmic-nuclear localization of PIKE via a positively charged nuclear localization sequence (NLS). Lipid membrane binding of the PH domain is further enhanced by the NLS79. Therefore, based on this model, it is conceivable that regulation of phosphatidylinositol phosphate (PIP) concentration at cytoplasmic membranes could possibly control the localization of PIKE, leading to alterations in its activity and cellular functions.
Modulating PIKE phosphorylation
It has been demonstrated that Akt1/PKBα regulates invasion of inflammatory breast cancer cells through Akt1 phosphorylation of Rho C GTPase80. Calmodulin binds to K-Ras B and inhibits phosphorylation of K-Ras B at Ser1-81, near to the membrane anchoring domain, modulating the signaling of oncogenic K-Ras B, which may be relevant to normal cell physiology, and opens new therapeutic perspectives for the inhibition of oncogenic K-Ras B signaling in tumors. PIKE can also be regulated by protein phosphorylation. It has been reported that PIKE-A can be phosphorylated by CDK5, Akt, and Fyn on Ser-27952, Ser-62923/Ser-47239, and Tyr-682/Tyr-77281, respectively. In addition, it has been shown that PIKE-A can also be phosphorylated upon growth factor stimulation82.
Phosphorylation of PIKE plays an important role in dictating its biological functions. To regulate the functions of PIKE through altering the levels of PIKE phosphorylation is rational. As described above, serine/threonine and tyrosine kinases are involved in the phosphorylation of PIKE; these kinases contribute to cancer progression and their cross-talk also plays a role in tumorigenesis83. Since PIKE-A acts as a proto-oncogene and contributes to tumorigenesis, it is reasonable to assume that suppressing activity of the kinases with inhibitors is a good approach for targeting PIKE in cancer therapy. It should be noted that there are many other putative phosphorylation sites on PIKE, which suggests that there are additional potential roles of PIKE in tumorigenesis, as well as more ways to impinge on PIKE function.
Disrupting protein-protein interactions
Protein-protein interactions (PPI) play an important role in tumorigenesis and disrupting PPI is considered a novel therapy for cancer treatment84. It has been reported that the ERK1/2-binding IQGAP1 (IQ motif-containing GTPase activating protein 1) WW domain peptide disrupts IQGAP1-ERK1/2 interactions and inhibits RAS- and RAF-driven tumorigenesis85. EHop-016, a novel small molecule, inhibits Rac GTPase activity by disrupting the interaction of Rac with its direct guanine nucleotide exchange factors86. It is now clear that PIKE interacts with numerous molecules to trigger multiple physiological functions potentially involved in tumorigenesis (Table 1). As cancer is a complex disease, the representation of a malignant cell as a protein-protein interaction network (PPIN) and its subsequent analysis can provide insight into the behavior of cancer cells and lead to the discovery of new biomarkers84. The feasibility of targeting PPIs for functional studies has been well established87 and targeting PPI is now considered a potential strategy for cancer therapy88. Therefore, disrupting the PIKE binding networks may provide an efficient strategy for regulating PIKE activity in tumorigenesis.
As such, we have demonstrated that the disruption of PIKE-A/Akt interaction leads to reduction of glioblastoma cell proliferation, survival, migration and invasion53. Moreover, peptides derived from the binding fragments of PIKE-A/Akt are able to inhibit this protein complex and sensitize glioblastoma cells to clinical anti-gloom chemotherapeutics53. Since PIKE-A/Akt association also plays an important role in prostate cancer progression23, it is reasonable to consider targeting this PPI for prostate cancer treatment. Among the binding partners of PIKE (Table 1), most of them are involved in tumorigenesis, such as UNC5B89, AMPK90, and Fyn37. Therefore, regulating PIKE-related PPI may serve as a good strategy for targeting PIKE in tumorigenesis.
Future perspectives
The role of PIKE GTPase in tumorigenesis is evident based on the studies performed in the past 10 years, yet we believe that the functional activity of PIKE in tumorigenesis has not been fully elucidated. With the availability of PIKE knockout animals, we will further delineate the authentic functions of PIKE in tumorigenesis. We and others have established that PIKE acts as a proto-oncogene. Thus, knockout mice may be resistant to the spontaneous or chemical-induced formation of cancers. However, experiments performed in PIKE knockout mice could not address the question convincingly as it is unknown if the high PIKE expression is the cause or consequence of cellular transformation. Another fundamental question regarding PIKE signaling is whether PIKE activation is growth factor specific, ie its activity can be triggered by NGF/EGF. In this regard, a parallel carcinogenic research study in PIKE transgenic and PIKE knockout mice may provide a more definitive answer. Additionally, the role of PIKE as an initiator or promoter of tumorigenesis still needs to be determined. Further study of PIKE signaling in tumorigenesis will not only enhance our knowledge about its physiological activities, but may also provide significant implications for cancer prevention and treatments.
