Abstract
A major priority in HIV vaccine research is the development of an immunogen to elicit broadly neutralizing antibodies (NAbs). Monoclonal antibody (mAb) b12 is one of now several broadly neutralizing mAbs that bind epitopes overlapping the CD4-binding site (CD4bs) on HIV-1 gp120 and that serve as templates to engineer effective immunogens. We are exploring a strategy whereby extra glycans are incorporated onto gp120 to occlude the epitopes of non-neutralizing mAbs while maintaining exposure of the b12 site. Immunizing with these so-called hyperglycosylated gp120s is hypothesized to preferentially elicit b12-like NAbs. Here, the effects of two adjuvants, monophosphoryl lipid A (MPL) and Quil A, on eliciting b12-like responses when formulated with a new hyperglycosylated mutant, ΔN2mCHO(Q105N), is presented. Sera from ΔN2mCHO(Q105N)_MPL immunized animals bound the homologous antigen ΔN2mCHO(Q105N) with greater preference than sera from ΔN2mCHO(Q105N) QuilA immunized animals, demonstrating the modulation of antibody fine specificity by these two adjuvants. We also found that sera from ΔN2mCHO(Q105N)_QuilA immunized animals bound best to a resurfaced HIV gp120 core protein on which non-CD4bs epitopes are substituted with non-HIV residues, suggesting that these sera contain a relatively larger fraction of CD4bs-specific antibodies. Consistent with these data, inhibition assays revealed epitope overlap with the binding sites of the CD4bs-specific antibodies b12, b13 and VRC03. Unexpectedly, these sera did not exhibit significant neutralizing activity against a set of HIV-1 primary strains. Our results show that although formulating mutant ΔN2mCHO(Q105N) with Quil A promotes the elicitation of CD4bs-directed antibodies relative to wild-type gp120, tweaking of the immunization regimen is needed to yield robust, CD4bs-focused NAbs.
Keywords: Protein engineering, b12, Hyperglycosylation, Immunofocusing
1. Introduction
The surface unit gp120 of the HIV envelope spike (Env) is a principal target for anti-viral neutralizing antibodies (NAbs) [1,2] and therefore of particular interest for the design of an HIV vaccine. However, the extensive antigenic diversity exhibited by gp120 poses significant challenges for formulating a vaccine immunogen capable of eliciting NAb responses that are effective against the many different HIV strains that a vaccinee might be exposed to. To address this challenge, novel vaccine design strategies are being explored. One major avenue of research has been made to delineate sites on Env that are most conserved among virus strains yet accessible for antibody recognition (reviewed in [1,3,4]). The CD4-binding site (CD4bs) on gp120 is one of the sites that is of considerable interest for vaccine design purposes because (a) its structure is relatively conserved as necessitated by its role in viral host entry [5], (b) of the occurrence of anti-CD4bs NAbs with exceptional activity in the sera of select long-term infected HIV+ individuals [6–19], and (c) of the results from passive antibody transfer studies in macaques demonstrating protection against chimeric simian–human immunodeficiency virus (SHIV) challenge upon administration of the broadly neutralizing CD4bs-specific monoclonal antibody (mAb) b12 [20–24]. Screening and mapping of sera from HIV-infected individuals has recently led to the discovery of a number of additional broadly neutralizing CD4bs-specific mAbs, including VRC01, VRC03, HJ16, VRC-PG04 and 3BCN60 [7,14–17,25]. Armed with an increasingly better understanding of how CD4bs antibodies structurally and functionally interact with gp120 [17,26–31], the focus has turned largely to the structure-based design of immunogens to elicit NAbs of equivalent activity (reviewed in [32,33]), supplemented by studies characterizing B cell responses at the cellular level[34–36].
Among the avenues under investigation are strategies to focus antibody responses on the epitopes of broadly neutralizing CD4bs antibodies such as b12 [37–39]. In earlier work, we reported on a panel of gp120 mutants engineered to elicit antibodies targeted to the b12 epitope [28,38,40]. The design of these mutants began with the identification of four residues – Gly473-Asp474-Met475-Arg476 (dubbed GDMR) – at the heart of the CD4bs that are generally important for the binding of several non-neutralizing CD4bs antibodies, but unnecessary for b12 binding [28,41]. Substitution of this string of 4 residues with alanines abrogated the binding of several non-neutralizing CD4bs antibodies; the design of this so-called GDMR mutant was meant to reduce the likelihood of eliciting non-neutralizing CD4bs antibodies upon immunization [28,42]. Binding of antibodies to other regions on monomeric gp120 was further eliminated by truncation of the N terminus and masking other unwanted epitopes via the incorporation of extra glycans [38,40]. We have dubbed this approach hyperglycosylation and hypothesize, as have others [43–46], that immunizing with these molecules will result in antibody responses preferentially directed at sites that remain accessible, most desirably the b12 binding site. While most mutants designed so far have not exhibited a reduction in immunogenicity relative to wild-type gp120, their antigenic design has not translated into the elicitation of high levels of CD4bs-specific NAbs ([42] and unpublished data). We have focused therefore on ‘fine tuning’ exposure of the CD4bs and are exploring immunization regimens that will promote the elicitation of the desired CD4bs-specific antibodies.
One factor that may impact the elicitation of CD4bs responses is the choice of adjuvant. Adjuvants are known to increase robustness of humoral responses to immunogens and have now become a key component in most vaccine formulations [47–50]. We therefore conducted a pilot study with wild-type gp120 (gp120wt) in mice using six different adjuvants and adjuvant mixtures – Alum, Quil A, CpG-ODN, monophosphoryl lipid A (MPL), Alum plus CpG, and Quil A plus MPL. Based on the results of this pilot study, which are reported here, Quil A and MPL were moved forward for closer investigation.
This report focuses on the head-to-head comparison of the ability of MPL and Quil A to promote CD4bs-specific antibody responses in mice immunized with the engineered mutant Q105N compared to gp120wt. To dissect the specificities of the elicited serum antibodies, three gp120 derivatives were used: (i) a truncated gp120 outer domain construct (XOD6) lacking the N-terminus, V1/V2 loops and half of the gp120 bridging sheet formed by the inner domain, (ii) a resurfaced gp120 core (RSC3) that specifically presents the neutralizing face of the CD4bs but largely eliminates other antigenic faces through non-HIV-1 residue substitution [25], and (iii) a mutant of RSC3 containing a deletion of Ile371 that eliminates binding of several CD4bs antibodies (RSC3Δ371I; also termed ΔRSC3 [25]). Overall, the data indicate that mutant Q105N elicits greater CD4bs-specific antibody responses when formulated with the adjuvant Quil A. Although the elicited antibodies did not exhibit significant neutralizing activity, this study shows the significance of adjuvant on the magnitude of CD4bs-directed responses and provides an incremental step in understanding molecular and physiological features that might promote antibody responses to the CD4bs.
