Summary
HIV-1 resulted from cross-species transmission of SIVcpz, a simian immunodeficiency virus that naturally infects chimpanzees. SIVcpz, in turn, is a recombinant between two SIV lineages from old world monkeys. Lentiviral inter-species transmissions are partly driven by the evolution and capacity of viral accessory genes, such as vpx, vpr or vif, to antagonize host antiviral factors, like SAMHD1 and the APOBEC3 proteins. We show that vpx, which, in other lentiviruses, antagonizes SAMHD1, was deleted during the creation of SIVcpz. This genomic deletion resulted in the reconstruction of the overlapping vif gene by “overprinting”, creating a unique vif that overlaps in its 3’end with the vpr gene and can antagonize hominid APOBEC3s. Moreover, passage of SIVs through chimpanzees facilitated subsequent adaptation of HIV-1 to humans. Thus, HIV-1 originated through a series of gene loss and adaptation events that generated its chimpanzee precursor and lowered the species barrier to human infection.
Introduction
The human immunodeficiency virus type 1 (HIV-1) is the result of cross-species transmissions of simian immunodeficiency viruses (SIVs) from African apes to humans. SIVcpz strains from chimpanzees (Pan troglodytes) were transmitted on at least two occasions to human, including the cross-species transmission of the precursor of HIV-1 group M that spawned the current AIDS pandemic (Keele et al., 2006; Sharp and Hahn, 2011). SIVcpz, in turn, originated from inter-species transmissions and recombination events involving the ancestors of at least two distant SIV lineages: SIVrcm from red-capped mangabeys (RCM) and SIVmus/mon/gsn from Cercopithecus monkeys (Bailes et al., 2003). While the adaptation of lentiviruses from chimpanzees to humans has been described (Kirchhoff, 2010), the transmission and adaptive processes of SIVs from monkeys to chimpanzees, which underlie the ultimate origin of HIV-1, are not well understood.
Host susceptibility to viral infections and the likelihood of lentiviral transmission from one primate species to another is partially governed by the antiviral proteins produced by the innate immune system of the host. These proteins, also called restriction factors, inhibit different stages of lentiviral replication and are usually counteracted in a species-specific manner by viral accessory proteins (Duggal and Emerman, 2012). One well-described antagonism is the degradation of the host cytidine deaminase APOBEC3G protein (A3G) by the viral infectivity protein Vif (Bishop et al., 2008; Mangeat et al., 2003; Sheehy et al., 2002). In the absence of Vif, A3G is packaged into assembling virions and transferred to target cells, where it produces hypermutation in the viral genome. However, during infection, Vif interacts with A3G, recruits it to an ubiquitin ligase complex to target A3G for proteasomal degradation, which prevents the encapsidation of A3G, and eventually allow viral replication in the new target cells. A more recently identified virus-host antagonism is the degradation of the host protein SAMHD1 by the accessory proteins Vpx and Vpr to allow the virus to efficiently infect myeloid and resting T cells (Baldauf et al., 2012; Hrecka et al., 2011; Laguette et al., 2011; Lim et al., 2012). The capacity to antagonize the host SAMHD1 was acquired by the vpr gene during primate lentiviral evolution. Subsequently, the recombination/duplication of the vpr gene led to the acquisition of a vpx gene in SIVrcm and SIVsmm from sooty mangabeys (Lim et al., 2012). The two viral lineages that recombined to give rise to SIVcpz have the capacity to degrade their respective SAMHD1 proteins: SIVrcm uses its Vpx protein, while members of the SIVmus/mon/gsn lineage use their Vpr protein to antagonize their hosts’ restriction factor (Lim et al., 2012). In contrast, neither HIV-1 nor SIVcpz, its immediate precursor, have the ability to degrade SAMHD1 since they do not encode a vpx gene, and their Vpr protein does not antagonize SAMHD1 (Laguette et al., 2011; Lim et al., 2012). Thus, one seemingly important function to antagonize a host restriction factor was lost during the process of primate lentivirus adaptation from monkeys to hominids, but the mechanism and the reason for this loss remain unknown.
Here, we analyzed the viral genomic reorganization and functional consequences that occurred during the transmission of lentiviruses from old world monkeys (OWMs) to hominids in order to understand the selective pressures leading to the ultimate origin of HIV-1 strains. We found that the vpx gene was entirely lost during the birth of SIVcpz and led to the absence of vpx in HIV-1. Furthermore, this loss was associated with the reconstruction of the overlapping vif gene. This Vif protein, unique to SIVcpz and its descendants, gained the function to fully antagonize the hominid A3G proteins. Finally, we found that chimpanzees represented a means for certain lentiviruses to adapt to hominids, which was likely essential for their subsequent transmission to humans. Our study elucidates how the HIV-1 lineage, leading to the emergence of a pandemic virus in the human population, had its ultimate origin in a monkey to hominid cross-species transmission that involved the loss of a viral gene, the creation of a distinctive 3’end region to an existing gene, and subsequent adaptation in chimpanzees.
