Abstract
Neuropeptide Y (NPY) is an appetite hormone that acts centrally to control feeding behavior. The 5’ and exon 2 regions of NPY2R, one of 5 NPY receptor genes, have been weakly and inconsistently implicated with obesity. With the ATG start site of the gene at the beginning of exon 2, SNPs across intron 1 may show stronger associations with obesity than expected. Two 5’ SNPs, three intron 1 SNPs, and one synonymous exon 2 SNP were genotyped on 2985 Caucasian Utah subjects. Previously associated FTO, NPY, NPY1R, MC4R, PPARGC1A, OR7D4, and four NPFFR2 SNPs were also genotyped and related to BMI. One NPY2R 5’ SNP (rs12649641, p=0.008), an exon 2 SNP (rs2880415, p=0.009), and an intron 1 SNP (rs17376826, p=7×10−6) were each significantly associated with BMI. All 3 SNPs, plus FTO (rs9939609, p=1.5×10−6) and two NPFFR2 SNPs (rs4129733, p=3.7×10−13 and rs11940196, 4.2×10−10) remained significant in a multiple regression additive model. Diplotypes using the estimated haplotypes of NPY2R, NPFFR2, and MC4R were significantly associated with BMI (p=1.0×10−10, 3.2×10−8, and 1.1×10−4, respectively). Haplotypes of NPY2R, NPFFR2, and MC4R, plus the FTO SNP, explained 9.6% of the BMI variance. SNP effect sizes per allele for the four genes ranged from 0.8 to 3.5 kg/m2. We conclude that haplotypes containing the rs17376826 SNP in intron 1 of NPY2R have strong associations with BMI, some NPFFR2 haplotypes are strongly protective against or increase risk of obesity, and both NPY2R and NPFFR2 play important roles in obesity predisposition independent of FTO and MC4R.
Large case-control studies using genome-wide association markers have been published identifying genes related to obesity (1). At least 17 genetic regions have been suggested by such studies. The best replicated of these genes are the FTO and MC4R genes (2–8). There is a long history of candidate gene studies relating common genetic variants to various measures of obesity (9). The gene with the most frequent prevalence of multiple mutations or polymorphisms related to obesity is the MC4R gene (10). Because obesity is an appetite-related condition, the genes related to appetite control have been studied in various populations, particularly the NPY and NPY receptor genes. Generally, there has been poor replication of variants in NPY, PYY, NPY1R and NPY5R being associated with obesity, while more positive studies have implicated NPY2R, a well-known candidate gene for obesity development and control of food intake (11–20). However, even the NPY2R studies have not been compelling because of their marginal significance levels and findings that occur most often in men rather than women (12–14). In addition, most of these studies have genotyped SNPs in the 5’ region of the NPY2R gene or one of the three synonymous change variants in exon 2 of the gene. The NPY2R gene has two exons, but the start codons for transcription occur at exon 2, not exon 1. Intron 1 is a region with low linkage disequilibrium (LD) that is not well tagged by the 5’ SNPs, which are located in one LD block, or the downstream end of intron 1 and exon 2 SNPs, which are in a second LD block. Therefore, we typed multiple markers in intron 1, along with some previously studied 5’ and exon 2 SNPs. The independence of the NPY2R association with BMI was assessed by also typing one or more SNPs in the NPY, NPY1R, NPFFR2 (a G-protein receptor that binds neuropeptides), PPARGC1A (a PPAR-γ coactivator of gene transcription including UCP1), OR7D4 (an olfactory receptor gene), and MC4R genes (14, 21–24). Four SNPs were genotyped in the NPFFR2 gene that formed a haplotype previously shown to be associated with leanness (25). The SNP rs9939609 in the FTO gene was previously genotyped in these subjects and was significantly associated with BMI (5). In addition to testing the independence of each of the above genes on BMI, the ability of haplotypes to improve the associations with BMI was assessed.
Methods
Subjects
A sample of 2985 Caucasian Utah subjects was studied, consisting of members of 107 severely obese (BMI≥35 kg/m2) ascertained pedigrees containing at least 5 severely obese subjects (N=882), members of pedigrees ascertained for two or more thin subjects (males<20 kg/m2, females<19 kg/m2) (N=220), and a case/control series of unrelated subjects (severely obese subjects (N=1056) and randomly-ascertained Utah subjects (N=827)). Selecting the extremes of BMI for both pedigree selection (severely obese and thin pedigrees) and the case/control series (severely obese and random subjects) should increase our power to detect both alleles increasing or decreasing BMI from average levels. Weight was measured by a Scaletronix scale (model 5100) (Scaletronix Corporation, Wheaton, IL) that has an 800 pound capacity and weighing accuracy of 0.1 kilogram. There was little skewness (0.49) or kurtosis (0.02) in the overall BMI distribution, indicating approximate normality. These groups of subjects are described in Table 1 and Table in more detail in a paper describing the association of the FTO gene with obesity (5). All subjects signed informed consent and the study was approved by the University of Utah Institutional Review Board.
Table 1.
Subject characteristics by ascertainment group.
| Severe Obesity Pedigrees |
Severely Obese Cases |
Random Subjects |
Thin Pedigrees |
|
|---|---|---|---|---|
| N | 882 | 1056 | 827 | 220 |
| % Females | 65 | 82 | 52 | 59 |
| Age±SD, y (min-max) |
43.8±17.0 (15–90) |
44.3±11.4 (18–72) |
52.6±8.6 (19–77) |
37.4±17.7 (15–90) |
| BMI±SD, kg/m2 (min-max) |
35.2±7.6 (17–64) |
46.0±7.5 (33–92) |
27.4±5.0 (17–51) |
21.1±3.6 (15–34) |
Genotyping. We genotyped six SNPs across the NPY2R gene for this study, as shown in Table 2
Table 2.
