Abstract
DNA damage and genetic rearrangements are hallmarks of cancer. However, gene fusions as driver mutations in cancer have classically been a distinction in leukemia and other rare instances until recently with the discovery of gene fusion events occurring in 50 to 75% of prostate cancer patients. The discovery of the TMPRSS2-ERG fusion sparked an onslaught of discovery and innovation resulting in a delineation of prostate cancer via a molecular signature of gene fusion events. The increased commonality of high-throughput sequencing data coupled with improved bioinformatics approaches not only elucidated the molecular underpinnings of prostate cancer progression, but the mechanisms of gene fusion biogenesis. Interestingly, the androgen receptor (AR), already known to play a significant role in prostate cancer tumorigenesis, has recently been implicated in the processes resulting in gene fusions by inducing the spatial proximity of genes involved in rearrangements, promoting the formation of double-strand DNA breaks (DSB), and facilitating the recruitment of proteins for non-homologous end-joining (NHEJ). Our increased understanding of the mechanisms inducing genomic instability may lead to improved diagnostic and therapeutic strategies. To date, the majority of prostate cancer patients can be molecularly stratified based on their gene fusion status thereby increasing the potential for tailoring more specific and effective therapies.
Keywords: Androgen receptor, gene fusions, genomic instability, prostate cancer
INTRODUCTION
The concept of genomic instability and rearrangements are hallmarks of cancer; however, until recently recurrent DNA rearrangements were largely undiscovered in solid tumors. Genomic rearrangements can be caused by a deletion, insertion, inversion, or translocation of distal intra- or inter-chromosomal sequences and ultimately result in a gene fusion. The consequences of these gene rearrangements include gene activation or repression thereby triggering oncogenic signaling programs. The classical example of gene rearrangements is the Philadelphia chromosome found in chronic myelogenous leukemia (CML) consisting of the translocation of chromosome 9 and 22 creating BCR-ABL, an activated tyrosine kinase that drives CML [1, 2]. As demonstrated by the discovery of the BCR-ABL gene fusion, genomic rearrangements provide key insights into tumor biology and have significant clinical impact by serving as potential diagnostic markers or therapeutic targets [3]. More recently, a bioinformatics strategy for processing cancer profiling data led to the discovery of the gene fusion between TMPRSS2 and the ETS oncogenic transcription factor ERG in 50 to 75% of prostate cancer patients. This discovery not only revealed that a gene fusion was a common event acquired early in prostate cancer progression, and represents a potential driver of disease progression, but also suggested that gene fusions represent an understudied class of mutations in solid tumors. More recently work has begun to show how androgen receptor is involved in the formation of gene fusions in prostate cancer. Here, we will review the research efforts since the initial discovery of ETS fusions in prostate cancer that have led to a comprehensive landscape of rearrangements, an improved understanding of the mechanisms driving genomic instability, and potential therapeutic strategies.
DISCOVERY OF GENE FUSION EVENTS IN PROSTATE CANCER
Whereas recurrent gene fusions had been widely characterized in leukemias, lymphomas and soft tissue tumors, the lack of known gene fusions in epithelial cancers had been attributed to clonal heterogeneity of the tumors as well as the technical limitations of earlier detection strategies such as cytogenetic analysis, spectral karyotyping, fluorescence in-situ hybridization (FISH), and microarray-based comparative genomic hybridization (aCGH). However, the use of microarrays, and more recently high throughput transcriptome sequencing (RNA-Seq) has helped circumvent these limitations. The first significant advancement involved a systematic analysis of cancer microarray data to nominate genes that are ‘highly’ deregulated in a subset of samples rather than focusing on those that are widely shared, a signature of a gene fusions [4]. The development of a data transformation algorithm, or cancer outlier profile analysis (COPA), led to the identification of such ‘outlier’ genes. Through the COPA transformation, a typical biomarker profile characterized by a general overexpression of genes is compressed and instead emphasizes ‘outlier gene’ profiles, characterized by general low expression with marked overexpression in a fraction of the samples [5]. The COPA transformation was applied to >10,000 microarray experiments across human cancers and identified several well-known cancer genes involved in recurrent chromosomal aberrations. Surprisingly, two genes known to be involved in gene fusions in Ewing’s sarcoma, namely ERG and ETV1, were scored as high ranking outliers in several independent prostate-cancer profiling studies. Interestingly, ERG and ETV1 overexpression was observed to be mutually exclusive in prostate cancer suggesting that elevated expression of ERG or ETV1 may be a key molecular event in a subset of prostate cancers. This in silico finding was further validated and characterized in prostate cancer cell lines and tissue leading to the discovery that the 5’ ends of ERG and ETV1 are replaced with the 5’ untranslated region of a prostate-specific, androgen responsive, transmembrane serine protease gene, TMPRSS2, in the outlier cases. Overall, this work revealed an androgen-regulated on-switch for activating oncogenic ETS transcription factor family members. Furthermore, it changed the preconceived notion that the majority of gene rearrangements were present in a small portion of selected cancer types. Instead, multiple independent research groups were able to confirm that 50 −75% of all prostate cancers harbor the TMPRSS2-ERG aberration, now the most common gene fusion found in any cancer.