Acknowledgments
This work is supported by the National Institutes of Health grant No RO1 CA127119, USA (Keqiang YE). We apologize for not citing original work of many colleagues because of space constraints. We would like to thank Obiamaka OBIANYO for proof reading the manuscript.
References
- Jackson TR, Kearns BG, Theibert AB. Cytohesins and centaurins: mediators of PI 3-kinase-regulated Arf signaling. Trends Biochem Sci. 2000;25:489–95. doi: 10.1016/s0968-0004(00)01644-3. [DOI] [PubMed] [Google Scholar]
- Ahn JY, Rong R, Kroll TG, Van Meir EG, Snyder SH, Ye K. PIKE (phosphatidylinositol 3-kinase enhancer)-A GTPase stimulates Akt activity and mediates cellular invasion. J Biol Chem. 2004;279:16441–51. doi: 10.1074/jbc.M312175200. [DOI] [PubMed] [Google Scholar]
- Rong R, Ahn JY, Huang H, Nagata E, Kalman D, Kapp JA, et al. PI3 kinase enhancer-Homer complex couples mGluRI to PI3 kinase, preventing neuronal apoptosis. Nat Neurosci. 2003;6:1153–61. doi: 10.1038/nn1134. [DOI] [PubMed] [Google Scholar]
- Ye K, Hurt KJ, Wu FY, Fang M, Luo HR, Hong JJ, et al. Pike. A nuclear gtpase that enhances PI3kinase activity and is regulated by protein 4.1N. Cell. 2000;103:919–30. doi: 10.1016/s0092-8674(00)00195-1. [DOI] [PubMed] [Google Scholar]
- Ahn JY, Hu Y, Kroll TG, Allard P, Ye K. PIKE-A is amplified in human cancers and prevents apoptosis by up-regulating Akt. Proc Natl Acad Sci U S A. 2004;101:6993–8. doi: 10.1073/pnas.0400921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkahloun AG, Krizman DB, Wang Z, Hofmann TA, Roe B, Meltzer PS. Transcript mapping in a 46-kb sequenced region at the core of 12q13.3 amplification in human cancers. Genomics. 1997;42:295–301. doi: 10.1006/geno.1997.4727. [DOI] [PubMed] [Google Scholar]
- Nagase T, Seki N, Ishikawa K, Tanaka A, Nomura N. Prediction of the coding sequences of unidentified human genes. V. The coding sequences of 40 new genes (KIAA0161-KIAA0200) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1996;3:17–24. doi: 10.1093/dnares/3.1.17. [DOI] [PubMed] [Google Scholar]
- Xia C, Ma W, Stafford LJ, Liu C, Gong L, Martin JF, et al. GGAPs, a new family of bifunctional GTP-binding and GTPase-activating proteins. Mol Cell Biol. 2003;23:2476–88. doi: 10.1128/MCB.23.7.2476-2488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CB, Ye K. PIKE GTPase are phosphoinositide-3-kinase enhancers, suppressing programmed cell death. J Cell Mol Med. 2007;11:39–53. doi: 10.1111/j.1582-4934.2007.00014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Jang SW, Liu X, Cheng D, Peng J, Yepes M, et al. Neuroprotective actions of PIKE-L by inhibition of SET proteolytic degradation by asparagine endopeptidase. Mol Cell. 2008;29:665–78. doi: 10.1016/j.molcel.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Jang SW, Okada M, Chan CB, Feng Y, Liu Y, et al. Netrin-1 mediates neuronal survival through PIKE-L interaction with the dependence receptor UNC5B. Nat Cell Biol. 2008;10:698–706. doi: 10.1038/ncb1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CB, Chen Y, Liu X, Papale L, Escayg A, Mei L, et al. Essential role of PIKE GTPases in neuronal protection against excitotoxic insults. Adv Biol Regul. 2011;52:66–76. doi: 10.1016/j.advenzreg.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CB, Liu X, Pradoldej S, Hao C, An J, Yepes M, et al. Phosphoinositide 3-kinase enhancer regulates neuronal dendritogenesis and survival in neocortex. J Neurosci. 2011;31:8083–92. doi: 10.1523/JNEUROSCI.1129-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CB, Liu X, Ensslin MA, Dillehay DL, Ormandy CJ, Sohn P, et al. PIKE-A is required for prolactin-mediated STAT5a activation in mammary gland development. EMBO J. 2010;29:956–68. doi: 10.1038/emboj.2009.