Results
The absence of vpx in HIV-1 results from the loss of the entire gene during the genesis of SIVcpz
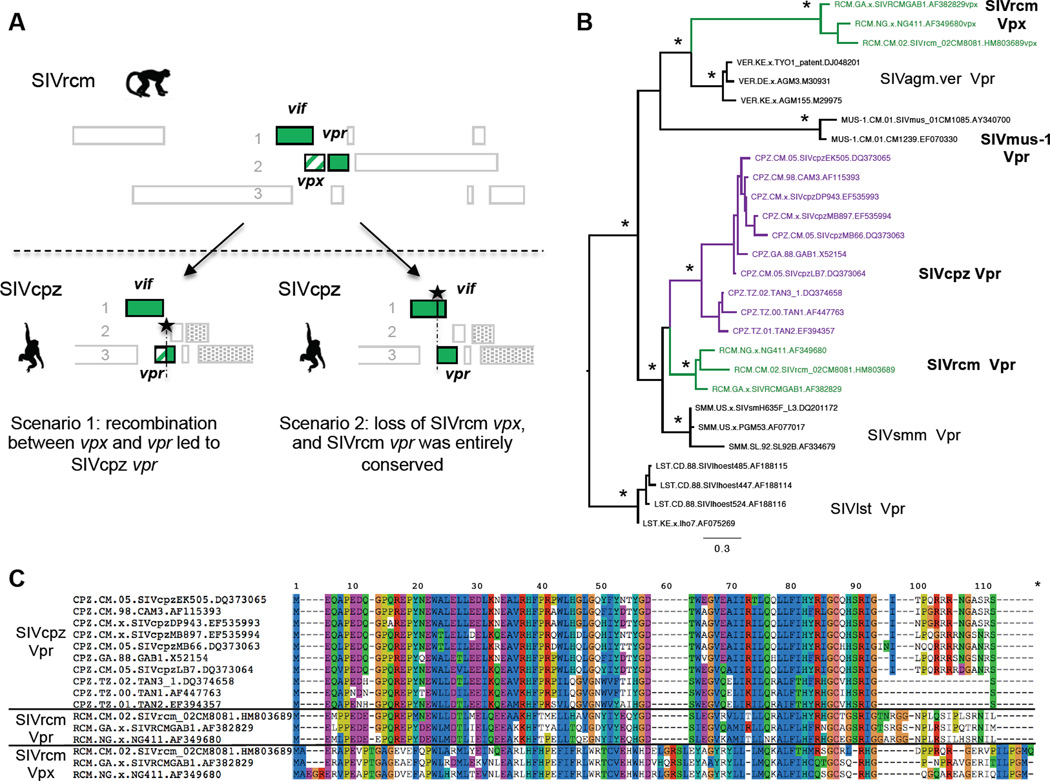
Phylogenetic analyses of the SIVcpz genome previously showed that vif and vpr, amongst other genes, originated from an ancient SIVrcm-like virus, while vpu and env genes came from an ancient SIVmus/mon/gsn-like strain (Sharp and Hahn, 2011), and these conclusions were confirmed by analyses including all the SIV lineages characterized to date (data not shown). However, SIVrcm encodes for a vpx gene between vif and vpr, while vpx is absent in SIVcpz (Figure 1A). Thus, we asked how and why the vpx gene is absent in SIVcpz. We first considered the possibility that the ancestor of the modern day SIVrcm lacked the vpx gene when it crossed to chimpanzees. Therefore, we analyzed an alignment of the surrounding regions of vpx to look for any evidence of a recent vpx gene transfer to SIVrcm (details of the hypotheses and the analyses are in Figure S1). The absence of recombination marks and the phylogenetic tree topologies revealed the lack of evidence for any vpx gene transfer between SIVsmm and SIVrcm, showing that vpx has been acquired by the SIVrcm and SIVsmm lineages before their divergence (Figure S1). This indicates that the SIVrcm-like strain, which recombined with a SIVmus-like strain to give rise to SIVcpz, encoded a vpx gene between vif and vpr. Thus, SIVcpz lost the vpx gene from the ancestral SIVrcm-like virus.
Figure 1. The entire vpx gene was lost in the lineage that gave rise to SIVcpz, SIVgor, and HIV-1. See also Figure S1.
A- Two possible scenarios leading to the absence of vpx in SIVcpz. Only the region of interest spanning vif, vpx, and vpr is detailed. The horizontal dashed line represents the transmission from red-capped mangabeys (top) to chimpanzees (bottom). The potential recombination breakpoints of interest are represented by plain stars and the vpx gene is highlighted by striped lines. In the SIVcpz genome, the dotted regions correspond to the vpu gene and the env gene that are originated from SIVmus/mon/gsn-like viruses. The numbers correspond to the three reading frames.
B- Vpr from SIVcpz is more closely related to SIVrcm Vpr than SIVrcm Vpx, or SIVmus Vpr. The phylogenetic analysis was performed from an alignment of full-length Vpr and Vpx from various SIVs (trimmed alignment of 76 amino acids). The name and the accession number of each SIV are at the tip of each branch. Sequences were retrieved from www.hiv.lanl.gov. The asterisks show bootstrap values superior to 85%.
C- The N-ter region of SIVcpz Vpr is closely related to the N-ter region of SIVrcm Vpr. Amino acid alignment of full-length SIVcpz Vpr, SIVrcm Vpr, and SIVrcm Vpx.
We then considered two main scenarios for how the vpx gene was lost during the origin of SIVcpz (Figure 1A). First, it was possible that recombination occurred between the paralogous genes vpr and vpx, leading to just one gene in SIVcpz (scenario 1) where the 5’end derives from SIVrcm vpx and the 3’end derives from SIVrcm vpr; or, second, that the entire vpx was deleted in SIVcpz while the vpr gene from SIVrcm remained intact in its entirety (scenario 2) (Figure 1A). The difference between these two scenarios is important because Vpr and Vpx perform different functions (Ayinde et al., 2010), and recombination between the genes (scenario 1) would imply that some of the functional domains of both genes were conserved, while direct deletion of vpx (scenario 2) would imply that only the functions of one gene, vpr, were conserved.
We found multiple lines of evidence that scenario 1 is not plausible, while scenario 2 is very likely. First, phylogenetic analysis of Vpr and Vpx from diverse SIVs showed that SIVcpz Vpr is closely related to SIVrcm Vpr and both cluster distantly from SIVrcm Vpx (Figure 1B). Second, if SIVcpz Vpr was a recombinant between SIVrcm Vpx and Vpr (Scenario 1), then one would expect the N-terminal (N-ter) region of SIVcpz Vpr to show more similarity with Vpx than Vpr (Figure 1A). However, when we aligned sequences of SIVcpz Vpr with sequences of SIVrcm Vpr and Vpx, we found that this was not true and that, instead, the N-ter of SIVrcm and SIVcpz Vpr shared higher similarity (the SIVcpz vpr gene has 54% identity with SIVrcm vpr vs. 39% with SIVrcm vpx) (Figure 1C). Third, we found no evidence of ancient recombination events within SIVcpz vpr, nor any remnant genomic region of SIVrcm vpx in SIVcpz (see experimental procedures). These results exclude the first scenario of recombination between vpr and vpx, and favor a scenario whereby the entire vpx gene was lost in SIVcpz.