SNPs Genotyped, Chromosomal Location, Minor Allele Frequency, Genotype BMI Means and P-Values from 2985 Utah subjects.
| SNP | Gene (chrom) |
Location (bpa) |
Minor allele freq. |
Genotype 1/1b |
Genotype 1/2 |
Genotype 2/2 |
P |
|---|---|---|---|---|---|---|---|
| rs12649641 | NPY2R (4q) | 156125333 | A: 0.38 | 35.1±0.33 | 35.9±0.32 | 36.8±0.58 | 0.008 |
| rs12507396 | NPY2R (4q) | 156129044 | T: 0.11 | 35.5±0.25 | 36.5±0.48 | 36.6±2.1 | 0.58 |
| rs17376826 | NPY2R (4q) | 156130948 | T: 0.04 | 35.4±0.24 | 38.9±0.71 | --- | 7.0 × 10−6 |
| rs10461238 | NPY2R (4q) | 156132216 | C: 0.44 | 35.8±0.37 | 35.6±0.31 | 35.7±0.43 | 0.79 |
| rs10461239 | NPY2R (4q) | 156132447 | C: 0.05 | 35.7±0.24 | 35.8±0.69 | --- | 0.90 |
| Rs2880415 | NPY2R (4q) | 156136027 | G: 0.45 | 35.1±0.37 | 35.7±0.30 | 36.6±0.44 | 0.009 |
| rs9939609 | FTO (16q) | 53820527 | A: 0.43 | 34.6±0.35 | 35.7±0.30 | 37.5±0.46 | 3.3 × 10−7 |
| rs12510838 | NPFFR2 (4q) | 72961548 | G: 0.18 | 35.5±0.26 | 36.0±0.38 | 36.2±1.17 | 0.56 |
| rs4129733 | NPFFR2 (4q) | 72963002 | G: 0.31 | 34.9±0.31 | 36.1±0.32 | 37.6±0.65 | 1.5 × 10−4 |
| rs9291171 | NPFFR2 (4q) | 72981626 | G: 0.29 | 35.6±0.29 | 35.8±0.33 | 36.2±0.73 | 0.39 |
| rs11940196 | NPFFR2 (4q) | 73003569 | G: 0.36 | 35.9±0.34 | 35.7±0.31 | 35.2±0.57 | 0.29 |
| rs17782313 | MC4R (18q) | 57851097 | C: 0.26 | 35.4±0.28 | 36.0±0.35 | 37.0±0.69 | 0.026 |
| rs477181 | MC4R (18q) | 57896038 | T: 0.36 | 35.4±0.33 | 35.8±0.33 | 36.3±0.52 | 0.13 |
| rs8192678 |
PPARGC1A (4p) |
23815662 | A: 0.34 | 35.6±0.31 | 35.7±0.31 | 36.4±0.59 | 0.21 |
| rs9764 | NPY1R (4q) | 164245404 | C: 0.28 | 36.0±0.30 | 35.3±0.32 | 35.7±0.70 | 0.61 |
| rs16139 | NPY (7p) | 24324879 | G: 0.04 | 35.7±0.24 | 36.2±0.72 | --- | 0.43 |
| rs2878329 | OR7D4 (19p) | 9325742 | T: 0.08 | 35.6±0.25 | 36.4±0.52 | 31.5±4.3 | 0.38 |
Basepair location using build 37.1.
BMI means (kg/m2±SE) and P-values were obtained after adjustment for age, age2, sex, and family relatedness. An additive genetic model was assumed. 1=major allele; 2=minor allele.
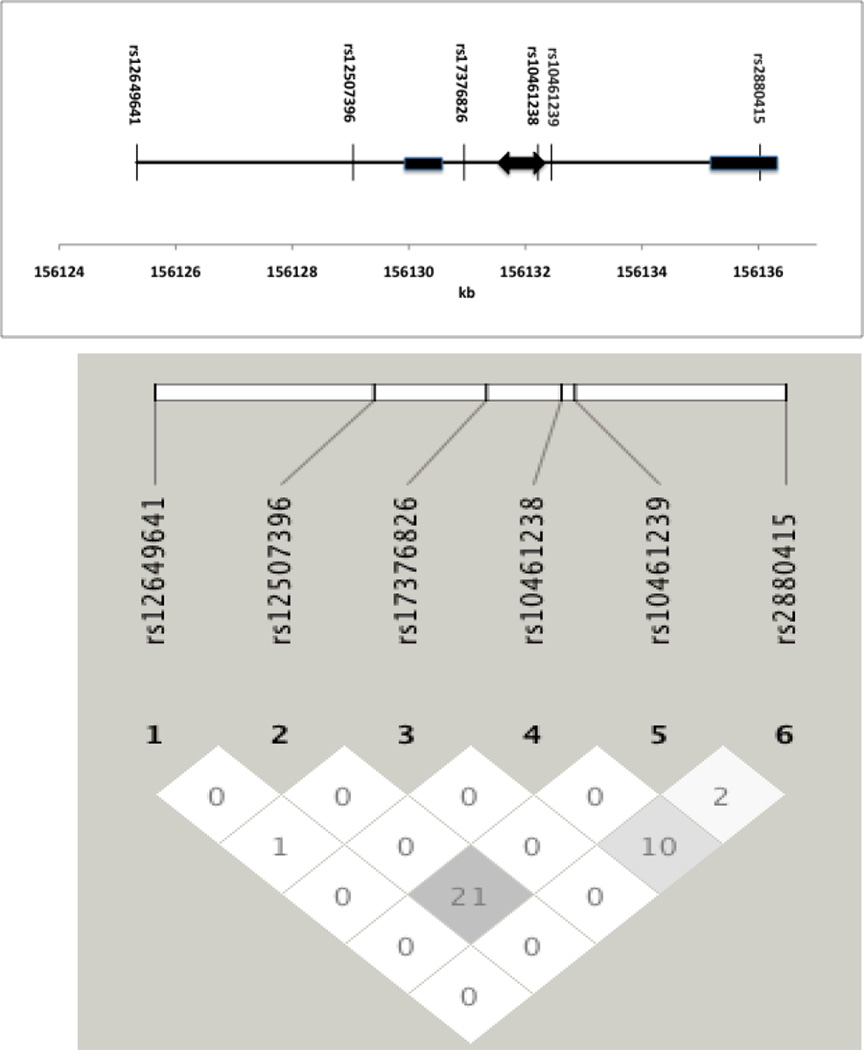
The NPY2R gene has two exons with the ATG start site occurring at the beginning of exon 2. From HapMap data, there is one LD block in the 5’ portion of NPY2R to exon 1 and a second LD block from the downstream portion of intron 1 to the 3’ end of the gene, which covers exon 2 (not shown). However, there appears to be low LD in the majority of NPY2R, especially intron 1, part of which is not covered by a block. As part of another study, one severely obese pedigree was selected for copy number variation (CNV) analysis performed using an Agilent 244k CGH array with further resequencing. Concordance of a small 500 bp deletion (156131730-156132230 bp on chromosome 4, NCBI build 37.1) in severely obese members of this pedigree was located in intron 1 of the NPY2R gene (unpublished data). Further resequencing in other pedigrees suggested that the deletion was a very rare or private mutation in this pedigree and therefore study of the CNV has not been further pursued at this point. However, the presence of this deletion was instrumental in selecting intron 1 SNPs that flanked or were within the deletion region.