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CB, Liu X, Jung DY, Jun JY, Luo HR, Kim JK, et al. Deficiency of phosphoinositide 3-kinase enhancer protects mice from diet-induced obesity and insulin resistance. Diabetes. 2010;59:883–93. doi: 10.2337/db09-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CB, Liu X, He K, Qi Q, Jung DY, Kim JK, et al. The association of phosphoinositide 3-kinase enhancer A with hepatic insulin receptor enhances its kinase activity. EMBO Reports. 2011;12:847–54. doi: 10.1038/embor.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hu Y, Hao C, Rempel SA, Ye K. PIKE-A is a proto-oncogene promoting cell growth, transformation and invasion. Oncogene. 2007;26:4918–27. doi: 10.1038/sj.onc.1210290. [DOI] [PubMed] [Google Scholar]
- Adamson ED. Oncogenes in development. Development. 1987;99:449–71. doi: 10.1242/dev.99.4.449. [DOI] [PubMed] [Google Scholar]
- Weinstein IB, Joe AK. Mechanisms of disease: Oncogene addiction — a rationale for molecular targeting in cancer therapy. Nat Clin Prac. 2006;3:448–57. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- Anderson MW, Reynolds SH, You M, Maronpot RM. Role of proto-oncogene activation in carcinogenesis. Environ Health Perspect. 1992;98:13–24. doi: 10.1289/ehp.929813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putzer BM, Drosten M. The RET proto-oncogene: a potential target for molecular cancer therapy. Trends Mol Med. 2004;10:351–7. doi: 10.1016/j.molmed.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Halpern M. Proto-oncogene products as target antigens for cancer vaccines. Int J Oncol. 1997;11:863–8. doi: 10.3892/ijo.11.4.863. [DOI] [PubMed] [Google Scholar]
- Cai Y, Wang J, Li R, Ayala G, Ittmann M, Liu M. GGAP2/PIKE-a directly activates both the Akt and nuclear factor-kappaB pathways and promotes prostate cancer progression. Cancer Res. 2009;69:819–27. doi: 10.1158/0008-5472.CAN-08-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobbe CB, Trampe-Kieslich A, Reifenberger G. Genetic alteration and expression of the phosphoinositol-3-kinase/Akt pathway genes PIK3CA and PIKE in human glioblastomas. Neuropathol Appl Neurobiol. 2005;31:486–90. doi: 10.1111/j.1365-2990.2005.00660.x. [DOI] [PubMed] [Google Scholar]
- Xie Z, Jiang Y, Liao EY, Chen Y, Pennypacker SD, Peng J, et al. PIKE mediates EGFR proliferative signaling in squamous cell carcinoma cells. Oncogene. 2012;31:5090–8. doi: 10.1038/onc.2012.10. [DOI] [PubMed] [Google Scholar]
- Reifenberger G, Ichimura K, Reifenberger J, Elkahloun AG, Meltzer PS, Collins VP. Refined mapping of 12q13-q15 amplicons in human malignant gliomas suggests CDK4/SAS and MDM2 as independent amplification targets. Cancer Res. 1996;56:5141–5. [PubMed] [Google Scholar]
- Reifenberger G, Reifenberger J, Ichimura K, Collins VP. Amplification at 12q13-14 in human malignant gliomas is frequently accompanied by loss of heterozygosity at loci proximal and distal to the amplification site. Cancer Res. 1995;55:731–4. [PubMed] [Google Scholar]
- Reifenberger G, Reifenberger J, Ichimura K, Meltzer PS, Collins VP. Amplification of multiple genes from chromosomal region 12q13-14 in human malignant gliomas: preliminary mapping of the amplicons shows preferential involvement of CDK4, SAS, and MDM2. Cancer Res. 1994;54:4299–303. [PubMed] [Google Scholar]
