The emerging use and influence of nanotechnology in the biomedical field has driven investigators to create innovative solutions for the targeted treatment of pathological conditions such as atherosclerosis,[1] cancer,[2–4] diabetes, [5] traumatic brain and spinal cord injury, [6] and thrombosis.[7, 8] The rapid expansion in the use of nanotechnology-based products evoked concerns related to the possible adverse effects associated with nanomaterials. However, disagreements on methodology to evaluate toxicity levels and on the choice of nanoparticles to use as a standard have troubled this field of research from its inception.[9]
The development of effective in vitro methods to understand the impact of nanomaterials on cells and tissues, must consider that exposure to toxic agents typically results in three possible outcomes: cells undergo a loss of membrane integrity resulting in rapid death (necrosis), or they become senescent and stop functioning and dividing, or they activate a sequence of proteolytic events resulting in a highly ordered and controlled cell death (apoptosis).[10, 11] Necrosis is characterized by the rapid swelling of the cell resulting in declined metabolism and rapid expulsion of proteins, metabolites, and organelles into the surrounding environment. Necrosis typically occurs as a consequence of acute toxicity. Cellular senescence normally occurs in response to cellular aging, however, low concentrations of a cytotoxic compound could cause cells to be unable to perform a particular function or to divide. In contrast, apoptosis is distinguished by an extremely ordered process, which produces a cascade of biochemical events resulting in specific morphological alterations that trigger the efficient removal of cells from tissues.
There are several accounts that demonstrate the capacity of nanoparticles to elicit the aforementioned cellular outcomes.[12–14] Metabolic activity of cells and their membrane integrity are commonly measured to evaluate the toxicity of nanoparticles. However, these approaches, even if tested in combination, often fail to reveal if the exposure to nanoparticles has affected or altered other cellular functions in cases of low, minimally acute toxicity.[15] Hence, appropriate end points must be chosen to evaluate if cell senescence has been induced, if apoptosis markers are expressed, or if biochemical pathways have been altered, in addition to determining if cells are damaged acutely.[9] Another concern is the selection of an assay that limits false positives, which can occur if the nanomaterials absorb or spontaneously interact with dyes. [9, 16, 17]
In this study, we tested our multistage mesoporous silicon nanovectors (MSV) designed and engineered to decouple the multiplicity of tasks required of nanovectors (mononuclear phagocyte system avoidance, margination, etc.,) and distribute them onto multiple stages.[18–22] We produced MSV through established photolithography techniques[23, 24] with well-defined shapes and well-controlled size distribution (Supplementary Information) and used conventional assays to investigate their effect on key cell functions. In particular, we examined the consequences of MSV internalization on cell proliferation, tube formation, differentiation, and migration of human umbilical vein endothelial cells (HUVEC)[25] and mouse adipose-derived mesenchymal stromal cells (ADMSC).[26, 27] The endothelial model was chosen because the vasculature is the first tissue to be encountered by a circulating particle upon systemic administration. The choice of ADMSC was based on the propensity of these cells for multipotent differentiation and migration[26, 27] thus providing a representative experimental model to study the influence of MSV on these processes. The impact on the cytoskeleton and ultrastructure were also studied to interpret potential effects on the cellular architecture, which is essential for maintaining normal cellular shape and size, motility, and intracellular transport. Using both cell types we showed that MSV neither induced any detectable adverse effects on cell architecture, viability or physiology nor affected the capacity of HUVEC to form networks and of ADMSC to differentiate and migrate toward cancer cells. Combined, these results show that MSV have minimal effects on fundamental cell functions.