2. Materials and methods
2.1. Construction, expression and purification of the truncated outer domain protein XOD6
XOD6 is a gp120 outer domain construct comprised of residues 249–485 of JR-CSF gp120. A plasmid encoding codon-optimized JR-CSF gp120 (kindly gifted by Maxygen) was used as template to amplify the relevant segment by standard PCR. The reverse primer encoded also a 6-HIS tag sequence to facilitate protein purification. The resulting fragment was cloned into a modified pRMHa Drosophila expression vector (Maxygen) and checked by DNA sequencing. To express the protein, the plasmid was transfected into Drosophila S2 cells along with plasmid pCoBlast (Invitrogen) at a 20:1 ratio. Stably transfected cells were selected by serially passaging the cells in S2 media containing 25 μg/ml blastidicin (Invivogen). Stably transfected clones were expanded in multi-level Cell Factories (Nunc) and allowed to grow until near full-confluency. CuSO4 (0.5 mM final concentration) was then added to induce protein expression. Supernatant was harvested 3–4 days later and stored at 4 °C until needed.
XOD6 was purified in a 2-step process. Culture supernatant was first passed over a Galanthus nivalis lectin (Vector Labs) column. Non-specifically bound protein was removed by washing and bound glycoproteins eluted with buffer supplemented with 1 M methyl mannoside. The eluate was then passed over a Ni2+-NTA agarose (Qiagen) column. After washing, HIS-tagged XOD6 was eluted with buffer containing a high concentration of imidazole (200–300 mM). The eluate was dialyzed against PBS and purity assessed by SDS-PAGE.
2.2. Construction, expression and purification of JR-FL gp120wt and Q105N
Mutant Q105N was generated by QuikChange mutagenesis (Agilent Technologies) using mutant ΔN2mCHO [40] as template. The mutagenesis primers were designed to insert the glycosylation signal sequence Asn-(X)-Thr at positions 105–107. The sequence of the Q105N mutant was verified by DNA sequencing. To facilitate recombinant protein purification, the sequences for JR-FL gp120wt [28] and mutant Q105N were appended with a C-terminal 8-HIS tag by standard PCR. Following digestion with EcoRI and XhoI, the PCR products were cloned into a previously described pCMV-tag4 expression vector (Agilent Technologies) containing a tissue plasminogen activator leader sequence for recombinant protein secretion [28]. After confirming addition of the HIS tag by DNA sequencing, CHO-K1 cells were stably transfected with each construct using FuGENE (Roche). Stably transfected clones were selected by two rounds of limiting dilution in selection media (Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (Invitrogen) supplemented with 5% FBS and 200 μg/ml G418). Clones producing the highest levels of protein from the limiting dilution rounds were sub-cultured to select the best-growing clone that produced the highest amount of protein. Cultures of the best clones were then expanded. To maximize production, stably transfected clones were cultured in multi-level Cell Factories (Nunc). Supernatants were normally harvested at 10-day intervals, filtered to remove cell debris and then purified on Ni2+-NTA columns (Qiagen) as described above for protein XOD6. Protein purity was determined as described above.
2.3. Immunizations
Groups of roughly six-week old female BALB/c mice (Jackson), with 8 animals per group, were used for immunizations. Each animal received 20 μg of antigen per injection formulated with adjuvant. For the pilot adjuvant study with gp120wt we used 6 different adjuvants and adjuvant combinations: (1) Alhydrogel (Accurate Chemical and Scientific Corp.; 50 μl per animal of 1.3% aluminum hydroxide formulation); (2) Quil A (Accurate Chemical and Scientific Corp.; 20 μg per animal); (3) CpG-ODN 2006 [51–53] (Invitrogen); (4) monophosphoryl lipid A (MPL; 50 Sigma Adjuvant System, 20 μg per animal); (5) Alhydrogel plus CpG; and (6) Quil A plus MPL. For the MPL formulations, adjuvant was pre-heated at 45 °C for 5 min and then mixed with antigen. For all other formulations, adjuvant solubilised in PBS was mixed directly with antigen. Ca2+/Mg2+-free PBS was used to make up a final volume of 200 μl per animal, and formulations were mixed for 1 h at RT. All formulations were prepared on the day of immunization. Each animal received two 100 μl subcutaneous injections of immunogen at each immunization time point. The animals received a first booster injection at 4 weeks after priming and a second boost 8 weeks later. Bleeds were taken 2 weeks after each injection. Sera from the final bleeds were used for all analyses reported here. All experiments were approved by and followed the guidelines set out by the University Animal Care Committee.
2.4. Enzyme-linked immunosorbent assay (ELISA)
ELISA’s were performed as described previously [28]. To assess the antigenic profile of Q105N, microtiter plates were coated with Q105N at 2 μg/ml (in PBS) and serially diluted mAbs (F105, b6, b12, b13, VRC01, VRC03, CD4-IgG2, B4e8 and 2G12) assessed for binding starting at 5 μg/ml. Plates were allowed to develop for 30 min in the dark and then read at 405 nm.
To determine serum antibody titres and to dissect serum antibody specificities, 96-well ELISA plates were coated with antigen as described above and serially diluted sera assessed for binding starting at 1:500. Serum antibody titres were defined as the serum dilution that yielded an OD405 signal > 0.2 after 30 min. This cutoff is twice above the average background OD405 signal (0.1) based upon binding of pre-bleed samples and corresponding blank controls. To determine the IgG subclasses of the elicited serum antibodies, assays were set up as described above and binding detected with subclass-specific secondary antibodies.
Inhibition assays were performed in 2 ways. For the analysis of sera from the pilot immunization with gp120wt, serially diluted sera were added to gp120-coated microtiter plates that had been pre-incubated for 1 h with F(ab’)2 b12. F(ab’)2 b12 was made by digestion of IgG b12 with pepsin (Sigma) following a previously described procedure [54]. Serum binding was quantified with a rabbit Fc-specific secondary antibody and ELISA titres determined relative to buffer-only control wells. For the analysis of sera from the head-to-head comparison between gp120wt and Q105N, sera were serially diluted (starting at 1:50) and incubated on plates for 1 h. MAbs (diluted to 10 μg/ml) were then added to the plates and these incubated for a further 1 h. MAb binding was quantified using AP-conjugated anti-human F(ab’)2-specific secondary antibody (Jackson ImmunoResearch). A F(ab’)2-specific secondary antibody was used to ensure specific detection of the mAbs. Relative changes in mAb binding were calculated by subtracting the OD values of no serum blank controls from the OD values of the corresponding serum samples.
2.5. Neutralization assay
Pseudovirus neutralization assays with luciferase read-out were performed with U87 CD4+ CCR5+ target cells essentially as described before [28,55]. Heat-inactivated (56 °C, 30 min) pooled sera from each immunization group were diluted 1:10 in media and pre-incubated with an equal volume of pseudovirus for 1 h at 37 °C. The sera/virus mixture was then added to U87 CD4+ CCR5+ cells. Luciferase activity in cell lysates was assessed on a Viktor X5 luminometer. Percentage neutralization was calculated relative to cell-only and virus-only controls.
2.6. Statistical analyses
All statistical analyses were performed using GraphPad Prism 5. Differences between groups were assessed using one-way ANOVA, with p < 0.05 being considered significant.
