Abstract
Following the completion of oogenesis and oocyte maturation, histone mRNAs are synthesized and stored in the sea urchin egg pronucleus. Histone mRNAs are the only mRNAs that are not polyadenylated but instead end in a stem–loop which has been conserved in evolution. The 3′ end binds the stem–loop-binding protein (SLBP), and SLBP is required for histone pre-mRNA processing as well as translation of the histone mRNAs. A cDNA encoding a 59 kDa sea urchin SLBP (suSLBP) has been cloned from an oocyte cDNA library. The suSLBP contains an RNA-binding domain that is similar to the RNA-binding domain found in SLBPs from other species, although there is no similarity between the rest of the suSLBP and other SLBPs. The suSLBP is present at constant levels in eggs and for the first 12 h of development. The levels of suSLBP then decline and remain at a low level for the rest of embryogenesis. The suSLBP is concentrated in the egg pronucleus and is released from the nucleus only when cells enter the first mitosis. SuSLBP expressed by in vitro translation does not bind the stem–loop RNA, suggesting that suSLBP is modified to activate RNA binding in sea urchin embryos.
INTRODUCTION
Histone mRNAs are the only cellular mRNAs in metazoans that are not polyadenylated (1). Instead, they end in a conserved stem–loop structure (2). The 3′ end of histone mRNAs is formed by an endonucleolytic cleavage (3) that requires the stem–loop and a purine-rich region, the histone downstream element (HDE) located 3′ to the stem–loop that binds the U7 snRNP (4–6). In most cells, histone mRNAs are expressed coordinately with DNA replication (7). An exception is in oocytes and many early embryos. Oocytes often store histone mRNAs that are not translated. Many early embryos (amphibians, insects, echinoderms) go through an initial period of rapid cell cycles that lack gap phases, but consist only of a series of alternating S phase and mitoses. During these cell cycles, the histone mRNAs are not cell cycle regulated (7).
The 3′ end of histone mRNA is the major element that determines cell cycle regulation of the mammalian histone mRNAs (8). The protein that binds the 3′ end of histone mRNA in mammalian cells, the stem–loop-binding protein (SLBP), is required for histone pre-mRNA processing (9) as well as for translation of the mRNA (10). SLBP is also cell cycle regulated, accounting for much of the cell cycle regulation of histone mRNA (11,12). There are two SLBPs present during oogenesis in Xenopus; xSLBP1 which is the ortholog of the mammalian SLBP (13) and xSLBP2 which is oocyte specific and is involved in storage of histone mRNA in an inactive form (14). XSLBP2 is destroyed during oocyte activation, allowing xSLBP1 to bind histone mRNA and activate translation. There is very little SLBP present in mouse oocytes, probably accounting for the lack of histone protein synthesis in oocytes. During oocyte maturation, the levels of SLBP increase dramatically, presumably resulting in activation of histone mRNA translation (15).
Sea urchins are unusual in that they store their female gametes as haploid eggs rather than oocytes. During early embryogenesis, the majority of the histone mRNAs are derived from a cluster of genes, the α-histone genes, that is tandemly repeated several hundred times (16). The sea urchin α-histone mRNAs and genes were the first metazoan genes cloned and sequenced (17). The U7 snRNA, which is required for 3′ processing of histone mRNA, was also first isolated from sea urchin embryos (5,18,19). The α-histone genes are first expressed in the haploid egg, and the histone mRNAs are retained in the pronucleus (20,21). Histone mRNAs are only released to the cytoplasm after fertilization, at the end of the first S phase when the nuclear envelope breaks down (22). The α-histone mRNAs are present throughout early embryogenesis, reaching a maximum at about the 100 cell stage, and then decline rapidly (23). Another set of histone mRNAs, transcribed from the late histone genes (24,25), then become the major histone mRNAs in the embryo. Both the α-histone mRNAs and the late histone mRNAs end in the same stem–loop and have similar HDEs.
We report here the characterization of a sea urchin SLBP (suSLBP) which is present in sea urchin eggs and embryos, and is the only SLBP detected in the sea urchin genome. This SLBP is present in the egg pronucleus and released at the end of the first S phase. SuSLBP is present at constant levels throughout early embryogenesis and at reduced levels after 24 h of embryogenesis. It is likely that suSLBP plays a role in the regulation of expression and metabolism of both the α-histone genes and late histone genes.
MATERIALS AND METHODS
Cloning of suSLBP
We prepared DNA from an oocyte phage cDNA library and used this cDNA as a template for PCR. The primers were from the conserved regions of the RNA-binding domain (RBD) chosen based on aligning the RBDs of the human, and two Xenopus SLBPs. The forward primer was CARAARC AGATHRAYTAYGGNAA [QKQI(E/D)YGK] and the reverse primer was CCANARYTTDATYTGYTGRTCCCA [WDQQIKLW]. PCR products of the appropriate size (175–200 nt) were cloned into the TA cloning vector (Invitrogen). Potential positive cDNA inserts of the correct size were tested by PCR using the same forward primer and the reverse primer YTTRTTNGGNGTNYKNGGRTG [HP(K/R)TPNK] and clones that gave PCR products of ∼110 nt were sequenced. They all had the same sequence.
The 186 nt insert containing the RBD was then used to screen the Strongylocentrotus purpuratus phage ovary cDNA library. Multiple phage were isolated that all encoded portions of the same cDNA, which contained a 525 amino acid open reading frame (ORF). The ORF was cloned into pGEM5. The 1575 nt ORF was used to screen an Lytechinus variegatus ovary cDNA library. Multiple clones were isolated which contained portions of the same cDNA, and these covered most of the ORF.
The sea urchin S.purpuratus genome is currently being sequenced (http://www.hgsc.bcm.tmc.edu/projects/seaurchin/). We analyzed the sequences from the shotgun reads (about 6× coverage) using the nucleotide sequence of the suSLBP cDNA. We located the exon–intron junctions, with each junction identified in at least three independent reads.