The loss of vpx led to the creation of a vif with a unique 3’ terminal region by “overprinting”
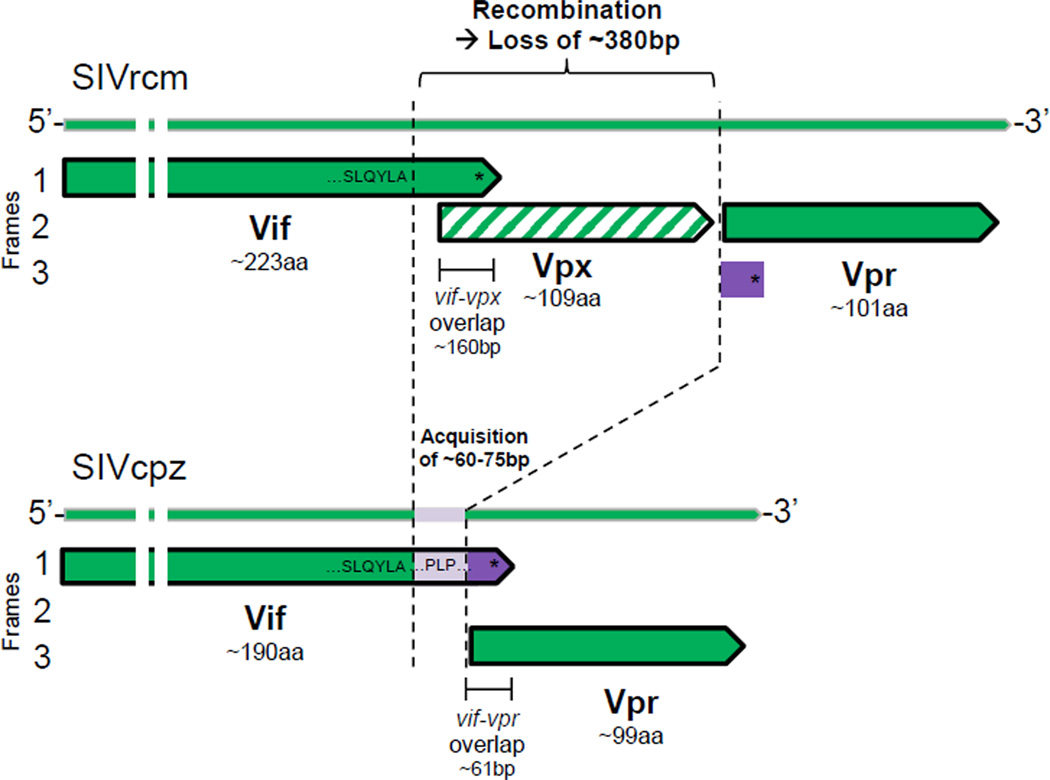
To determine the extent of the deletion that led to the loss of vpx, the impact on the overlapping genes, as well as the associated genomic modifications, we analyzed the region spanning vif, vpx, and vpr. In SIVrcm, the 5’end of vpx overlaps with the 3’end of vif by approximately 160 bp (Figure 2). However, in SIVcpz, we found that the loss of vpx led to the loss of this entire overlapping region including the stop codon for vif (Figure 2). Thus, we speculated that the 3’end truncation of the vif gene led to its reconstruction in an alternate reading frame of the vpr gene by a mechanism called “overprinting”. Overprinting is the process by which a sequence, which originally encodes for only one protein, undergoes modifications leading to an additional second open reading frame (ORF) (Keese and Gibbs, 1992). Indeed, we found that during the recombination events that generated SIVcpz, the 3’end of vif was reconstituted by overprinting of the 5’end of SIVrcm vpr (ORF 2) (Figure 2, frame 3 in SIVrcm, fragment in purple). Here, in frame 3 of SIVrcm, a preexisting stop codon within the SIVrcm vpr gene (but not in-frame with SIVrcm vpr), served as the stop codon for SIVcpz vif after the deletion of vpx from SIVrcm (Figure 2 and S2). Hence, in SIVcpz, the 3’end of vif (overprinting region) overlaps with the 5’end of vpr by approximately 61 bp in an alternative reading frame (dark purple, Figure 2). Furthermore, upstream of the overprinted region of SIVcpz vif, a sequence of 60–75 bp in SIVcpz was created (light purple, Figure 2) for which we could find no homology with any genomic region of SIVrcm, nor homology to any other nucleotide sequence present in any sequenced primate genome or lentivirus (see experimental procedures). These Vif sequences are unique to SIVcpz and its related strains, SIVgor and HIV-1. Importantly, this fragment harbors the “cullin box”, which includes the PPLP motif and surrounding residues that are highly conserved in HIV-1 and are necessary for HIV-1 Vif to efficiently degrade human A3G (Bergeron et al., 2010; Donahue et al., 2008; Walker et al., 2010). Thus, SIVcpz Vif and therefore HIV-1 Vif, acquired a unique C-ter domain with sequences important for their protein function during the loss of Vpx.
Figure 2. The loss of vpx had major consequences for vif. See also Figure S2.
The 3’end of SIVrcm vif was reconstructed when SIVrcm vpx was lost during the birth of SIVcpz. Representations of SIVrcm (top) and SIVcpz (bottom) genomes in the region spanning the vif, vpx (in stripped lines), and vpr open reading frames (ORFs). The approximate length of each protein is given (aa, amino acids). The reading frames are given on the left (1 to 3) and the proteins are represented by large plain arrows. Green stands for proteins and nucleotides related to SIVrcm; purple stands for sequences that were not expressed in SIVrcm but were in an ORF in SIVcpz (overprinting region); light purple stands for new amino acids in SIVcpz. Asterisks are stop codons. Important nucleotide and amino acid motifs or regions are given as well as gene overlaps. The dashed lines represent breakpoints. The white lines in the 5’ region of the arrows indicate regions that were cut for representation purposes only.