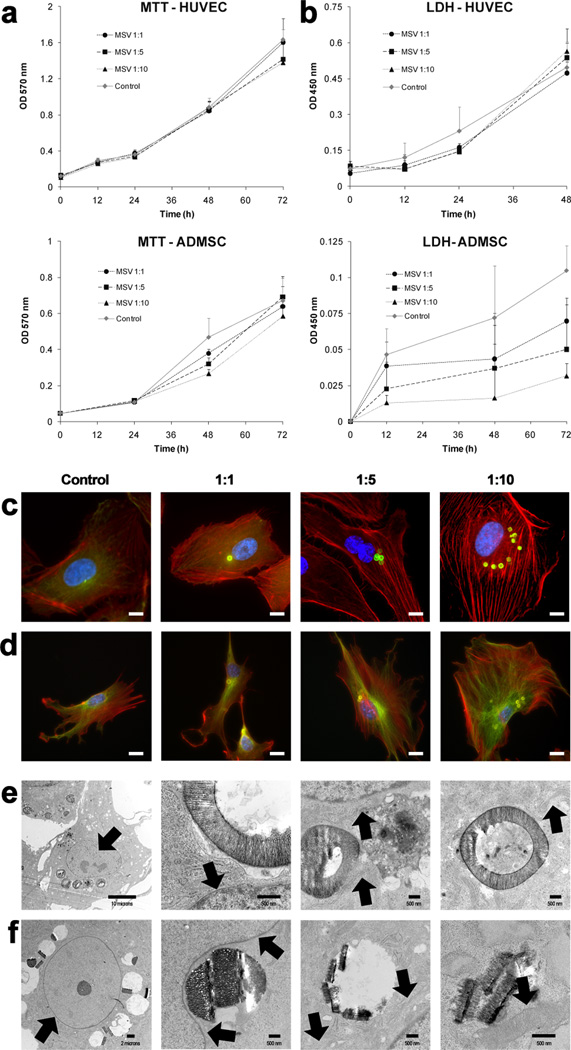
Previous investigations demonstrated that HUVEC, when seeded as a monolayer, quickly captured and internalized MSV by endocytosis[28] and resulted in partitioning of MSV into daughter cells upon division.[29] Cells (HUVEC, ADMSC) were seeded at high confluence and treated with MSV (Cell:MSV ratio of 1:1, 1:5, and 1:10) resulting in nearly 100% of cells containing at least one MSV per cell. Conventional cellular viability (MTT, 3-[4,5–Dimethylthiazol-2-yl]-2,5-Diphenyltetrazolium Bromide) and toxicity (LDH, lactate dehydrogenase) assays were conducted on both cell lines comparing different ratios in order to investigate if there was a dose dependent effect of MSV on cells. Both cell types showed a linear increase in MTT incorporation over 72 hours with minor difference between treated and control cells (Figure 1a). This result indicated that the exposure of MSV at all concentrations did not significantly alter the proliferation of cells and that MSV internalization appears to be well tolerated by both cell types. Also the release of LDH was followed in both cell types, and it confirmed the lack of a cellular cytotoxic stress due to MSV internalization.
Figure 1.
Cellular proliferation, toxicity, and architecture upon internalization of MSV. A) MTT assay was used to determine the effect MSV internalization on proliferation of HUVEC (top) and ADMSC (bottom). B) LDH assay was used to assess cellular toxicity and membrane damage that could have developed within HUVEC (top) and ADMSC (bottom) after the internalization of MSV. C, D) Cytoskeleton staining of HUVEC (C) and ADMSC (D) at increasing doses (1:1, 1:5, 1:10) of MSV demonstrated conservation of cellular structure. In C & D: microfilaments (f-actin) are in red, microtubules (α-tubulin) in green, nuclei are in blue, and MSV are in green (C) or yellow (D), scale bar = 10 µm. E, F) TEM images of HUVEC (E) and ADMSC (F), 24 hours after internalization of MSV. Starting from the left, images were selected to demonstrate the effect of MSV internalization on the nucleus/nucleolus, nuclear envelope, rough endoplasmic reticulum, and mitochondria, indicated by black arrows. Scale bars are 500 nm, except for left most images where it is 10 µm (E) and 2 µm (F).
To understand the effect of MSV internalization on cell architecture, the cytoskeletal arrangement and the ultrastructure of the cell were investigated. HUVEC (Figure 1c) and ADMSC (Figure 1d) were exposed at several ratios and stained with fluorescent markers to label f-actin microfilaments (red), alpha tubulin microtubules (green), and nuclei (blue). Previous results have demonstrated that, upon internalization by endocytosis, MSV are found in the lysosomal compartment within a few hours.[30] Here, we show that after internalization, MSV accumulate in the peri-nuclear region for both cells lines. The increased number of MSV internalized did not affect their ability to undergo peri-nuclear localization. Microfilaments of both cell types were well organized and filamentous, with parallel stress fibers distributed throughout the cytoplasm. These cytoskeletal components displayed characteristic endothelial and fibroblastoid phenotypes for HUVEC and ADMSC, with microtubules radiating from the peri-nuclear area throughout the cell body as previously observed.[31, 32] These observations indicated that cytoskeletal elements were largely unaffected by MSV incorporation. The ultrastructure of cells was also investigated using transmission electron microscopy (TEM) to understand the effect of MSV internalization at the organelle scale. HUVEC (Figure 1e) and ADMSC (Figure 1f) were exposed to MSV at a ratio of 1:10 and inspected for the integrity of several organelles in close proximity to internalized MSV. The analysis of TEM micrographs confirmed that the nucleus, nucleolus, and nuclear envelope of both cells were unaffected. The proximity to the nuclear envelope did not display any visible damage, and that the organization of heterochromatin and euchromatin within the nucleus was comparable with that of untreated cells, even when internalized MSV appeared to modify the curvature of the nuclear membrane (Figure 1f). As in untreated cells (supplementary information), rough endoplasmic reticulum (RER) studded with more electron dense areas indicative of ribosomes was found adjacent to the nuclear envelope and mitochondria with defined matrix and membranes were observed and appeared intact and healthy upon MSV uptake. Hence, upon MSV internalization, both cell types retained normal organization of cytoskeletal elements and ultrastructure and appeared unaffected by the presence of MSV.