The accession numbers for the S.purpuratus and L.variegatus SLBP sequences are AY495699 and AY495700, respectively.
Antibodies to the suSLBP
A peptide, CFNLDQSFLKDEELIL, corresponding to the C-terminal 15 amino acids with C516 changed to serine was synthesized and coupled to keyhole limpet hemocyanin (KLH). Antisera were prepared by injecting into rabbits. The IgG was purified by chromatography on protein A–Sepharose. The antibody was also affinity purified using a peptide affinity column (Pierce Sulfolink).
The immunofluorescence on early embryos was performed as previously described for cyclin E (26). Western blotting was performed on sea urchin extracts as previously described using the affinity-purified anti-SLBP antibody (26).
Preparation of sea urchin extracts
Sea urchin egg and embryo extracts were prepared as previously described (26), and used for western blotting. Egg extracts for mobility shift assays were prepared as described (26), adjusted to 0.35 M KCl and passed over a DEAE–cellulose column, which retained much of the yolk and all the cellular RNAs. The flow-through from the column was used for the mobility shift assays. Nuclear extracts were prepared as previously described from hatching blastula embryos (27).
Analysis of RNA
Total cell RNA was prepared from embryos as previously described (26) and fractionated into poly(A)+ and poly(A)– RNA by chromatography on oligo(dT) cellulose. The RNAs were denatured with formaldehyde and resolved by agarose gel electrophoresis. The suSLBP mRNA was detected by northern blotting as previously described (26).
Affinity purification of SLBP
Extracts were prepared from sea urchin embryos as previously described. Nuclear extracts were prepared from highly purified nuclei from 16 h embryos (28), by extraction with 0.23 M NaCl as described for mammalian cell extracts (29). The procedure used to partially purify proteins from whole-cell extracts using a biotinylated stem–loop RNA has been described (30). The extracts were incubated with a biotinylated stem–loop RNA and then streptavidin–agarose beads were added. The beads were washed with an excess of the same buffer used to prepare the extracts. The beads were boiled in SDS loading buffer, resolved by SDS–gel electrophoresis and the suSLBP detected by western blotting using anti-SLBP.
Construction of domain-swapping mutants
We constructed a series of mutants with the portions of the RBD from human SLBP interchanged with the suSLBP RBD. These were made by using PCR to amplify the appropriate fragment introducing the appropriate restriction site for cloning.
RESULTS
Control of histone protein synthesis is important in the early development of all organisms, as the rapid cell cycles in early embryos require a constant supply of histones to package the newly replicated DNA. The early sea urchin histone genes (the α-histone genes) are one of the best studied systems of gene expression in early embryogenesis. Expression of this tandemly repeated gene set is restricted to early embryogenesis (23). While there are histone mRNAs present in the sea urchin egg, these all accumulate after germinal vesicle breakdown and are restricted to the female pronucleus (20,21). The 3′ end of histone mRNA plays an important role not only in cell cycle regulation in mammalian cells (31) but also in regulation of translation of histone mRNA during oogenesis and early development (10,14), and these regulatory events are mediated by proteins that bind the stem–loop, SLBPs.
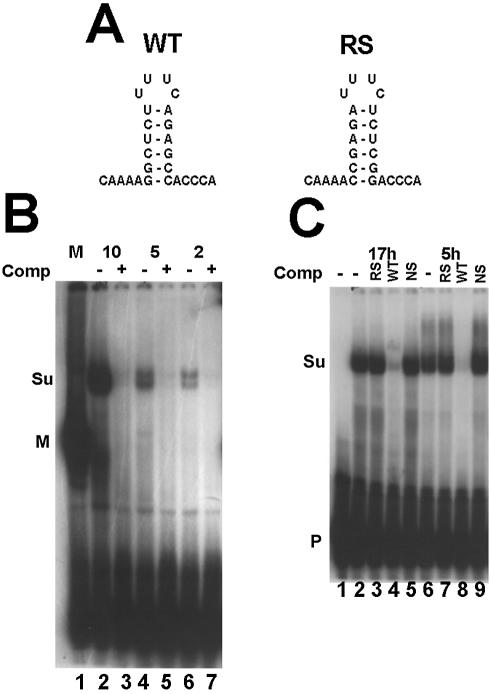
To define the SLBP(s) present in the sea urchin embryo, we prepared extracts from sea urchin eggs and embryos and analyzed these using a mobility shift assay. The sequences of the 3′ end of histone mRNA and the reverse-stem competitor used are shown in Figure 1A. In sea urchin eggs, we observed a complex which specifically binds the stem–loop (Fig. 1B, lane 2). This complex, which often resolved into a doublet, migrated more slowly than any of the SLBP–stem–loop complexes previously observed in mammals (Fig. 1B, lane 1) or frogs. The amount of the complex increased with increasing amounts of egg extract (Fig. 1B, lanes 2, 4 and 6), and formation of this complex was competed by the stem–loop, demonstrating that it was a specific complex (Fig. 1B, lanes 3, 5 and 7). We prepared total cell extracts from 5 and 17 h embryos and carried out mobility shift assays. The same major complex found in sea urchin eggs was also present at both stages of embryogenesis (Fig. 1C). Formation of the complex was competed by the unlabeled stem–loop but not by the reverse-stem or a non-specific oligoribonucleotide (Fig. 1C).
Figure 1.
