Abstract
Exon repetition describes the presence of tandemly repeated exons in mRNA in the absence of duplications in the genome. Its existence challenges our understanding of gene expression, because the linear organization of sequences in apparently normal genes must be subverted during RNA synthesis or processing. It is restricted to a small number of genes in some of which over half of the mRNA contains specific patterns of repetition. Although it is sometimes assumed to arise by trans-splicing, there is no evidence of this and the efficiency is very much higher than for examples of bona fide trans-splicing in mammals. Furthermore, a potentially ubiquitous reaction such as trans-splicing is not consistent with a phenomenon that involves such a high proportion of the products of so few genes. Instead, it seems more probable that exon repetition is caused by a specific trans-acting factor. We have tested this and demonstrate for the two best characterized examples that the property is restricted to specific alleles of the affected genes and is determined in cis. It is not determined by exonic splicing signals, as had been suggested previously. In heterozygotes, RNA transcribed from the two alleles of an affected gene can have fundamentally different fates.
INTRODUCTION
The discovery of exon repetition suggested that something extraordinary can happen during RNA processing (1,2). Tandem repeats of specific exons were found originally in a majority of the mRNA from two rat genes, COT (carnitine octanoyl transferase) and Sa (a medium-chain acyl-CoA synthetase) (1,2), although in the case of Sa the repeats were seen only in mRNA from the kidney of specific rat strains (2). Since these reports, exon repetition has been observed in mRNA from a small number of other rat and human genes (3–7). In most cases, the repeats were detected by RT–PCR and confirmed by direct analysis of the mRNA (1,2,4,5,7). In principle, exon repetition might arise from duplications in the genome or unusual RNA processing reactions. Duplications of the repeated exons in the genome have been excluded (1,2,4,5,7). Tandem duplication of the entire gene might allow run-through transcription and splicing, but this too was excluded in the case of Sa (2). Hence, it was inferred that exon repetition must take place at the level of the RNA, and it was the first example of heterogeneity in natural mammalian mRNA that did not arise from the use of alternative signals in linear, contiguous sequences.
The only reaction known at present that might explain exon repetition is intragenic trans-splicing. However, the precedents in mammals do not support this. Most examples of trans-splicing in mammalian cells have involved pairs of partial genes in which transcripts began or ended within the introns (which therefore became ‘outruns’), such that there were unpaired splice sites (8–10). There is no evidence of appropriate internal promoters in the Sa or COT genes. Trans-splicing of intact genes has been reported very rarely, perhaps in part because it could be detected only if it were interallelic, meaning that the mRNA contains sequences originating from two distinguishable alleles, or intergenic, containing sequences from two different genes. A comprehensive analysis of protocadherin gene expression excluded the existence of trans-splicing between transcripts originating from different alleles, although there was evidence of a very low level of probable trans-splicing between tandem gene clusters on the same chromosome (11). The only precedent for efficient trans-splicing between transcripts from one intact gene comes from Drosophila, in which splicing involves alleles of lola on paired homologous chromosomes (12). However, it seems that lola splicing might depend upon the use of alternative promoters to produce transcripts with partial terminal introns (12). We infer that, even if trans-splicing of intact genes were a ubiquitous side-reaction, it could account for neither the high level of exon repetition nor its restriction to so few genes. Exon repetition is clearly special.
If exon repetition is not an occasional aberration of RNA processing, but efficient, it is unclear why it should be restricted to so few genes. The absence of exon repetition in Sa mRNA from one strain of rat suggests that there is a strain-specific trans-acting factor that promotes exon repetition in the mRNA from specific genes. The existence of such a factor would be a strong indication that exon repetition had a biological function. We have investigated exon repetition of the Sa and COT genes, and conclude that it is tightly restricted and not determined by a common trans-acting factor; furthermore, we show that it is a unique property of specific alleles acting in cis.
MATERIALS AND METHODS
Analysis of exon repetition by RT–PCR
RNA extraction, reverse transcription and PCR were as described (2). In all figures, the mRNA was extracted from kidneys. RT–PCR showed that the proportion of COT mRNA with exon repetition was similar in all tissues tested. This proportion was quantified by amplification in the presence of [α-32P]dCTP and measurement of the intensities of the signals after various numbers of cycles. The efficiencies of amplification for each product were determined by fitting a straight line to each plot of log10signal versus cycle number, and used to calculate the starting ratios after cycle 1 (13). The ratios were corrected for the numbers of dCMP moieties incorporated on each strand during amplification. Reverse transcription was done with 2 µg of total RNA and oligo(dT) primers. PCR products were isolated for sequence analysis by electrophoresis on agarose gels, followed by elution using centrifugation through Spin-X filters (Costar). Sequencing was done using Big Dye terminators (Applied BioSystems). PCR conditions were as follows:
Figure 2 panel A: 94°C, 2 min, then 25 cycles of 94°C, 10 s, 55°C, 30 s and 72°C, 1 min 30 s. 72°C, 10 min.
