Abstract
Background:
It is believed that paraoxonase-2 gene polymorphism is associated with type 2 diabetes. This study is aimed to investigate the association between paraoxonase-2 gene polymorphism and type 2 diabetes in an Iranian population.
Methods:
This study was performed on 200 individuals including 100 diabetics and 100 healthy subjects. Genomic DNA was extracted from peripheral blood leukocytes, and PCR-RFLP was carried out. Palindromic sequence in PON2 gene was recognized by Dde1 restriction endonuclease. In order to visualize restriction products, electrophoresis was carried out using polyacrylamide gel (8%) and ethidium bromide staining.
Results:
The expected PCR product of 331 bp was obtained. Digestion of this product with DdeI showed four Ser homozygotes, three Cys homozygotes, and five Ser311 Cys heterozygotes. The gene frequency of Cys (C) in diabetic subjects was significantly higher than in healthy subjects.
Conclusions:
This study suggests that an association exists between Ser311 Cys polymorphism and type 2 diabetes mellitus.
Keywords: Iranian population, paraxonase-2 gene, polymorphism, type 2 diabetes
INTRODUCTION
Type 2 diabetes (T2D) has been a common, serious, and costly public health problem. Millions of people have been diagnosed with T2D, and many more are unaware that they are exposed to T2D. Either the body does not make enough insulin or the cells ignore the insulin in T2D. Despite the impact of environmental components upon T2D, this disease clusters in families. To date, no specific genes causing T2D have been conclusively identified. Nonetheless, it is believed that gene families such as paraoxonase (PON) are associated with the high risk of T2D. PON gene family is located on the long arm of chromosome 7 (7q21-22) close to glucose absorption genes.[1] The paraoxonase gene family includes three known members as follows: PON1, PON2, and PON3. These genes encode enzymes, which are responsible for breaking acetylcholinesterase inhibitors.[2] PON1 is synthesized in the liver and released along with HDL secretion in the plasma. It also poses antioxidant properties and prevents LDL oxidation. Inflammatory changes and levels of serum oxidized-LDL affect its concentration. PON2 protects cells against oxidative damage.[3] PON3 shares nearly 60% homology in the amino acid sequence with PON1 and its proteins are found on HDL. Effect of PON3 upon lipoprotein oxidation, which can retard aforesaid processes, has been demonstrated.[4,5] The effect of PON genes upon various disorders is well-documented in the literature. For instance, an association between the paraoxonase 192 Arg allele and coronary artery disease (CAD) was reported in French and Japanese patients with type 2 diabetes.[6,7] Paraoxonase polymorphism (Gln192-Arg) was found to be associated with coronary heart disease in Japanese non-insulin-dependent diabetes mellitus.[8] An association between paraoxonase-2 G148 variant with non-insulin-dependent diabetes was seen in aboriginal Canadians elsewhere.[9] Sureerut et al. stated that the role of PON genes in metabolic syndromes and cancers heightened the need for more investigation.[10] A correlation between paraoxonase and diabetes mellitus was found in Pima Indians.[11] The PON2 was associated with increased risk of T2D in Chinese population.[12] Sanghera et al. demonstrated that a variation at codon 311 (substitution of serine [S] for cysteine [C]) in the PON2 gene was associated with CAD in an Asian–Indian population.[13] A relationship between low serum PON activity and diabetes mellitus was reported elsewhere.[14] In spite of the fact that a large body of literature was published on the involvement of PON genes in T2D development in different populations, little is known about this association in Iranian population. Therefore, this study was designed to investigate the association between T2D and Ser 311 Cys variation of PON2 gene (rs17876171) in an Iranian population.
