Abstract
Introduction
Gene expression signatures have been identified for epithelial ovarian cancer survival (TCGA) and intrinsic subtypes (Tothill et al.). One obstacle to clinical translation is these signatures were developed using frozen tissue, whereas usually only formalin-fixed, paraffin embedded (FFPE) tissue is available. The aim of this study was to determine if gene expression signatures can be translated to fixed archival tissues.
Methods
RNA extracted from FFPE sections from 240 primary ovarian cancers were analyzed by DASL on Illumina BeadChip arrays. Concordance of expression at the individual gene level was assessed by comparing array data from the same cancers (30 frozen samples analyzed on Affymetrix arrays versus FFPE DASL).
Results
The correlation between FFPE and frozen survival signature estimates was 0.774. The TCGA signature using DASL was predictive of survival in 106 advanced stage high grade serous ovarian cancers (median survival 33 versus 60 months, estimated hazard ratio for death 2.30, p=0.0007). Similar to Tothill, we found using DASL that most high grade serous ovarian cancers (102/110, 93%) were assigned to subtypes 1, 2, 4 and 5, whereas most endometrioid, clear cell, mucinous and low grade serous cases (39/57, 68%) were assigned to subtypes 3 and 6 (p<10e-15).
Conclusions
Although individual probe estimates of microarrays may be weakly correlated between FFPE and frozen samples, combinations of probes have robust ability to predict survival and subtype. This suggests that it may be possible to use these signatures for prognostic and predictive purposes as we seek to individualize the treatment of ovarian cancer.
Keywords: ovarian cancer, gene expression, microarray, survival, histological subtype
Introduction
Epithelial ovarian cancer is a leading cause of cancer death in US women because most cases present at an advanced stage. Clinically useful molecular markers that are both prognostic and predict response to specific therapies have been discovered for some cancer types. In this regard, the multi-marker OncotypeDx test has become widely adopted to predict recurrence risk in breast cancer [1] and assessment of HER-2/neu amplification is used to guide the use of trastuzumab [2]. Similar tests do not exist for ovarian cancer and this has been a limitation in the management of the disease.
Our group and others have sought to use expression microarrays to identify signatures that are predictive of outcome and other phenotypes in ovarian cancer [3–9]. As has been noted in other cancer types, there is little overlap in the gene lists that comprise the ovarian cancer survival signatures derived by various groups. Possible explanations for the lack of overlap include use of different microarray platforms as well as false-discovery due to multiple comparisons inherent in large data sets. Alternatively, the differences in gene lists may reflect the complexity of discoverable pathways involved in determining ovarian cancer outcome. More recently, The Cancer Genome Atlas Project (TCGA) has embarked on a genomic analysis of various types of human cancers. High grade serous ovarian cancer was among the first cancers characterized by TCGA. This histologic subtype comprises about 60% of epithelial ovarian cancers and is responsible for most disease-related deaths. Data from 489 tumors analyzed by TCGA was published and made publically available in 2011[10]. In the TCGA ovarian cancer project, a robust gene expression signature for overall survival was developed and then validated in several other existing data sets. The TCGA also identified four gene expression “intrinsic subtypes” of high grade serous ovarian cancer based on patterns of gene expression (mesenchymal, immunoreactive, differentiated, proliferative) that are highly congruent with the subtypes described by Tothill et al. [9]. These patterns appear to reflect basic biologic heterogeneity within high grade serous cancers.
Nearly all prior studies that have developed gene expression signatures of ovarian cancer phenotypes have used fresh frozen tumor samples, including the TCGA. This may represent a significant obstacle to clinical application, since only formalin fixed, paraffin embedded (FFPE) tissue is typically available. In this regard, both OncotypeDx and HER-2/neu testing generally are performed on FFPE tissue blocks. The aim of the present study was to determine if the TCGA high grade serous survival signature and Tothill ovarian cancer subtype expression signatures can be reliably detected in archival FFPE tissue using the cDNA-mediated annealing, selection, extension and ligation (DASL) assay [11]. We chose this method as it was the most widely used approach for whole genome profiling from FFPE at the time of the analysis.
Methods
Study subjects
Samples were obtained from 240 separate invasive epithelial ovarian cancer cases. These included a set of 43 invasive high grade (moderately or poorly differentiated) serous ovarian cancer cases from the Duke University IRB approved frozen gynecologic tumor bank and 197 cases of various histologic types from the North Carolina Ovarian Cancer Study (NCOCS). The NCOCS is an IRB approved population-based case-control study initiated in 1999 using rapid case ascertainment to identify incident ovarian cancers in 50 counties in North Carolina [12]. Among the 197 NCOCS cases, 128 were serous, 26 endometrioid, 9 clear cell, 7 mucinous and 27 mixed/other histologic types. Grade distribution among the NCOCS cases was 29 well differentiated (15%), 68 moderately differentiated (35%) and 99 poorly differentiated (51%). Of the 108 advanced stage (III/IV) high grade serous cancers (54 moderately differentiated, 54 poorly differentiated and 54 were optimally debulked), 106 of these were used to validate the TCGA survival signature. Two cases were eliminated as technical outliers from our QC analysis. All cases were reviewed by a study pathologist at Duke University to confirm these histologic designations.