Since the initial discovery of ETS gene fusions in prostate cancer, there has been a steady unraveling of gene fusions in prostate cancer, and more recently additional solid tumors. One of the most significant technological contributions towards the discovery of gene fusions in human cancers has been the ongoing development of high-throughput sequencing approaches. An initial study using transcriptome sequencing, or RNA-Seq, leveraged a hybrid strategy of long-read sequences (Roche 454), to accurately capture a putative fusion junction, and short-read sequences (Illumina Genome Analyzer), to obtain deep sequence read coverage supporting true positive fusion transcripts [6]. This approach was able to “rediscover” the BCR-ABL1 and TMPRSS2-ERG in a chronic myeloid leukemia cell line, K562, and a prostate cancer cell line, VCaP, respectively. However, despite the success of this methodology, it required substantial overhead to leverage two sequencing platforms. Subsequent studies have since demonstrated the effectiveness of paired-end tran-scriptome sequencing to systematically identify gene fusions and chimeric transcripts in cancers. While transcriptome sequencing offers an unbiased view of expressed transcripts, and therefore enables the detection of gene fusions, the vast quantities of data still require highly sensitive computational approaches for accurately detecting gene fusions while minimizing false positives. To address this challenge, a number of software packages have been developed, and applied to prostate cancer cohorts, including ChimeraScan, FusionSeq, dRanger [7–9]. Overall, new tools are continually being implemented in the field and applied to improve our ability to comprehensively detect gene fusions [9, 10].
GENE FUSION STRATIFICATION OF PROSTATE CANCERS
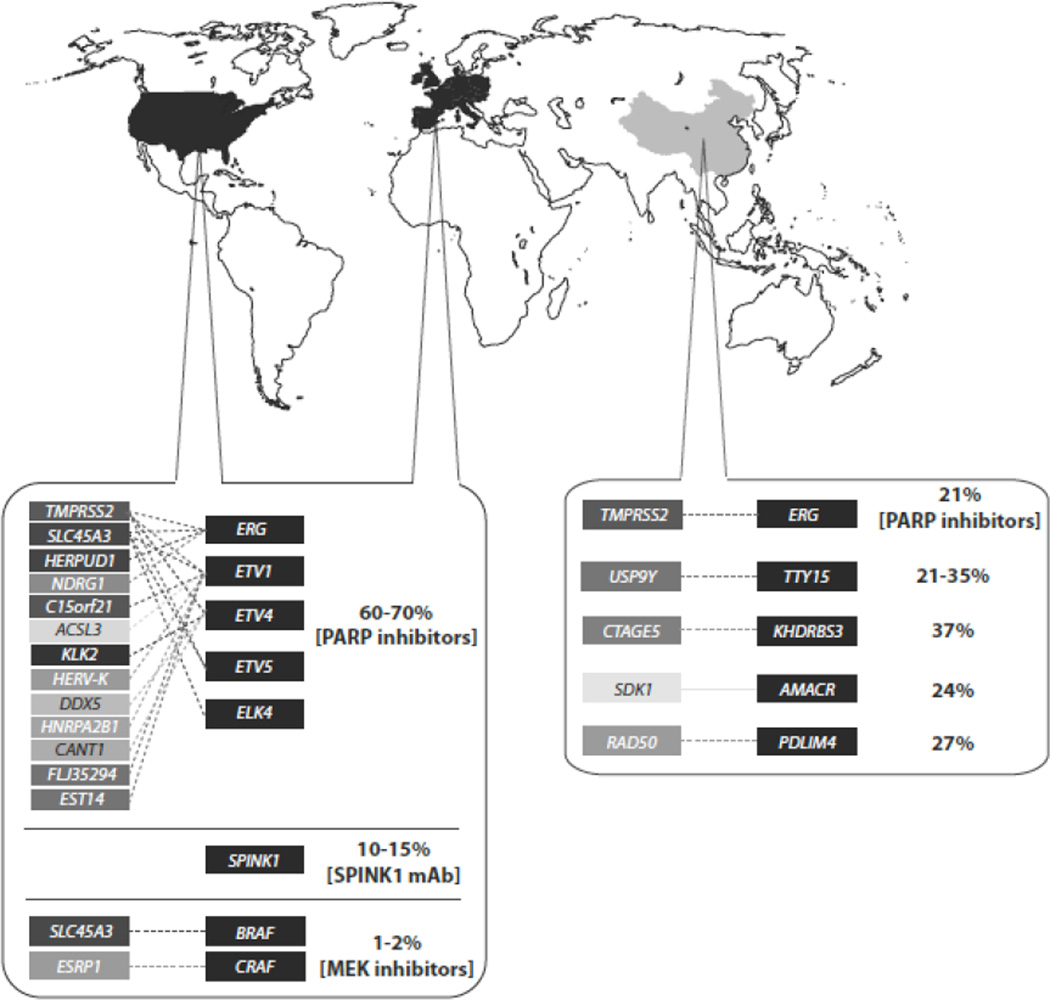
Since the initial discovery of ETS fusions in prostate cancer, bioinformatics approaches using outlier expression analysis or unbiased sequencing of whole genomes and transcriptomes have revealed a more comprehensive and complex mutational spectrum in prostate cancer (Fig. 1). Interestingly, closer inspection of the gene fusion landscape in prostate cancer revealed three virtually mutually exclusive aberrations, as initially proposed by Rubin et al. [11], including ETS gene fusions, RAF kinase gene fusions, and SPINK1 outlier expression. While two of these categories involve genomic rearrangements, the combination of all three molecular subtypes offer a mechanism for stratifying prostate cancer patients.