SIVrcm vif adapted to efficiently antagonize chimpanzee A3G and A3D, while SAMHD1-antagonism was not retained
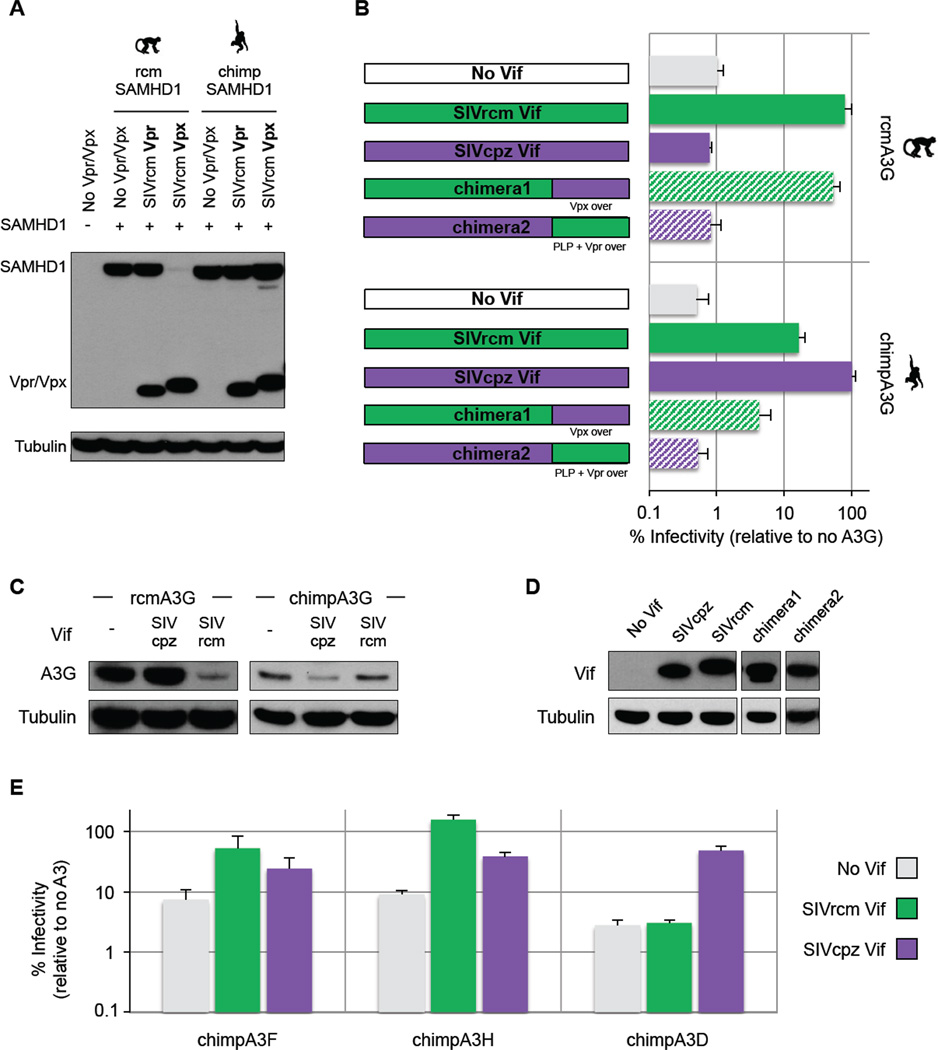
The capacity of Vif to antagonize host A3G is a feature conserved throughout SIVs and primates, however this antagonism is usually species-specific (Compton and Emerman, 2013). We hypothesized that the loss of vpx during the birth of SIVcpz was driven by selection for changes in the overlapping reading frame that encodes Vif, i.e. the ability of Vif to antagonize chimpanzee A3G was of greater importance than the need to retain Vpx for the purpose of SAMHD1 antagonism. In support of this hypothesis we found that neither SIVrcm Vpx nor SIVrcm Vpr were able to degrade the chimpanzee SAMHD1 (Figure 3A), although, as expected (Lim et al., 2012), SIVrcm Vpx, but not SIVrcm Vpr, had the capacity to degrade SAMHD1 from RCM (Figure 3A). Thus, the absence of SAMHD1-antagonism presumably had little consequence for the initial transmission of SIV from monkeys to chimpanzees.
Figure 3. SIVrcm Vif adaptation to the hominid A3G. See also Figure S3.
A- Vpx and Vpr from SIVrcm do not degrade chimpanzee SAMHD1. The ability of SIVrcm to degrade SAMHD1 was assayed by western-blot analyses of HA-tag SAMHD1 from red-capped mangabey (rcm) or chimpanzee (chimp) (+, presence; -, empty plasmid) cotransfected with or without 3xFLAG-tag Vpr or Vpx from SIVrcmNG411. Tubulin was probed as a loading control.
B- SIVrcm Vif has some activity against chimpanzee A3G to rescue viral infection, but adaptions were needed for full antagonism. Single-round infectivity assays were performed in the presence or absence of A3G; infectivity in the absence of A3G was normalized to 100%. Infectivity values in percentage are the average of six infections; error bars indicate the standard deviation from the mean of these replicates. Infectivity of HI V-1Δ Vif (light grey) and HIV-1 expressing SIVrcmCM8081 Vif (green), SIVcpzTan3 Vif (purple), or chimeric Vifs were tested against red-capped mangabey A3G (top) or chimpanzee A3G (bottom). Constructs are depicted on the left, with SIVrcm and SIVcpz Vif fragments in green and purple, respectively, and with the PLP motif and the region of Vpr or Vpx overlap shown (“Vpr over” or “Vpx over”) for chimera1 and chimera2.
C- SIVrcm Vif does not degrade chimpanzee A3G. Western-blot analysis against HA-tag A3G (red-capped mangabey, left; chimpanzee, right) was performed from 293T cells cotransfected with HIV-1Δ Vif, and HIV-1 expressing SIVcpzTan3Vif and SIVrcmCM8081Vif Tubulin was probed as a loading control.