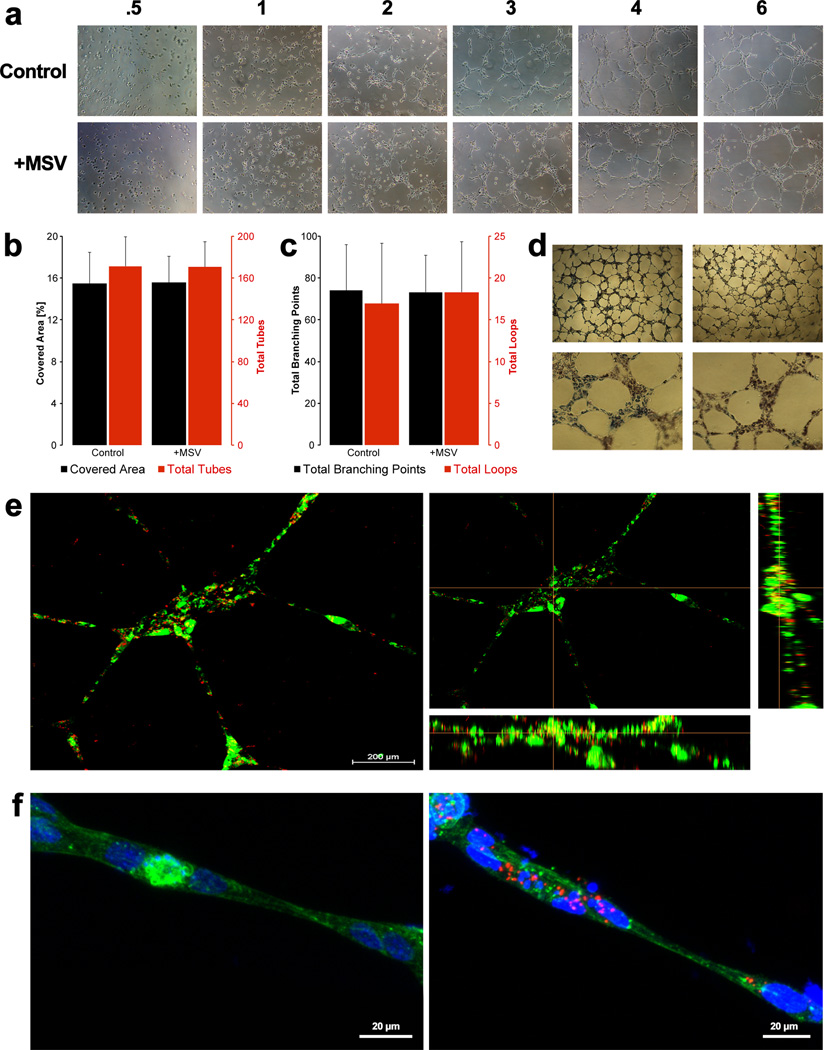
Understanding the potential effects that MSV may have on the function of endothelial cells is vital for the development of a drug delivery system with no impact per se on cell physiology. For this reason, we chose to investigate if HUVEC retained the ability to form tubular networks, a conventional ex vivo angiogenesis assay.[33] HUVEC were exposed overnight to 1000 × 200 nm discoidal MSV at a ratio of 1:40. The following day, HUVEC containing MSV were seeded onto matrigel and assayed for tube formation (Figure 2). Initially, HUVEC were seen as single cells (Figure 2a), however over time cells migrated and elongated towards other cells and reorganized in space to begin the formation of vessel-like structures. Upon MSV internalization, HUVEC assembled and formed tubes in a similar fashion, indicating that MSV did not alter this process. Quantitative assessment of tube formation was performed using Wimasis Image Analysis software and comparing the percentage of area covered by tubes, and the total number of tubes, branching points, and loops in four separate 4× images taken at 24 hours (Figure 2b,c). Compared to control HUVEC, cells containing MSV covered equivalent surface and formed a comparable total number of tubes. Furthermore, the final count of branching points and loops between different treatment groups was similar. In the MTT assay, exposed cells retained the ability to actively reduce the dye, demonstrating their viability post tube-formation (Figure 2d). In order to confirm MSV retention (Figure 2e) and to analyze cytoskeletal organization (Figure 2f), tubes were imaged with a laser scanning confocal system. MSV were discovered predominantly distributed within the center of the tubes and mostly co-localized with nuclei, while fewer particles could be found in the tube projections (Figure 2e,f). Internalization was confirmed by inspecting a cross-section displaying x and y plane profiles that localized MSV within the cellular boundaries (Figure 2e, right). Lastly, the cytoskeletal organization was inspected by staining f-actin. Consistent with results from monolayer HUVEC culture (Figure 1c), tubes formed by both control HUVEC and HUVEC containing MSV exhibited similar parallel microfilaments, indicating minimal actin reorganization (Figure 2f).
Figure 2.