RNA isolation
From hematoxylin and eosin stained sections, percent tumor nuclei was quantified and ranged from 10 – 90% with a median of 70%. RNA was extracted from two slides containing 5µm-thick sections of tumor. Total RNA was isolated using the Ambion Recover All Total Nucleic Acid Isolation kit (Cat. No.AM1975) according to the manufacturer’s protocol. RNA concentration was determined with a ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA). The normal metric of RNA quality, 28S:18S rRNA ratio, is not useful for the degraded RNA derived from FFPE tissue. Therefore to obtain a quantitative metric of RNA quality real time RT-PCR was performed to amplify small regions of two abundant mRNAs, the 18s rRNA (F 5’CGAAGACGATCAGATACCGT, R 5’GGTCATGGGAATAACGCCG; 77bp amplicon) and gACTB (F 5’CAGCAGATGTGGATCAGCAAG, R 5’GCATTTGCGGTGGACGAT; 65bp amplicon).
Whole genome DASL microarray analysis
In brief, a total of 200ng of RNA was analyzed using the Illumina Whole-Genome DASL (cDNA-mediated annealing, selection, extension, and ligation) assay with the HumanRef-8 Bead Chip (Cat. No. DA-903-1024) corresponding to 24,526 gene transcripts or 18,631 unique genes. Total RNA was converted to cDNA using biotinylated oligo-dT18 and random nonamer primers, followed by immobilization to a streptavidin-coated solid support. The biotinylated cDNAs were then simultaneously annealed to a set of assay-specific oligonucleotides designed to correspond to 24526 probes (18626 unique genes), based on content derived from the National Center for Biotechnology Information Reference Sequence Database (Build 36.2, Release 22). Extension and ligation of the annealed oligonucleotides generated PCR templates that were amplified using fluorescence-labeled and biotinylated universal primers. The labeled PCR products were then captured on streptavidin paramagnetic beads, washed, and denatured to yield single-stranded fluorescent molecules which were hybridized to Whole Genome gene expression BeadChips (HumanRef-8, Illumina) for 16 h at 58 °C. Images were extracted and fluorescence intensities were read on a BeadArray Reader and scanned data were uploaded into BeadStudio for further analysis.
Expression analysis
In the 43 cases from our institutional bank, expression data from fresh frozen tissue was derived from Affymetrix U133A arrays. The justRMA R/Bioconductor implementation of RMA [13] was used at its default settings to normalize, background correct and estimate expression on the log2 scale for the Affymetrix data. The DASL data for 13 of the 43 paired samples and four of the 197 NCOCS samples failed to meet laboratory quality control standards and were removed from the analysis. We used neqc from the limma package [14] using its default settings to background correct and normalize the remaining DASL data after removing probes not expressed in all samples following Shi et al. [15]. Finally, we used limma’s removeBatchEffect to adjust for batch. Affymetrix and Illumina probes were matched on the basis of the RefSeq ID(s) assigned to each. Illumina provides a single RefSeq ID in its annotation data. The 'hgu133aREFSEQ' data set distributed with the R/Bioconductor package hgu133a.db was used to associate RefSeq IDs with the Affymetrix probes. DASL probes were mapped to Affymetrix probes by gene symbol. For each Affymetrix probe that matched one or more DASL probes, the median expression across those DASL probes was used to represent the DASL estimate corresponding to that Affymetrix probe. The subtype signatures are defined in terms of probe-level expression estimates while the TCGA survival signature is defined in terms of gene-level estimates. For purposes of the subtype analysis, we normalized the Affymetrix and DASL data for the remaining probes separately, so that across samples in each set the probe means are all zero and the probe SDs are all one; for purposes of the survival signature, we calculated gene level estimates of expression by taking the median probe level estimate of expression for each gene prior to normalization.
Statistical analysis
The TCGA survival signature gene set comprises 193 genes, 85 with increased expression related to a "good" prognosis and 108 with increased expression related to a "poor" prognosis [10]. We identified 166 genes in the normalized gene-level expression sets that matched a gene in the TCGA gene set. The TCGA reported the log hazard estimate for each gene used in the TCGA survival analysis (Supplementary Table 6.1) [10]. We defined the survival signature as the weighted sum of the normalized gene-level DASL expression values where each gene in the signature was weighted by the product of its published log hazard estimate and its squared Affymetrix/DASL correlation in the 30 paired samples. We normalized the survival signature to have mean zero and standard deviation one in the 193 cases from the North Carolina Ovarian Cancer study.