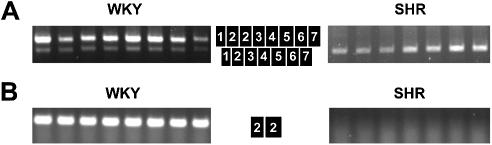
Figure 2.
Exon repetition of COT mRNA in WKY and SHR strains of rat. The presence or absence of exon repetition in COT mRNA was evaluated by RT–PCR followed by separation on agarose gel. Eight WKY and seven SHR animals were used in the experiment. (A) Analysis by amplification with primers COT E1F and COT E7R. This set of primers amplified both the normal COT mRNA and mRNA containing repeated exons, depicted by the adjacent black boxes with white exon numbers. (B) Analysis of COT mRNA by amplification with primers COT E2F and COT E2R, which amplify only a tandem repeat of exon 2.
Figure 2 panel B: 94°C, 2 min, then 28 cycles of 94°C, 45 s, 55°C, 45 s and 72°C, 45 s. 72°C, 10 min.
Figure 3 panels A and B: 94°C, 2 min, then 28 cycles of 94°C, 10 s, 55°C, 30 s, 72°C, 1 min. 72°C, 10 min.
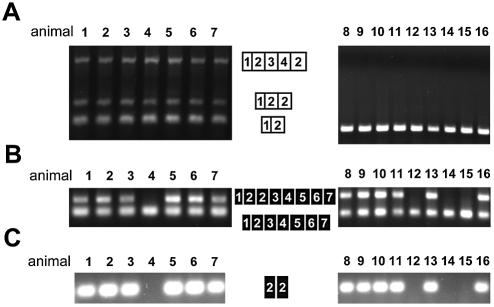
Figure 3.
Segregation of exon repetition in Sa and COT mRNA in an F2 population. Animals from an F2 population derived by crossing F1 individuals (WKY:SHR) were divided into two groups: WKY Sa homozygotes (animals 1–7) and SHR Sa homozygotes (animals 8–16). (A) Analysis of exon repetition in Sa mRNA by RT–PCR with primers SA E1F and SA E2R. The exon organization of the Sa PCR products is shown by the white boxes alongside each PCR product. (B) Analysis of exon repetition in COT mRNA by RT–PCR with primers COT E1F and COT E7R. The exon organization of the COT PCR products is shown by the black boxes. (C) Confirmation of exon repetition in COT mRNA by PCR with primers COT E2F and COT E2R.
Figure 3 panel C: 94°C, 2 min, then 30 cycles of 94°C, 45 s, 55°C, 45 s, 72°C, 45 s. 72°C, 10 min.
Figure 4 panel A and C: 94°C, 2 min, then 30 cycles of 94°C, 10 s, 55°C, 30 s, 72°C, 1 min. 72°C, 10 min.
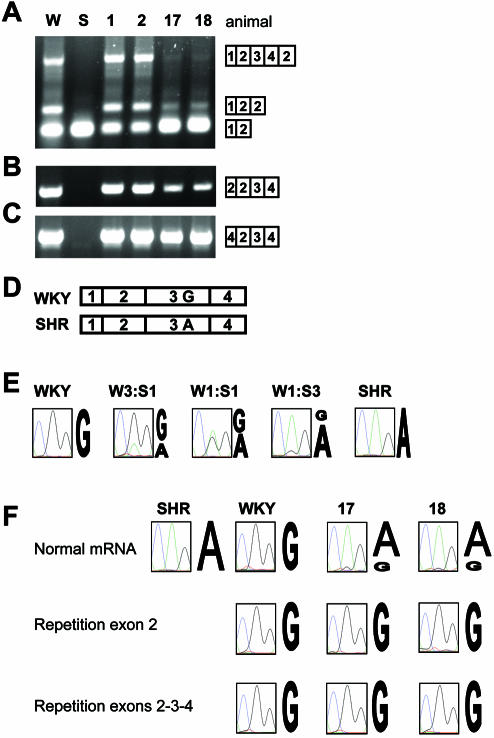
Figure 4.