HUVEC network formation upon internalization of MSV. A) Images were taken at increasing times (starting from left, 0.5, 1, 2, 3, 4, and 6 hours) to investigate the effect of MSV on migration of tube network assembly. B) Quantitative assessment of the percentage of area covered and total tubes formed. Left axis (black): displays the quantity of the percent covered area and right axis (red) is for the total tubes formed comparing control and MSV treated HUVEC. C) Quantitative assessment of the total branching points and total loops detected during tube formation. Left axis (black): plots the total number of branching points. Right axis (red) plots the total number of loops. E, F) Confocal imaging of tubes formed at 24 hours demonstrating retention of MSV after tube formation, with HUVEC in green and MSV in red (E). Maintained cytoskeletal structure after tube formation, microfilaments (f-actin) in green, MSV in red, and nucleus in blue (F). Scale bar = 200 µm (E) and 20 µm (F).
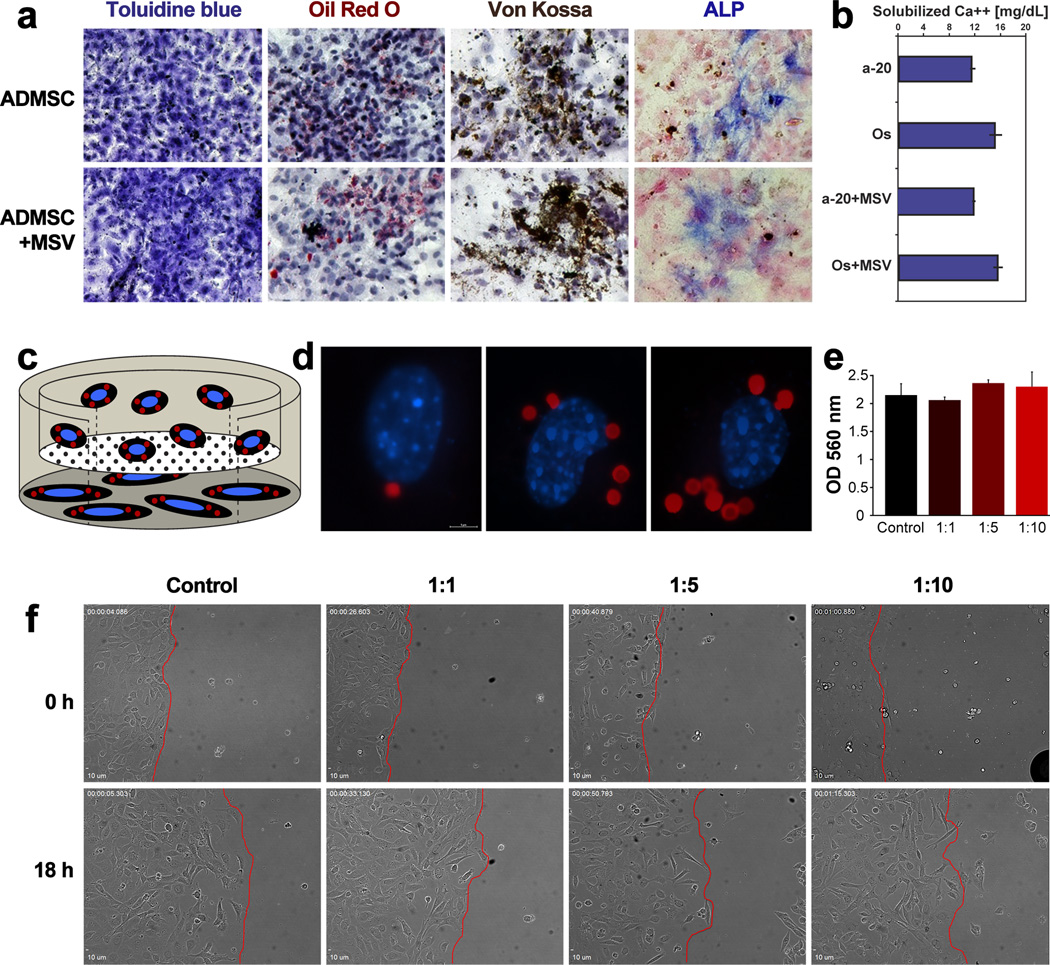
Differentiation is integral for efficient tissue repair and maintaining homeostasis, hence conservation of this function is critical and can be assayed as an indicator of toxicity. Stromal cells, initially isolated as plastic-adherent colony-forming fibroblasts,[34] have a marked capacity to differentiate into mesenchymal cell types, such as adipocytes and osteoblasts.[35] Similar properties have been demonstrated for ADMSC,[36] and here we tested whether MSV affect cell multipotency. As shown above (Figure 1), upon internalization (at 1:5 cell:MSV ratio), ADMSC retained the ability to adhere to plastic. Under induction media, ADMSC containing MSV maintained morphology similar to that of control cells, as evident from toluidine blue staining (Figure 3a). Oil Red O, a lipophilic dye that stains lipid droplets, was used to identify adipocytes upon adipogenesis induction. ADMSC containing internalized MSV displayed similar staining for Oil Red O as control cells indicating that ADMSC retained the ability to differentiate into adipocytes. Upon osteoblast differentiation, ADMSC were assessed by Von Kossa, alkaline phosphatase (ALP), and alizarin red staining to determine mineralization, osteoblast activity, and calcium deposition, respectfully. ADMSC with internalized MSV demonstrated comparable staining with all three techniques. Furthermore, a quantitative calcium assay was used to compare ADMSC containing MSV with control cells incubated with inductive media or regular media (Figure 3b). ADMSC with internalized MSV produced an equivalent amount of calcium as control cells indicating an unchanged capacity of ADMSC to differentiate into osteoblasts upon internalization of MSV.