3. Results
3.1. Hyperglycosylated mutant Q105N limits access of select CD4bs antibodies
The design of previous hyperglycosylated mutants has focused mostly on masking the epitopes of antibodies to non-CD4bs epitopes, in particular the V1/V2 and V3 regions, through the introduction of extra glycans at those locations [38,40]. Although these mutants also contained a 4-string alanine substitution of residues 473–476 (the GDMR region) that prevented the binding of non-neutralizing CD4bs antibodies, access to the CD4bs was not specifically constrained by glycans. We reasoned that elicitation of CD4bs-focused responses might be improved by replacing the GDMR/AAAA mutation with a glycan that would restrict access to the target site. Combining insight from the b12:gp120 complex structure [56] and alanine mutagenesis data [28], we inserted a glycan at position 105 (Gln) on JR-FL gp120 (Fig. 1). Residues at position 105 are highly variable [28,57] and thus it seemed likely that our mutation would not be substantially disruptive. Furthermore, we reasoned that a glycan on the left perimeter of the b12 epitope near the non-neutralizing face/inner domain of gp120 [58] (Fig. 1) would limit antibody access to the CD4bs from that unwanted angle. The resulting mutant, ΔN2mCHO(Q105N), has a total of 11 extra glycans on gp120 relative to the wild-type sequence.
Fig. 1.
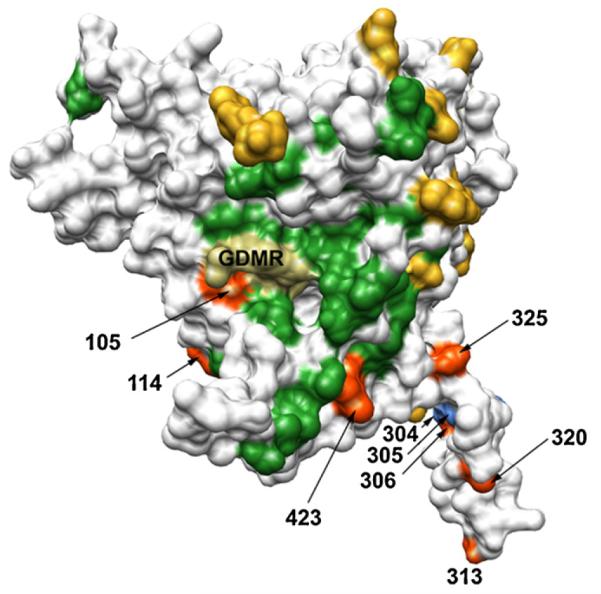
Locations of modifications on HIV-1 gp120 to focus CD4bs antibody responses: Structure of the JR-FL gp120 core (PDB ID 2B4C) denoting the locations of glycan attachment sites (naturally occurring (yellow) and those inserted for hyperglycosylation (orange)) and engineered Ala substitutions (light blue). The location of residues that upon alanine substitution result in ≥50% reduction in b12 binding to gp120 (ref. [28]) are shown in green. The location of Gly-Asp-Met-Arg (GDMR) residues at the center of the CD4bs that are necessary for binding of several non-NAbs, but not b12 (ref. [28]), is also marked (dark khaki).
To assess CD4bs exposure on mutant Q105N, the binding of a panel of mAbs was evaluated relative to wild-type JR-FL gp120. Neutralizing and non-neutralizing antibodies targeting three areas on gp120 – the CD4bs (F105, b6, b12, b13, VRC01, VRC03 and CD4-IgG2), the glycosylated ‘silent face’ (2G12) and the V3 loop (B4e8) – were assessed for binding. As expected, the mAbs bound at high levels and with high affinity to gp120wt (Fig. 2A). The only exception was VRC03. BIAcore measurements show that VRC03 binds with reasonable affinity but at poor saturation levels to monomeric gp120 [25], congruent with the results obtained here. The antibodies b6, b12, b13, VRC01 and 2G12 bound best to mutant Q105N, albeit with lower affinities than to gp120wt (Fig. 2B). Retention of b6 and b13 binding was not expected, but can be explained by their very similar mode of interaction with the CD4bs compared to b12[25,27,28,56]. CD4-IgG2, which used here as a surrogate for CD4, bound poorly to Q105N relative to gp120wt and mAbs F105 and VRC03 did not bind Q105N at all. The V3-specific antibody B4e8 did not bind to Q105N as expected given its inability to bind the parental mutant ΔN2mCHO [40].
Fig. 2.
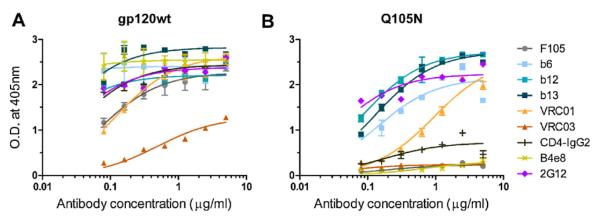
Mutant Q105N limits exposure of antibody epitopes overlapping the CD4bs. The antigenicity of hyperglycosylated mutant Q105N relative to gp120wt (JR-FL) was determined using mAbs F105, b6, VRC01, b13, B4e8, b12, 2G12, CD4-IgG2 and VRC0. CD4-IgG2 was used as a surrogate for CD4. Error bars denote the signal ranges from replicate wells.
To assess whether the changes in mAb binding were due to a glycan insertion, we also generated ΔN2mCHO variants with only an Asn at position 105 or only a Thr at position 107. The binding of b12, F105 and CD4-IgG2 was no different between the mutant ΔN2mCHO and these 2 variants (Fig. S1). From these results we concluded that it is likely indeed the specific insertion of a glycan at position 105 on gp120 that restricts access to the CD4bs. However, unlike the ΔN2mCHO-GDMR mutant, Q105N does not eliminate binding of all non-neutralizing CD4bs mAbs [40].
3.2. Adjuvants Quil A and MPL promote the elicitation of CD4bs-directed responses
Given the antigenicity of Q105N, we wished to assess the impact of adjuvant formulation on the magnitude of antibody responses. We first conducted a pilot study with gp120wt to compare the adjuvanticity of Alum (Alhydrogel) to formulations of Quil A, CpG-ODN, MPL, Alum + CpG and Quil A + MPL. As a source of MPL we used the Sigma Adjuvant System, a squalene oil-in-water emulsion containing MPL and synthetic trehalose dicorynomycolate. The composition of the Sigma Adjuvant System is analogous to that of the more commonly known Ribi Adjuvant System. The adjuvant activity of these systems is likely mediated primarily by the interaction of MPL with TLR4 [59]. Quil A is a complex mixture of chemically related saponins extracted from the bark of the Quillaja saponaria tree [60]. The adjuvant activity of Quil A, and its purified derivative QS-21, is thought to be mediated through a chemical reaction between an aldehyde group on the saponin and amino groups on certain T cell receptor molecules [61]. Except for CpG-ODN, all adjuvant formulations elicited more robust antibody responses than Alum (Fig. S2); Quil A and a mixture of Quil A plus MPL gave the most robust antibody responses. However, animals that received the latter mixture also developed swelling at the injection sites (data not shown) and thus this mixture was not considered further. We next assessed which of the adjuvants might best promote the elicitation of CD4bs antibodies by performing inhibition ELISAs with F(ab’)2 b12. The largest (and statistically significant) reduction in serum binding in the presence of F(ab’)2 b12 was observed with sera from the Quil A-only formulation (Fig. S3). Of the remaining sera, those from animals immunized with MPL showed the greatest reduction in binding in the presence of F(ab’)2 b12. However, the level of diminution was not statistically significant.