Identification of the Sa gene alleles producing mRNA with tandemly repeated exons in heterozygotes. Exon repetition in Sa mRNA was evaluated by RT–PCR in the parental rat strains WKY (W), SHR (S), and in four F2 animals, two of which were homozygous for the WKY Sa allele (animals 1 and 2, the same as in Fig. 2); the other two were heterozygotes, having one copy of the Sa gene from each parent (animals 17 and 18). (A) RT–PCR with primers SA E1F and SA E2R. (B) RT–PCR with primer SA 2–2 jct (which amplifies only Sa isoforms that contain a tandem repeat of exon 2) and primer SA E4R. (C) RT–PCR with primers SA 4–2 jct and SA E4R, to specifically amplify Sa cDNA molecules in which exons 2, 3 and 4 are repeated. (D) An alignment of the first four exons of the WKY and SHR Sa cDNA with the SNP in exon 3 (WKY → G, SHR → A). (E) DNA sequence analysis of Sa exon 3 SNP in PCR products from WKY (W) and SHR (S) alleles mixed in the molar ratios shown. (F) Sequence analysis of RT–PCR products from Sa heterozygotes. PCR products corresponding to the normal Sa mRNA (top), exon 2 repetition (middle) and repetition of exons 2, 3 and 4 (bottom) were produced in separate reactions, purified by agarose gel electrophoresis and analyzed by sequencing. Incorporation of A (SHR allele) is green; G (WKY allele) is black.
Figure 4 panel B: 94°C, 2 min, then 30 cycles of 94°C, 10 s, 60°C, 30 s, 72°C, 1 min. 72°C, 10 min.
Figure 5 panel A: 94°C, 2 min, then 35 cycles of 94°C, 10 s, 55°C, 30 s, 72°C, 1 min. 72°C, 10 min.
Figure 5.
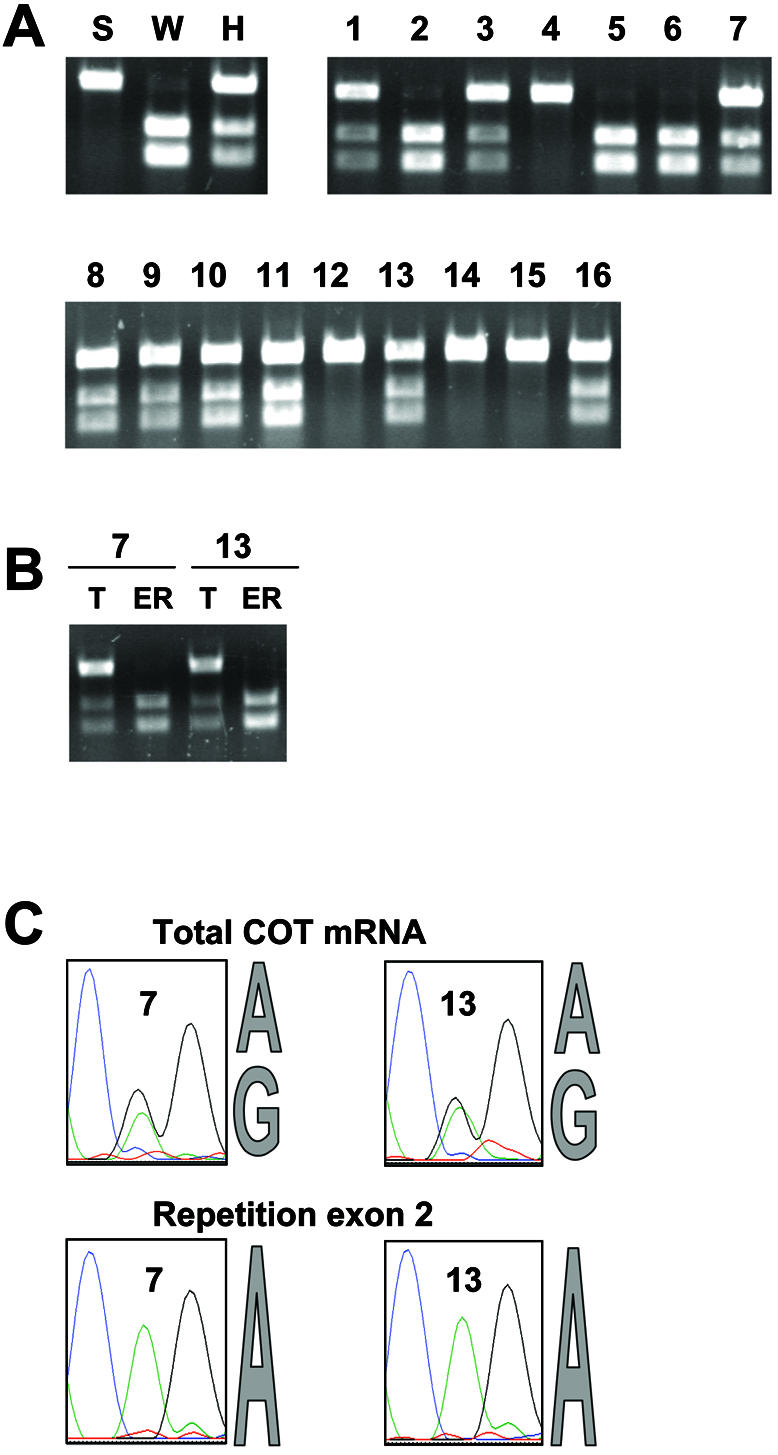
Analysis of COT gene alleles involved in exon repetition in heterozygotes. The segregation of the WKY and SHR COT alleles was studied in the F2 population of Figure 3. A SNP in the COT 3′UTR was used to analyze the distribution of the COT alleles in the F2 population. The point mutation creates a recognition sequence for the restriction endonuclease AflII in the WKY COT allele. Thus, amplification products from the COT 3′UTR can be cleaved into two fragments by AflII only if they contain the WKY allele. (A) Identification