- Su WT, Alaminos M, Mora J, Cheung NK, La Quaglia MP, Gerald WL. Positional gene expression analysis identifies 12q overexpression and amplification in a subset of neuroblastomas. Cancer Genet Cytogenet. 2004;154:131–7. doi: 10.1016/j.cancergencyto.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PloS one. 2010;5:e8918. doi: 10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GX, Poo MM. Requirement of TRPC channels in netrin-1-induced chemotropic turning of nerve growth cones. Nature. 2005;434:898–904. doi: 10.1038/nature03478. [DOI] [PubMed] [Google Scholar]
- Ellezam B, Selles-Navarro I, Manitt C, Kennedy TE, McKerracher L. Expression of netrin-1 and its receptors DCC and UNC-5H2 after axotomy and during regeneration of adult rat retinal ganglion cells. Exp Neurol. 2001;168:105–15. doi: 10.1006/exnr.2000.7589. [DOI] [PubMed] [Google Scholar]
- Arakawa H. Netrin-1 and its receptors in tumorigenesis. Nat Rev Cancer. 2004;4:978–87. doi: 10.1038/nrc1504. [DOI] [PubMed] [Google Scholar]
- Lee HK, Seo IA, Seo E, Seo SY, Lee HJ, Park HT. Netrin-1 induces proliferation of Schwann cells through Unc5b receptor. Biochem Biophys Res Commun. 2007;362:1057–62. doi: 10.1016/j.bbrc.2007.08.143. [DOI] [PubMed] [Google Scholar]
- Guenebeaud C, Goldschneider D, Castets M, Guix C, Chazot G, Delloye-Bourgeois C, et al. The dependence receptor UNC5H2/B triggers apoptosis via PP2A-mediated dephosphorylation of DAP kinase. Mol cell. 2012;40:863–76. doi: 10.1016/j.molcel.2010.11.021. [DOI] [PubMed] [Google Scholar]
- Llambi F, Lourenco FC, Gozuacik D, Guix C, Pays L, Del Rio G, et al. The dependence receptor UNC5H2 mediates apoptosis through DAP-kinase. EMBO J. 2005;24:1192–201. doi: 10.1038/sj.emboj.7600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito YD, Jensen AR, Salgia R, Posadas EM. Fyn: a novel molecular target in cancer. Cancer. 2010;116:1629–37. doi: 10.1002/cncr.24879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mille F, Llambi F, Guix C, Delloye-Bourgeois C, Guenebeaud C, Castro-Obregon S, et al. Interfering with multimerization of netrin-1 receptors triggers tumor cell death. Cell Death Differ. 2009;16:1344–51. doi: 10.1038/cdd.2009.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Jang SW, Joshi J, Yoo MH, Ye K. Akt-phosphorylated PIKE-A inhibits UNC5B-induced apoptosis in cancer cell lines in a p53-dependent manner. Mol Biol Cell. 2011;22:1943–54. doi: 10.1091/mbc.E10-11-0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A, Matsuda F, Goto Y, Cheng Y, Gonda H, Inoue N, et al. Molecular cloning of a porcine (Sus scrofa) apoptosis inhibitory ligand, netrin-1, and its receptor, p53RDL1. J Reprod Dev. 2008;54:275–80. doi: 10.1262/jrd.20044. [DOI] [PubMed] [Google Scholar]
- Tanikawa C, Matsuda K, Fukuda S, Nakamura Y, Arakawa H. p53RDL1 regulates p53-dependent apoptosis. Nat Cell Biol. 2003;5:216–23. doi: 10.1038/ncb943. [DOI] [PubMed] [Google Scholar]
- Gross JM, Yee D. The type-1 insulin-like growth factor receptor tyrosine kinase and breast cancer: biology and therapeutic relevance. Cancer Metastasis Rev. 2003;22:327–36. doi: 10.1023/a:1023720928680. [DOI] [PubMed] [Google Scholar]
- Hopfner M, Sutter AP, Huether A, Baradari V, Scherubl H. Tyrosine kinase of insulin-like growth factor receptor as target for novel treatment and prevention strategies of colorectal cancer. World J Gastroenterol. 2006;12:5635–43. doi: 10.3748/wjg.v12.i35.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi AB, Hara T, Ramesh V, Gao J, Klein-Szanto AJ, Morin F, et al. Mutations in transcript isoforms of the neurofibromatosis 2 gene in multiple human tumour types. Nat Genet. 1994;6:185–92. doi: 10.1038/ng0294-185. [DOI] [PubMed] [Google Scholar]