The pilot study thus favoured MPL and Quil A for improving CD4bs responses. We therefore conducted a head-to-head comparison of MPL and Quil A. We found that both gp120wt and Q105N formulated with Quil A elicited higher titres on average than when formulated with MPL (Fig. 3). However, these differences were not statistically significant. Serum antibodies elicited by gp120wt formulated in MPL as well as in Quil A bound preferentially to gp120wt (Fig. 3A), suggesting that the elicited antibodies recognize predominantly epitopes exclusive to this antigen. Anti-Q105N MPL sera also bound its homologous antigen (ΔN2mCHO(Q105N)) preferentially, suggesting the possible creation of neo-epitopes when Q105N is mixed with MPL. However, this preference was less pronounced with Q105N_QuilA sera (Fig. 3B).
Fig. 3.
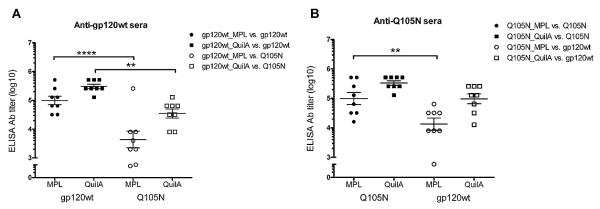
Serum antibody binding to JR-FL gp120wt and Q105N antigens. Antibodies elicited by gp120wt or Q105N formulated in adjuvant MPL bind preferentially to their respective homologous antigen. Sera from gp120wt_QuilA animals also bind preferentially to JR-FL gp120wt. Filled symbols indicate serum binding to the respective homologous antigen whereas open symbols indicate binding to heterologous antigen. Error bars denote the standard error of mean from replicate wells. Statistical analyses show that Quil A induces higher overall antibody titres regardless of antigen. **, p < 0.01; ****, p < 0.0001.
In addition to overall titres we also assessed IgG subclass composition. Sera from both Quil A-adjuvanted groups elicited strong IgG1, IgG2a and IgG2b titres whereas most MPL-adjuvanted animals only produced IgG1 antibodies (Fig. 4). These results are in accordance with Quil A’s ability to stimulate both Th1 and Th2 responses [61].
Fig. 4.
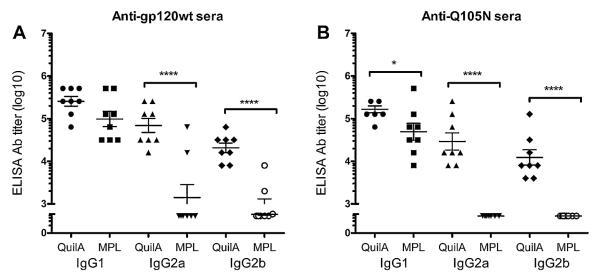
Subclass profile of immune sera. Binding of sera from mice immunized with gp120wt or Q105N was detected with noted subclass-specific secondary antibodies. The results show that Quil A elicited higher levels of IgG2 when formulated with either gp120wt or Q105N. Error bars denote the standard error of mean from replicate wells. *, p < 0.05; ****, p < 0.0001.
3.3. Q105N in Quil A adjuvant elicits higher levels of CD4bs-specific antibodies
To determine the proportion of CD4bs-specific antibodies elicited by each immunogen, we assessed serum binding to XOD6, RSC3 and RSC3Δ371I. XOD6 is a gp120 outer domain construct comprised of residues 249–485 of JR-CSF gp120; it is bound by mAbs VRC01 and b13 with moderate affinity but is not bound at all by mAbs VRC03 or b12 (Fig. S4). RSC3 is an antigenically resurfaced, conformationally stabilized gp120 core protein that exclusively presents the structurally most conserved part of the CD4bs [25]. Variant RSC3Δ371I contains a residue deletion (Ile371) that knocks out the binding of most CD4bs antibodies capable of binding RSC3 [25].
Because these three gp120 derivatives present the CD4bs in different ways, we reasoned that they would allow us to gain insight into how the elicited antibodies might be interacting with their epitopes. We found that sera from all groups bound XOD6 with reasonable affinity (Fig. 5A), though sera from Q105N_QuilA animals bound slightly better relative to sera from the 3 other groups, suggesting the somewhat greater presence of antibodies with epitopes overlapping the gp120 outer domain in this group of immunized animals. Most notably, Q105N_QuilA sera bound significantly better to RSC3 than sera from the other 3 serum groups (Fig. 5B), indicating that a comparatively larger fraction of CD4bs-specific antibodies had been elicited by specifically combining mutant Q105N with Quil A. No substantial diminution of serum binding to mutant RSC3Δ371I relative to RSC3 was observed (Fig. 5C), suggesting that the elicited antibodies do not require the presence of Ile371 for efficient binding.
Fig. 5.
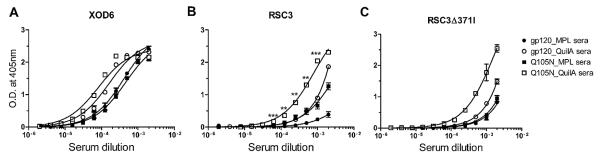
Sera from animals immunized with hyperglycosylated mutant Q105N mixed with Quil A elicit greater CD4bs-directed responses. (A) All sera bind fairly equally to construct XOD6, indicating that all contain antibodies to the outer domain portion of gp120. (B) The Q105N_QuilA formulation elicited a significantly higher proportion of antibodies to epitopes overlapping the CD4bs relative to gp120wt mixed with the same adjuvant as judged by stronger affinity for the RSC3 mutant. **, p < 0.01; ***, p < 0.001. (C) None of the sera bound significantly less to mutant RSC3Δ371I compared to RSC3, suggesting that the elicited CD4bs-specific antibodies do not require Ile371 for binding. Error bars denote the signal ranges from replicate wells.
3.4. Serum antibodies elicited by Q105N bind epitopes that partially overlap the CD4bs on gp120
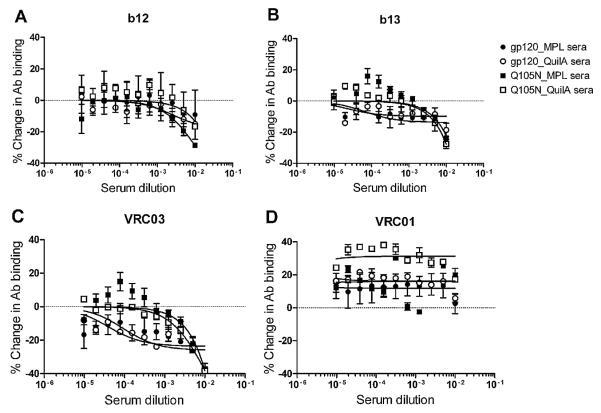
To obtain further insight into the specificity of the elicited CD4bs-specific antibodies we conducted inhibition ELISAs with CD4bs-specific mAbs. The percentage change in the binding of mAbs b12, b13, VRC01, and VRC03 to RSC3 in the presence of pooled sera was used as a measure of the antibody specificities contained in the sera. Sera from all 4 animal groups inhibited b12 binding to roughly the same extent, with the level of inhibition by the Q105N_MPL sera somewhat greater than the other 3 sera. Sera from both the Q105N_MPL and Q105N_QuilA animals inhibited the binding of mAbs b13 and VRC03 to a greater extent (30–40%) than sera from the gp120wt groups (10–20%) (Fig. 6), suggesting that the Q105N-immunized animals contain a greater abundance of antibodies to epitopes overlapping the epitopes of those two mAbs. Unexpectedly, no inhibition of VRC01 binding was observed, despite significant overlap in the binding footprint of VRC01 and VRC03 on gp120 [17]. This result is not explained by differences in affinity for RSC3 between VRC01 and VRC03; VRC01 only binds with 3-fold better affinity to RSC3 than VRC03 (5.6 nM vs. 16.1 nM), and only 5-fold better than b12 (22.4 nM) [25]. We interpret the results therefore as suggesting that the epitopes of elicited serum antibodies only partially overlap the CD4bs. MAbs binding to epitopes that partially overlap the CD4bs can inhibit the binding of some CD4bs antibodies but enhance the binding of others [62] and similar observations have been made recently with mAbs from macaques immunized with gp140 [63].