of COT alleles expressed in total mRNA from F2 animals derived from a WKY × SHR cross. The animals are numbered as in Figure 3. The results are also shown for the parental lines (S, W) and an F1 heterozygote (H). (B) Contributions of the two COT alleles in heterozygotes (animals 7 and 13) to mRNA isoforms. T, COT mRNA was amplified between exons 1 and 17; it was re-amplified by PCR between exons 16 and 17, and analyzed by digestion with AflII and agarose gel electrophoresis. This procedure is similar to that in (A), with prior amplification between exon 1 and 17. ER, exon repetition-containing COT mRNA was amplified between the exon 2–2 jct and exon 17, and then re-amplified and analyzed as for T. (C) Sequence analysis of products of amplification between exons 16 and 17. As in (B), PCR products derived initially by RT–PCR between exons 1 and 17 represent the alleles present in total mRNA, whereas those derived initially by RT–PCR with a primer specific for the tandem repetition of exon 2 represent the alleles present in exon repetition isoforms.
Figure 5 panel B: first round PCR with primers COT E1F and COT E17R: 94°C, 2 min, then 38 cycles of 94°C, 10 s, 55°C, 30 s, 72°C, 3 min. 72°C, 10 min.
First round PCR, primers COT 2–2 jct and COT E17R: 94°C, 2 min, then five cycles of 94°C, 10 s, 65°C, 30 s, 72°C, 3 min, followed by five cycles of 94°C, 10 s, 63°C, 30 s, 72°C, 3 min, then 25 cycles of 94°C, 10 s, 60°C, 30 s, 72°C, 3 min. 72°C, 10 min.
Second round of PCR, primers COT E16F and COT E17R: 94°C, 2 min, then 30 cycles of 94°C, 10 s, 55°C, 30 s, 72°C, 1 min. 72°C, 10 min.
Primer sequences
COT E1F, GAGTGCAGAGAGCCAAGCCGGGTGCA; COT E3R, TGATGCAATGTCTTGCCAAC; COT E7R, GGCCCAACAGGTTCATTCCAG; COT E2F, GGACTCTC TTCCGCCCTTGC; COT E2R, GGAATGTTCGTTCTT CAATTGAC; COT 2–2 jct, CTTGAGTCAGTTTTCTT ACT; COT E16F, TTCCCATGGTACATAATGGATAC; COT E17R, CATGAAATCTCATAGGCTTG; SA E1F, GGCTTTCTCTCCCATTAAGCAGGTCTA; SA E2R, ATG GATTTGGTGGTTAGGCATCG; SA 2–2 jct, GCCTAA CCACCAAATCCATGTGAAAAACAA; SA 4–2 jct, AAATGTAGCCTGTCTGCGAACAGGTGAAAAAC; SA E4R, CACCACTCTGGGATCTTGGGCAGAATCACC.