D- Vif expression. Western-blot analyses against FLAG-tag Vifs from the BruΔVifΔEnvLuc backbone with corresponded inserted Vif (SIV strains and chimeras correspond to panel B). Similar Vif expression relative to tubulin.
E- SIVrcm was pre-equipped to antagonize chimpanzee A3F and A3H, but not A3D. Single round infectivity assays were performed against various chimpanzee APOBEC3 family proteins, as described in panel B. Left, chimpanzee A3F; middle, A3H; right, A3D. Infectivity values in percentage are the average of six infections; error bars indicate the standard deviation from the mean of these replicates.
We therefore tested if the deletion in vpx was important for the adaptation of vif to antagonize chimpanzee A3G. First, we tested the ability of SIVrcm Vif to antagonize chimpanzee A3G. We found that, while Vif from SIVrcm was potent at antagonizing its own host A3G (Figure 3B, top in plain green), it was only able to partially counteract the antiviral effects of chimpanzee A3G compared to SIVcpz Vif ability (16% of rescue vs. 100%; Figure 3B, bottom plain green vs. purple). Degradation assays further confirmed that SIVrcm and SIVcpz Vifs efficiently degraded RCM and chimpanzee A3Gs, respectively, whereas neither Vif degraded the heterologous A3G (Figure 3C and 3D). Thus, SIVrcm Vif needed to adapt to efficiently antagonize chimpanzee A3G and gain full infectivity.
We found that, alone, the unique C-ter part of SIVcpz Vif that was reconstructed upon the loss of vpx was not sufficient to rescue the infection in the presence of chimpanzee A3G (Figure 3B, chimera1). However, this C-ter domain of SIVcpz Vif was essential for the protein function, as its replacement with the C-ter part of SIVrcm Vif led to a chimera that had no activity against A3G (Figure 3B, chimera2). Changes in SIVrcm Vif to any of the conserved N-ter motifs from SIVcpz/HIV-1 Vif essential for Vif function (reviewed in (Malim and Bieniasz, 2012)) (Figure S3A) led to chimeric Vifs that lost their activity against chimpanzee A3G, while retaining their activity against RCM A3G (Figure S3B). Thus, the domains in SIVrcm Vif required for the degradation of chimpanzee A3G and RCM A3G are distinct and non linear and changes at both the N-and C-ter of SIVrcm Vif were needed to fully gain chimpanzee A3G antagonism.
We also investigated if SIVrcm Vif needed to adapt to antagonize other genes from the chimpanzee APOBEC3 family. We found that SIVrcm Vif has the capacity to antagonize both chimpanzee A3F and A3H. Therefore, chimpanzee A3F and A3H did not represent barriers for SIVrcm-like strains to jump to chimpanzees. On the other hand, SIVrcm Vif did not antagonize chimpanzee A3D (Figure 3E). Chimpanzee A3D is particularly active relative to other primate A3D proteins due to positive selection in this gene in the chimpanzee-bonobo lineage (Duggal et al., 2011). Thus, SIVrcm Vif needed to adapt to fully antagonize chimpanzee A3G and A3D and the deletion of vpx may have been required for these changes to take place.
Chimpanzee as a “passage” for lentiviruses to human infection
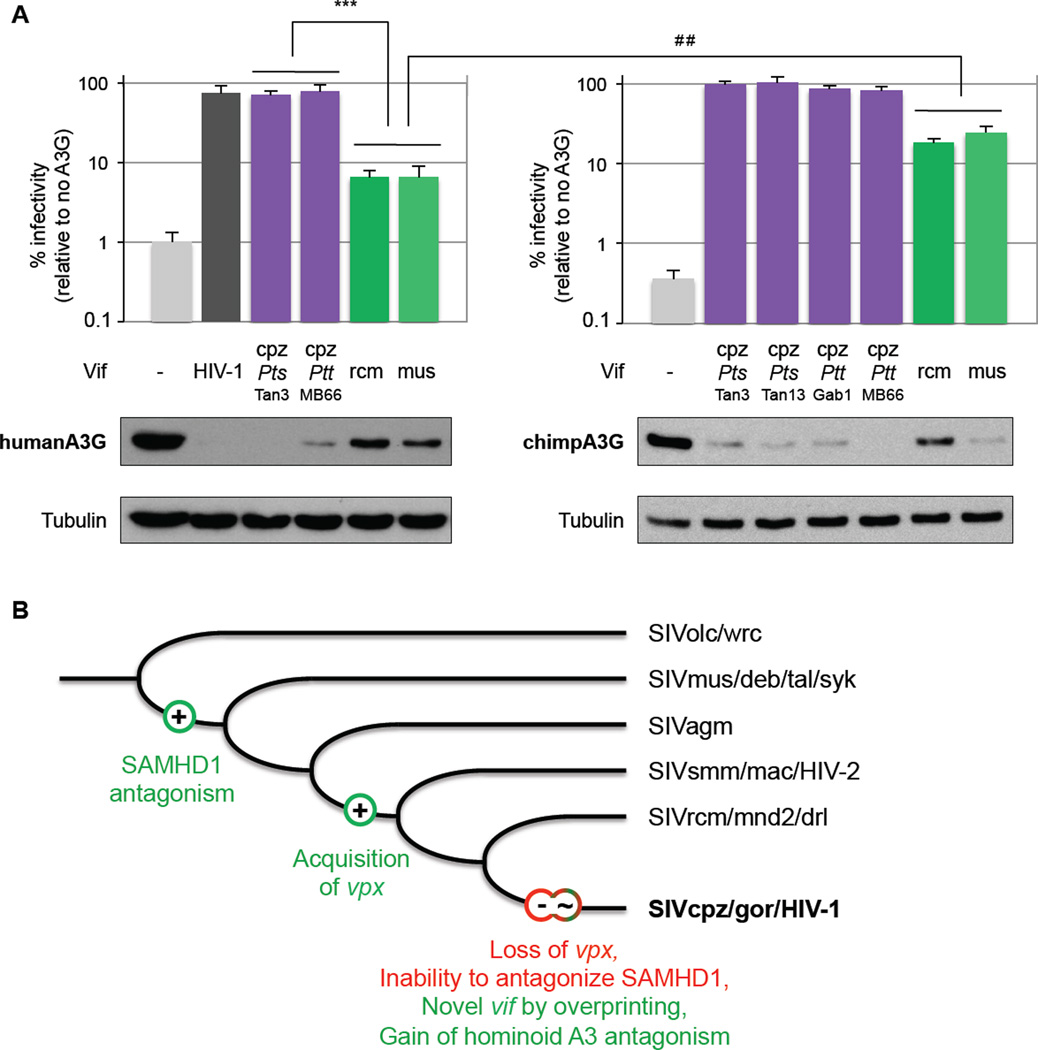
The only known lentiviruses that directly transferred from OWMs to humans are strains of SIVsmm from sooty mangabeys, which gave rise to HIV-2 (Gao et al., 1992; Santiago et al., 2005). Interestingly, Vif from SIVsmm is pre-equipped to counteract human A3G (Compton and Emerman, 2013). As SIVrcm and SIVmus never crossed to humans as opposed to their recombinant progeny, SIVcpz, we hypothesized that passage through chimpanzees was a determinant step for vif to adapt to humans. Thus, we determined if adaptation of SIV Vif to human A3G antagonism occurred during the transfer from chimpanzees to humans, or if it occurred during the adaptation to chimpanzees. We found that SIVcpz Vif rescued the infection to levels similar to HIV-1 Vif in the presence of human A3G and that it could efficiently degrade the host protein (Figure 4A, left panel), consistent with previous studies (Gaddis et al., 2004). On the other hand, we found that SIVrcm Vif, as well as SIVmus Vif, had very little capacity to rescue the infection (less than 10%) in the presence of human A3G (Figure 4A, left panel) as compared to SIVcpz or HIV-1 Vifs (p<0.0001). Hence, human A3G is a hurdle for SIVrcm and SIVmus, but not for SIVcpz. Furthermore, SIVrcm and SIVmus Vifs were better at antagonizing chimpanzee than human A3G (Figure 4A, comparing right and left panels; 3-fold difference in infectivity; p<0.005). Thus, the adaptive hurdle from monkeys to chimpanzees appears to be lower than the barrier directly from monkeys to humans for SIVs, such as SIVrcm or SIVmus, to overcome A3G antagonism. Therefore, the chimpanzee-adapted Vif pre-equipped to antagonize human A3G may have been a path to transmission to humans.