Detection of suSLBP using a mobility shift assay. (A) The sequence of the 3′ end of histone mRNA used in the mobility shift assay is shown as well as the reverse-stem RNA used as a non-specific competitor. (B) An extract (10 mg/ml) was prepared from sea urchin eggs as described in Materials and Methods. Varying amounts (indicated above each lane) of the extract were incubated with the radiolabeled stem–loop probe. In lanes 3, 5 and 7, a 100-fold excess of unlabeled stem–loop (SL) was mixed with the probe prior to addition of the extract. In lane 1, 10 µg of extract from mouse myeloma cell nuclei was used. The complexes were resolved by native gel electrophoresis in a 10% polyacrylamide gel and detected by autoradiography. M is the complex with mouse SLBP and Su the complex with sea urchin SLBP. P indicates the unbound probe. (C) Extracts were prepared from 5 h embryos, or from purified nuclei from 17 h embryos. The extracts were incubated with the radiolabeled stem–loop (lanes 2 and 6). A 100-fold excess of the reverse-stem RNA (RS, lanes 3 and 7), the stem–loop RNA (SL, lanes 4 and 8) or a non-specific oligoribonucleotide (lanes 4 and 9) was added to the probe before addition of the extract. Lane 1 is the probe incubated in buffer. The complexes were resolved as in (B).
To clone the suSLBP, we purified DNA from an ovary phage cDNA library (32) and used PCR with primers designed to amplify the region of the cDNA encoding the RBD, which has been conserved from Caenorhabditis elegans to human (1,7). We obtained clones of the expected size for the RBD, and all these clones encoded the same sequence, suggesting that there was a single major SLBP in this library. We used this clone to screen the cDNA library and isolated multiple clones, which overlapped and contained a complete ORF of 525 amino acids (59 kDa; Fig. 2A), significantly larger than the 250–290 amino acid SLBPs found in vertebrates, Ciona and Drosophila. We used the S.purpuratus cDNA as a probe to isolate clones containing most of the SLBP cDNA from the sea urchin L.variegatus, which diverged from S.purpuratus ∼35 million years ago (33). Strikingly, these two proteins were <50% identical in the first 130 amino acids (56/130) with several insertions and deletions. Although we did not obtain a clone with the complete C-terminus of the L.variegatus cDNA, the suSLBPs from the two species were 70% identical in the 486 amino acids from the L.variegatus cDNA, with the greatest identity in the 190 amino acid region encompassing the RBD and including 60 amino acids before and 60 amino acids after the RBD (164/190, 87% identity).
Figure 2.
The sequence of suSLBP. (A) The predicted amino acid sequence from the cDNA sequence of S.purpuratus suSLBP is shown. The underlined sequences are the translation activation region required for translation of histone mRNA (WAVQVEE) and the RBD. The alternating bold blocks of amino acids represents the boundaries of the seven exons. (B) Differences between the S.purpuratus (top) and L.variegatus (bottom) suSLBP are indicated. Only the first 500 amino acids of the L.variegatus cDNA were isolated. The RBD is in bold. The sequence similar to the region required for translation of histone mRNA is in bold and underlined. (C) The RBDs of a variety of metazoan SLBPs are compared. The sequence of the mammalian SLBP (35), Xenopus xSLBP1 and xSLBP2 (14), Drosophila (34), C.elegans (38) and Ciona (39). There were multiple cDNAs for a single SLBP from the Ciona EST collection. The region around the conserved TPNK in the suSLBPs is highlighted. The amino acids in bold are invariant in the metazoan SLBPs.
The RBD of the suSLBP is located in the middle of the protein, starting about 240 amino acids from the N-terminus, and there are about 200 amino acids after the RBD. The RBDs of known SLBPs from a variety of metazoans are compared in Figure 2C. The suSLBP contains most of the invariant residues implicated in RNA binding that are found in the vertebrate and invertebrate SLBPs. The other portions of the protein were not similar to any known protein domains and did not show extensive similarity to other SLBPs. There was a short region from amino acids 190 to 197, which is similar in location and sequence to the core region of vertebrate SLBP required to stimulate translation of histone mRNA (10) (underlined in Fig. 2A). We have previously observed the same properties for the mammalian, Xenopus xSLBP2 (14) and Drosophila SLBPs (34); there are no obvious common domains in the multiple SLBPs outside of the RBD and no recognizable protein domains in the rest of the molecule.
The S.purpuratus genome is currently being sequenced. Searching the preliminary sequence reads with the suSLBP nucleotide sequence allowed us to deduce the exon–intron structure of the S.purpuratus suSLBP, which is indicated in Figure 2A. There are seven exons in the suSLBP, and the position of intron 5 (between exons 5 and 6) in the RBD is at the exact same site as an intron in the mammalian SLBP (35). Exons 5 and 6 in suSLBP are the same size as the corresponding exons in human SLBP, further supporting the interpretation that these two genes are orthologous. Since the two proteins have diverged outside of the RBD, it is not possible to precisely compare the exon–intron structure of the remainder of the protein, although the two genes contain the same number up to the RBD. There is one less exon in the suSLBP, probably due to the loss of the last intron found in the mammalian SLBP.
We also searched the sea urchin genome using the protein sequences of the RBD from the two frog SLBPs, dSLBP and Ciona SLBP. Each of these identified only a single gene, the suSLBP that we have cloned. Thus we conclude that it is likely that there is only a single SLBP in the sea urchin genome.