- Lasota J, Fetsch JF, Wozniak A, Wasag B, Sciot R, Miettinen M. The neurofibromatosis type 2 gene is mutated in perineurial cell tumors: a molecular genetic study of eight cases. Am J Pathol. 2001;158:1223–9. doi: 10.1016/S0002-9440(10)64072-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau P, Marchio A, Nagamori S, Seki S, Tiollais P, Dejean A. Homozygous deletion scanning in hepatobiliary tumor cell lines reveals alternative pathways for liver carcinogenesis. Hepatology. 2003;37:852–61. doi: 10.1053/jhep.2003.50138. [DOI] [PubMed] [Google Scholar]
- Sheikh HA, Tometsko M, Niehouse L, Aldeeb D, Swalsky P, Finkelstein S, et al. Molecular genotyping of medullary thyroid carcinoma can predict tumor recurrence. Am J Surg Pathol. 2004;28:101–6. doi: 10.1097/00000478-200401000-00012. [DOI] [PubMed] [Google Scholar]
- Rong R, Tang X, Gutmann DH, Ye K. Neurofibromatosis 2 (NF2) tumor suppressor merlin inhibits phosphatidylinositol 3-kinase through binding to PIKE-L. Proc Natl Acad Sci U S A. 2004;101:18200–5. doi: 10.1073/pnas.0405971102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui H, Kirken RA, Farrar WL. Activation of receptor-associated tyrosine kinase JAK2 by prolactin. J Biol Chem. 1994;269:5364–8. [PubMed] [Google Scholar]
- Britschgi A, Andraos R, Brinkhaus H, Klebba I, Romanet V, Muller U, et al. JAK2/STAT5 inhibition circumvents resistance to PI3K/mTOR blockade: a rationale for cotargeting these pathways in metastatic breast cancer. Cancer Cell. 2012;22:796–811. doi: 10.1016/j.ccr.2012.10.023. [DOI] [PubMed] [Google Scholar]
- Wagner KU, Rui H. Jak2/Stat5 signaling in mammogenesis, breast cancer initiation and progression. J Mammary Gland Biol Neoplasia. 2008;13:93–103. doi: 10.1007/s10911-008-9062-z. [DOI] [PubMed] [Google Scholar]
- Liu R, Tian B, Gearing M, Hunter S, Ye K, Mao Z. Cdk5-mediated regulation of the PIKE-A-Akt pathway and glioblastoma cell invasion. Proc Natl Acad Sci U S A. 2008;105:7570–5. doi: 10.1073/pnas.0712306105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q, He K, Liu X, Pham C, Meyerkord C, Fu H, et al. Disrupting the PIKE-A/Akt interaction inhibits glioblastoma cell survival, migration, invasion and colony formation. Oncogene. 2013;32:1030–40. doi: 10.1038/onc.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandilands E, Serrels B, McEwan DG, Morton JP, Macagno JP, McLeod K, et al. Autophagic targeting of Src promotes cancer cell survival following reduced FAK signalling. Nat Cell Biol. 2011;14:51–60. doi: 10.1038/ncb2386. [DOI] [PubMed] [Google Scholar]
- Sawhney RS, Liu W, Brattain MG. A novel role of ERK5 in integrin-mediated cell adhesion and motility in cancer cells via Fak signaling. J Cell Physiol. 2009;219:152–61. doi: 10.1002/jcp.21662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Slambrouck S, Grijelmo C, De Wever O, Bruyneel E, Emami S, Gespach C, et al. Activation of the FAK-src molecular scaffolds and p130Cas-JNK signaling cascades by alpha1-integrins during colon cancer cell invasion. Int J Oncol. 2007;31:1501–8. [PubMed] [Google Scholar]
- Zhu Y, Wu Y, Kim JI, Wang Z, Daaka Y, Nie Z. Arf GTPase-activating protein AGAP2 regulates focal adhesion kinase activity and focal adhesion remodeling. J Biol Chem. 2009;284:13489–96. doi: 10.1074/jbc.M900469200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieg DJ, Hauck CR, Schlaepfer DD. Required role of focal adhesion kinase (FAK) for integrin-stimulated cell migration. J Cell Sci. 1999;112:2677–91. doi: 10.1242/jcs.112.16.2677. [DOI] [PubMed] [Google Scholar]
- Yeo MG, Sung BH, Oh HJ, Park ZY, Marcantonio EE, Song WK. Focal adhesion targeting of v-Crk is essential for FAK phosphorylation and cell migration in mouse embryo fibroblasts deficient src family kinases or p130CAS. J Cell Physiol. 2008;214:604–13. doi: 10.1002/jcp.21247. [DOI] [PubMed] [Google Scholar]