Figure 3.
ADMSC differentiation and migration upon internalization of MSV. A) Control and MSV-containing ADMSC were induced to differentiate into adipocytes and osteoblasts. Toluidine blue demonstrates similar cellular morphology, while Oil Red O reveal lipid droplets upon adipogenic differentiation. Von Kossa and ALP staining demonstrate osteogenic differentiation. B) Quantitative determination of solubile calcium upon osteogenic differentiation. C) Schematic of Boyden chamber illustrating the set-up used to verify migration of ADMSC toward MDA-231 cells. D) Sample images of migrated ADMSC containing different ratios of internalized MSV (from left: 1:1, 1:5, and 1:10). E) Quantitative assessment using Boyden chambers, comparing control ADMSC with ADMSC containing different amounts of MSV. F) Scratch assay still frame images from time lapse microscopy at 0 and 18 hours, demonstrating conserved migration at the different ratios of MSV compared to the leading edge (red-line) within each sample.
Finally, a potentially useful property of mesenchymal cells, including ADMSC, is their trafficking toward tumors[37, 38] Here, we tested the ability of ADMSC to traffic towards breast cancer cells (MDA-MB-231) using modified Boyden chamber (transwell) and wound-healing (scratch) assays. The transwell assay was established as depicted in Figure 3c, where MDA-MB-231 cells were seeded below the insert within the well while ADMSC (Cell:MSV ratio of 1:1, 1:5, and 1:10) were seeded on the top of the inserts. In each group, ADMSC were found to transverse the membrane, as illustrated by representative images (Figure 3d). Quantitation was performed using crystal violet staining and comparing absorbance of each sample (Figure 3e) and indicated that ADMSC migration was not significantly affected by internalization of MSV. In the wound-healing assay, ADMSC and cancer cells were seeded into inserts that separated the cells by an average of 500 µm (Figure 3f). Snapshots shown at 0 and 18 hours of migration demonstrated that the leading edge (red line) within the different groups was equivalent, suggesting that the incorporation of MSV did not affect migration. Furthermore, inspection of time-lapse microscopy movies (supplementary information) confirmed that ADMSC containing MSV displayed comparable migration kinetics.
In summary, we applied multiple biochemical and functional assays to evaluate the ability of primary cells to preserve their key biological functions upon MSV internalization. HUVEC and ADMSC were chosen to evaluate whether our novel multistage delivery vector[39] has an effect on the ability of cells to proliferate, form vessel-like structures, differentiate, or migrate. Analysis of cell ultrastructure, cytoskeleton, and viability did not reveal detectable changes indicating that MSV had no detrimental effects. Internalization of MSV by endothelial cells did not interfere with their assembly into endothelial networks on matrigel. The formation of tubes, their number, structure and features were also comparable to those of untreated control cells. Upon MSV internalization, ADMSC retained the capacity to differentiate into osteoblasts and adipocytes and the ability to traffic toward cancer cells. In conclusion, we demonstrated that MSV elicited minor responses resulting in minimal toxicity that did not compromise key cellular functions. On account of cells maintaining these functions while containing up to ten MSV, our data demonstrate a remarkable tolerance toward this type of nanomaterial. In conjunction with providing a methodological platform for analyzing nanoparticle toxicity, this study provides a rationale for using MSV as a nanomaterial whose compatibility and benign nature may streamline its biomedical applications.
Experimental Section
MSV Fabrication
MSV were fabricated in the Microelectronics Research Center at The University of Texas at Austin using previously described protocols.[23, 24] Briefly, a heavily doped p-typed silicon wafer was deposited with a 100 nm silicon nitride in a low pressure chemical vapor deposition furnace. Standard photolithography was performed to pattern 2 µm circle arrays over the wafer using photoresist. Patterned silicon nitride was removed by CF4 reactive ion etching, followed by etching of 600 nm trenches into patterns. Photoresist was removed by incubation in a piranha solution and the wafer was carefully assembled into a Teflon cell for two-step electrochemical etching. After etching, the nitride layer was removed in hydrofluoric acid and porous silicon particles were released by ultrasonication in alcohol for one minute.
Cell Culture
HUVEC were purchased from LONZA and maintained using endothelial basal media-2 supplemented with EGM-2 SingleQuots (Lonza). ADMSC were isolated from fat, as previously described.[37, 40] Briefly, cells from isolated using a mixed population of subcutaneous and intraperitoneal white adipose tissue from C57BL/6 mice by enzymatic digestion. MDA-MB-231 were purchased from American Type Culture Collection. ADMSC and MDA cells were maintained using dulbecco’s modified eagle medium (DMEM) with high glucose, L-glutamine (Invitrogen, GIBCO) and supplemented with 10% fetal bovine serum.