Tothill et al. published (Supplementary Table S2) the 4,809 Affymetrix U133A probes that were at least 2-fold over or under expressed in each of six subtype clusters (5). The 1806 of these that mapped directly to an Affymetrix U133A probe were used to create a 'signature' associated with each of the 6 original Tothill subtypes. In the 193 cases from the North Carolina Ovarian Cancer study, the signature for each subtype was defined as a weighted sum of the normalized expression values for the over-expressed probes minus a weighted sum of the normalized expression values for the under-expressed probes. We weighted each probe in the signature functions by the square of its correlation between the Affymetrix and DASL log2 expression estimates calculated in the 30 paired samples. This was to ensure that the transcripts measured most accurately by DASL had the most influence. Thus, the value of a subtype signature is large and positive when the case is likely to belong in that subtype and small when it is not.
Results
Evaluating RNA and DASL quality
A challenge of gene expression array studies is to understand and control for unwanted variables that are not related to RNA levels in the tissues. From frozen or fresh material the quality of the extracted RNA should not be a major source of variation but variably degraded and modified RNA in FFPE is typical, and therefore samples cannot be excluded based on the standard metric i.e., the 28S:18S ratio. The majority of samples in this study were derived from a North Carolina population based case control study of ovarian cancer. Tissue blocks were obtained from participating institutions, sectioned, and then returned. FFPE sections were stored at 4°C without desiccation from 179 to 3399 days (median of 1660 days) before RNA was extracted for this current study. The number of days stored did not correlate with the amount of RNA obtained or its quality as assessed by qRT-PCR for actin and the 18S rRNA indicating that time of storage under these conditions did not adversely affect the RNA in these sections (supplementary Figure S1).
The DASL assay itself produced a series of quality control metrics related to the final hybridization and detection on the bead array. Of these metrics, the percentage of positive probes is a primary measure of the overall success of the assay. Increased time of storage was positively correlated with percentage of positive probes (Pearson r = 0.22, P= 0.0017), again indicating that there was no degradation of signal based on extensive storage of the sections.
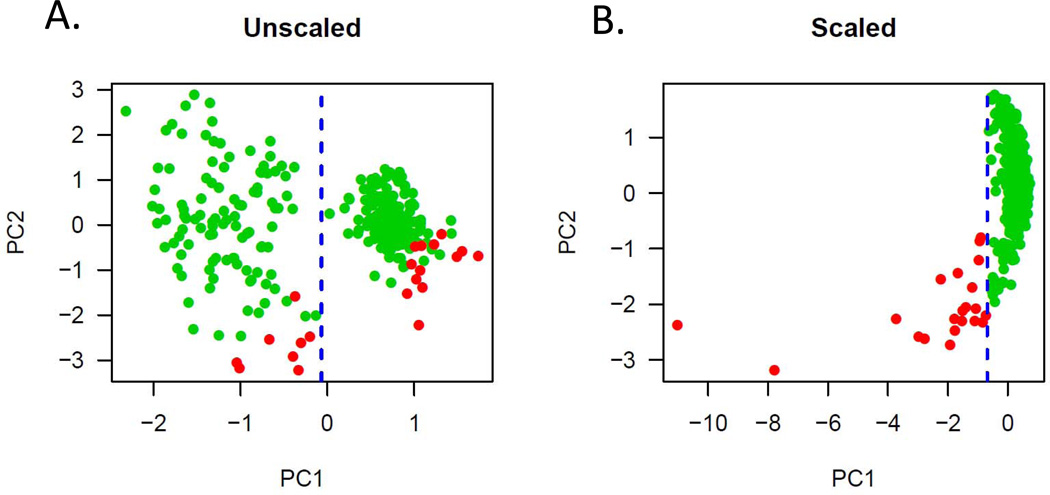
While age of the specimen was not a major factor, principal component (PC) analysis of the quality metrics and the DASL expression data revealed significant structure in the data inconsistent with true biologic variation. The first PC (PC1), separating samples into two distinct groups, was found to be correlated with the specific BeadChip (8 assays per chip or batch) on which each specific sample was run (Figure 1A). Once this structure was revealed, scaling of the expression data based on this variable (batch) produced a uni-modal distribution (Figure 1B). Importantly, the values of PC1 after this correction correlated very strongly with both our metrics (qRT-PCR for actin: Pearson r = −0.50, P<10e-15; qRT-PCR for 18s rRNA: Pearson r = −0.52, P<10e-15) and Illumina BeadChip metrics (percent positive probes: Pearson r = 0.74, P<10e-15). Finally, based on this PC grouping, we eliminated outliers (PC1< −0.69) for the analyses described below (outliers indicated in red, Figure 1B). Only through analysis of all available quality control data could we logically and confidently remove specific samples. Further, these data indicate that qRT-PCR is an accurate predictor for the utility of degraded RNA samples for high throughput expression analysis.
Figure 1.
Principal component analysis of the DASL expression data on all samples.