Intron 1 contains only one polymorphic SNP that is found in HapMap. Therefore we selected the one HapMap SNP (rs10461238) that occurs within the deletion and a second non-HapMap SNP (rs10461239) in intron 1 that was 3’ of the deletion, both of which were located at the beginning of block 2. We genotyped a SNP in intron 1 on the 5’ side of the deletion (rs17376826), a synonymous SNP in exon 2 (rs2880415) and two SNPs genotyped by other studies in the 5’ region of the gene in block 1 (rs12649641 and rs12507396). The NPY2R SNPs showed low r2 LD estimates in our data (Figure 1), similar to those seen from HapMap
Figure 1.
NPY2R gene structure on chromosome 4 with two exons (rectangles) and 6 SNPs genotyped for this study. Exon 1 is a noncoding exon. The double arrow is a 500 base pair deletion that segregates with severe obesity in only one pedigree with severe obesity. NPY2R is transcribed from left to right. Below is the linkage disequilibrium structure in terms of r2 of the six SNPs genotyped in this study.
Four NPFFR2 SNPs were genotyped (Table 2) and had r2 LD values ranging from 0.29 to 0.75. These SNPs were the four SNPs that formed a protective haplotype in the study of Dahlman, et al (25). One FTO SNP, two MC4R SNPs, and one SNP in PPARGC1A, NPY1R, NPY, and OR7D4 were genotyped (Table 2), Genotype calling was greater than 95% for all SNPs, and the sample size of 2985 subjects represents the subjects who had complete genotyping for all SNPs. All SNPs were in Hardy-Weinberg equilibrium (p>0.05) in the randomly ascertained sample except for OR7D4 rs2878329 (p=0.02). For the pedigree samples, misinheritances were zeroed out. Since the goal of the study was to look at all SNPs using multiple regression models, any subject with a missing genotype was removed from the study, leaving a sample size of 2985 subjects (Table 1).
SNP genotyping was performed using a LightScanner instrument (Roche Applied Science, USA). A single probe (Roche Applied Science, USA) labeled with fluorescent dye was used. Primers and probes for each SNP were designed using LightTyper SNP design software (Idaho Technologies, Inc.). The probe primer sets for each SNP are available from the authors. The PCR cocktail was mixed by adding 2.5ul sterilized distilled water, 5ul PCR premix (containing dNTP, EPICENTRE), 1µl (10µM) forward primer, 0.2µl (2µM) reverse primer, 0.1µl (0.5U) Taq (KlenTaq), 0.1µl (20µM) probe and 1µl (15µg) DNA, making total volume of 10µl. PCR cocktails were aliquoted into a 384 well PCR white plate (BioRad), followed by aliquoting 10µl light mineral oil to each well on the top of the PCR cocktail. The plate was sealed by clear adhesive film (Applied Biosystems). PCR was performed using a 9700 PCR apparatus (Applied Biosystems, USA). The thermal cycling conditions were: initial denaturation of 95ο C for 5 minutes, followed by 40 cycles of 95ο C 30 seconds, 58 – 65ο C 20 seconds, 72ο C 30 seconds and final denaturation of 95ο C for 10 minutes. It was then soaked in 4οC. After PCR, the 384- well plate was put into the LightScanner instrument to measure the probe melting curve. Computerized genotyping is carried out using melting curves analysis by LightScanner analysis software (Idaho Technologies, Inc., USA). Each SNP nucleotide was aligned with the + strand from NCBI.