The cloned suSLBP is the SLBP detected by mobility shift in egg extracts
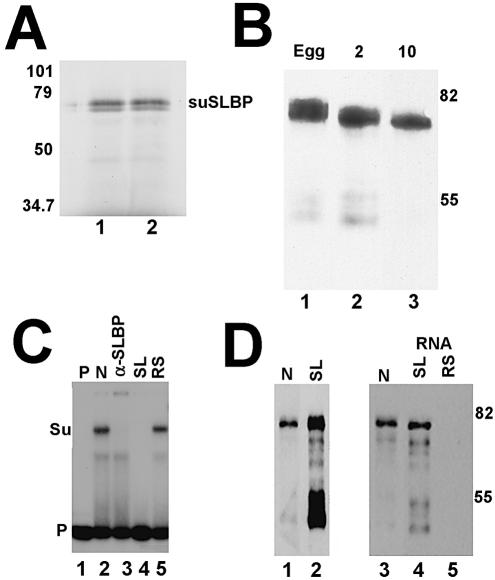
We estimated the size of the suSLBP by subjecting a partially purified preparation of SLBP from sea urchin eggs to gel filtration, followed by mobility shift assay. The suSLBP eluted from the gel filtration column with an apparent mol. wt of 65 kDa, consistent with the calculated size of the suSLBP assuming the suSLBP binding to the stem–loop as a monomer (not shown). Antibodies were prepared against a synthetic peptide corresponding to the C-terminus of the suSLBP from S.purpuratus. The antibody reacted with a single protein in S.purpuratus egg and embryo extracts on western blots (Fig. 3B and C), and the protein detected by western blotting had the same mobility (∼65 kDa) as the in vitro translated suSLBP (Fig. 3A). To demonstrate conclusively that the cloned suSLBP was the protein responsible for the observed complex with the radiolabeled stem–loop, we added the antibody to a mobility shift assay. The complex was shifted to the top of the gel (Fig. 3C, lane 3).
Figure 3.
The cloned suSLBP is expressed in sea urchin eggs and embryos. (A) The suSLBP was expressed by in vitro translation in a rabbit reticulocyte lysate in the presence of [35S]methionine. The proteins were resolved by gel electrophoresis and detected by autoradiography. (B) Extracts were prepared from sea urchin eggs, 2 (2-cell) and 10 h (60–100 cells) embryos in SDS. Proteins from an equal number of embryos were resolved by gel electrophoresis, transferred to nitrocellulose and the suSLBP detected by western blotting. (C) A nuclear extract from 17 h embryos was incubated with the radiolabeled stem–loop (lane 2) and the complexes resolved by native gel electrophoresis. In lane 3, affinity-purified anti-suSLBP was added to the reaction prior to electrophoresis. In lanes 4 and 5, excess stem–loop (SL) or reverse-stem (RS) was added to the probe prior to addition of the extract. Lane 1 is the probe incubated in buffer. Su is the complex formed with sea urchin SLBP. (D) A biotinylated stem–loop RNA was incubated with an extract from 5 h embryos. The proteins bound to the stem–loop (lane 2) were resolved by gel electrophoresis, and transferred to nitrocellulose. The suSLBP was detected by western blotting. Lane 1 is a nuclear extract from 17 h embryos. There was some proteolysis of the suSLBP during the purification. A nuclear extract from 17 h embryos (lane 3) was incubated with the biotinylated stem–loop (lane 4) or reverse-stem (lane 5) and the bound proteins resolved by gel electrophoresis, transferred to nitrocellulose and the suSLBP detected by western blotting.
The suSLBP was enriched from sea urchin extracts using a biotinylated stem–loop RNA. We incubated either a 5 h embryo extract (Fig. 3D, lanes 1 and 2) or a nuclear extract from 17 h embryos (Fig. 3D, lanes 3–6) with a biotinylated stem–loop RNA or a biotinylated reverse-stem RNA which does not bind SLBP, recovered the bound proteins on streptavidin–agarose and tested them for the presence of suSLBP by western blotting. A 65 kDa band was detected in both egg and embryo extracts, confirming that the suSLBP we cloned binds to the stem–loop sequence. No suSLBP bound to the biotinylated reverse-stem RNA (Fig. 3D, lane 5). There was significant proteolysis during the incubation of the embryo extract, resulting in the formation of a 53 kDa fragment which bound the stem–loop and reacts with the antibody, presumably resulting from cleavage of the SLBP on the N-terminal side of the RBD. This fragment was not present in the starting 5 h embryo extract analyzed by western blotting (not shown), and resulted from proteolysis during the purification procedure.
The suSLBP is present at constant levels in eggs and early embryos
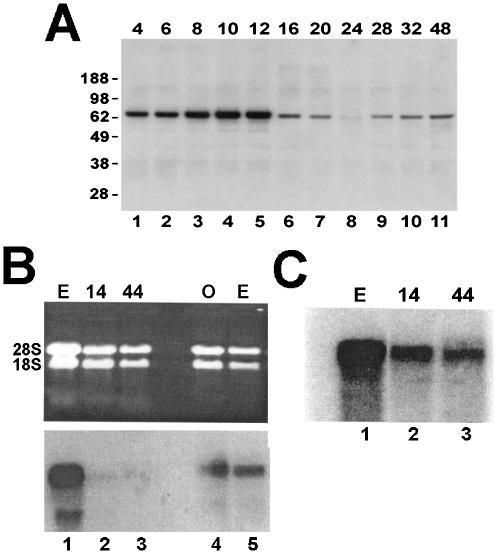
Western blots across development demonstrated that the suSLBP was present in sea urchin eggs and early embryos (Figs 3B and 4A). The levels of suSLBP were essentially constant in eggs and for the first 12–16 h of development, and then dropped dramatically (Figs 3B and 4A). The dropoff occurs at about the same time as the silencing of the α-histone genes (23), consistent with a role for suSLBP in α-histone gene expression. The levels of suSLBP were relatively constant throughout the next 48 h of embryogenesis (Fig. 4A, lanes 6–11). We also measured the levels of suSLBP mRNA by northern blotting. The suSLBP mRNA was detected in oocytes and eggs but much lower levels of suSLBP mRNA were present in embryos. The suSLBP mRNA was detected in blastula and gastrula stage (44 h) embryos by northern blotting of poly(A)+ mRNA (Fig. 4C, lanes 2 and 3), and similar results were obtained by RT–PCR (not shown).
Figure 4.