Figure 4. The deep origin of HIV-1 lies in the passage of OWM SIVs in the chimpanzee host and in the evolution of vif and vpx.
A- Chimpanzee as a passage to human infection for OWM SIVs. Top panel, single-round infectivity assays were performed in the presence or absence of A3G as described in Figure 3. The graphs show the result for the average of six infections; error bars indicate the standard deviation from the mean of these replicates. Infectivity of HIV-1Δ Vif (grey, negative control), and HIV-1 plasmid with inserted HIV-1 LAI Vif (black), SIVcpzPtsTan3 and Tan13, SIVcpzPttMB66 and Gab1 Vifs (purple), SIVrcmCM8081 and SIVmus1CM1085 Vifs (green) were tested against human (left) or chimpanzee (right) A3Gs. HIV-1 and SIVcpz Vifs serve as positive controls against human and chimpanzee A3G, respectively. All Vifs were expressed at a level sufficient for their anti-A3G activity. *** and ##, statistically different under the Mann-Whitney test, p<0.0001 and p<0.005, respectively.
Lower panel: Western-blot analyses against HA-tag A3G (human, left; chimpanzee, right) were performed from 293T cells cotransfected with the corresponding Vif constructs.
B- The deep origin of HIV-1 is associated with the evolution of the accessory genes vpx and vif as a result of host antiviral gene selective pressure. Representation of the primate lentiviral evolution highlighting major events that ultimately played a role in the origin of HIV-1. The main genetic events associated with vpx and vif are depicted as well as SAMHD1 and A3G antagonism gain and loss. Green and positive signs are associated with a gain of function or a gene acquisition/evolution, while red and negative signs are associated with the loss of function or gene loss.
Discussion
We have studied the deep origins of HIV-1 through an investigation of the gene loss and the adaptations that occurred during and after the transfer of lentiviruses from monkeys to chimpanzees to create SIVcpz. Specifically, we found that (1) the SIVrcm vpx gene was lost in its entirety upon adaptation to chimpanzee; (2) the loss of vpx was associated with the creation of a unique vif region by overprinting; (3) Vif adapted to antagonize chimpanzee APOBEC3 proteins including A3G and A3D; (4) the chimpanzee-adapted lentivirus was more efficient than its monkey ancestors at antagonizing human restriction factors such as A3G. Thus, lentiviral gene loss and adaptations in the chimpanzee host were at the origin of the human HIV-1 pandemic.
Although SAMHD1-antagonism by the vpx or the vpr genes is conserved in many lentiviruses (Lim et al., 2012), we found that this function is not strictly necessary for viral adaptation to hominids. It had previously been hypothesized that the ancestral SIVcpz had a vpx gene and was transmitted from chimpanzee to RCM and mandrills giving rise to SIVrcm(Vpx+) and SIVmnd2(Vpx+), respectively, and that subsequently SIVcpz lost its vpx gene in its natural host (Zhang et al., 2012). However, this scenario is not plausible because the ancestor of SIVrcm and SIVsmm acquired the vpx gene prior to their divergence and prior to the jump of SIVs from RCM to chimpanzee (Figure S1).
It is remarkable that a viral antagonist to a host protein could be lost from the viral genome, since one would have expected adaptation of Vpx to counteract the new host SAMHD1 (Lim et al., 2012). However, the selective pressure during the cross-species jump from OWMs to chimpanzees may have favored the maintenance of a virus in which a poorly active gene was lost by recombination, but a critical function in an overlapping gene was restored/gained. Vif adaptation to chimpanzee APOBEC3s may have been more critical than Vpx adaptation to chimpanzee SAMHD1. This suggests that the A3 proteins are a more potent selective force in the transmission of lentiviruses than SAMHD1. Finally, it is possible that SIVcpz acquired functions to balance for the absence of Vpx-driven antagonism of SAMHD1 and that these adaptations have increased pathogenicity in the HIV-1 lineage.