- Xie Z, Sanada K, Samuels BA, Shih H, Tsai LH. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell. 2003;114:469–82. doi: 10.1016/s0092-8674(03)00605-6. [DOI] [PubMed] [Google Scholar]
- Ilic D, Damsky CH, Yamamoto T.Focal adhesion kinase: at the crossroads of signal transduction J Cell Sci 1997110(Pt 4): 401–7. [DOI] [PubMed] [Google Scholar]
- Martelli AM, Evangelist C, Billi AM, Manzoli L, Papa V, Cocco L. Intranuclear 3′-phosphoinositide metabolism and apoptosis protection in PC12 cells. Acta Biomed. 2007;78:113–9. [PubMed] [Google Scholar]
- Martelli AM, Faenza I, Billi AM, Manzoli L, Evangelisti C, Fala F, et al. Intranuclear 3′-phosphoinositide metabolism and Akt signaling: new mechanisms for tumorigenesis and protection against apoptosis. Cell Signal. 2006;18:1101–7. doi: 10.1016/j.cellsig.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Matteucci A, Faenza I, Gilmour RS, Manzoli L, Billi AM, Peruzzi D, et al. Nuclear but not cytoplasmic phospholipase C beta 1 inhibits differentiation of erythroleukemia cells. Cancer Res. 1998;58:5057–60. [PubMed] [Google Scholar]
- York JD, Majerus PW. Nuclear phosphatidylinositols decrease during S-phase of the cell cycle in HeLa cells. J Biol Chem. 1994;269:7847–50. [PubMed] [Google Scholar]
- Wang Z, Gluck S, Zhang L, Moran MF. Requirement for phospholipase C-gamma1 enzymatic activity in growth factor-induced mitogenesis. Mol Cell Biol. 1998;18:590–7. doi: 10.1128/mcb.18.1.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diakonova M, Chilov D, Arnaoutov A, Alexeyev V, Nikolsky N, Medvedeva N. Intracellular distribution of phospholipase C gamma 1 in cell lines with different levels of transformation. Eur J Cell Biol. 1997;73:360–7. [PubMed] [Google Scholar]
- Neri LM, Borgatti P, Capitani S, Martelli AM. Nuclear diacylglycerol produced by phosphoinositide-specific phospholipase C is responsible for nuclear translocation of protein kinase C-alpha. J Biol Chem. 1998;273:29738–44. doi: 10.1074/jbc.273.45.29738. [DOI] [PubMed] [Google Scholar]
- Ye K, Snyder SH.PIKE GTPase: a novel mediator of phosphoinositide signaling J Cell Sci 2004117(Pt 2): 155–61. [DOI] [PubMed] [Google Scholar]
- Kim H, Huang W, Jiang X, Pennicooke B, Park PJ, Johnson MD. Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proc Natl Acad Sci U S A. 2010;107:2183–8. doi: 10.1073/pnas.0909896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Chen Y, Liao EY, Jiang Y, Liu FY, Pennypacker SD. Phospholipase C-gamma1 is required for the epidermal growth factor receptor-induced squamous cell carcinoma cell mitogenesis. Biochem Biophys Res Commun. 2010;397:296–300. doi: 10.1016/j.bbrc.2010.05.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Chen Y, Pennypacker SD, Zhou Z, Peng D. The SH3 domain, but not the catalytic domain, is required for phospholipase C-gamma1 to mediate epidermal growth factor-induced mitogenesis. Biochem Biophys Res Commun. 2010;398:719–22. doi: 10.1016/j.bbrc.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Aghdasi B, Luo HR, Moriarity JL, Wu FY, Hong JJ, et al. Phospholipase C gamma 1 is a physiological guanine nucleotide exchange factor for the nuclear GTPase PIKE. Nature. 2002;415:541–4. doi: 10.1038/415541a. [DOI] [PubMed] [Google Scholar]