Fig. 6.
Dissection of the CD4bs specificities of serum antibodies. Sera were serially diluted (starting at 1:50) and incubated on plates for 1 h. The indicated mAbs (diluted to 10 μg/ml) were then added to the plates and these incubated for a further 1 h. MAb binding was quantified using AP-conjugated anti-human F(ab’)2-specific secondary antibody (Jackson ImmunoResearch). Sera from animals immunized with Q105N inhibited the binding of mAbs b12, b13 and VRC03 to a greater extent than gp120wt animals, suggesting that the epitopes of the serum antibodies overlap most with the epitopes of these CD4bs mAbs. Error bars denote the signal ranges from replicate wells.
3.5. Serum antibodies elicited by immunization with Q105N do not show neutralizing activity
Having determined that some level of CD4bs-directed responses was elicited with Q105N formulated with Quil A, we assessed the neutralizing activity of sera from all 4 immunization groups by testing them on tier 1B and 2 subtype B viruses, SS1196 and JR-FL, and the tier 1B subtype C virus ZM197M. All 3 viruses are sensitive, albeit at different levels, to neutralization by b12 and other CD4bs antibodies. SIVmac239 was used as a negative control. We observed low levels of neutralization with all sera against all three viruses (Fig. 7). However, neutralizing activity was also observed against the SIVmac239 control, with no statistical difference between test viruses and control. These results suggest that despite the presence of a fraction of CD4bs-like antibodies in Q105N_QuilA sera, the elicited antibodies may not be sufficiently abundant to neutralize virus effectively.
Fig. 7.
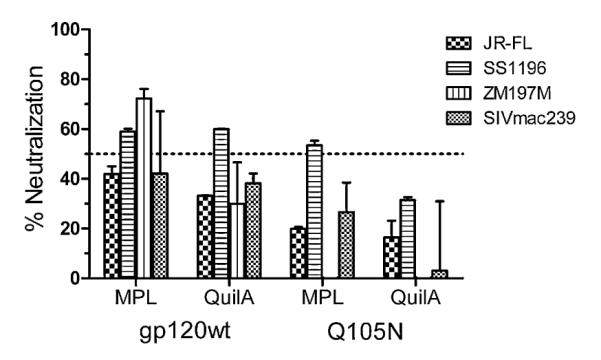
Sera from gp120wt and Q105N animals exhibit poor neutralizing activity. The neutralizing activity of the immune sera was assessed using the tier 1B viruses SS1196 (subtype B) and ZM197M (subtype C) and the tier 2 virus JR-FL (subtype B). SIVmac239 was used as a control for non-specific neutralizing activity. The sera (1:10) were mixed with an equal volume of virus and incubated for 1 h at 37 °C before adding to target cells. The dashed line denotes 50% neutralization. Error bars denote the signal ranges from replicate wells.
4. Discussion
In this study we have investigated the influence of adjuvant on the elicitation of CD4bs-specific antibody responses in combination with an immunogen hyperglycosylation strategy to focus the response. Our results show that the hyperglycosylated gp120 mutant Q105N selectively limits exposure of the CD4bs without loss of immunogenicity. Formulating this mutant with adjuvant Quil A, but not the adjuvant MPL, elicited greater CD4bs-directed antibody responses than wild-type gp120 formulated in either of the two adjuvants. However, this apparently greater response did not translate effectively into better neutralizing activity.
We should note that there is some uncertainty around the use of mice for immunization studies, as they may not be able to make the desired type of antibodies. Human and mouse antibody germline repertoires share broad similarities [64,65], providing some rationale for the use of mice. On the other hand, a significant difference between mice and human antibodies is the length of the third complementary determining region (CDR H3). Mice are known to generate antibodies with CDR H3s that are shorter relative to those characteristic of human antibodies [64,66], including those of most anti-HIV antibodies [67]. The b12:gp120 structure shows that it is the base, not the tip, of b12’s CDR H3 that makes extensive contacts with gp120, specifically with the CD4-binding loop [56]. The CDR H3 of murine antibodies, with a mean length of 12 residues [66], should be sufficiently long to make contacts with the vulnerable CD4-binding loop. Notably, the CDR H3’s of more recently isolated CD4bs antibodies such as VRC01 and 3BNC60 also exhibit lengths (12 and 10 residues, respectively) that easily fall within the capacity of the mouse antibody repertoire, demonstrating that a long CDR H3 is not an absolute requirement for a CD4bs antibody to gain effective access to gp120 on the virus. Other laboratories exploring HIV immunogen design have favoured the use of rabbits, in large part probably because rabbits can produce antibodies with longer CDR H3s [68]. However, rabbits have a limited heavy chain VHDHJH repertoire due to preferential usage of their VH1 gene segment [69–71], which may equally hamper the elicitation of the desired NAb response. A good alternative would be to immunize monkeys given that their antibody repertoire is very similar, though not identical, to that of humans [72,73]. However, larger scale immunization of monkeys is clearly impractical from a cost and availability perspective.
Our data are in general agreement with the noted influence of adjuvant on the quality of antibody responses to HIV immunogens. For example, Li et al. [74] found that certain GlaxoSmithKline adjuvants elicited higher NAb responses relative to formulations with Ribi adjuvant, which is similar in composition to the MPL adjuvant used here. Along with adjuvant, the glycosylation pattern of gp120 has been reported to impact immunogenicity [75,76]. Grundner et al. [76] showed that gp120 expressed from HEK293 cells, which impart a mixture of complex/hybrid and high-mannose glycans [77,78], elicited higher anti-gp120 antibody titres when formulated with Ribi adjuvant than gp120 from insect cells that impart mainly paucimannosidic glycans. Kong et al. [75] however observed the opposite effect, with insect cell-derived gp120 being more immunogenic than HEK293-gp120, with the notable difference that the Kong et al. study used a different set of adjuvants (Freund’s and CpG) compared to the adjuvant used in the Grunder et al. study (Ribi). We did not observe any difference in the overall level of antibody titres between gp120wt and Q105N (Fig. 3), regardless of whether formulated in MPL or Quil A adjuvant, when expressed in CHO-K1 cells. These cells impart a higher abundance of high-mannose glycans on gp120 than complex/hybrid glycans [77]. Whether the glycosylation pattern of Q105N played a pivotal role in the quality of the antibody responses elicited here remains to be determined.