Statistical analysis
Genetic association was investigated by two methods. First we utilized multiple linear regression using BMI as a continuous dependent variable and sex, age, and age2 as covariates with each SNP as the independent predictor variable. All subjects were analyzed together. Relatedness among subjects in the pedigrees was handled by using generalized estimating equations with pedigree ID as the repeated measure variable and an exchangeable correlation matrix. Following use of each SNP individually, we included all SNPs in the same model with backwards stepwise elimination to test for independence. The six SNPs that remained significant were then used in a forwards stepwise regression model to estimate the increases in R2 as each SNP was entered into the model. Because we combined both pedigree-ascertained and case/control samples into a pooled analysis despite their different ascertainment criteria, we also performed a meta analysis of the two groups (N=1102 and 1883, respectively) by combining the group-specific p-values using Fisher’s method (26).
Second we estimated haplotypes for the three genes with more than one SNP typed so that within gene interactions need not be modeled and to better subset subjects carrying untyped causal variants. The three significant SNPs at the NYP2R locus were first used to infer haplotypes for each chromosome by using the Pedigree Analysis Package (PAP, version 5.0), which uses the pedigree structure for maximum likelihood inference in the pedigrees or standard likelihood-based analysis using the SNP frequency data for the unrelated subjects. Therefore, all 2985 subjects were included in all analyses and tables. Probabilities of each possible haplotype for each chromosome are obtained for each subject and the haplotype with the highest probability for each chromosome was assigned to that subject. All three combinations of 4-SNP haplotypes and the full 6-SNP haplotypes were subsequently estimated, diplotypes for each subject formed, and a global F-test from the above general linear model was used to assess significance among the diplotypes. Haplotypes with frequencies less than 1% were combined into one combined haplotype labeled as ‘rare’ before the diplotypes were formed or tested. NPFFR2 haplotypes from the four genotyped SNPs and MC4R haplotypes from the two genotyped SNPs were similarly estimated. The haplotypes from the three genes plus the FTO locus SNP were included in a final regression model. Interaction terms were added to the final SNP or diplotype models to test whether or not the effects of each gene on BMI were independent.
Results
Table 1 shows the age, gender and BMI distributions for each of the four ascertainment groups. Age, age2, and gender were included in all regression models and were always significant at p<1 × 10−4. Without any genotypes in the model, the beta coefficients for age, age2, and gender were 0.37±0.065, −0.005±0.0006, and 3.40±0.40, respectively. Figure 1 shows the location of the six genotyped SNPs at the NPY2R locus, the location of the deletion found in one pedigree, and the LD structure in our data for the six SNPs. Table 2 lists all 17 SNPs that were genotyped, their physical positions, and the minor allele frequencies as estimated from our data. Table 2 also shows that three SNPs at the NPY2R locus were significantly associated with BMI. rs12649641 was in the 5’ region, rs17376826 was in intron 1 and rs2880415 was in exon 2, with the intron 1 SNP rs17376826 having the highest significance level (p=7.0×10−6). As we had shown previously (5), the FTO locus SNP rs9939609 was also highly significant (p=3.3×10−7). One of the two MC4R locus SNPs, rs17782313, was significant, but if adjusted for 17 multiple comparisons, would no longer be significant. NPFFR2 locus SNP rs4129733 was significant at p=1.5×10−4. SNPs at the NPY, NPY1R, OR7D4 and PPARGC1A loci did not show associations with BMI.
Including all SNPs in a backwards stepwise elimination model resulted in six SNPs in three genes (NPY2R, NPFFR2, and FTO) being independently predictive of BMI (Table 3). In this model, NPFFR2 became the most strongly associated locus with BMI rather than NPY2R. The percent of variance explained in BMI after age and sex adjustment was 4.3% (R2 from a forward stepwise inclusion shown for the 6 SNPs). The BMI effect sizes (beta coefficients) for each SNP allele in the additive model ranged from 0.8 to 3.5 kg/m2, although note the minor allele of rs11940196 had a protective effect on BMI (Table 3). Because some studies have suggested association mostly in males, the analyses were repeated for males and females. Four of the six SNPs in Table 3 were significant for both males and females (rs4129733, rs11940196, rs9939609, and rs17376826). At the NPY2R locus, rs12649641 was only significant in females (p=0.025) and rs2880415 was significant only in males (p=0.006). We also subdivided the sample into pedigree (N=1102) and case/control subsets (N=1883) and performed a meta analysis. The effect sizes were slightly smaller in the pedigrees compared with the case/control samples, but the meta-analysis p-values all remained significant when the two groups were combined (last column of Table 3). For example, rs17376826 had an effect size and p-value of 2.0+1.2 kg/m2 and 0.09, respectively, in the pedigree sample compared with 2.4+0.8 kg/m2 and 0.004, respectively, in the case/control sample (results not shown). Two SNPs located in intron1 of the NPY2R gene, rs17376826 and rs2880415, showed significant interactions on BMI levels (p=0.0008), However, there were no significant interactions between pairs of SNPs selected from two different genes. Therefore, the SNPs from the four genes appeared to have independent associations with BMI.
Table 3.