Cellular toxicity and Proliferation
LDH and MTT assays were purchased from Abcam and Invitrogen, respectfully, and preformed simultaneously following manufacturers instructions. Briefly, cells were seeded into 96 well plates. At pre-determined times, the media was removed for LDH and adherent cells were treated with 0.5 mg mL−1 of MTT dye for two hours in the incubator. LDH samples were measured on a Synergy H4 (BioTek) plate reader at OD of 450 nm. After incubation with MTT dye, cells were solubilized with DMSO for 30 minutes and then measured at OD of 570 nm.
TEM
Cells were grown to 90% confluency in six well plates. Cells were then treated with MSV overnight in incubator and then fixed in a 2% paraformaldehyde (Electron Microscopy Sciences) and 3% glutaraldehyde (Sigma-Aldrich) in PBS. Samples were then processed and imaged for TEM as previously described.[20, 41]
Tube Formation
8-chambered slides (LabTek) were pre-coated with 100 µL of Matrigel (BD Biosciences) at 37 °C for 30–45 minutes. Upon uniform coverage of well, HUVEC (40,000) were added to each well. Images were taken at 0.5, 1, 2, 3, 4, 6, and 24 hours using a Nikon TS100 equipped with a DS-Fi1. 24-hour images at 4× were sent to Wimasis for image analysis. In addition, HUVEC were labeled with Cell Tracker® Red (Invitrogen) and imaged at 24 hours for tube formation using a Nikon A-1 confocal microscope.
ADMSC Differentiation
ADMSC (80,000) were seeded in 24 well plates and allowed to adhere overnight. The following day, plates were divided for osteogenic and adipogenic differentiation and media was replaced with their respected differentiation media. Media was replaced every 3 days for 3 weeks, after cells were stained with toluidine blue, oil red o, von kossa, and ALP. Furthermore, a quantitative calcium assay (QuantiChrom) was used to compare the production of calcium in the mineral produced in the cells. Solubilized cells were measured at OD 610 nm on a plate reader.
ADMSC Migration
Transwell: Thincerts™ (Greiner Bio-One) in 24 well plates assessed the migration affinity of ADMSC towards breast cancer cells. Transwell were established by seeding cancer cells (20,000) in complete media on the bottom of the well, inserted a 8.0 pore transwell, and ADMSC (25,000, with and without MSV) were seeded in serum-free media within the transwell. ADMSC were allowed to migrate for 48 hours after which transwells were stained with crystal violent. Inserts were transferred to a new plate and measured on a plate reader at OD 560 nm. Adapted Wound-healing Assay: Ibidi® culture inserts within an ibidi® µ-Dish (35 mm, high) were used to investigate the 2D migration affinity of ADMSC towards breast cancer cells. Here, ADMSC (10,000) were seeded into one compartment while cancer cells (30,000) were seeded in the other. The following day, inserts were removed and migration was assessed using time-lapse imaging. Images were acquired in four z-planes every 4 minutes using an DSU IX81 Olympus microscope with a 20× objective for 18 continuous hours. Still shots were taken with a 10× objective before and after acquisition of time-lapse and used within the figure, while movies were composed of projection images of all captured z-planes per time.
Supplementary Material
Acknowledgments
The authors would like to thank Kenneth Dunner Jr. for TEM sample preparation and analysis at the High-Resolution Imaging Facility at The University of Texas M.D. Anderson Cancer Center (UT-MDACC), Jared K. Burks for time-lapse imaging preparation and acquisition performed at the Flow Cytometry and Cellular Imaging Core Facility within UT-MDACC. We wish to thank Kemi Cui of The Methodist Hospital Research Institute (TMHRI) Advanced Cellular and Tissue Microscope Core Facility for Laser Scanning Confocal microscope access. In the Department of Nanomedicine at TMHRI, we wish to thank Daniel Blashki for assistance on running ADMSC differentiation, Michael Evangelopoulous for assistance in acquiring images for tube formation, and Matt Landry for assistance with image preparation. This research was supported by The Alliance for NanoHealth, for DOD TATRC grants W81XWH-09-2-0139 and W81XWH-10-2-0125; MDACC Institutional Core Grant #CA-016672. In addition, JOM was supported by NIH fellowships TL1RR024147 and 1F31CA154119-01A1 and AP was supported from the Bianca Garavaglia Association, (Busto Arsizio, Varese, Italy).
Footnotes
Supporting Information is available on the WWW under http://www.small-journal.com or from the author.
Contributor Information
Jonathan O. Martinez, Department of Nanomedicine, The Methodist Hospital Research Institute, 6670 Bertner Ave. MS R7-414 Houston, TX 77030 (USA) Graduate School of Biomedical Sciences, University of Texas Health Science Center at Houston, Houston, TX USA.