The loss of vpx together with additional modifications led to a unique viral vif gene. However, the origin of the region including the cullin box in SIVcpz/HIV-1 Vif is unknown. It is possible that these sequences arose during the vif reconstruction associated with the species-jump or it may have also been acquired later. While SIVcpz, SIVgor, and HIV-1 Vifs harbor a cullin box with a highly conserved PLP motif, most SIV Vifs harbor a very divergent cullin box that has a distinct evolutionary history, which explains why HIV-1 has a distinctive way to antagonize human A3G compared to HIV-2 and other SIVs (Barraud et al., 2008; Gaur and Strebel, 2012). Although SIVsmm, which crossed to humans, harbors a Vif with such divergent C-ter domain, its Vif and Vpx were pre-equipped to antagonize human A3G and SAMHD1, respectively (Compton and Emerman, 2013; Hrecka et al., 2011). Whether the unique cullin box from the SIVcpz/HIV-1 lineage has a distinctive role still needs to be addressed, but in any case, the passage of the SIVrcm/SIVmus-like recombinant to hominids was not as easy as the passage of SIVsmm to humans. Furthermore, we found that mutations in both the N- and the C-ter domains of Vif were necessary to gain full chimpanzee A3G-antagonism. Since an SIVrcm Vif with only the C-ter reconstruction of SIVcpz Vif (analogous to chimera1 in Figure 3) is poorly active against chimpanzee APOBEC3 proteins, the deletion of vpx may have put the intermediate virus into a fitness valley from which it could have recovered only with additional N-ter mutations in Vif. However, it is also possible that an ancestral version of SIVrcm Vif already harbored a Vif with an N-ter domain that directly provided full activity to Vif after its C-ter reconstruction in SIVcpz.
Due to species specificities in virus-host interactions and antagonisms, lentiviruses need to adapt to the new host proteins to efficiently infect a new species. This jump is easier when the virus is pre-equipped to antagonize the recipient species’ restriction factors, e.g. human A3G is not a barrier for SIVcpz. However, we further confirmed that human A3G cannot be counteracted by SIVs from most OWMs, except by SIVsmm, which crossed to humans on multiple occasions (Compton and Emerman, 2013; Gao et al., 1992; Santiago et al., 2005). Thus, human A3G appears to pose a major hurdle for SIVs from most OWMs, limiting their transfer to humans. On the other hand, viruses such as SIVrcm and SIVmus have some activity against chimpanzee A3G. This may have been a key component of their jump to the ape by allowing first for a poorly efficient viral infection, followed by subsequent viral adaptation. Since A3G has been under strong positive selection in the human genome after the divergence of Pan and Homo from a yet unknown selective pressure (Sawyer et al., 2004), it is possible that this evolution led to variation in human A3G that is more poorly recognized than chimpanzee A3G by monkey SIVs. Therefore, the chimpanzee host may have constituted an intermediary in the adaptive processes allowing for certain OWM lentiviruses to infect humans. While adaptation in the chimpanzee host does not provide adaptation to all human proteins, e.g. SIVcpz is not pre-equipped to antagonize human Tetherin (Sauter et al., 2009), we propose that SIV adaptation to chimpanzee restriction factors reduced both the number and the size of the hurdles for cross-species transmission to humans, which favored a successful viral emergence in the human population.
In summary, it is possible to trace back many of the ancient genetic events in the evolution of primate lentiviruses that ultimately lead up to the emergence of HIV-1 (Figure 4B), and the adaptations during the cross-species transmissions leading to SIVcpz in chimpanzee reveal the functional origins of the pandemic HIV-1 in humans.
Experimental procedures
Plasmids
SAMHD1, Vpr and Vpx expression plasmids are as previously reported (Lim et al., 2012). Apobec3 and recombinant HIV-1 proviral plasmids were constructed as detailed in the Supplemental Information.
Transfection and western-blot analysis for the SAMHD1-Vpr/Vpx study
293T cells were transfected with 100 ng of SAMHD1 expression plasmid with or without 100 ng of Vpr/Vpx constructs using TransIT-LT1 transfection reagent (Mirus Bio). The amount of Vpr/Vpx and SAMHD1 expression plasmid transfected was normalized for similar protein expression, but the total quantity of DNA transfected was maintained constant. Cells were harvested 48 h post-transfection for western-blot analysis, as previously described (Lim et al., 2012). Details of the antibodies are Supplemental Information.
Single-round viral infectivity assay and western-blot analysis for the A3-Vif study
293T cells were transfected with 400 ng of A3 plasmid or an empty pcDNA3.1 vector, 200 ng of L-VSV-G (the fusogenic envelope G glycoprotein of the vesicular stomatitis virus used for pseudotyping), and 600 ng of proviral plasmid with various Vif genes, using TransIT-LT1. The virus supernatants and the cells were harvested 48 h post-transfection. Each transfection condition was performed in duplicate in independent experiments. The total amount of virus in the supernatant was quantified by p24 Gag enzyme-linked immunosorbent assay (Advanced Bioscience Laboratories). SupT1 cells plated at 0.4 M cells/mL in the presence of 20 µg/mL DEAE-Dextran were infected with 2 ng of virus. Infections were performed in triplicate for 72 h and luciferase activity was measured with the Bright-Glo Luciferase Assay Reagent (Promega). Statistical analyses were performed with the Mann-Whitney test. The harvested cells were used for western-blot analyses.