Associations of SNPs in a single multiple regression model versus BMI (kg/m2) in 2985 Utah subjects.
| Gene, SNP, major/minor allele |
Genotype 1/1a |
Genotype 1/2 |
Genotype 2/2 |
Allele Effect Size (β, kg/m2) |
P-Value | Δ R2 from stepwise model |
Fisher’s meta- analysis P-Value |
|---|---|---|---|---|---|---|---|
| Sex, Age, Age2 | 5.55% | ||||||
| NPFFR2 rs4129733 T/G | 34.6 | 37.8 | 42.0 | 3.5 | 3.7 × 10−13 | 0.76% | 9 × 10−5 |
| NPFFR2 rs11940196 A/G | 41.0 | 38.4 | 35.0 | −2.8 | 4.2 × 10−10 | 1.47% | 8 × 10−7 |
| FTO rs9939609 T/A | 37.0 | 37.9 | 39.6 | 1.3 | 1.5 × 10−6 | 0.77% | 6 × 10−5 |
| NPY2R rs17376826 C/T | 36.5 | 39.7 | --- | 3.1 | 3.5 × 10−5 | 0.73% | 0.0034 |
| NPY2R rs2880415 A/G | 37.3 | 38.1 | 38.9 | 0.8 | 0.003 | 0.30% | 0.022 |
| NPY2R rs12649641 C/A | 37.3 | 38.3 | 38.8 | 0.8 | 0.017 | 0.29% | 0.041 |
1=major allele, 2=minor allele; see Table 2 for the minor allele; P-value is from an additive model using a test for trend. Δ R2 is from a forward stepwise regression model with the first row entered first and the last row entered last. See text for a description of the meta-analysis.
Mean BMI was estimated for each diplotype combination of the 3-SNP NPY2R haplotypes (1–3–6) consisting of the three individually significant SNPs (rs12649641, rs17376826, and rs2880415). The association in Table 4 was approximately as significant as rs17376826 alone (Table 3). However, diplotype differences in mean BMI using the 4-SNP haplotypes (3–4–5–6) showed much stronger associations with BMI than any of the other haplotype definitions, including the full 6-SNP haplotype. Using the NPY2R 3–4–5–6 haplotype definition to form diplotypes, Table 5 shows the full model using diplotypes for NPY2R, NPFFR2 and MC4R and the FTO SNP. In this full diplotype model adjusting for the three other significant loci, the NPY2R association (p=3.6 × 10−11) was stronger than in the single SNP model (p=3.5 × 10−5 for rs17376826 in Table 3). NPFFR2 association was weaker than in the single SNP model but still highly significant (p=2.8 × 10−8). While the two MCR4 SNPs did not remain significant in the SNP multiple regression model of Table 3, the haplotypes became highly significant in the haplotype model (both with and without the other genes in the model). The r2 between the two MC4R SNPs was 0.60. None of the SNPs at the NPY, NPY1R, OR7D4 or PPARGC1A loci were significant when added to the haplotype model.
Table 4.
Comparison of diplotype associations with BMI using various haplotype definitions across NPY2R in 2985 Utah subjects.
| Diplotypes formed from NPY2R haplotypes below |
P-Value |
|---|---|
| SNPs 1-3-6 | 1.1 × 10−5 |
| SNPs 1-2-3-4 | 1.4 × 10−5 |
| SNPs 2-3-4-5 | 1.6 × 10−3 |
| SNPs 3-4-5-6 | 3.6 × 10−11 |
| SNPs 1-2-3-4-5-6 | 6.5 × 10−9 |
Adjusted for sex, age, age2, relatedness, FTO, MC4R diplotypes, and NPFFR2 diplotypes. Global F-test on BMI means.
SNP 1: rs12649641; SNP 2: rs12507396; SNP 3: rs17376826;
SNP 4: rs10461238; SNP 5: rs10461239; SNP 6: rs2880415
Table 5.
Associations of diplotypes in a single multiple regression model versus BMI (kg/m2) in 2985 Utah subjects.
| Haplotype | P-Value | Δ R2 (%) | Degrees of Freedom |
|---|---|---|---|
| Sex, Age, Age2 | 5.55 | 3 | |
| NPY2R (hap 3-4-5-6) | 3.6 × 10−11 | 4.44 | 30 |
| NPFFR2 | 2.8 × 10−8 | 3.11 | 34 |
| MC4R | 2.0 × 10−4 | 1.10 | 9 |
| FTO | 8.4 × 10−5 | 0.57 | 2 |
No genetic model specified. Total R2 for all SNPs is 4.3% from table 3 while for diplotypes above it is 9.2%.
Δ R2 is from a forward stepwise regression model with the first row entered first and the last row entered last.
Table 6 shows the frequency and mean BMI for each haplotype with the haplotypes with frequencies less than 1% grouped into a rare haplotype definition. Compared to a common NPY2R haplotype that had the lowest mean BMI (CCGA), there were three common haplotypes with 0.5, 1.0 and 4.2 kg/m2 increases in mean BMI. The combined rare haplotype at 1.9% frequency showed the highest mean BMI, with a 5.8 kg/m2 mean BMI increase over the BMI of the CCGA haplotype. There were common NPFFR2 haplotypes with both lower BMI (ATAG, BMI=31.7) and higher BMI (AGAG, BMI=36.8) than the most common haplotype (ATAA, BMI=35.6 kg/m2).
Table 6.