Expression of suSLBP during embryogenesis. (A) Total cell extracts were prepared at the indicated times after fertilization. The suSLBP was detected by western blotting. The sample in the 24 h lane is probably underloaded. (B) Total RNA from 14 and 44 h embryos (lanes 1–3) and a separate preparation of egg and oocyte mRNA (lanes 4 and 5) were resolved by agarose gel electrophoresis and analyzed by northern blotting using the suSLBP coding region as a probe. (C) Total RNA from the same preparation of eggs, 14 and 44 h embryos as in (B) were fractionated on oligo(dT) cellulose and the poly(A) RNA was resolved by gel electrophoresis and analyzed by northern blotting using the suSLBP as a probe. Equal amounts of total RNA from each sample were also analyzed.
SuSLBP is present in the female pronucleus and is released at the first mitosis
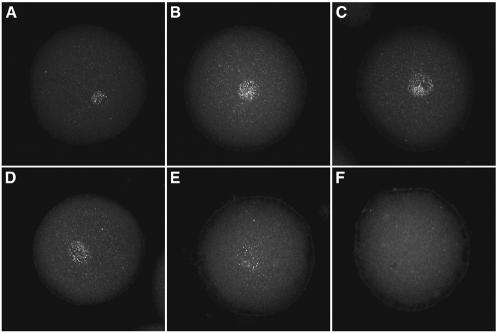
The α-histone mRNAs are synthesized in the female pronucleus, but are not exported to the cytoplasm. They only leave the nucleus at nuclear envelope breakdown during the first mitosis. We used immunofluorescence to localize the suSLBP in the sea urchin egg. SuSLBP is concentrated in the pronucleus (Fig. 5A), the same site as the α-histone mRNA (20). During the first cell cycle, the suSLBP remains in the pronucleus after pronuclear fusion and throughout the first S phase (Fig. 5B and C). However, at nuclear envelope breakdown, the suSLBP is released from the nucleus (Fig. 5D–F), as is the histone mRNA (20), consistent with the possibility that the suSLBP is associated with the α-histone mRNA.
Figure 5.
Localization of suSLBP during the first cell cycle. Unfertilized eggs (A) and fertilized eggs 30 (B), 45 (C), 60 (D), 75 (E), 90 (F) and 105 min (G) after fertilization were fixed and suSLBP detected by immunofluorescence using affinity-purified SLBP. Pronuclear fusion had occurred at 30 min, the cells had entered mitosis by 90 min and were in metaphase at 105 min. suSLBP is concentrated in the egg pronucleus and remains in the zygotic nucleus during S phase. At mitosis, the suSLBP is dispersed throughout the zygote.
Recombinant suSLBP does not bind RNA
We expressed the suSLBP in a reticulocyte lysate (Fig. 3A), and it was readily detected by western blotting. However, when we incubated the reticulocyte extract with a radiolabeled stem–loop and carried out a mobility shift assay, there was no detectable binding activity (Fig. 6B, lane 2). Similar results were obtained when suSLBP was expressed in Xenopus oocytes (not shown). Clearly the suSLBP is active in binding the stem–loop in vivo from the experiments in Figures 3 and 4, but not when expressed in a heterologous system.
Figure 6.
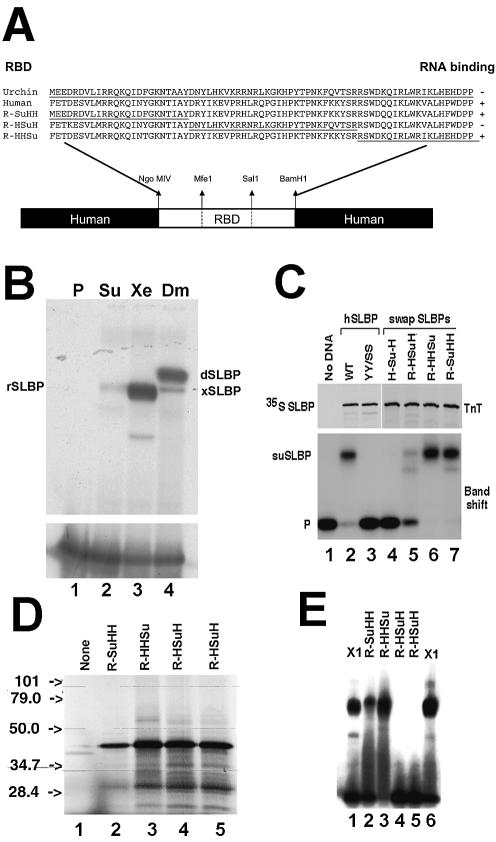
Recombinant suSLBP does not bind the stem–loop. (A) Structure of the chimeric clones, with portions of the suSLBP RBD interchanged with the human RBD. (B) The suSLBP, Xenopus SLBP1 and Drosophila SLBP were expressed in a reticulocyte lysate. Aliquots of the lysate were then tested for binding to the stem–loop in a mobility shift assay. Lane 1 is the probe, lane 2 is the suSLBP, lane 3 is xSLBP1 and lane 4 is dSLBP. There is a small amount of rabbit SLBP in the lysate (35), indicated by rSLBP. (C) The hSLBP (lane 2), a mutant of hSLBP (Y24S,Y27S; lane 3) which does not bind the stem–loop (40), and the indicated hybrid human–sea urchin SLBPs (lanes 4–7) were expressed in the reticulocyte lysate in the presence of [35S]methionine (top). The lysates were incubated with the radiolabeled stem–loop and the complexes formed were resolved by native gel electrophoresis (bottom). (D) An independent preparation of the indicated hybrid SLBPs (lanes 2–5) was translated in the reticulocyte lysate in the presence of [35S]methionine, and the proteins detected by autoradiography. (E) The lysates from (D) (lanes 2–5), together with a lysate expressing xSLBP1 (lanes 1 and 6), were incubated with the radiolabeled stem–loop and the complexes formed were resolved by native gel electrophoresis.