Molecular sequence analyses
Alignments were performed using Muscle (Edgar, 2004) or FSA (Bradley et al., 2009), and minor adjustments were done where necessary. Maximum likelihood (ML) trees were constructed using PhyML (Guindon et al., 2010) with 1,000 bootstrap replicates and the GTR model with four gamma rate categories. Recombination analyses were performed using GARD from Datamonkey (Kosakovsky Pond et al., 2006) and the PHI test (Bruen et al., 2006) implemented in SplitsTree (Huson and Bryant, 2006). The p-value cutoff for any evidence of recombination was set at 0.1. We used BLAST (Altschul et al., 1990), hmmer amino acid HMM search and nhmmer (Eddy, 2011; Finn et al., 2011) to search for the origin of the 60–75 bp fragment in vif that is only present in the SIVcpz/HIV-1 lineage; first, by looking in all sequences available in the NCBI database, and then more finely in primate and SIV genomes (excluding this short fragment of the SIVcpz/HIV-1 lineage).
Supplementary Material
Highlights.
-
-
The entire SIVrcm vpx gene was lost upon the adaptation of SIV to chimpanzee.
-
-
The loss of vpx led to the creation of a unique vif region/gene by overprinting.
-
-
SIVcpz Vif adapted to counteract hominid APOBEC3 proteins.
-
-
Adaptation of SIV to chimpanzees facilitated virus adaptation to human APOBEC3G.
Acknowledgements
We thank Oliver Fregoso and Maulik Patel for comments on the manuscript. We thank Connor McCoy for bioinformatic analyses to search for the origin of the 3’end Vif fragment in SIVcpz, and Efrem Lim for helpful discussion on the loss of vpx in SIVcpz. The work was supported by NIH grant AI30937 (M.E.), amfAR Mathilde Krim Fellowship in Basic Biomedical Research # (108499–53-RKGN) to L.E., and by the Fred Hutchinson Cancer Research Center Interdisciplinary Fellowship (L.E.). L.E., F.A.M. and M.E. designed the experiments and the phylogenetic analyses. L.E. performed the experiments and the phylogenetic analyses. L.E. and M.E. wrote the paper. All authors participated in the study concept and gave comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ayinde D, Maudet C, Transy C, Margottin-Goguet F. Limelight on two HIV/SIV accessory proteins in macrophage infection: is Vpx overshadowing Vpr? Retrovirology. 2010;7:35. doi: 10.1186/1742-4690-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, Hahn BH, Sharp PM. Hybrid origin of SIV in chimpanzees. Science. 2003;300:1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med. 2012;18:1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud P, Paillart JC, Marquet R, Tisne C. Advances in the structural understanding of Vif proteins. Curr HIV Res. 2008;6:91–99. doi: 10.2174/157016208783885056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron JR, Huthoff H, Veselkov DA, Beavil RL, Simpson PJ, Matthews SJ, Malim MH, Sanderson MR. The SOCS-box of HIV-1 Vif interacts with ElonginBC by induced-folding to recruit its Cul5-containing ubiquitin ligase complex. PLoS Pathog. 2010;6:e1000925. doi: 10.1371/journal.ppat.1000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley RK, Roberts A, Smoot M, Juvekar S, Do J, Dewey C, Holmes I, Pachter L. Fast statistical alignment. PLoS Comput Biol. 2009;5:e1000392. doi: 10.1371/journal.pcbi.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton AA, Emerman M. Convergence and Divergence in the Evolution of the APOBEC3G-Vif Interaction Reveal Ancient Origins of Simian Immunodeficiency Viruses. PLoS Pathog. 2013;9:e1003135. doi: 10.1371/journal.ppat.1003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue JP, Vetter ML, Mukhtar NA, D’Aquila RT. The HIV-1 Vif PPLP motif is necessary for human APOBEC3G binding and degradation. Virology. 2008;377:49–53. doi: 10.1016/j.virol.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal NK, Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol. 2012;12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal NK, Malik HS, Emerman M. The breadth of antiviral activity of Apobec3DE in chimpanzees has been driven by positive selection. J Virol. 2011;85:11361–11371. doi: 10.1128/JVI.05046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddis NC, Sheehy AM, Ahmad KM, Swanson CM, Bishop KN, Beer BE, Marx PA, Gao F, Bibollet-Ruche F, Hahn BH, et al. Further investigation of simian immunodeficiency virus Vif function in human cells. J Virol. 2004;78:12041–12046. doi: 10.1128/JVI.78.21.12041-12046.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Yue L, White AT, Pappas PG, Barchue J, Hanson AP, Greene BM, Sharp PM, Shaw GM, Hahn BH. Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature. 1992;358:495–499. doi: 10.1038/358495a0. [DOI] [PubMed] [Google Scholar]
- Gaur R, Strebel K. Insights into the dual activity of SIVmac239 Vif against human and African green monkey APOBEC3G. PLoS ONE. 2012;7:e48850. doi: 10.1371/journal.pone.0048850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keese PK, Gibbs A. Origins of genes: "big bang" or continuous creation? Proc Natl Acad Sci U S A. 1992;89:9489–9493. doi: 10.1073/pnas.89.20.9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe. 2010;8:55–67. doi: 10.1016/j.chom.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. GARD: a genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ES, Fregoso OI, McCoy CO, Matsen FA, Malik HS, Emerman M. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe. 2012;11:194–204. doi: 10.1016/j.chom.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Bieniasz PD. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harbor perspectives in medicine. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Santiago ML, Range F, Keele BF, Li Y, Bailes E, Bibollet-Ruche F, Fruteau C, Noe R, Peeters M, Brookfield JF, et al. Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Tai Forest, Cote d'Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2. J Virol. 2005;79:12515–12527. doi: 10.1128/JVI.79.19.12515-12527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, et al. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe. 2009;6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Hahn BH. Origins of HIV and the AIDS Pandemic. Cold Spring Harbor perspectives in medicine. 2011;1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Walker RC, Jr, Khan MA, Kao S, Goila-Gaur R, Miyagi E, Strebel K. Identification of dominant negative human immunodeficiency virus type 1 Vif mutants that interfere with the functional inactivation of APOBEC3G by virus-encoded Vif. J Virol. 2010;84:5201–5211. doi: 10.1128/JVI.02318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, de Silva S, Wang JH, Wu L. Co-evolution of primate SAMHD1 and lentivirus Vpx leads to the loss of the vpx gene in HIV-1 ancestor. PLoS ONE. 2012;7:e37477. doi: 10.1371/journal.pone.0037477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.