Haplotype frequencies and sex-, age-, age2-adjusted BMI in 2985 Utah subjects.
| Haplotype | Frequency (%) |
Mean±SD (kg/m2) |
Haplotype | Frequency (%) |
Mean±SD (kg/m2) |
|---|---|---|---|---|---|
| NPY2R | NPFFR2 | ||||
| Rare | 1.9 | 40.7±11.3 | AGAA | 2.5 | 37.4±9.3 |
| CCGG | 7.9 | 39.1±9.4 | AGAG | 27.2 | 36.8±10.5 |
| TCGA | 1.2 | 38.7±8.4 | GTGA | 15.7 | 36.1±11.2 |
| TGGA | 1.5 | 37.3±11.5 | ATGA | 11.2 | 35.9±10.2 |
| CGGA | 16.3 | 35.9±10.6 | ATAA | 33.5 | 35.6±10.4 |
| CCCA | 3.8 | 35.5±11.4 | Rare | 2.9 | 35.5±8.5 |
| CGGG | 36.4 | 35.4±10.5 | ATAG | 7.1 | 31.7±10.0 |
| CCGA | 30.9 | 34.9±10.3 | MC4R | ||
| CG | 4.9 | 39.0±9.4 | |||
| FTO | TT | 14.3 | 36.6±10.0 | ||
| A | 42.5 | 37.9±0.45 | CT | 21.1 | 35.9±10.7 |
| T | 57.5 | 36.8±0.46 | TG | 59.6 | 35.3±10.6 |
SNP order: NPY2R (3-4-5-6): rs17376826, rs10461238, rs10461239, rs2880415
NPFFR2: rs12510838, rs4129733, rs9291171, rs11940196
MC4R: rs17782313, rs477181.
Discussion
The main purpose of this study was to look at the association of the NPY2R locus with BMI, since it was the identification of a small intron 1 deletion in severely obese members of a large pedigree that led us to this well-known candidate gene for obesity. We selected two SNPs on either side of the deletion in intron 1 plus one SNP that was within the deletion region (rs10461238). We then tested whether the associations found for NPY2R were independent of the other obesity-related genes that we had genotyped.
We were able to confirm weak findings of a previously studied 5’ SNP (rs12649641) near NPY2R associated with BMI. Like previous studies (12, 13, 16, 27), our association of the exon 2 synonymous SNP rs2880415 was borderline when both genders were combined, but significant when analyzing men only. It should be noted that some previous studies genotyped rs1047214 instead of rs2880415, but the estimated r2 is nearly 1.0, indicating that the results using either SNP would be the same. Although the NPY2R locus was not implicated in the GIANT consortium meta analysis (28), a recent candidate gene study from CARDIA found that the only significantly associated SNP with BMI in whites was the NPY2R intron 1 SNP rs17376826 (29). The four-SNP haplotypes of rs17376826, rs10461238, rs10461239, and rs2880415 showed highly significant results, suggesting that genetic variation at multiple sites across the NPY2R locus combine to increase the risk of obesity. In fact, two of the SNPs at the NPY2R locus showed a significant interaction, rs17376826 and rs2880415, which helps explain the much higher significance level found for the diplotype analysis compared to the single SNP analysis. BMI was significantly higher in the rs2880415 G/G genotype group than the other two genotypes when rs17376826 was C/T rather than C/C.
The association of the NPY2R locus was independent of three other loci that showed significance with BMI, namely, NPFFR2, FTO, and MC4R. There were no interactions among SNPs between these genetic loci. The percent of variance of BMI explained by the six best SNPs was 4.3%, while the diplotypes of the 3 genes and FTO explained 9.6% of the variance of BMI. However, the percent of variance explained is related to the degrees of freedom, so that analyzing diplotypes with their corresponding degrees of freedom inflates this latter estimate. While some authors have posited that some of the missing heritability in BMI from GWA studies would be found in interactions, this limited study of eight loci does not suggest this to be the case. The independent effects of multiple loci appear to be the better supported explanation, along with possible interactions with diet, physical activity, or psychosocial factors. The missing heritability from any one study more likely comes from the specific characteristics or size of the sample, preventing the identification of all relevant obesity genes. This possibility is suggested by the non-overlap of most of the GWAS findings and the non-overlap of many replicated candidate genes with the GWAS findings. Part of the non-overlap is probably caused from the severe statistical penalty of multiple comparisons required in the GWA studies. Further in silico lookups of particular findings may increase the consistency among studies. Both MC4R and FTO loci are strongly associated with BMI in GIANT (28).
Another reason for non-overlap in study findings might be that adjustment for particular obesity genes or the formation of haplotypes of multiple SNPS across a gene of interest may be required before strong and significant signals may be uncovered. This seems to be the case for the MC4R gene in this study, as it required the haplotypes using two MC4R SNPs before strong signals were found after adjusting for other genes. The intronic SNP rs17376826 with an allele frequency of only 4% was critical to include for both single SNP and haplotype models.
As reported in other studies (12, 13, 16, 27), NPY2R SNP rs2880415 (or its equivalent rs1047214) had the strongest association with BMI in men. While a couple of these studies only included men, those that included women did not see associations with rs2880415. Also, three of these studies showed higher BMI with the minor allele, and one showed lower BMI with the minor allele. Four other studies did not find association of rs2880415 in exon 2 with obesity (14, 15, 17, 18). None of the studies of NPY2R looked at haplotypes consisting of SNPs in other regions of the gene. Our data appear to suggest that the intron 1 SNP rs17376826 and the exon 2 SNP greatly increase the risk of higher BMI. Others have looked at haplotypes in the 5’ area of the gene, but haplotypes were not dramatically more significant than individual SNPs (14). Campbell et al. also looked at two of the SNPs we typed in intron 1, rs10461238 and rs10461239. These two SNPs were not significant in their study, nor were they significant in our study, even though rs10461238 was located within the deletion region we found in one pedigree. rs10461238 was not significant in the CARDIA study (29). However rs10461238 is a common genetic variant, suggesting it does not mark the deletion we found.