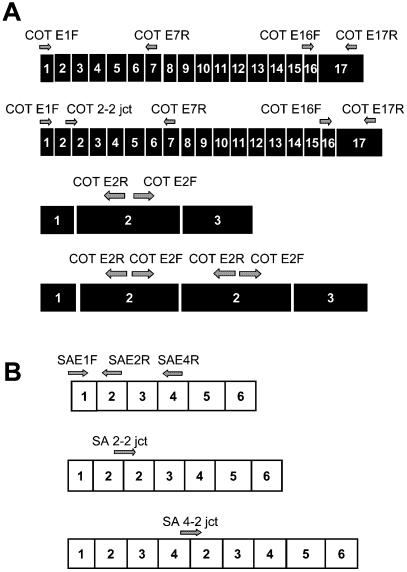
Figure 1 shows the sites of annealing of the primers on COT and SA mRNA.
Figure 1.
Schematic representation of the mRNA isoforms expressed by the rat COT and Sa genes, and of the primers used in the PCR reactions. (A) Analysis of exon repetition in the COT mRNA. A normal COT mRNA and a COT mRNA in which exon 2 is repeated are illustrated, with (below) an expanded view of exons 1–3 in each case. Primers specific for the COT gene are shown as arrows (5′ to 3′) and named. The COT 2–2 jct primer was designed to amplify a PCR product only if exon 2 is repeated. (B) Analysis of exon repetition in the Sa mRNA. A portion of Sa cDNA is shown (exons 1–6). Three Sa cDNA molecules are depicted. One corresponds to the normal cDNA. The second one contains a repetition of exon 2. Finally, in the third Sa cDNA, exons 2, 3 and 4 are repeated. The primer referred to as SA 2–2 jct specifically amplified transcripts containing a repetition of exon 2, while primer SA 4–2 jct detected the simultaneous repetition of exons 2, 3 and 4.
Gene sequences
The sequences of cloned Sa genes from WKY and SHR rats have been deposited with accession numbers AY455861 and AY456695, respectively.
RESULTS
The rat Sa gene is expressed in the liver and in kidney proximal tubules, and it is expressed more abundantly in the spontaneously hypertensive (SHR) rat than in the normotensive Wistar–Kyoto (WKY) rat (14,15). However, exon repetition in Sa is restricted to the WKY rat, in which either exon 2 or exons 2, 3 and 4 are tandemly repeated (2). Exon repetition in COT mRNA has been investigated in Sprague–Dawley (SD) rats, in which either exon 2 or exons 2 and 3 are tandemly repeated, and the detection of a protein product suggested that repetition might have a biological role (1). If exon repetition was determined by a common trans-acting factor, then it would be predicted that SHR rats would not show repetition in COT mRNA. To test this, COT mRNA from WKY and SHR rats was examined by RT–PCR. Amplification between exons 1 and 7 (Fig. 2A) in mRNA from eight WKY rats showed a very strong signal from exon repetition-containing mRNA compared with normal mRNA, confirmed by RT–PCR between the two copies of exon 2 (Fig. 2B). Quantitative PCR showed that the fraction of cDNA molecules with repetition of exon 2 was 54% (see Supplementary Material). However, the parallel experiments with mRNA from seven SHR rats showed no evidence of exon repetition. This shows that COT exon repetition occurs in the same rats as Sa exon repetition, consistent with the hypothesis of a common trans-acting factor. The results also show that the exon repetition of COT mRNA is non-essential, even though at least one exon repetition isoform appears to be translated (1).
It has been suggested previously that exon repetition in COT mRNA depended on a purine-rich enhancer sequence in exon 2, based on splicing reactions with exogenous RNA and experiments with transfected mini-genes (16). To examine whether the failure of the SHR COT mRNA to produce repetition isoforms was caused by a difference in the exon sequence, COT cDNA was amplified by PCR from WKY, SD and SHR rat strains and sequenced. Exon 2 is identical in all three strains. We conclude that, even if the purine-rich sequence in exon 2 is required for exon repetition, it is not sufficient. Furthermore, all of the exons in SD and SHR rats are identical, even though a high proportion of COT mRNA from the SD rats contains repeated exons whereas mRNA from SHR contains no repetition. Thus, exonic sequences do not determine the property of exon repetition in the SD rat strain.