Compared with other SLBPs, suSLBP contains significant differences in the RBD before and after the invariant TPNK sequence (highlighted in Fig. 2C). The TPNK in other SLBPs is preceded by a basic amino acid and followed by four amino acids that contain one or two basic amino acids and end in an aromatic residue. The sea urchin SLBPs differ in this conserved region, with a hydrophobic amino acid before the TPNK and an uncharged region ending in a T after the TPNK. Also there are additional basic residues 4–6 amino acids before the TPNK not present in other SLBPs. To confirm that these changes were not a result of a mutation introduced during cloning, we isolated other clones from the S.purpuratus library as well as cDNA clones from a L.variegatus library. The sequence of the RBD was identical in all the S.purpuratus clones, and in the recently available genome sequence. The L.variegatus cDNA was clearly the ortholog of the S.purpuratus cDNA, with similar properties (although a slightly different sequence) around the TPNK (highlighted in Fig. 2C).
To determine the basis for the failure of the suSLBP to bind RNA, we constructed a number of chimeric SLBPs, interchanging regions of the human SLBP with the suSLBP (Fig. 6A). One possibility is that the sequences flanking the RBD were inhibiting the binding of the in vitro translated SLBP. The sea urchin RBD was inserted in place of the human RBD. The resulting HSuH protein was expressed well, but did not bind RNA (Fig. 6C, lane 4), indicating that the defect was in the sea urchin RBD and not due to an inhibitory effect of the N- or C-terminal region of the suSLBP (Fig. 6C, lane 4).
We then constructed three different chimeras of the RBD in the context of the human protein. We divided the RBD into three pieces shown in Figure 6A and interchanged different parts of the sea urchin and human RBDs. Each protein was expressed by in vitro translation in a reticulocyte lysate and assayed for its ability to bind the stem–loop RNA. Placing the N- or C-terminal third of the RBD in the context of the human protein resulted in a protein that bound the stem–loop as well as the human SLBP (Fig. 6C, lanes 6 and 7). However, when the central fragment of the suSLBP RBD was inserted into the human SLBP, the resulting protein, R-HSuH, did not bind the stem–loop. Again with two additional preparations of the R-HSuH protein, we did not observe any binding to the stem–loop (Fig. 6E, lanes 4 and 5) although the other chimeras bound to the stem–loop (Fig. 6E, lanes 2 and 3). We repeated this experiment with separate preparations of the chimeric proteins (Fig. 6D and E). Thus the region of the suSLBP which is defective in stem–loop binding is in the central region of the RBD, possibly around the conserved HPXTPNK. This suggests that the suSLBP requires a post-translational modification to bind the stem–loop which is only found on the suSLBP present in sea urchin embryo extracts.
DISCUSSION
In organisms that have large eggs and which undergo rapid cleavage without concomitant growth of the embryo, there is a very high rate of DNA replication and chromatin assembly. Under these conditions, the synthesis of histone protein and replication of DNA are not tightly coupled. In addition, the cell cycles are controlled differently from the situation in ‘normal’ cells, often lacking gap phases and consisting of alternating S and M phases. As the embryo transitions into a slower rate of cell division, there is a change to cell cycles which have G1 and G2 phases, with a tight coupling of histone protein synthesis to DNA replication.
Different organisms have different strategies for providing the large amounts of histone proteins required during the rapid cleavage stages. Mammals have a single SLBP, which is required for histone pre-mRNA processing and translation of histone mRNA. In the initial cell cycle, prior to activation of the zygotic genome in mouse embryos, histone synthesis is controlled by the maternal stores of histone mRNA and SLBP. SLBP mRNA is stored in the mouse oocyte and translation activated after treatment with progesterone, resulting in synthesis of high levels of SLBP (15), which in turn activates accumulation of histone protein for remodeling the sperm chromatin and the initial cell cycle. In frogs, histone mRNAs are stored in the oocyte, as are histone proteins. There are two SLBPs, one of which, xSLBP2, is involved in silencing the oocyte histone mRNA (14). At oocyte maturation, the xSLBP2 is destroyed, allowing xSLBP1, the ortholog of mammalian SLBP, to bind and activate translation of histone mRNA (14). In the Drosophila genome, there is only one SLBP (34). However, although there is histone mRNA stored in the egg that persists through the syncytial cell cycles, there is no SLBP (36), and no translation of histone mRNA during the syncytial stage. At the completion of the syncytial cell cycles, maternal histone and SLBP mRNA is destroyed and the synthesis of both SLBP and histone mRNA from the zygotic genome is activated (36). All of these organisms use a single set of histone genes to provide the core histones in all cells, including the early embryo.
Sea urchins have two sets of ‘replication-dependent’ histone genes: the α-histone genes are tandemly repeated about 400 times, with each repeat unit containing one copy of each of the five histone genes. These genes are transcribed in the haploid egg, and throughout the cleavage stage. Transcription of these multiple gene copies is probably necessary to provide the histone mRNAs needed during the rapid cleavage cycles. They are then silenced and are not expressed again during development or in adult tissues. The ‘late’ histone genes are expressed at low levels in early embryogenesis; their expression is activated at the blastula stage (24) and they provide the histone proteins after the cleavage stage.
We have cloned suSLBP from sea urchin embryos. The suSLBP is present in eggs and throughout embryogenesis. Only one SLBP is present in the sea urchin genome, based on searches for the RBD sequence in the current genome sequence, using the RBD from human, Xenopus xSLBP2, Ciona and Drosophila to search the urchin genome. Each of these RBDs identifed the cloned suSLBP but no other SLBPs in the sea urchin genome. It is likely that suSLBP is the ortholog of the mammalian SLBP, since the position of the intron in the RBD is identical in the suSLBP and the mammalian SLBP.