The highly significant SNP at the NPY2R locus, rs17376826, had a minor allele frequency of only 0.04 in our data and 0.06 in the CARDIA whites (29). When creating haplotypes, the combined ‘rare’ category of NPY2R haplotypes with frequencies less than 1% showed the highest mean BMI levels of all haplotypes. This may suggest that these haplotypes tag multiple rare variants in intron 1 that affect gene transcription and the development of severe obesity. The only other study we could find that typed an intron 1 SNP was in the Pima Indians (13). This SNP, rs10461257, is also a common variant and showed marginal significance across the different genetic models.
NPFFR2 is a gene that shows 33% amino acid identity with the NPY2R gene and is expressed at high levels in the brain and heart (30). Dahlman et al. have typed four SNPs at this locus and showed that one particular haplotype was protective against obesity (25). The protective haplotype was associated with higher adipocyte lipid mobilization. We typed those same four SNPs, estimated haplotypes, and confirmed that the ATAG haplotype, with a frequency of 7%, was associated with significantly lower BMI than the most common haplotype (a 3.9 kg/m2 lower BMI). Other haplotypes in NPFFR2 appeared to have increased risk of obesity compared to the most common haplotype. Thus, another NPY-family genetic locus is strongly associated with obesity and is independent of the NPY2R locus association.
We previously did not find significant associations with obesity for the GAD2 and INSIG2 loci in a subset of the subjects in this study (31,31, 32). Other genetic loci that we tested in this study (NPY, NPY1R, PPARGC1A, and OR7D4) were not associated with obesity. However, only one SNP at each locus that had been suggested by prior studies to possibly be associated with obesity was genotyped. Therefore, the lack of significance for these loci should not be interpreted as ruling out involvement with obesity, as additional SNPs and haplotype analysis may provide evidence for an association with obesity. In addition, none of the p-values were adjusted for multiple comparisons in the tables. There were 17 tests of the initial SNPs, following by creation of 5 different NPY2R haplotype definitions (which are highly correlated). Even if the most conservative adjustment is used for multiple comparisons, the results remain highly significant and our conclusions are not changed.
In summary, four genetic loci were strongly and independently associated with obesity, NPY2R, NPFFR2, MC4R, and FTO. Haplotype type estimation greatly increased the level of association for these loci. The NPY2R locus association appeared to be mostly explained by an intron 1 SNP located between two LD blocks covering the two exons of the NPY2R gene. The four loci explained a large proportion of variance in BMI (9.7%), which may have been larger than other studies because of the wide range of BMI in our study and the significant portion of the sample that were severely obese. Adjustment of future studies for these genes may allow greater power to detect additional genes associated with obesity and further explain its polygenic heritability. Future studies investigating how subjects with protective and risk haplotypes respond to different dietary, exercise, and stress environments will hopefully provide further insights into the mechanisms leading to obesity.
Acknowledgments
This research was supported by NIH grants DK073550, DK055006, HD17463, and the National Center for Research Resources (MO1-RR00064). The Huntsman Cancer Institute Microarray Core Facility is partially supported by NCI Cancer Center Support Grant P30CA042014 and by institutional support provided through the Huntsman Cancer Institute.
References
- 1.Hinney A, Vogel CI, Hebebrand J. From monogenic to polygenic obesity: recent advances. European Child & Adolescent Psychiatry. 2010;19:297–310. doi: 10.1007/s00787-010-0096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 3.Frayling TM, Timpson NJ, Weedon MN, et al. A Common Variant in the FTO Gene Is Associated with Body Mass Index and Predisposes to Childhood and Adult Obesity. Science. 2007;316:889–994. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinney A, Nguyen TT, Scherag A, et al. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS One. 2007;2:e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunt SC, Stone S, Xin Y, et al. Association of the FTO Gene With BMI. Obesity. 2008;16:902–904. doi: 10.1038/oby.2007.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grant SF, Li M, Bradfield JP, et al. Association analysis of the FTO gene with obesity in children of Caucasian and African ancestry reveals a common tagging SNP. PLoS One. 2008;3:e1746. doi: 10.1371/journal.pone.0001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hotta K, Nakata Y, Matsuo T, et al. Variations in the FTO gene are associated with severe obesity in the Japanese. J Hum Genet. 2008;53:546–553. doi: 10.1007/s10038-008-0283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loos RJ, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rankinen T, Zuberi A, Chagnon YC, et al. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 10.Lubrano-Berthelier C, Cavazos M, Dubern B, et al. Molecular genetics of human obesity-associated MC4R mutations. Ann N Y Acad Sci. 2003;994:49–57. doi: 10.1111/j.1749-6632.2003.tb03161.x. [DOI] [PubMed] [Google Scholar]