- Niemann HH, Knetsch ML, Scherer A, Manstein DJ, Kull FJ. Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms. EMBO J. 2001;20:5813–21. doi: 10.1093/emboj/20.21.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razidlo GL, Wang Y, Chen J, Krueger EW, Billadeau DD, McNiven MA. Dynamin 2 potentiates invasive migration of pancreatic tumor cells through stabilization of the Rac1 GEF Vav1. Dev Cell. 2013;24:573–85. doi: 10.1016/j.devcel.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Haber C, Kazanietz MG. Cucurbitacin I inhibits rac1 activation in breast cancer cells by a reactive oxygen species-mediated mechanism and independently of janus tyrosine kinase 2 and p-rex1. Mol Pharmacol. 2013;83:1141–54. doi: 10.1124/mol.112.084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Pleckstrin homology (PH) domains and phosphoinositides. Biochem Soc Symp. 2007;74:81–93. doi: 10.1042/BSS0740081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Liu Z, Ye K. Phosphoinositol lipids bind to phosphatidylinositol 3 (PI3)-kinase enhancer GTPase and mediate its stimulatory effect on PI3-kinase and Akt signalings. Proc Natl Acad Sci U S A. 2005;102:16853–8. doi: 10.1073/pnas.0507365102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Wen W, Chan LN, Zhang M. Split pleckstrin homology domain-mediated cytoplasmic-nuclear localization of PI3-kinase enhancer GTPase. J Mol Biol. 2008;378:425–35. doi: 10.1016/j.jmb.2008.02.052. [DOI] [PubMed] [Google Scholar]
- Lehman HL, Van Laere SJ, van Golen CM, Vermeulen PB, Dirix LY, van Golen KL. Regulation of inflammatory breast cancer cell invasion through Akt1/PKBalpha phosphorylation of RhoC GTPase. Mol Cancer Res. 2012;10:1306–18. doi: 10.1158/1541-7786.MCR-12-0173. [DOI] [PubMed] [Google Scholar]
- Tang X, Feng Y, Ye K. Src-family tyrosine kinase fyn phosphorylates phosphatidylinositol 3-kinase enhancer-activating Akt, preventing its apoptotic cleavage and promoting cell survival. Cell Death Differ. 2007;14:368–77. doi: 10.1038/sj.cdd.4402011. [DOI] [PubMed] [Google Scholar]
- Tang X, Ye K. Pike tyrosine phosphorylation regulates its apoptotic cleavage during programmed cell death. Adv Enzyme Regul. 2006;46:289–300. doi: 10.1016/j.advenzreg.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Yadav V, Denning MF. Fyn is induced by Ras/PI3K/Akt signaling and is required for enhanced invasion/migration. Mol Carcinog. 2011;50:346–52. doi: 10.1002/mc.20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Pamplona R, Berenguer A, Sole X, Cordero D, Crous-Bou M, Serra-Musach J, et al. Tools for protein-protein interaction network analysis in cancer research. Clin Transl Oncol. 2012;14:3–14. doi: 10.1007/s12094-012-0755-9. [DOI] [PubMed] [Google Scholar]
- Jameson KL, Mazur PK, Zehnder AM, Zhang J, Zarnegar B, Sage J, et al. IQGAP1 scaffold-kinase interaction blockade selectively targets RAS-MAP kinase-driven tumors. Nat Med. 2013;19:626–30. doi: 10.1038/nm.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalvo-Ortiz BL, Castillo-Pichardo L, Hernandez E, Humphries-Bickley T, De la Mota-Peynado A, Cubano LA, et al. Characterization of EHop-016, novel small molecule inhibitor of Rac GTPase. J Biol Chem. 2012;287:13228–38. doi: 10.1074/jbc.M111.334524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Rivas J, Fontanillo C. Protein-protein interaction networks: unraveling the wiring of molecular machines within the cell. Brief Funct Genomics. 2012;11:489–96. doi: 10.1093/bfgp/els036. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Osada H. Phosphorylation-dependent protein-protein interaction modules as potential molecular targets for cancer therapy. Curr Drug Targets. 2012;13:1654–8. doi: 10.2174/138945012803530035. [DOI] [PubMed] [Google Scholar]
- Navankasattusas S, Whitehead KJ, Suli A, Sorensen LK, Lim AH, Zhao J, et al. The netrin receptor UNC5B promotes angiogenesis in specific vascular beds. Development. 2008;135:659–67. doi: 10.1242/dev.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. The LKB1-AMPK pathway-friend or Foe in cancer. Cancer Cell. 2013;23:131–2. doi: 10.1016/j.ccr.2013.01.009. [DOI] [PubMed] [Google Scholar]