Alessandro Parodi, Department of Nanomedicine, The Methodist Hospital Research Institute, 6670 Bertner Ave. MS R7-414 Houston, TX 77030 (USA); Department of Experimental Oncology and Molecular Medicine, Fondazione IRCCS Istituto Nazionale Tumori, Milan 20133, Italy.
Xuewu Liu, Department of Nanomedicine, The Methodist Hospital Research Institute, 6670 Bertner Ave. MS R7-414 Houston, TX 77030 (USA).
Mikhail G. Kolonin, The Brown Foundation Institute of Molecular Medicine, University of Texas Health Science Center at Houston, Houston, TX USA
Mauro Ferrari, Department of Nanomedicine, The Methodist Hospital Research Institute, 6670 Bertner Ave. MS R7-414 Houston, TX 77030 (USA).
Ennio Tasciotti, Email: etasciotti@tmhs.org, Department of Nanomedicine, The Methodist Hospital Research Institute, 6670 Bertner Ave. MS R7-414 Houston, TX 77030 (USA).
References
- 1.Lobatto ME, Fuster V, Fayad ZA, Mulder WJ. Nat Rev Drug Discov. 2011;10(11):835–852. doi: 10.1038/nrd3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrari M. Nat Rev Cancer. 2005;5(3):161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 3.Jain RK, Stylianopoulos T. Nat Rev Clin Oncol. 2010;7(11):653–664. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nature nanotechnology. 2007;2(12):751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 5.Krol S, Ellis-Behnke R, Marchetti P. Nanomedicine. 2012 doi: 10.1016/j.nano.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Cho Y, Borgens RB. Exp Neurol. 2012;233(1):126–144. doi: 10.1016/j.expneurol.2011.09.028. [DOI] [PubMed] [Google Scholar]
- 7.Korin N, Kanapathipillai M, Matthews BD, Crescente M, Brill A, Mammoto T, Ghosh K, Jurek S, Bencherif SA, Bhatta D, Coskun AU, Feldman CL, Wagner DD, Ingber DE. Science. 2012 [Google Scholar]
- 8.Holme MN, Fedotenko IA, Abegg D, Althaus J, Babel L, Favarger F, Reiter R, Tanasescu R, Zaffalon PL, Ziegler A, Muller B, Saxer T, Zumbuehl A. Nature nanotechnology. 2012 [Google Scholar]
- 9.Lewinski N, Colvin V, Drezek R. Small. 2008;4(1):26–49. doi: 10.1002/smll.200700595. [DOI] [PubMed] [Google Scholar]
- 10.Soti C, Sreedhar AS, Csermely P. Aging Cell. 2003;2(1):39–45. doi: 10.1046/j.1474-9728.2003.00031.x. [DOI] [PubMed] [Google Scholar]
- 11.Kanduc D, Mittelman A, Serpico R, Sinigaglia E, Sinha AA, Natale C, Santacroce R, Di Corcia MG, Lucchese A, Dini L, Pani P, Santacroce S, Simone S, Bucci R, Farber E. Int J Oncol. 2002;21(1):165–170. [PubMed] [Google Scholar]
- 12.Kumari M, Rajak S, Singh SP, Kumari SI, Kumar PU, Murty US, Mahboob M, Grover P, Rahman MF. J Nanosci Nanotechnol. 2012;12(3):2149–2159. doi: 10.1166/jnn.2012.5796. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Zhou G, Liu H, Niu X, Han J, Zheng L, Fan Y. Nanoscale. 2012;4(9):2894–2899. doi: 10.1039/c2nr00044j. [DOI] [PubMed] [Google Scholar]
- 14.Gao J, Wang HL, Shreve A, Iyer R. Toxicol Appl Pharmacol. 2010;244(2):130–143. doi: 10.1016/j.taap.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 15.Monteiro-Riviere NA, Inman AO, Zhang LW. Toxicol Appl Pharmacol. 2009;234(2):222–235. doi: 10.1016/j.taap.2008.09.030. [DOI] [PubMed] [Google Scholar]
- 16.Albini A, Mussi V, Parodi A, Ventura A, Principi E, Tegami S, Rocchia M, Francheschi E, Sogno I, Cammarota R, Finzi G, Sessa F, Noonan DM, Valbusa U. Nanomedicine. 2010;6(2):277–288. doi: 10.1016/j.nano.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Laaksonen T, Santos H, Vihola H, Salonen J, Riikonen J, Heikkila T, Peltonen L, Kumar N, Murzin DY, Lehto VP, Hirvonen J. Chem Res Toxicol. 2007;20(12):1913–1918. doi: 10.1021/tx700326b. [DOI] [PubMed] [Google Scholar]
- 18.Shen H, You J, Zhang G, Ziemys A, Li Q, Bai L, Deng X, Erm DR, Liu X, Li C, Ferrari M. Advanced Healthcare Materials. 2012;1(1):84–89. doi: 10.1002/adhm.201100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tasciotti E, Liu X, Bhavane R, Plant K, Leonard AD, Price BK, Cheng MM, Decuzzi P, Tour JM, Robertson F, Ferrari M. Nat Nanotechnol. 2008;3(3):151–157. doi: 10.1038/nnano.2008.34. 10.1038/nnano.2008.34. [DOI] [PubMed] [Google Scholar]