The possibility that a trans-acting factor might predispose specific lines of rats to exon repetition was tested further by examining whether the property of exon repetition in the Sa and COT genes co-segregates when WKY and SHR rats are crossed. First, the Sa genotypes of rats in the second filial (F2) generation from the cross were determined, and RT–PCR was used to determine their pattern of exon repetition. Rats homozygous for the WKY allele of the Sa gene all expressed mRNA with repetition of either exon 2 or exons 2, 3 and 4 (animals 1–7; Fig. 3A); rats homozygous for the SHR allele showed no exon repetition (animals 8–16; Fig. 3A). In both cases, the result matched the phenotype of the parental rats with the same Sa genotype. If exon repetition of the Sa mRNA depended upon a trans-acting factor, then it is likely to be linked to the Sa WKY allele.
When the same rats were screened by RT–PCR of COT RNA, it was evident that exon repetition in COT mRNA did not co-segregate with exon repetition in Sa mRNA (animal 4 showed exon repetition in Sa but not COT mRNA, and animals 8, 9, 10, 11, 13 and 16 showed exon repetition in COT but not Sa mRNA; Fig. 3B and C, cf. Fig. 3A). The level of exon repetition appeared to vary somewhat in the positive animals (Fig. 3B; see below). We conclude that Sa and COT exon repetition are not determined by a common trans-acting factor but are caused either by separate trans-acting factors or by cis-acting properties of the WKY and SHR alleles of each gene.
The contributions of trans-acting and cis-acting factors in exon repetition were tested by determining which alleles produced the mRNA isoforms containing repeated exons in Sa heterozygotes. The allele from which the RNA originates can be identified via a polymorphism in exon 3. Genotype analysis of the F2 population revealed a number of rats that contained the WKY and SHR alleles of the Sa gene. Two of these were analyzed. Amplification between exons 1 and 2 of Sa mRNA produced bands indicative of exon repetition (animals 17 and 18; Fig. 4A), but they were weaker than in rats homozygous for the WKY allele (animals 1 and 2). This conclusion was confirmed by two further analyses with different sets of primers (Fig. 4B and C). The contribution of each allele to the different mRNA isoforms was determined by sequence analysis of the PCR products. To test whether sequence analysis would give semi-quantitative data, PCR products from normal mRNA from the two homozygous parent lines were derived by amplification between exons 1 and 4 (Fig. 4D), mixed in various ratios and subjected to sequencing. Figure 4E shows that the heights of the peaks at the position of the single nucleotide polymorphism (SNP) in exon 3 were correlated with the starting proportions of the alleles in the mixture of PCR products, although the signal from the WKY allele was a little weaker. The mRNA isoforms in the F2 population were analyzed by sequencing three RT–PCR products: normal mRNA amplified by primers in exons 1 and 4, exon 2 repetition mRNA amplified by a primer specific for the junction of exon 2 with exon 2 and a reverse primer in exon 4 (as in Fig. 4B), and exon 2-3-4 repetition mRNA amplified by a primer specific for the junction of exon 4 with exon 2 and a reverse primer in exon 4 (as in Fig. 4C). Sequencing across the SNP in exon 3 showed that in the two heterozygotes tested (Fig. 4F, animals 17 and 18), the normal mRNA derived predominantly from the SHR allele (Fig. 4F, normal mRNA), whereas mRNA containing repetition of exon 2 (Fig. 4F, repetition exon 2) and mRNA containing the exon 2-3-4 repetition (Fig. 4F, repetition exons 2, 3 and 4) gave signals identical to those from these isoforms in the WKY homozygote (Fig. 4F, W). We conclude that mRNA isoforms with exon repetition in the heterozygote are derived from the WKY allele alone, and therefore that exon repetition is a cis-acting property of that allele. The lack of a contribution to the repetition isoforms from the SHR allele is consistent with the relatively faint level of exon repetition in animals 17 and 18 in Figure 4A.
The analysis was repeated for COT mRNA. To characterize the COT genotype in the F2 population, mRNA from the F2 animals was tested for the presence of a WKY-specific AflII site in exon 17. The assignments were confirmed by genomic PCR (not shown). The results in Figure 5A show that the digestion products (lower bands) associated with the presence of a WKY allele are seen in animals 1, 2, 3, 5, 6, 7, 8, 9, 10, 11, 13 and 16, all of which showed exon repetition in COT mRNA in Figure 3. We conclude that COT exon repetition segregates with the presence of a WKY allele of the COT gene. Hence, exon repetition is a cis-acting property of the allele or it is determined by a trans-acting factor closely linked to the WKY allele.