METHODS
A total of 200 unrelated people including 100 diabetics and 100 nondiabetic individuals (average age of 51 years) referring to Sedigheh Tahereh Research Center (Isfahan, Iran) were selected. T2D was clinically diagnosed (polyuria, polydipsia, and polyphagia) and confirmed by criteria of American Diabetes Association (ADA). More precisely, T2D was determined by (1) a fasting plasma glucose level of more than 7.0 mmol/L after a minimum 12 h fast or (2) a 2 h postglucose level [2 h oral glucose tolerance test (OGTT)] of more than 11.1 mmol/L. Impaired glucose tolerance (IGT) was determined by a fasting plasma glucose level of between 5.6 and 7.0 mmol/L or a 2 h OGTT of between 7.8 and 11.1 mmol/L. Normo-glycemic subjects were diagnosed by a fasting glycemia of less than 5.6 mmol/L or a 2 h glucose of less than 7.8 mmol/L. Biochemical and anthropometric parameters of participants are summarized in Table 1. Cholesterol, triglycerides, HDL, and glucose levels in blood were measured by standard enzymatic assays. LDL cholesterol level in blood was also derived by means of Friedewald equation.[15] Blood HbA1c level was measured using ion-exchange high performance liquid chromatography (normal reference range of 4.1-6.4%). Owing to having misleading results in analyses of the control group, people with positive family history of T2D and impaired glucose tolerance were excluded. Furthermore, individuals with impaired renal and hepatic function, previous diagnosis of blood disease, and those receiving pharmacological treatment for T2D, hypercholesterolemia, or hypertension were excluded to eliminate interference in biological variables. A written informed consent was obtained from all the subjects participated in the study.
Table 1.
Biochemical and anthropometric parameters of the type 2 diabetes group compared to the control group
A blood sample (5 cc) was collected from each participant after an overnight fasting for PCR-RFLP. Extraction of genomic DNA was performed using a high pure PCR template preparation kit (Roche, Germany) following the manufacturer's instructions. PCR-RFLP was then carried out. A segment of PON2 gene (331 bp fragment) was PCR-amplified using the following primers: Forward primer: 5-TGGAAAACAGGGCTTATTGATGA-3 and reverse primer: 5-CTGGGTCAATGTTGCTGGT TAAA-3. Primers were selected according to a preceding study[12] and then were ordered (BIORON). The PCR mixture contained 100-200 ng DNA template, 0.2 mmol/L dNTPs, 1.5 mmol/L MgCl2, 0.3 mmol/L of each primer, and 1.25 IU Taq DNA polymerase with 5% dimethyl sulfoxide in a final volume of 50 μL. For warming up, an initial denaturation at 94°C for 3 min was carried out. Thereafter, denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min were performed for 30 cycles. It was then followed by a postextension at 72°C for 10 min. In order to confirm the PCR amplification, gel electrophoresis (8% polyacrylamide gel and ethidium bromide staining) was carried out. The amplified products were then digested by DdeI restriction endonuclease enzyme at 37°C for 1 h. In order to visualize restriction products, electrophoresis was carried out using polyacrylamide gel (8%) and ethidium bromide stain. Four fragments were expected to see as follows: Ser 311 homozygotes (128, 105, 67, and 31 bp) and three fragments for Cys 311 homozygotes (172, 128, and 31 bp).[12] Therefore, Ser Cys heterozygotes had to yield five bands of 172, 128, 105, 67, and 31 bp. Inasmuch as the lower bands of 67 and 31 bp could not be separated from the bands of primer dimers, the presence of 172, 128, and 105 bp bands represented the polymorphism.
Data were analyzed using SPSS 17.0. More precisely, Student's t-test and χ2-test were applied for analyses of the variables. A P < 0.05 was considered to be statistically significant. Odds ratio with 95% confidence interval was applied for determination of the association of the type of PON2 polymorphism with the risk of T2DM. Using the χ2 results, the test for Hardy–Weinberg equilibrium and comparison of genotype and allele frequencies in the diabetic and nondiabetic subjects was carried out.
RESULTS
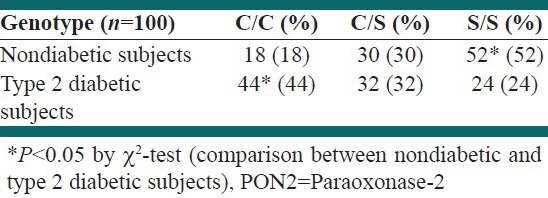
Patients’ biochemical and anthropometric blood parameters are summarized in Table 1. PCR amplification of PON2 gene produced the desired 331 bp band [Figure 1]. This product was subjected to digestion with DdeI restriction enzyme. As shown in Figure 2, different band patterns were observed due to the polymorphism of PON2 gene. As mentioned above, 67 and 31 bp bands were difficult to be observed owing to the presence of primer dimer bands. However, larger bands were sufficient for determination of the presence of Ser311 Cys polymorphism. Gene frequencies of PON2 gene are illustrated in Table 2. The obtained data showed that the frequency of C allele in diabetic subjects was significantly higher than in healthy subjects. The frequency of S allele in the healthy subjects was significantly high as compared to the C allele. Therefore, a correlation between diabetes and the presence of C allele was demonstrated (P < 0.001). Additionally, the genotype frequency of Ser/Ser was significantly associated with low frequency of T2D in diabetic subjects and genotype frequency of Cys/Cys and Cys/Ser was associated with high frequency of this disease [Table 3]. No significant difference was observed between the Cys311Cys and Cys311Ser genotypes in the healthy and diabetic groups.