Although our goal was to design an antigen that prevents binding of all poorly neutralizing CD4bs mAbs, the ability of mAbs b6 and b13, both of which neutralize poorly, to recognize mutant Q105N suggests that future studies may need to explore strategies such as heterologous prime-boosting to selectively promote antibody responses to the b12 site. Two recent reports using a prime-boosting strategy with a set of 2F5 epitope scaffolds show that such an approach can help to focus antibodies to the target epitope [79,80]. Guenaga et al. [80] also showed that attaching a promiscuous T-helper epitope to their epitope scaffolds improved titres against the target epitope, demonstrating how weak immunogenicity of a target epitope may be enhanced with certain immunopotentiators. Similar strategies may prove beneficial in promoting CD4bs-focused antibody responses.
Despite the suboptimal antigenic profile of mutant Q105N, Q105N_QuilA sera did bind strongest overall to RSC3, providing support for the use of Q105N to preferentially stimulate CD4bs responses. The observation that antibodies elicited with Q105N_QuilA bound the RSC3Δ371I mutant to similar levels as the parent molecule RSC3 was unexpected given that this residue is normally a critical binding residue for CD4bs antibodies [28]. However, we have found that the recently described broadly neutralizing CD4bs antibody HJ16 [7] also is not sensitive to the I371 deletion, demonstrating that not all CD4bs antibodies necessarily interact with this residue. Unfortunately, due to serum volume limitations we were not able to assess whether HJ16 binding was also blocked by the Q105N_QuilA serum antibodies.
Disappointingly, we observed no significant neutralizing activity with the Q105N_QuilA sera or any of the other 3 sera against the tier 1 and 2 subtype B and C pseudoviruses tested. The lack of significant neutralizing activity may be due to the relatively low abundance of CD4bs-specific antibodies that were elicited, as suggested by the shallow slope of the binding curves for the resurfaced core protein RSC3 (Fig. 5). Our analyses were confounded by apparent neutralizing activity against the control virus SIVmac239. Due to volume limitations we were unable to investigate the possible reason(s) for this phenomenon, including purification of serum IgG for follow-up experiments. However, it has been suggested that complement activation may lead to lysis and inactivation of virus, including SIV, particularly with sera from animals such as mice and rabbits [81,82]. Longer heat-inactivation times (1 h rather than the more common 30 min as used here) may therefore be necessary to ensure complete inactivation of complement prior to performing neutralization assays.
In summary, we have shown that hyperglycosylated gp120 mutant Q105N can be used to induce relatively greater responses to the CD4bs, particularly when formulated with adjuvant Quil A. However, the level of CD4bs-directed antibodies was not as high as would be desired. Future studies will therefore explore additional immunostimulants and immunogen formulations to boost the desired CD4bs-specific antibody response.
Supplementary Material
Acknowledgements
Financial support for this study was provided by CIHR operating grant 200100 and a CIHR New Investigator Award (to R.P.) and infrastructure development funding from the Canada Foundation for Innovation and the British Columbia Knowledge Development Fund (to R.P.). Financial support for some of the pilot studies was provided by the IAVI Neutralizing Antibody Consortium and by NIH grants AI33292 and AI60425 (to D.R.B.). MAbs VRC01, VRC03, CD4-IgG2, and HJ16 as well as antigens RSC3 and RSC3Δ371I were obtained via the NIH AIDS Research and Reference Reagent Program from John Mascola (VRC01 and 03), Progenics Pharmaceuticals (CD4-IgG2), Antonio Lanzavecchia (HJ16), and Zhi-Yong Yang, Peter Kwong, and Gary Nabel (RSC3 and Δ371I). We thank Peter Kwong and John Mascola (Vaccine Research Center, NIH) for providing additional VRC01, Lisa Cavacini (Beth Israel Deaconess Medical Center) for mAbs B4e8 and F105, and Dietmar and Hermann Katinger (Polymun Scientific GmbH, Austria) for the generous gift of 2G12. We also thank Kate Auyeung for assistance with immunogen preparation, James Kwon, Jose Tam-Huang and Dennis Chau for assistance with cloning, plasmid preparation and protein purification, Savrina Manhas for assistance with neutralization assays and Kevin Henry for critical review of the manuscript during the draft stage.
Footnotes
Appendix A. Supplementary data Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.vaccine.2011.11.089.
References
- [1].Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu Rev Immunol. 2010;28:413–44. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- [2].McElrath MJ, Haynes BF. Induction of immunity to human immunodeficiency virus Type-1 by vaccination. Immunity. 2010;33:542–54. doi: 10.1016/j.immuni.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine. Nat Med. 2009;15:866–70. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- [4].Verkoczy L, Kelsoe G, Moody MA, Haynes BF. Role of immune mechanisms in induction of HIV-1 broadly neutralizing antibodies. Curr Opin Immunol. 2011;23:383–90. doi: 10.1016/j.coi.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–8. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- [6].Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, et al. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82:11651–68. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Corti D, Langedijk JPM, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One. 2010;5:e8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13:1032–4. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li Y, Svehla K, Louder MK, Wycuff D, Phogat S, Tang M, et al. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J Virol. 2009;83:1045–59. doi: 10.1128/JVI.01992-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol. 2009;83:757–69. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sather DN, Stamatatos L. Epitope specificities of broadly neutralizing plasmas from HIV-1 infected subjects. Vaccine. 2010;28:B8–12. doi: 10.1016/j.vaccine.2009.07.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, et al. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dhillon AK, Donners H, Pantophlet R, Johnson WE, Decker JM, Shaw GM, et al. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J Virol. 2007;81:6548–62. doi: 10.1128/JVI.02749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tomaras GD, Binley JM, Gray ES, Crooks ET, Osawa K, Moore PL, et al. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J Virol. 2011;85:11502–19. doi: 10.1128/JVI.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Olivera TY, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;16:1633–7. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–70. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gray ES, Taylor N, Wycuff D, Moore PL, Tomaras GD, Wibmer CK, et al. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J Virol. 2009;83:8925–37. doi: 10.1128/JVI.00758-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nandi A, Lavine CL, Wang P, Lipchina I, Goepfert PA, Shaw GM, et al. Epitopes for broad and potent neutralizing antibody responses during chronic infection with human immunodeficiency virus type 1. Virology. 2010;396:339–48. doi: 10.1016/j.virol.2009.