A) Unscaled data demonstrating an obvious bi-modal distribution along the PC1 space. This distribution was found to correlate with batch and presents a systematic variable in the data not related to the type of cancer.
B) After batch correction, PC1 now distinguishes outliers that correlate with RNA quality (by PCR) and percent positive probes on the array. These outliers in red were excluded from subsequent analyses.
Testing the TCGA survival signature in FFPE samples
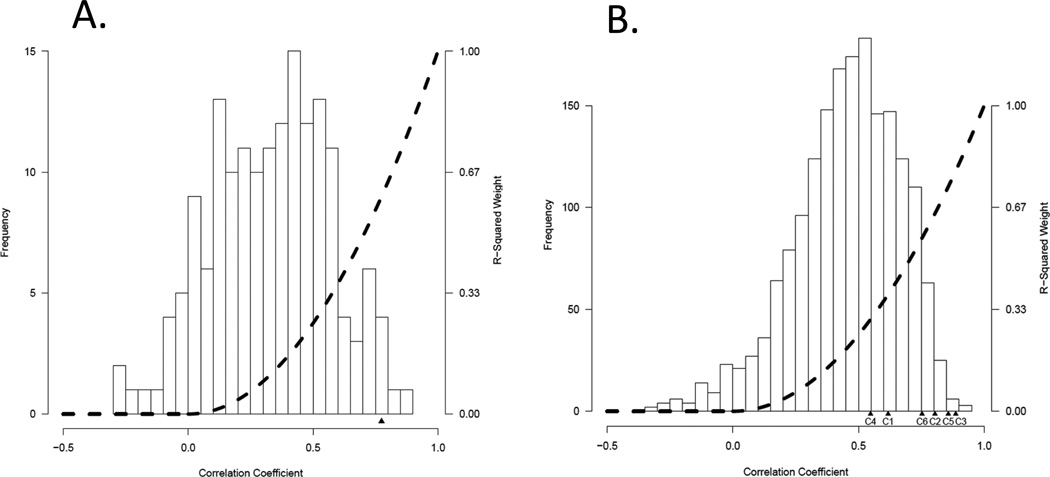
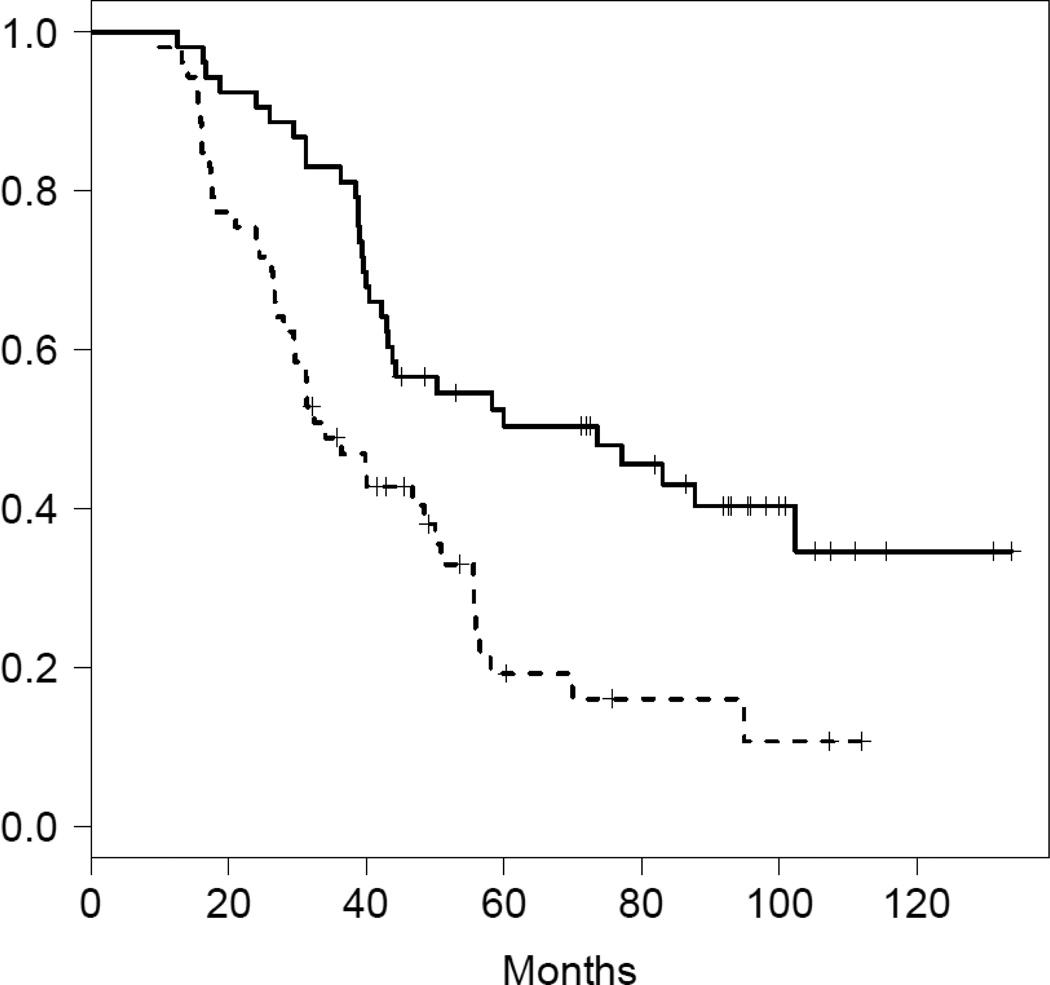
To assess the relationship between gene expression measures derived from optimal RNA (flash frozen tissue) compared to the degraded material from the FFPE sections, we analyzed data from 30 paired frozen (Affymetrix U133A) and paraffin samples (that passed the quality control described above). The TCGA survival signature consists of 194 genes, 166 of which map back to the DASL array. Linear correlations of these 166 genes between DASL and Affymetrix expression estimates in the 30 paired samples ranged from −0.265 to 0.868 (mean = 0.331), and included 14 genes with negative correlations and 43 genes with a linear correlation >0.50 (Figure 2A). After weighting individual probes based on their squared correlation between the two platforms, we arrive at highly concordant estimates of the TCGA survival between frozen and FFPE in these 30 cases (Pearson r=0.774, P=5.2e-7). The survival signature was developed on high grade serous cancers specifically. Therefore we next applied this algorithm to the DASL data in the subset of 106 advanced stage high grade serous ovarian cancers from the North Carolina Ovarian Cancer Study. When cases were divided into two groups based on whether their TCGA survival signature scores were above or below the median value, the median survival of the two groups was 33 months versus 60 months (Figure 3). In a Cox proportional hazards model adjusted for age and grade the estimated hazard ratio for death was 2.30 (p=0.0007). These results indicate that the survival signature is robust and retains prognostic information in FFPE expression data.
Figure 2.
Correlation between Affymetrix and DASL gene expression signatures.