The suSLBP we have described here has the properties expected of a protein that is involved in the expression of both the α-histone genes and the late histone genes. SuSLBP is expressed in early embryogenesis, and is concentrated in the egg pronucleus, where the α-histone mRNAs are sequestered (21). Like the histone mRNA, the suSLBP is dispersed throughout the cell at the first mitosis. SuSLBP contains a sequence similar to that found in vertebrate SLBPs, that is required for stimulation of translation of histone mRNA (10), suggesting that suSLBP is involved in translation of the α-histone mRNAs during the cleavage stage. The levels of suSLBP remain constant during early embryogenesis, and then decline to a new steady-state level at the time the α-histone genes are turned off. These data are consistent with the suSLBP being required for expression of both the α-histone and late histone genes.
The suSLBP is larger than the SLBPs from vertebrates, insects and the urochrodate Ciona. Urochordates are a sister group of vertebrates, and are now widely considered to be in the phylum chordata (37). The only SLBP in the Ciona genome is similar in size to the vertebrate SLBPs. The extended N-terminus of suSLBP is very different between L.variegatus and S.purpuratus suSLBP, suggesting that this region of the protein does not have a critical function. The sequence around the RBD has been highly conserved between the two sea urchin species, including a region similar to the sequence in vertebrate SLBPs required for translation of histone mRNA.
The most striking property of the suSLBP is that the recombinant protein expressed either in Xenopus oocytes or rabbit reticulocyte lysates does not bind the stem–loop, although the suSLBP in the sea urchin embryo is active in binding the 3′ end of histone mRNA. We were also unsuccessful in our attempts to clone the suSLBP using the three-hybrid system, consistent with the possibility that the protein expressed in yeast also does not bind the 3′ end of histone mRNA. A likely explanation for this observation is that there is a covalent modification in the RBD of the suSLBP in sea urchins that is essential for RNA binding and that does not occur when the suSLBP is expressed in organisms other than sea urchins.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Hemant Kelkar of the UNC Bioinformatics Center for assistance in the sea urchin genome analysis. This work was supported by NIH grants GM58961 to W.F.M. and F32GM20151 to B.J.S., and by the Stowers Foundation for Medical Research (J.A.C.).
REFERENCES
- 1.Dominski Z. and Marzluff,W.F. (1999) Formation of the 3′ end of histone mRNA. Gene, 239, 1–14. [DOI] [PubMed] [Google Scholar]
- 2.Birnstiel M.L., Busslinger,M. and Strub,K. (1985) Transcription termination and 3′ processing: the end is in site! Cell, 41, 349–359. [DOI] [PubMed] [Google Scholar]
- 3.Gick O., Krämer,A., Vasserot,A. and Birnstiel,M.L. (1987) Heat-labile regulatory factor is required for 3′ processing of histone precursor mRNAs. Proc. Natl Acad. Sci. USA, 84, 8937–8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilmartin G.M., Schaufele,F., Schaffner,G. and Birnstiel,M.L. (1988) Functional analysis of the sea urchin U7 small nuclear RNA. Mol. Cell. Biol., 8, 1076–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strub K. and Birnstiel,M.L. (1986) Genetic complementation in the Xenopus oocyte: co-expression of sea urchin histone and U7 RNAs restores 3′ processing of H3 pre-mRNA in the oocyte. EMBO J., 5, 1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mowry K.L. and Steitz,J.A. (1987) Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNAs. Science, 238, 1682–1687. [DOI] [PubMed] [Google Scholar]
- 7.Marzluff W.F. and Duronio,R.J. (2002) Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol., 14, 692–699. [DOI] [PubMed] [Google Scholar]
- 8.Lüscher B., Stauber,C., Schindler,R. and Schümperli,D. (1985) Faithful cell-cycle regulation of a recombinant mouse histone H4 gene is controlled by sequences in the 3′-terminal part of the gene. Proc. Natl Acad. Sci. USA, 82, 4389–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominski Z., Zheng,L.-X., Sanchez,R. and Marzluff,W.F. (1999) The stem–loop binding protein facilitates 3′ end formation by stabilizing U7 snRNP binding to the histone pre-mRNA. Mol. Cell. Biol., 19, 3561–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez R. and Marzluff,W.F. (2002) The stem–loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol., 22, 7093–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng L.-X., Dominski,Z., Yang,X., Elms,P., Raska,C.S., Borchers,C.H. and Marzluff,W.F. (2003) Phosphorylation of SLBP on two threonines triggers degradation of SLBP, the sole cell-cycle regulated factor required for regulation of histone mRNA processing, at the end of S-phase. Mol. Cell. Biol., 23, 1590–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitfield M.L., Zheng,L.-X., Baldwin,A., Ohta,T., Hurt,M.M. and Marzluff,W.F. (2000) Stem–loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol., 20, 4188–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingledue T.C., Dominski,Z., Sanchez,R., Erkmann,J.A. and Marzluff,W.F. (2000) Dual role for the RNA binding domain of Xenopus laevis SLBP1 in histone pre-mRNA processing. RNA, 6, 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z.-F., Ingledue,T.C., Dominski,Z., Sanchez,R. and Marzluff,W.F. (1999) Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol. Cell. Biol., 19, 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allard P., Champigny,M.J., Skoggard,S., Erkmann,J.A., Whitfield,M.L., Marzluff,W.F. and Clarke,H.J. (2002) Stem loop binding protein accumulates during oocyte maturation and is not cell cycle regulated in the early mouse embryo. J. Cell Sci., 115, 4577–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxson R., Mohun,T.J., Cohn,R. and Kedes,L. (1983) Expression and organization of histone genes. Annu. Rev. Genet., 17, 239–277. [DOI] [PubMed] [Google Scholar]