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15:951–4. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hessell AJ, Hangartner L, Hunter M, Havenith CE, Beurskens FJ, Bakker JM, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–4. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- [22].Parren PW, Marx PA, Hessell AJ, Luckay A, Harouse J, Cheng-Mayer C, et al. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J Virol. 2001;75:8340–7. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Burton DR, Hessell AJ, Keele BF, Klasse PJ, Ketas TA, Moldt B, et al. Limited or no protection by weakly or nonneutralizing antibodies against vaginal SHIV challenge of macaques compared with a strongly neutralizing antibody. Proc Natl Acad Sci U S A. 2011;108:11181–6. doi: 10.1073/pnas.1103012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ng CT, Jaworski JP, Jayaraman P, Sutton WF, Delio P, Kuller L, et al. Passive neutralizing antibody controls SHIV viremia and enhances B cell responses in infant macaques. Nat Med. 2010;16:1117–9. doi: 10.1038/nm.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, Seaman MS, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–61. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhou T, Georgiev I, Wu X, Yang Z-Y, Dai K, Finzi A, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;29:811–7. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen L, Do Kwon Y, Zhou T, Wu X, O’Dell S, Cavacini L, et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–7. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pantophlet R, Ollmann Saphire E, Poignard P, Parren PW, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J Virol. 2003;77:642–58. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zwick MB, Parren PW, Saphire EO, Church S, Wang M, Scott JK, et al. Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. J Virol. 2003;77:5863–76. doi: 10.1128/JVI.77.10.5863-5876.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wu X, Zhou T, O’Dell S, Wyatt RT, Kwong PD, Mascola JR. Mechanism of human immunodeficiency virus type 1 resistance to monoclonal antibody b12 that effectively targets the site of CD4 attachment. J Virol. 2009;83:10892–907. doi: 10.1128/JVI.01142-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Klein JS, Gnanapragasam PNP, Galimidi RP, Foglesong CP, West AP, Bjorkman PJ. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc Natl Acad Sci U S A. 2009;106:7385–90. doi: 10.1073/pnas.0811427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Walker LM, Burton DR. Rational antibody-based HIV-1 vaccine design: current approaches and future directions. Curr Opin Immunol. 2010;22:358–66. doi: 10.1016/j.coi.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Phogat S, Wyatt R. Rational modifications of HIV-1 envelope glycoproteins for immunogen design. Curr Pharm Des. 2007;13:213–27. doi: 10.2174/138161207779313632. [DOI] [PubMed] [Google Scholar]
- [34].Dosenovic P, Chakrabarti B, Soldemo M, Douagi I, Forsell MN, Li Y, et al. Selective expansion of HIV-1 envelope glycoprotein-specific B cell subsets recognizing distinct structural elements following immunization. J Immunol. 2009;183:3373–82. doi: 10.4049/jimmunol.0900407. [DOI] [PubMed] [Google Scholar]
- [35].Douagi I, Forsell MNE, Sundling C, O’Dell S, Feng Y, Dosenovic P, et al. Influence of novel CD4 binding-defective HIV-1 envelope glycoprotein immunogens on neutralizing antibody and T-cell responses in nonhuman primates. J Virol. 2010;84:1683–95. doi: 10.1128/JVI.01896-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sundling C, Forsell MNE, O’Dell S, Feng Y, Chakrabarti B, Rao SS, et al. Soluble HIV-1 Env trimers in adjuvant elicit potent and diverse functional B cell responses in primates. J Exp Med. 2010;207:2003–17. doi: 10.1084/jem.20100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wu L, Zhou T, Yang Z-y, Svehla K, O’Dell S, Louder MK, et al. Enhanced exposure of the CD4-binding site to neutralizing antibodies by structural design of a membrane-anchored human immunodeficiency virus type 1 gp120 domain. J Virol. 2009;83:5077–86. doi: 10.1128/JVI.02600-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pantophlet R, Wilson IA, Burton DR. Hyperglycosylated mutants of human immunodeficiency virus (HIV) type 1 monomeric gp120 as novel antigens for HIV vaccine design. J Virol. 2003;77:5889–901. doi: 10.1128/JVI.77.10.5889-5901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bhattacharyya S, Rajan RE, Swarupa Y, Rathore U, Verma A, Udaykumar R, et al. Design of a non-glycosylated outer domain-derived HIV-1 gp120 immunogen that binds to cd4 and induces neutralizing antibodies. J Biol Chem. 2010;285:27100–10. doi: 10.1074/jbc.M110.152272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pantophlet R, Wilson IA, Burton DR. Improved design of an antigen with enhanced specificity for the broadly HIV-neutralizing antibody b12. Protein Eng Des Sel. 2004;17:749–58. doi: 10.1093/protein/gzh085. [DOI] [PubMed] [Google Scholar]
- [41].Pietzsch J, Scheid JF, Mouquet H, Klein F, Seaman MS, Jankovic M, et al. Human anti-HIV-neutralizing antibodies frequently target a conserved epitope essential for viral fitness. J Exp Med. 2010;207:1995–2002. doi: 10.1084/jem.20101176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Selvarajah S, Puffer B, Pantophlet R, Law M, Doms RW, Burton DR. Comparing antigenicity and immunogenicity of engineered gp120. J Virol. 2005;79:12148–63. doi: 10.1128/JVI.79.19.12148-12163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Delves PJ, Lund T, Roitt IM. Can epitope-focused vaccines select advantageous immune responses. Mol Med Today. 1997;3:55–60. doi: 10.1016/s1357-4310(96)20036-x. [DOI] [PubMed] [Google Scholar]
- [44].Garrity RR, Rimmelzwaan G, Minassian A, Tsai WP, Lin G, de Jong JJ, et al. Refocusing neutralizing antibody response by targeted dampening of an immunodominant epitope. J Immunol. 1997;159:279–89. [PubMed] [Google Scholar]
- [45].Tobin GJ, Trujillo JD, Bushnell RV, Lin G, Chaudhuri AR, Long J, et al. Deceptive imprinting and immune refocusing in vaccine design. Vaccine. 2008;26:6189–99. doi: 10.1016/j.vaccine.2008.09.080. [DOI] [PubMed] [Google Scholar]
- [46].Nara PL, Tobin GJ, Chaudhuri AR, Trujillo JD, Lin G, Cho MW, et al. How can vaccines against influenza and other viral diseases be made more effective? PLoS Biol. 2010;8:e1000571. doi: 10.1371/journal.pbio.1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Dey AK, Srivastava IK. Novel adjuvants and delivery systems for enhancing immune responses induced by immunogens. Expert Rev Vaccines. 2011;10:227–51. doi: 10.1586/erv.10.142. [DOI] [PubMed] [Google Scholar]
- [48].Mbow ML, De Gregorio E, Valiante NM, Rappuoli R. New adjuvants for human vaccines. Curr Opin Immunol. 2010;22:411–6. doi: 10.1016/j.coi.2010.04.004. [DOI] [PubMed] [Google Scholar]