The two possibilities could be distinguished by determining whether in heterozygotes the RNA showing exon repetition was derived from both alleles or only the WKY allele, as was the case for Sa mRNA. However, the analysis of exon repetition in heterozygotes could not be done directly, because the SNP was distant from the repeated exons. Instead, mRNA with a repetition of exon 2 was amplified by a primer specific for the junction of exon 2 with exon 2 and a reverse primer in exon 17, and re-amplified in a second round of PCR between exons 16 and 17, as above. Both digestion with AflII (Fig. 5B) and sequence analysis of a second SNP in exon 17 (Fig. 5C) showed that the exon repetition-containing mRNA was derived from the WKY allele. As a control, total COT mRNA was amplified between exons 1 and 17, and then analyzed by re-amplification and both digestion and sequencing. The two alleles were present in almost equal concentrations. Hence, just as with the Sa gene, the property of exon repetition in the COT gene is restricted to the WKY allele even in a heterozygote. This explains the variation in the apparent level of exon repetition seen in Figure 3B: in the left-hand panel, animals 2, 5 and 6 were homozygous for the WKY allele, whereas animals 1, 3 and 7 were heterozygotes and the SHR allele would not have contributed to the exon repetition signal. The positive animals in the right-hand panel were all heterozygotes and gave a more uniform apparent level of repetition.
DISCUSSION
Exon repetition was an extraordinary and unexpected phenomenon that appeared to subvert our understanding that genes were expressed via a linear series of signals on contiguous sequences of DNA or RNA. However, it has never been clear whether it was essential. It had been shown that mRNA containing repetition of exons 2 and 3 of COT is translated to produce a longer protein, from which it was inferred that exon repetition might have functional significance (1), whereas we had found that the exon repetition in Sa mRNA from WKY rats was absent in SHR rats (2). In this work, we have demonstrated that SHR rats do not show exon repetition in COT mRNA either. Thus, the best characterized and most efficient examples of exon repetition are inessential. In addition, we have examined the effect of the repetition of COT exon 2 (which contains the initiation codon) upon expression of a reporter function, and the results show that the upstream, out of frame initiation codon reduces expression substantially (R.Rigatti, unpublished data). Since this is the major isoform in WKY rats, the implication is that exon repetition is actually detrimental to COT expression in these rats. These results only deepen the mystery surrounding exon repetition. There is no obvious reason why such an extraordinary property should have arisen, one that does not arise from a background level of imprecision in splicing but is emphatically specific and efficient.
The allele specificity of exon repetition in the heterozygotes demonstrates that it is not determined by diffusible factors, whether encoded on the same chromosome or elsewhere; the determinants are allele-specific. Allele-specific expression is usually associated with quantitative considerations of transcription efficiency or the proportion of two different splicing isoforms. In the case of exon repetition, it appears that allelic differences acting strictly in cis determine whether or not a transcript will undergo qualitatively different and extraordinary reactions.
The differences in sequence between the two alleles of the Sa gene suggest few candidate determinants. Within the 1.2 kb preceding exon 1, there are six single nucleotide differences (SNPs) between the WKY and SHR alleles, and a difference of five nucleotides in the length of an oligo(A) tract. The first five exons differ at only one position. Most of the introns contain only a few differences, but intron 1 contains 39 differences in 5.3 kb (25 SNPs and differences in tracts of di- or tri-nucleotide repeats), and there is a LINE element of 1.4 kb in SHR that is missing in the WKY allele. Since the LINE element is found only in the strain that does not display exon repetition, its only potential role would be as a suppressor rather than a determinant of exon repetition. In the case of the COT gene, our analysis of the genomic sequences is incomplete; we have not yet found any differences between the SHR allele and the SD allele, and we cannot exclude the possibility that the determinants of exon repetition are epigenetic.
Allele specificity excludes a number of possible explanations for exon repetition, including the simplest form of the idea that it arises by trans-splicing. It is not consistent with the idea that freely diffusing pre-mRNA sequences undergoing processing are brought into close proximity by specific RNA-binding proteins and that trans-splicing takes place. There is evidence from studies of intergenic trans-splicing that chromosomal proximity is required. For example, it was observed in protocadherin that there was no trans-splicing between alleles, but there was an extremely low level (less than 0.5% of mRNA) of trans-splicing between gene clusters on the same chromosome (11). In the lola gene of Drosophila, there is a high level of interallelic trans-splicing (possibly resulting from internal promoters), but this requires the proper pairing of chromosome homologues (12). This suggests that intragenic trans-splicing might be highly efficient.