- 20.Serda RE, Mack A, van de Ven AL, Ferrati S, Dunner K, Jr, Godin B, Chiappini C, Landry M, Brousseau L, Liu X, Bean AJ, Ferrari M. Small. 2010;6(23):2691–2700. doi: 10.1002/smll.201000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ananta JS, Godin B, Sethi R, Moriggi L, Liu X, Serda RE, Krishnamurthy R, Muthupillai R, Bolskar RD, Helm L, Ferrari M, Wilson LJ, Decuzzi P. Nature nanotechnology. 2010;5(11):815–821. doi: 10.1038/nnano.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakamoto J, Annapragada A, Decuzzi P, Ferrari M. Expert Opin Drug Deliv. 2007;4(4):359–369. doi: 10.1517/17425247.4.4.359. [DOI] [PubMed] [Google Scholar]
- 23.Chiappini C, Tasciotti E, Fakhoury J, Fine D, Pullan L, Wang Y, Fu L, Liu X, Ferrari M. Chem Phys Chem. 2010 doi: 10.1002/cphc.200900914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godin B, Chiappini C, Srinivasan S, Alexander JF, Yokoi K, Ferrari M, Decuzzi P, Liu X. Advanced Functional Materials. 2012 doi: 10.1002/adfm.201200869. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donovan D, Brown NJ, Bishop ET, Lewis CE. Angiogenesis. 2001;4(2):113–121. doi: 10.1023/a:1012218401036. [DOI] [PubMed] [Google Scholar]
- 26.Studeny M, Marini FC, Champlin RE, Zompetta C, Fidler IJ, Andreeff M. Cancer Res. 2002;62(13):3603–3608. [PubMed] [Google Scholar]
- 27.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Science. 1999;284(5411):143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 28.Serda RE, Gu J, Bhavane RC, Liu X, Chiappini C, Decuzzi P, Ferrari M. Biomaterials. 2009;30(13):2440–2448. doi: 10.1016/j.biomaterials.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 29.Serda RE, Ferrati S, Godin B, Tasciotti E, Liu X, Ferrari M. Nanoscale. 2009;1(2):250–259. doi: 10.1039/b9nr00138g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrati S, Mack A, Chiappini C, Liu X, Bean AJ, Ferrari M, Serda RE. Nanoscale. 2010;2(8):1512–1520. doi: 10.1039/c0nr00227e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez JP, Gonzalez M, Rios S, Cambiazo V. J Cell Biochem. 2004;93(4):721–731. doi: 10.1002/jcb.20234. [DOI] [PubMed] [Google Scholar]
- 32.Logie JJ, Ali S, Marshall KM, Heck MM, Walker BR, Hadoke PW. PLoS One. 2010;5(12):e14476. doi: 10.1371/journal.pone.0014476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arnaoutova I, Kleinman HK. Nat Protoc. 2010;5(4):628–635. doi: 10.1038/nprot.2010.6. [DOI] [PubMed] [Google Scholar]
- 34.Kolonin MG, Evans KW, Mani SA, Gomer RH. Stem Cell Res. 2012;8(2):312–323. doi: 10.1016/j.scr.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E. Cytotherapy. 2006;8(4):315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 36.Traktuev DO, Merfeld-Clauss S, Li J, Kolonin M, Arap W, Pasqualini R, Johnstone BH, March KL. Circ Res. 2008;102(1):77–85. doi: 10.1161/CIRCRESAHA.107.159475. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Daquinag A, Traktuev DO, Amaya-Manzanares F, Simmons PJ, March KL, Pasqualini R, Arap W, Kolonin MG. Cancer Res. 2009;69(12):5259–5266. doi: 10.1158/0008-5472.CAN-08-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Studeny M, Marini FC, Dembinski JL, Zompetta C, Cabreira-Hansen M, Bekele BN, Champlin RE, Andreeff M. J Natl Cancer Inst. 2004;96(21):1593–1603. doi: 10.1093/jnci/djh299. [DOI] [PubMed] [Google Scholar]
- 39.Godin B, Tasciotti E, Liu X, Serda RE, Ferrari M. Acc Chem Res. 2011;44(10):979–989. doi: 10.1021/ar200077p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daquinag AC, Zhang Y, Amaya-Manzanares F, Simmons PJ, Kolonin MG. Cell Stem Cell. 2011;9(1):74–86. doi: 10.1016/j.stem.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 41.Serda RE, Mack A, Pulikkathara M, Zaske AM, Chiappini C, Fakhoury JR, Webb D, Godin B, Conyers JL, Liu XW, Bankson JA, Ferrari M. Small. 2010;6(12):1329–1340. doi: 10.1002/smll.201000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.