A) TCGA survival signature: Linear correlations between the DASL and Affymetrix gene expression estimates in the 30 paired samples ranged from −0.265 to 0.868 (mean = 0.331). The dotted line indicates the relative weighting of probes at each correlation level. After this weighting, the correlation between the DASL and Affymetrix signature estimates was 0.774.
B) Tothill et al. gene expression subtype signature: Linear correlations between the DASL and Affymetrix gene expression estimates in the 30 paired samples ranged from −0.333 to 0.914 (mean = 0.453). The dotted line indicates the relative weighting of probes at each correlation level. After this weighting, correlation levels for the subtype signatures C1 through C6 are denoted under the X axis.
Figure 3.
Validation of the Cancer Genome Atlas survival predictor using DASL in 106 advanced stage high grade serous ovarian cancers from the North Carolina Ovarian Cancer Study. When cases were divided into two groups based on whether their TCGA survival signature scores were above or below the median score, the median survival of the two groups was 33 months versus 60. In a Cox proportional hazards model adjusted for age and grade the estimated hazard ratio for death was 2.30 (p=0.0007).
Testing signatures of ovarian cancer subtypes in FFPE samples
Tothill et al. developed a classification of epithelial ovarian cancer based on unsupervised hierarchical clustering that grouped these diseases into 6 gene expression subtypes [5]. Their analysis included low malignant potential serous and some endometrioid cancers accounting for two of the 6 groups. The great majority of high grade serous cancers clustered into 4 groups. These 4 subtypes that capture the expression heterogeneity of high grade serous cancers were recapitulated closely in the TCGA study [10]. These were named mesenchymal, immunoreactive, differentiated and proliferative by TCGA based on the identity of the genes that were overexpressed in each cluster. Over the 1806 probes in these signatures that map to the BeadChip array, the 30 paired frozen and FFPE samples linear demonstrated correlations between the DASL and Affymetrix gene expression estimates ranging from −0.333 to 0.914 (mean = 0.45) including 62 probes with negative correlations and 807 probes with a linear correlation >0.50 (Figure 2B).