Figure 1.
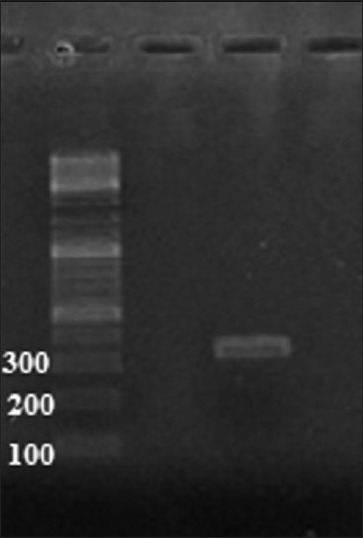
Confirmation of PCR amplification using gel electrophoresis. Lane 1: 100 bp DNA ladder, lane 2: PCR products from PON2 gene using forward and reverse primers mentioned in the “Methods” section. Extracted DNA from peripheral blood was used as a template
Figure 2.
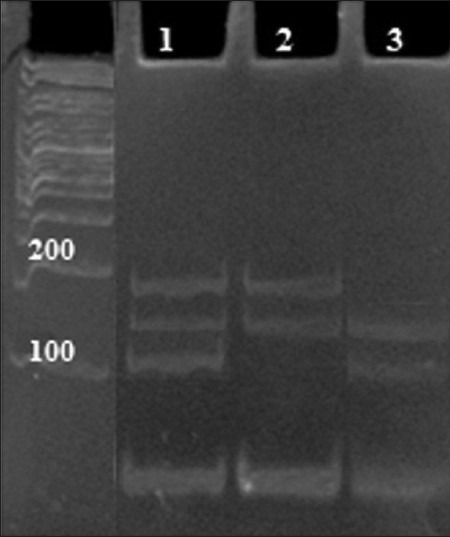
PCR-RFLP detection of the Ser/Cys polymorphism of PON2 gene using gel electrophoresis. Lane 1: Ser/Cys heterozygotes, lane 2: Cys 311 homozygotes, lane 3: Ser 311 homozygote. Extracted DNA from peripheral blood was used as a template
Table 2.
Gene frequency of the PON2 gene in nondiabetic and type 2 diabetic subjects
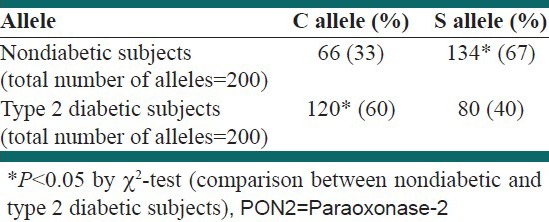
Table 3.
Genotype frequency of PON2 gene in nondiabetic and type 2 diabetic subjects
Finally, observed genotype frequencies of the Cys311Ser polymorphism of paraoxonase-2 in diabetic and nondiabetic subjects were in accordance with the Hardy–Weinberg equilibrium (data not shown).