If trans-splicing is involved in exon repetition, then the most plausible explanation of allele specificity is that the reaction is required to take place while the transcripts are associated with the gene. The differences between the alleles may well affect signals involved in transcription, with very remarkable consequences, and it will be of great interest to determine what the signals are and why the vast majority of genes lack them.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the Medical Research Council, UK, and the Wellcome Trust, UK.
DDBJ/EMBL/GenBank accession nos AY455861, AY456695
REFERENCES
- 1.Caudevilla C., Serra,D., Miliar,A., Codony,C., Asins,G., Bach,M. and Hegardt,F.G. (1998) Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. Proc. Natl Acad. Sci. USA, 95, 12185–12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frantz S.A., Thiara,A.S., Lodwick,D., Ng,L.L., Eperon,I.C. and Samani,N.J. (1999) Exon repetition in mRNA. Proc. Natl Acad. Sci. USA, 96, 5400–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hide W.A., Babenko,V.N., van Heusden,P.A., Seoighe,C. and Kelso,J.F. (2001) The contribution of exon-skipping events on chromosome 22 to protein coding diversity. Genome Res., 11, 1848–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flouriot G., Brand,H., Seraphin,B. and Gannon,F. (2002) Natural trans-spliced mRNAs are generated from the human estrogen receptor-alpha (hER alpha) gene. J. Biol. Chem., 277, 26244–26251. [DOI] [PubMed] [Google Scholar]
- 5.Takahara T., Kanazu,S.I., Yanagisawa,S. and Akanuma,H. (2000) Heterogeneous Sp1 mRNAs in human HepG2 cells include a product of homotypic trans-splicing. J. Biol. Chem., 275, 38067–38072. [DOI] [PubMed] [Google Scholar]
- 6.Finta C. and Zaphiropoulos,P.G. (2000) The human CYP2C locus: a prototype for intergenic and exon repetition splicing events. Genomics, 63, 433–438. [DOI] [PubMed] [Google Scholar]
- 7.Akopian A.N., Okuse,K., Souslova,V., England,S., Ogata,N. and Wood,J.N. (1999) Trans-splicing of a voltage-gated sodium channel is regulated by nerve growth factor. FEBS Lett., 445, 177–182. [DOI] [PubMed] [Google Scholar]
- 8.Eul J., Graessmann,M. and Graessmann,A. (1996) Trans-splicing and alternative-tandem-cis-splicing: two ways by which mammalian cells generate a truncated SV40 T-antigen. Nucleic Acids Res., 24, 1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eul J., Graessmann,M. and Graessmann,A. (1995) Experimental evidence for RNA trans-splicing in mammalian cells. EMBO J., 14, 3226–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X., Su,H. and Bradley,A. (2002) Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev., 16, 1890–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tasic B., Nabholz,C.E., Baldwin,K.K., Kim,Y., Rueckert,E.H., Ribich,S.A., Cramer,P., Wu,Q., Axel,R. and Maniatis,T. (2002) Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol. Cell, 10, 21–33. [DOI] [PubMed] [Google Scholar]
- 12.Horiuchi T., Giniger,E. and Aigaki,T. (2003) Alternative trans-splicing of constant and variable exons of a Drosophila axon guidance gene, lola. Genes Dev., 17, 2496–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eperon I.C. and Krainer,A.R. (1994) Splicing of mRNA precursors in mammalian cells. In Higgins,S.J. and Hames,B.D. (eds), RNA Processing: A Practical Approach. IRL Press, Oxford, Vol. I, pp. 57–101. [Google Scholar]
- 14.Kaiser M.A., Lodwick,D. and Samani,N.J. (1994) The rat SA gene shows genotype-dependent tissue-specific expression. Clin. Sci. (Lond.), 87, 1–4. [DOI] [PubMed] [Google Scholar]
- 15.Iwai N. and Inagami,T. (1991) Isolation of preferentially expressed genes in the kidneys of hypertensive rats. Hypertension, 17, 161–169. [DOI] [PubMed] [Google Scholar]
- 16.Caudevilla C., Codony,C., Serra,D., Plasencia,G., Roman,R., Graessmann,A., Asins,G., Bach-Elias,M. and Hegardt,F.G. (2001) Localization of an exonic splicing enhancer responsible for mammalian natural trans-splicing. Nucleic Acids Res., 29, 3108–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.