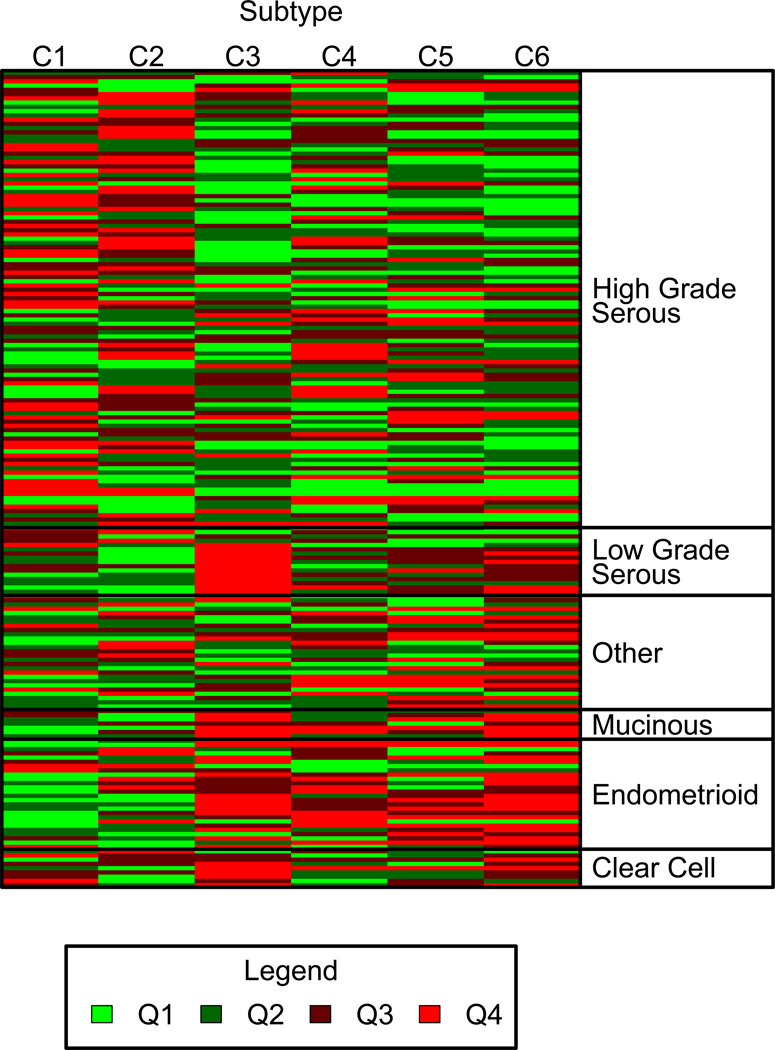
After applying corrections based on these correlations as above, the correlations between the Affymetrix and DASL versions of these 6 signatures in the 30 paired samples were 0.618 (cluster 1; P=2.8e-4), 0.804 (cluster 2; P=8.7e-8), 0.887 (cluster 3; P=7.9e-11), 0.548 (cluster 4; P=1.7e-3), 0.857 (cluster 5; P=1.5e-9) and 0.753 (cluster 6; P=1.6e-6). As reported by Tothill et al. (5), we found that most high grade serous ovarian cancers (102/110, 93%) were assigned to subtypes 1, 2, 4 and 5, whereas most endometrioid, clear cell, mucinous and low grade serous cases (39/57, 68%) were assigned to subtypes 3 and 6 (Table 1, Figure 4) (correct classification, p<10e-15). These data indicate that the intrinsic subtype signatures can also be discerned accurately in the majority of cases.
Table 1.
Predicted subtypes of epithelial ovarian cancer using DASL
| Tothill Expression Subtypes* | ||||||
|---|---|---|---|---|---|---|
| C1 | C2 | C3 | C4 | C5 | C6 | |
| Serous high grade | 38 | 26 | 8 | 16 | 22 | 0 |
| Serous low grade | 2 | 1 | 13 | 0 | 0 | 0 |
| Endometrioid | 2 | 3 | 1 | 4 | 3 | 12 |
| Clear cell | 2 | 1 | 4 | 0 | 0 | 2 |
| Mucinous | 0 | 0 | 1 | 0 | 0 | 6 |
Shaded areas represent correctly classified cases: 102/110 (93%) high grade serous cases were correctly assigned to subtypes 1, 2, 4 and 5, (Corresponding TCGA subtypes are C1=mesenchymal, C2=immunoreactive, C3=differentiated, C4=proliferative), while 39/57 (68%) clear cell, endometrioid, mucinous and low grade serous cases were correctly assigned to subtypes 3 and 6 (p<10e-15). 26 samples with mixed or other histological types are not included in this table.
Figure 4.
Heat map of the probability of being assigned to each of the six Tothill et al. gene expression subtype classes arranged by tumor histology. The predictions for each cancer (rows) for the six expression subtype signatures (columns) are demonstrated in this heat map. Probability predictions are grouped by quartiles from the lowest probability of class membership (Q1, light green) to the highest probability (Q4, red).