DISCUSSION
Complex diseases, such as T2D, arise from metabolic disruptions with genetic and environmental components. Multiple genes are believed to be associated with T2D. Paraoxonase has been found to exert antioxidant and protecting effects upon lipid peroxidation as well as its role in controlling oxidative stress[11,16] resulting in several human diseases.[10] PON2 polymorphisms include Ala148Gly and Cys311Ser which are associated with different pathological disorders.[17] It is believed that PON2 polymorphism is associated with the high risk of T2D. Some research studied this relationship in different countries. Thus, this study aimed to assess the association between PON2 polymorphism and T2D. This study showed that an association exists between Ser311Cys polymorphism with T2D; even so, a study investigating this association in a northern Chinese population was in agreement with our results.[12] In addition, a correlation between paraoxonase and T2D was demonstrated in Pima Indians elsewhere.[11] Hegele et al. stated that a relationship between PON2 and glycemic control existed inasmuch as PON1 is adjacent to PON2 gene on chromosome 7 and associated with complications of diabetes and glycemic control in T2D and insulin sensitivity.[9] Mackness et al. reported a relationship between PON2 variant and microvascular complications in diabetic patients.[18] Ikeda et al.[19] and Robertson et al.[20] demonstrated that patients with PON1 polymorphism encounter oxidative stress damage, which led to CAD and metabolic syndrome. Jalilian et al. showed that a variation in PON2 gene is associated with the risk of CAD in Iranian population.[21] Leus et al. showed an association between PON2 gene variants and clinical symptoms of cardiovascular disease in familial hypercholesterolemia patients.[22]
CONCLUSION
We suggest that a Ser311Cys variation of PON2 gene is associated with type 2 diabetes. This change in amino acid may occur at an active site of PON2.[12] This can impair its antioxidation activity and change the metabolic potency of the cell.[12] Finally, it should be mentioned that the most important limitation of this study is the relatively small sample size. This was due to difficulty of obtaining blood samples. In fact, the sample was regionally representative of center of Iran and thus would tend to miss people who were in marginal regions. Moreover, further investigation and experimentation into Ser311Cys variation of PON2 are strongly recommended to better understand the actual relationship between the risk of diabetes mellitus and PON2 polymorphism in Iranian populations.
ACKNOWLEDGMENT
The authors would like to acknowledge the financial support of Isfahan Pharmaceutical Sciences Research Center and other supportive aids. There is no conflict of interest.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared
REFERENCES
- 1.Prochazka M, Thompson DB, Scherer SW. Linkage and association of insulin resistance and NIDDM with markers at 7q21.3q22.1 in the pimaindians. Diabetes. 1995;44:42. [Google Scholar]
- 2.Primo-Parma SL. The human serum paraoxonase/arylesterase gene (PON1) is one member of a multigene family. Genomics. 1996;33:498–509. doi: 10.1006/geno.1996.0225. [DOI] [PubMed] [Google Scholar]
- 3.Ng CJ, Wadleigh DJ, Gangopadhyay A, Hama S, Grijalva VR, Navab M, et al. Paraoxonase-2 is a ubiquitously expressed protein with antioxidant properties and is capable of preventing cell-mediated oxidative modification of low density lipoprotein. J Biol Chem. 2001;276:44444–9. doi: 10.1074/jbc.M105660200. [DOI] [PubMed] [Google Scholar]
- 4.Draganov DI, Stetson PL, Watson CE, Billecke SS, La Du BN. Rabbit serum paraoxonase 3 (PON3) is a high density lipoprotein-associated lactonase and protects low density lipoprotein against oxidation. J Biol Chem. 2000;275:33435–42. doi: 10.1074/jbc.M004543200. [DOI] [PubMed] [Google Scholar]
- 5.Reddy ST, Wadleigh DJ, Grijalva V, Ng C, Hama S, Gangopadhyay A, et al. Human paraoxonase-3 is an HDL-associated enzyme with biological activity similar to paraoxonase-1 protein but is not regulated by oxidized lipids. Arterioscler Thromb Vasc Biol. 2001;21:542–7. doi: 10.1161/01.atv.21.4.542. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz J, Blanche H, James RW, Garin MC, Vaisse C, Charpentier G, et al. Gln-Arg192 polymorphism of paraoxonase and coronary heart disease in type 2 diabetes. Lancet. 1995;346:869–72. doi: 10.1016/s0140-6736(95)92709-3. [DOI] [PubMed] [Google Scholar]