- 17.Cohn R.H., Lowry,J.C. and Kedes,L.H. (1976) Histone genes of the sea urchin (S.purpuratus) cloned in E.coli; order, polarity and strandedness of the five histone coding and spacer regions. Cell, 9, 147–161. [DOI] [PubMed] [Google Scholar]
- 18.Birchmeier C., Schümperli,D., Sconzo,G. and Birnstiel,M.L. (1984) 3′ editing of mRNAs: sequence requirements and involvement of a 60-nucleotide RNA in maturation of histone mRNA precursors. Proc. Natl Acad. Sci. USA, 81, 1057–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLorenzi M., Rohrer,U. and Birnstiel,M.L. (1986) Analysis of a sea urchin gene cluster coding for the small nuclear U7 RNA, a rare RNA species implicated in the 3′ editing of histone precursor mRNAs. Proc. Natl Acad. Sci. USA, 83, 3243–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venezky D.L., Angerer,L.M. and Angerer,R.C. (1981) Accumulation of histone repeat transcripts in the sea urchin egg pronucleus. Cell, 24, 385–391. [DOI] [PubMed] [Google Scholar]
- 21.Angerer L.M., DeLeon,D.V., Angerer,R.C., Showman,R.M., Wells,D.E. and Raff,R.A. (1984) Delayed accumulation of maternal histone mRNA during sea urchin oogenesis. Dev. Biol., 101, 477–484. [DOI] [PubMed] [Google Scholar]
- 22.Wells D.E., Showman,R.M., Klein,W.H. and Raff,R.A. (1981) Delayed recruitment of maternal mRNA in sea urchin embryos. Nature, 292, 477–478. [DOI] [PubMed] [Google Scholar]
- 23.Maxson R.E. Jr and Wilt,F.H. (1981) The rate of synthesis of histone mRNA during the development of sea urchin embryos (Strongylocentrotus purpuratus). Dev. Biol., 83, 380–386. [DOI] [PubMed] [Google Scholar]
- 24.Knowles J.A. and Childs,G.J. (1984) Temporal expression of late histone messenger RNA in the sea urchin Lytechinus pictus. Proc. Natl Acad. Sci. USA, 81, 2411–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Childs G., Nocente-McGrath,C., Lieber,T., Holt,C. and Knowles,J. (1982) Sea urchin (L.pictus) late-stage H3 and H4 genes: characterization and mapping of a clustered but non-tandemly linked multigene family. Cell, 31, 383–393. [DOI] [PubMed] [Google Scholar]
- 26.Sumerel J.L., Moore,J.C., Schnackenberg,B.J., Nichols,J.A., Canman,J.C., Wessel,G.M. and Marzluff,W.F. (2001) Cyclin E and its associated cdk activity do not cycle during early embryogenesis of the sea urchin. Dev. Biol., 234, 425–440. [DOI] [PubMed] [Google Scholar]
- 27.Morris G.F., Price,D.H. and Marzluff,W.F. (1986) Synthesis of U1 RNA in a DNA-dependent system from sea urchin embryos. Proc. Natl Acad. Sci. USA, 83, 3674–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris G.F. and Marzluff,W.F. (1985) Synthesis of U1 RNA in isolated nuclei from sea urchin embryos: U1 RNA is initiated at the first nucleotide of the RNA. Mol. Cell. Biol., 5, 1143–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dominski Z., Sumerel,J., Hanson,R.J. and Marzluff,W.F. (1995) The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA, 1, 915–923. [PMC free article] [PubMed] [Google Scholar]
- 30.Dominski Z., Yang,X., Kaygun,H. and Marzluff,W.F. (2003) A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell, 12, 295–305. [DOI] [PubMed] [Google Scholar]
- 31.Harris M.E., Böhni,R., Schneiderman,M.H., Ramamurthy,L., Schümperli,D. and Marzluff,W.F. (1991) Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol., 11, 2416–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berg L.K., Chen,S.W. and Wessel,G.M. (1996) An extracellular matrix molecule that is selectively expressed during development is important for gastrulation in the sea urchin embryo. Development, 122, 703–713. [DOI] [PubMed] [Google Scholar]
- 33.Smith A.B. (1988) Phylogenetic relationships, divergence times and rates of molecular evolution for camarodent sea urchins. Mol. Biol. Evol., 5, 345–365. [Google Scholar]
- 34.Sullivan E., Santiago,C., Parker,E.D., Dominski,Z., Yang,X., Lanzotti,D.J., Ingledue,T.C., Marzluff,W.F. and Duronio,R.J. (2001) Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev., 15, 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z.-F., Whitfield,M.L., Ingledue,T.I., Dominski,Z. and Marzluff,W.F. (1996) The protein which binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev., 10, 3028–3040. [DOI] [PubMed] [Google Scholar]
- 36.Lanzotti D.J., Kaygun,H., Yang,X., Duronio,R.J. and Marzluff,W.F. (2002) Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3′ processing in vivo. Mol. Cell. Biol., 22, 2267–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cameron C.B., Garey,J.R. and Swalla,B.J. (2000) Evolution of the chordate body plan: new insights from phylogenetic analyses of deuterostome phyla. Proc. Natl Acad. Sci. USA, 97, 4469–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin F., Schaller,A., Eglite,S., Schümperli,D. and Müller,B. (1997) The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J., 16, 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dehal P., Satou,Y., Campbell,R.K., Chapman,J., Degnan,B., De Tomaso,A., Davidson,B., Di Gregorio,A., Gelpke,M., Goodstein,D.M. et al. (2002) The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science, 298, 2157–2167. [DOI] [PubMed] [Google Scholar]
- 40.Dominski Z., Erkmann,J.A., Greenland,J.A. and Marzluff,W.F. (2001) Mutations in the RNA binding domain of stem–loop binding protein define separable requirements for RNA binding and histone pre-mRNA processing. Mol. Cell. Biol., 21, 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]