Discussion
Most prior expression microarray work in ovarian and other cancers has been done using fresh frozen tissues where high quality RNA is reliably obtained. FFPE tissues represent the bulk of all clinical specimens (both retrospective and prospective), therefore translation of ovarian cancer genomic signatures to this platform is an important step towards potential clinical application. Although RNA in FFPE tissues is degraded and chemically altered, methods have been developed to perform whole genome expression analysis from this type of degraded RNA. The use of the DASL assay to develop a cancer prognosis signature using FFPE tissues was reported in 2008 in a study of hepatocellular carcinoma [11]. Samples from 90% of patients yielded interpretable data, including samples that had been archived for more than 24 years. In this study, the patterns of gene expression in the normal liver surrounding the tumors, rather than from the tumors, was prognostic. Subsequently, Xiang et al. used the DASL Human Cancer Panel Assay that targets 502 cancer related genes to develop a signature correlating with lymph node metastasis in hepatocellular carcinoma [16]. The Affymetrix U133 plus 2.0 array also has been used to develop a gene expression signature in FFPE that predicts prognosis of non-small-cell lung cancer patients [17]. The 59 gene lung cancer prognosis signature was validated successfully in independent data sets and was useful in refining the prognosis of stage I cases. A commercial disease specific array has also been successfully employed in developing a prognostic signature from archival FFPE stage II colon cancers [18]. These studies indicate that there is no intrinsic barrier to deriving useful array based expression data from archival FFPE specimens.
In the past decade a number of groups have developed gene expression signatures correlating with clinical features and having prognostic value in epithelial ovarian cancer [3–5]. The TCGA has developed and validated a gene expression signature that predicts survival in high grade serous cancers. Bowtell’s group in Australia was the first to classify ovarian cancers into subtypes based on gene expression signatures (5) and four of these subtypes comprising high grade serous cancers were validated in the TCGA study. In view of this prior work with signatures developed and validated in multiple data sets and across platforms, we attempted to validate these existing signatures using the DASL platform as opposed to developing new signatures based on FFPE derived data. This strategy allowed us to directly test the quality of the DASL data using established metrics and to provide further support for the strength of these signatures across platforms.
The only prior study using DASL in ovarian cancer was performed by Chien et al. and involved a comparison of 5 early stage invasive serous ovarian cancers and 5 serous borderline tumors [19]. Using a differentially expressed gene set, class comparison prediction analysis correctly identified serous carcinomas from serous borderline tumors in 3 independent data sets at over 80% accuracy.
A systematic issue with FFPE material is the inconsistent quality of RNA that can be extracted. The usual metric for RNA integrity, the 28S:18S ratio, is of little practical value in samples where these peaks cannot even be clearly discerned. To explore this problem, we employed quantitative RT-PCR analysis of two abundant mRNA’s. After discovering and correcting for systematic batch variation in the BeadChip DASL data, we were able to show that the qRT-PCR values directly and strongly correlated with the overall quality of the assay and could be used to eliminate samples before costly analysis.
We were able to directly evaluate the DASL assay by analyzing data from 30 matched frozen (Affymetrix U133A) and FFPE samples using the survival (166 genes on DASL) and Tothill classification (1806 genes). On a gene by gene basis, we observed a broad range of correlation from FFPE (Illumina BeadChip) to frozen samples (Affymetrix U133). This analysis led us to use the squared correlations as a means of weighting the contribution of individual probes to the overall signatures leading to improved classifications. A similar study on 20 matched frozen and FFPE breast cancers, also using the whole genome DASL platform, arrived at similar results and found that with appropriate data processing, the MammaPrint 70 gene prognostic signature could be recapitulated from archival specimens [20].
For assigning categories in the FFPE samples, we found that 93% of high grade serous cases were assigned to subtypes 1, 2, 4 and 5 (mesenchymal, differentiated, proliferative, immunoreactive high grade serous). In contrast, 68% of epithelial ovarian cancers with other histologies were assigned to subtypes 3 and 6. These results are similar to those reported by Tothill et al, who defined these six expression subtypes using frozen tissue samples [9]. This confirms that microarray expression data derived from FFPE samples can discriminate between histological subtypes of epithelial ovarian cancer and can define the 4 consistent subtypes of high grade serous ovarian cancer that are characterized by distinct patterns of gene expression. Since these subtypes may represent different etiologies, pathway activations, and potentially response to therapy the ability to go back into the FFPE archives and identify cancer subtype opens many possible avenues of investigation.
The TCGA developed a survival signature using data from four gene expression platforms and validated this in several independent data sets, including one from our institution [21]. We recapitulated the TCGA survival signature using DASL data and found that among high grade serous ovarian cancers the median survival of the poor versus favorable survival groups was 33 months versus 60 months. This was similar to what was seen in the TCGA study. Although a 27 month difference highlights the importance of gene expression in determining outcome, this information does not have clinical utility at present in view of the lack of predictive information to guide specific treatments.
The ability to assess the gene expression and survival subtypes in FFPE tissue samples now makes it possible to evaluate their predictive potential in the context of large clinical trials. Despite the weak correlation of individual probes between FFPE and frozen samples, these multi-gene signatures appear to be robust enough to withstand the significant technical variability introduced by fixation and expression platform. This is essential if we hope to use signatures such as these for prognostic and predictive purposes as we seek to improve and individualize the treatment of ovarian cancer.
Supplementary Material
Acknowledgments
Grant support
This work was supported by the American Cancer Society grant #120368-SIOP-06-090-06-COUN to AB and NCI CA084955 to JRM.