- 7.Zama T, Murata M, Matsubara Y, Kawano K, Aoki N, Yoshino H, et al. A192Arg variant of the human paraoxonase (HUMPONA) gene polymorphism is associated with an increased risk for coronary artery disease in the Japanese. Arterioscler Thromb Vasc Biol. 1997;17:3565–9. doi: 10.1161/01.atv.17.12.3565. [DOI] [PubMed] [Google Scholar]
- 8.Masato O, Yoichi T, Kamejiro Y. Paraoxonase polymorphism (Gln192-Arg) is associated with coronary heart disease. J Clin Endocrinol Metab. 1997;82:2257–60. doi: 10.1210/jcem.82.7.4096. [DOI] [PubMed] [Google Scholar]
- 9.Hegele RA, Connelly PW, Scherer SW, Hanley AJ, Harris AB, Tsui LC, et al. Paraoxonase-2 gene (PON2) G148 variant associated with elevated fasting plasma glucose in noninsulin-dependent diabetes mellitus. J Clin Endocrino lMetab. 1997;82:3373–7. doi: 10.1210/jcem.82.10.4289. [DOI] [PubMed] [Google Scholar]
- 10.Sureerut P, Thinnakorn P, Wanida S. Human Paraoxanase 2. Excli J. 2010;9:159–72. [PMC free article] [PubMed] [Google Scholar]
- 11.Horke S, Witte I, Wilgenbus P, Altenhöfer S, Krüger M, Li H, et al. Protective effect of paraoxonase-2 against endoplasmic reticulumstress-induced apoptosis is lost upon disturbance of calcium homoeostasis. Biochem J. 2008;416:395–405. doi: 10.1042/BJ20080775. [DOI] [PubMed] [Google Scholar]
- 12.Qu Y, Yang Z, Jin F, Sun L, Zhang C, Ji L, et al. The Ser311Cys variation in the paraoxonase 2 gene increases the risk of type 2 diabetes in northern Chinese. J Genet. 2008;87:165–9. doi: 10.1007/s12041-008-0025-3. [DOI] [PubMed] [Google Scholar]
- 13.Sanghera DK, Aston CE, Saha N, Kamboh MI. DNA polymorphisms in two paraoxonase genes (PON1 and PON2) is associated with the risk of coronary heart disease. Am J Hum Genet. 1998;62:36–44. doi: 10.1086/301669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackness B, Durrington PN, Abuashia B, Boulton AJ, Mackness MI. Low paraoxonase activity in type II diabetes mellitus complicated by retinopathy. Clin Sci (Lond) 2000;98:355–63. [PubMed] [Google Scholar]
- 15.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 16.Altenhöfer S, Witte I, Teiber JF, Wilgenbus P, Pautz A, Li H, et al. One enzyme, two functions: PON2 prevents mitochondrial superoxide formation and apoptosis independent from its lactonase activity. J Biol Chem. 2010;285:398–403. doi: 10.1074/jbc.M110.118604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mochizuki H, Scherer SW, Xi T, Nickle DC, Majer M, Huizenga JJ, et al. Human PON2 gene at 7q21.3: Cloning, multiplem RNA forms, and missense polymorphisms in the coding sequence. Gene. 1998;213:149–57. doi: 10.1016/s0378-1119(98)00193-0. [DOI] [PubMed] [Google Scholar]
- 18.Mackness B, Mcelduff P, Mackness MI. The paraoxonase-2-310 polymorphism is associated with the presence of microvascular complications in diabetes mellitus. J Intern Med. 2005;258:363–8. doi: 10.1111/j.1365-2796.2005.01554.x. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda Y, Suehiro T, Ohsaki F, Arii K, Kumon Y, Hashimoto K. Relationship between polymorphisms of the human serum paraoxonase gene and insulin sensivity in Japanese patients with type 2 diabetes. Diabetes Res Clin Pract. 2003;60:79–85. doi: 10.1016/s0168-8227(02)00280-2. [DOI] [PubMed] [Google Scholar]
- 20.Robertson KS, Hawe E, Miller GJ, Talmud PJ, Humphries SE. Human paraoxonase gene cluster polymorphisms as predictors of coronary heart disease risk in the prospective Northwick Park Heart Study II. Biochim BiophysActa. 2003;1639:203–12. doi: 10.1016/j.bbadis.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Jalilian A, Javadi E, Akrami M, Fakhrzadeh H, Heshmat R, Rahmani M, et al. Association of Cys 311 Ser polymorphism of Paraoxonase-2 gene with the risk of coronary artery disease. Arch Iran Med. 2008;11:544–9. [PubMed] [Google Scholar]
- 22.Leus FR, Zwart M, Kastelein JJ, Voorbij HA. PON2 gene variants are associated with clinical manifestations of cardiovascular disease in familial hypercholesterolemia patients. Atherosclerosis. 2001;154:641–9. doi: 10.1016/s0021-9150(00)00440-8. [DOI] [PubMed] [Google Scholar]