- [49].Steinhagen F, Kinjo T, Bode C, Klinman DM. TLR-based immune adjuvants. Vaccine. 2011;29:3341–55. doi: 10.1016/j.vaccine.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Coller B-AG, Clements DE, Bett AJ, Sagar SL, Ter Meulen JH. The development of recombinant subunit envelope-based vaccines to protect against dengue virus induced disease. Vaccine. 2011;29:7267–75. doi: 10.1016/j.vaccine.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Qian F, Rausch KM, Muratova O, Zhou H, Song G, Diouf A, et al. Addition of CpG ODN to recombinant Pseudomonas aeruginosa ExoProtein A conjugates of AMA1 and Pfs25 greatly increases the number of responders. Vaccine. 2008;26:2521–7. doi: 10.1016/j.vaccine.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mullen GE, Aebig JA, Dobrescu G, Rausch K, Lambert L, Long CA, et al. Enhanced antibody production in mice to the malaria antigen AMA1 by CPG 7909 requires physical association of CpG and antigen. Vaccine. 2007;25:5343–7. doi: 10.1016/j.vaccine.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Aebig JA, Mullen GE, Dobrescu G, Rausch K, Lambert L, Ajose-Popoola O, et al. Formulation of vaccines containing CpG oligonucleotides and alum. J Immunol Methods. 2007;323:139–46. doi: 10.1016/j.jim.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bell CH, Pantophlet R, Schiefner A, Cavacini LA, Stanfield RL, Burton DR, et al. Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization. J Mol Biol. 2008;375:969–78. doi: 10.1016/j.jmb.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zwick MB, Wang M, Poignard P, Stiegler G, Katinger H, Burton DR, et al. Neutralization synergy of human immunodeficiency virus type 1 primary isolates by cocktails of broadly neutralizing antibodies. J Virol. 2001;75:12198–208. doi: 10.1128/JVI.75.24.12198-12208.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–7. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–59. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–11. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- [59].Evans JT, Cluff CW, Johnson DA, Lacy MJ, Persing DH, Baldridge JR. Enhancement of antigen-specific immunity via the TLR4 ligands MPL adjuvant and Ribi.529. Expert Rev Vaccines. 2003;2:219–29. doi: 10.1586/14760584.2.2.219. [DOI] [PubMed] [Google Scholar]
- [60].Kensil CR. Saponins as vaccine adjuvants. Crit Rev Ther Drug Carrier Syst. 1996;13:1–55. [PubMed] [Google Scholar]
- [61].Sun HX, Xie Y, Ye YP. Advances in saponin-based adjuvants. Vaccine. 2009;27:1787–96. doi: 10.1016/j.vaccine.2009.01.091. [DOI] [PubMed] [Google Scholar]
- [62].Moore JP, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70:1863–72. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Sundling C, Li Y, Huynh N, Wilson R, O’Dell S, Mascola J, et al. Abstract OA04.04 – characterization of CD4-binding site directed monoclonal antibodies isolated from HIV-1 gp140 Env immunized rhesus macaques. Abstracts from AIDS Vaccine 2011 Bangkok, Thailand, 12–15 September, 2011. AIDS Res Hum Retroviruses. 2011;27:A-1–148. [Google Scholar]
- [64].Schroeder JHW. Similarity and divergence in the development and expression of the mouse and human antibody repertoires. Dev Comp Immunol. 2006;30:119–35. doi: 10.1016/j.dci.2005.06.006. [DOI] [PubMed] [Google Scholar]
- [65].Collis AVJ, Brouwer AP, Martin ACR. Analysis of the antigen combining site: correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J Mol Biol. 2003;325:337–54. doi: 10.1016/s0022-2836(02)01222-6. [DOI] [PubMed] [Google Scholar]
- [66].Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, et al. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J Mol Biol. 2003;334:733–49. doi: 10.1016/j.jmb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- [67].Breden F, Lepik C, Longo NS, Montero M, Lipsky PE, Scott JK. Comparison of antibody repertoires produced by HIV-1 infection, other chronic and acute infections, and systemic autoimmune disease. PLoS One. 2011;6:e16857. doi: 10.1371/journal.pone.0016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Popkov M, Mage RG, Alexander CB, Thundivalappil S, Barbas CF, 3rd, Rader C. Rabbit immune repertoires as sources for therapeutic monoclonal antibodies: the impact of kappa allotype-correlated variation in cysteine content on antibody libraries selected by phage display. J Mol Biol. 2003;325:325–35. doi: 10.1016/s0022-2836(02)01232-9. [DOI] [PubMed] [Google Scholar]
- [69].Mage RG, Lanning D, Knight KL. B cell and antibody repertoire development in rabbits: the requirement of gut-associated lymphoid tissues. Dev Comp Immunol. 2006;30:137–53. doi: 10.1016/j.dci.2005.06.017. [DOI] [PubMed] [Google Scholar]
- [70].Mehr R, Edelman H, Sehgal D, Mage R. Analysis of mutational lineage trees from sites of primary and secondary Ig gene diversification in rabbits and chickens. J Immunol. 2004;172:4790–6. doi: 10.4049/jimmunol.172.8.4790. [DOI] [PubMed] [Google Scholar]
- [71].Dennis Lanning XZ, Zhai S-K, Katherine LK. Development of the antibody repertoire in rabbit: gut-associated lymphoid tissue, microbes, and selection. Immunol Rev. 2000;175:214–28. [PubMed] [Google Scholar]
- [72].Yuan T, Li J, Zhang Y, Wang Y, Streaker E, Dimitrov DS, et al. Putative rhesus macaque germline predecessors of human broadly HIV-neutralizing antibodies: Differences from the human counterparts and implications for HIV-1 vaccine development. Vaccine. 2011;29:6903–10. doi: 10.1016/j.vaccine.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Link JM, Larson JE, Schroeder HW. Despite extensive similarity in germline DH and JH sequence, the adult Rhesus macaque CDR-H3 repertoire differs from human. Mol Immunol. 2005;42:943–55. doi: 10.1016/j.molimm.2004.09.027. [DOI] [PubMed] [Google Scholar]
- [74].Li Y, Svehla K, Mathy NL, Voss G, Mascola JR, Wyatt R. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J Virol. 2006;80:1414–26. doi: 10.1128/JVI.80.3.1414-1426.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Kong L, Sheppard NC, Stewart-Jones GB, Robson CL, Chen H, Xu X, et al. Expression-system-dependent modulation of HIV-1 envelope glycoprotein antigenicity and immunogenicity. J Mol Biol. 2010;403:131–47. doi: 10.1016/j.jmb.2010.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Grundner C, Pancera M, Kang JM, Koch M, Sodroski J, Wyatt R. Factors limiting the immunogenicity of HIV-1 gp120 envelope glycoproteins. Virology. 2004;330:233–48. doi: 10.1016/j.virol.2004.08.037. [DOI] [PubMed] [Google Scholar]
- [77].Raska M, Takahashi K, Czernekova L, Zachova K, Hall S, Moldoveanu Z, et al. Glycosylation patterns of HIV-1 gp120 depend on the type of expressing cells and affect antibody recognition. J Biol Chem. 2010;285:20860–9. doi: 10.1074/jbc.M109.085472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, Burton DR, et al. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci U S A. 2010;107:13800–5. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Ofek G, Guenaga FJ, Schief WR, Skinner J, Baker D, Wyatt R, et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci U S A. 2010;107:17880–7. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Guenaga J, Dosenovic P, Ofek G, Baker D, Schief WR, Kwong PD, et al. Heterologous epitope-scaffold prime-boosting immuno-focuses B cell responses to the HIV-1 gp41 2F5 neutralization determinant. PLoS One. 2011;6:e16074. doi: 10.1371/journal.pone.0016074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Montefiori D. Evaluating neutralizing antibodies against HIV, SIV and SHIV in luciferase reporter gene assays. In: Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, Coico R, editors. Current protocol in immunology. John Wiley & Sons; New York: 2004. pp. 12.1.1–5. [DOI] [PubMed] [Google Scholar]
- [82].Hosoi S, Borsos T, Dunlop N, Heat-Labile NPL. Complement-like factor(s) of animal sera prevent(s) HIV-1 infectivity in vitro. J Acquir Immune Defic Syndr. 1990;3:366–71. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.