References
- 1.Sparano JA, Paik S. Development of the 21-gene assay and its application in clinical practice and clinical trials. J Clin Oncol. 2008;26:721–728. doi: 10.1200/JCO.2007.15.1068. [DOI] [PubMed] [Google Scholar]
- 2.Press MF, Sauter G, Bernstein L, Villalobos IE, Mirlacher M, Zhou JY, et al. Diagnostic evaluation of HER-2 as a molecular target: an assessment of accuracy and reproducibility of laboratory testing in large, prospective, randomized clinical trials. Clin Cancer Res. 2005;11:6598–6607. doi: 10.1158/1078-0432.CCR-05-0636. [DOI] [PubMed] [Google Scholar]
- 3.Berchuck A, Iversen ES, Luo J, Clarke JP, Horne H, Levine DA, et al. Microarray analysis of early stage serous ovarian cancers shows profiles predictive of favorable outcome. Clin Cancer Res. 2009;15:2448–2455. doi: 10.1158/1078-0432.CCR-08-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonome T, Lee JY, Park DC, Radonovich M, Pise-Masison C, Brady J, et al. Expression profiling of serous low malignant potential, low-grade, and high-grade tumors of the ovary. Cancer Res. 2005;65:10602–10612. doi: 10.1158/0008-5472.CAN-05-2240. [DOI] [PubMed] [Google Scholar]
- 5.Spentzos D, Levine DA, Kolia S, Otu H, Boyd J, Libermann TA, Cannistra SA. Unique gene expression profile based on pathologic response in epithelial ovarian cancer. J Clin Oncol. 2005;23:7911–7918. doi: 10.1200/JCO.2005.02.9363. [DOI] [PubMed] [Google Scholar]
- 6.Shridhar V, Lee J, Pandita A, Iturria S, Avula R, Staub J, et al. Genetic analysis of early-versus late-stage ovarian tumors. Cancer Res. 2001;61:5895–5904. [PubMed] [Google Scholar]
- 7.Spentzos D, Levine DA, Ramoni MF, Joseph M, Gu X, Boyd J, et al. Gene expression signature with independent prognostic significance in epithelial ovarian cancer. J Clin Oncol. 2004;22:4648–4658. doi: 10.1200/JCO.2004.04.070. [DOI] [PubMed] [Google Scholar]
- 8.Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–5486. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schildkraut JM, Iversen ES, Wilson MA, Clyde MA, Moorman PG, Palmieri RT, et al. Association between DNA damage response and repair genes and risk of invasive serous ovarian cancer. PLoS One. 2010;5:e10061. doi: 10.1371/journal.pone.0010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–260. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 14.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and BIoconductor. New York: Springer; 2005. pp. 397–394. [Google Scholar]
- 15.Shi W, Oshlack A, Smyth GK. Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res. 2010;38:e204. doi: 10.1093/nar/gkq871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang ZL, Zeng ZC, Fan J, Tang ZY, Zeng HY, Gao DM. Gene expression profiling of fixed tissues identified hypoxia-inducible factor-1alpha, VEGF, and matrix metalloproteinase-2 as biomarkers of lymph node metastasis in hepatocellular carcinoma. Clin Cancer Res. 2011;17:5463–5472. doi: 10.1158/1078-0432.CCR-10-3096. [DOI] [PubMed] [Google Scholar]
- 17.Xie Y, Xiao G, Coombes KR, Behrens C, Solis LM, Raso G, et al. Robust gene expression signature from formalin-fixed paraffin-embedded samples predicts prognosis of non-small-cell lung cancer patients. Clin Cancer Res. 2011;17:5705–5714. doi: 10.1158/1078-0432.CCR-11-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy RD, Bylesjo M, Kerr P, Davison T, Black JM, Kay EW, et al. Development and independent validation of a prognostic assay for stage II colon cancer using formalin-fixed paraffin-embedded tissue. J Clin Oncol. 2011;29:4620–4626. doi: 10.1200/JCO.2011.35.4498. [DOI] [PubMed] [Google Scholar]
- 19.Chien J, Fan JB, Bell DA, April C, Klotzle B, Ota T, et al. Analysis of gene expression in stage I serous tumors identifies critical pathways altered in ovarian cancer. Gynecol Oncol. 2009;114:3–11. doi: 10.1016/j.ygyno.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Mittempergher L, de Ronde JJ, Nieuwland M, Kerkhoven RM, Simon I, Rutgers EJ, et al. Gene expression profiles from formalin fixed paraffin embedded breast cancer tissue are largely comparable to fresh frozen matched tissue. PLoS One. 2011;6:e17163. doi: 10.1371/journal.pone.0017163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berchuck A, Iversen ES, Lancaster JM, Pittman J, Luo J, Lee P, et al. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin Cancer Res. 2005;11:3686–3696. doi: 10.1158/1078-0432.CCR-04-2398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.