- 11.Bray MS, Boerwinkle E, Hanis CL. Sequence variation within the neuropeptide Y gene and obesity in Mexican Americans. Obes Res. 2000;8:219–226. doi: 10.1038/oby.2000.25. [DOI] [PubMed] [Google Scholar]
- 12.Hung CC, Pirie F, Luan J, et al. Studies of the peptide YY and neuropeptide Y2 receptor genes in relation to human obesity and obesity-related traits. Diabetes. 2004;53:2461–2466. doi: 10.2337/diabetes.53.9.2461. [DOI] [PubMed] [Google Scholar]
- 13.Ma L, Tataranni PA, Hanson RL, et al. Variations in peptide YY and Y2 receptor genes are associated with severe obesity in Pima Indian men. Diabetes. 2005;54:1598–1602. doi: 10.2337/diabetes.54.5.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell CD, Lyon HN, Nemesh J, et al. Association studies of BMI and type 2 diabetes in the neuropeptide Y pathway: a possible role for NPY2R as a candidate gene for type 2 diabetes in men. Diabetes. 2007;56:1460–1467. doi: 10.2337/db06-1051. [DOI] [PubMed] [Google Scholar]
- 15.Torekov SS, Larsen LH, Andersen G, et al. Variants in the 5' region of the neuropeptide Y receptor Y2 gene (NPY2R) are associated with obesity in 5,971 white subjects. Diabetologia. 2006;49:2653–2658. doi: 10.1007/s00125-006-0425-y. [DOI] [PubMed] [Google Scholar]
- 16.Siddiq A, Gueorguiev M, Samson C, et al. Single nucleotide polymorphisms in the neuropeptide Y2 receptor (NPY2R) gene and association with severe obesity in French white subjects. Diabetologia. 2007;50:574–584. doi: 10.1007/s00125-006-0555-2. [DOI] [PubMed] [Google Scholar]
- 17.Wang HJ, Wermter AK, Nguyen TT, et al. No association of sequence variants in the neuropeptide Y2 receptor (NPY2R) gene with early onset obesity in Germans. Horm Metab Res. 2007;39:840–844. doi: 10.1055/s-2007-992127. [DOI] [PubMed] [Google Scholar]
- 18.Santoro N, Del Giudice EM, Grandone A, et al. Y2 receptor gene variants reduce the risk of hypertension in obese children and adolescents. J Hypertens. 2008;26:1590–1594. doi: 10.1097/HJH.0b013e32830413ed. [DOI] [PubMed] [Google Scholar]
- 19.Kuo LE, Kitlinska JB, Tilan JU, et al. Neuropeptide Y acts directly in the periphery on fat tissue and mediates stress-induced obesity and metabolic syndrome. Nat Med. 2007;13:803–811. doi: 10.1038/nm1611. [DOI] [PubMed] [Google Scholar]
- 20.Beck B. Neuropeptide Y in normal eating and in genetic and dietary-induced obesity. Philos Trans R Soc Lond B Biol Sci. 2006;361:1159–1185. doi: 10.1098/rstb.2006.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez-Alvarez MI, Chiellini C, Manco M, et al. Genes involved in mitochondrial biogenesis/function are induced in response to bilio-pancreatic diversion in morbidly obese individuals with normal glucose tolerance but not in type 2 diabetic patients. Diabetologia. 2009;52:1618–1627. doi: 10.1007/s00125-009-1403-y. [DOI] [PubMed] [Google Scholar]
- 22.Goyenechea E, Crujeiras AB, Abete I, Parra D, Martinez JA. Enhanced short-term improvement of insulin response to a low-caloric diet in obese carriers the Gly482Ser variant of the PGC-1alpha gene. Diabetes Res Clin Pract. 2008;82:190–196. doi: 10.1016/j.diabres.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Cauchi S, Stutzmann F, Cavalcanti-Proenca C, et al. Combined effects of MC4R and FTO common genetic variants on obesity in European general populations. J Mol Med. 2009;87:537–546. doi: 10.1007/s00109-009-0451-6. [DOI] [PubMed] [Google Scholar]
- 24.Choquette AC, Bouchard L, rapeau V, et al. Evidence of association between a human olfactory receptor gene (OR7D4) and traits related to eating behavior and body fatness: Results from the Quebec Family Study (QFS) Obesity. 2009;17:S294. (abstract) [Google Scholar]
- 25.Dahlman I, Dicker A, Jiao H, et al. A common haplotype in the G-protein-coupled receptor gene GPR74 is associated with leanness and increased lipolysis. Am J Hum Genet. 2007;80:1115–1124. doi: 10.1086/518445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher RA. Statistical methods for research workers. 13th ed. Edinburgh and London: Oliver & Boyd; 1925. [Google Scholar]
- 27.Lavebratt C, Alpman A, Persson B, Arner P, Hoffstedt J. Common neuropeptide Y2 receptor gene variant is protective against obesity among Swedish men. Int J Obes (Lond) 2006;30:453–459. doi: 10.1038/sj.ijo.0803188. [DOI] [PubMed] [Google Scholar]
- 28.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedlander Y, Li G, Fornage M, et al. Candidate molecular pathway genes related to appetite regulatory neural network, adipocyte homeostasis and obesity: results from the CARDIA Study. Annals of Human Genetics. 2010;74:387–398. doi: 10.1111/j.1469-1809.2010.00596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker RM, Copeland NG, Eyre HJ, et al. Molecular cloning and characterisation of GPR74 a novel G-protein coupled receptor closest related to the Y-receptor family. Brain Res Mol Brain Res. 2000;77:199–208. doi: 10.1016/s0169-328x(00)00052-8. [DOI] [PubMed] [Google Scholar]
- 31.Hunt SC, Xin Y, Wu LL, Hopkins PN, Adams TD. Lack of association of glutamate decarboxylase 2 gene polymorphisms with severe obesity in Utah. Obesity. 2006;14:650–655. doi: 10.1038/oby.2006.73. [DOI] [PubMed] [Google Scholar]
- 32.Boes E, Kollerits B, Heid IM, et al. INSIG2 polymorphism is neither associated with BMI nor with phenotypes of lipoprotein metabolism. Obesity. 2008;16:827–833. doi: 10.1038/oby.2007.132. [DOI] [PubMed] [Google Scholar]