Fig. 1.
Prostate cancer gene fusion molecular stratification. This schematic highlights all of the gene fusion discoveries in both western countries and China to date indicating the ETS, SPINK1 overexpression, and RAF kinase classifications as well as incidence rates. Where applicable, current potential therapies are provided.
ETS subtype gene fusions
Similar to Ewing sarcoma rearrangements [12], gene fusions involving ERG and ETV1 have been found to be mutually exclusive events [4]. However, TMPRSS2-ERG is the most prevalent rearrangement having been observed in approximately 50–75% of prostate cancer patients. The most common TMPRSS2-ERG fusion involves the first or second exon of TMPRSS2 fused to exon 2, 3, 4, or 5 of ERG [13]. Interestingly, transcriptome sequencing from 14 matched controls and tumors from patients at Shanghai Changhai Hospital in China determined a lower frequency of TMPRSS2-ERG prevalence (approximately 20%) than reported in western countries [14]. This is in agreement with previous work that has shown variability in the prevalence of the TMPRSS2-ERG gene fusion across ethnicities with the highest prevalence occurring in Caucasians (∼50%) followed by both African Americans (∼31%) and Asians (16%–30%) [15, 16]. However, the most common TMPRSS2-ERG transcript isoform observed in the Chinese population corresponds to the same isoform observed in western countries involving exon 1 of TMPRSS2 and exon 4 of ERG. This finding was validated by RT-PCR and Sanger sequencing in this cohort and also in an independent cohort of 54 prostate tumors collected from patients across three different cities in China. This study highlights the different molecular signaling in prostate cancer from other ethnicities and suggests the necessity to understand the genomic complexity in prostate cancer across different racial groups. In addition to TMPRSS2, other genes have been shown to activate ERG including the androgen responsive SLC45A3 (Solute carrier family 45 member 3) [17]. This observation was then confirmed in a large prostatectomy cohort [18]. Since then HERPUD1 and NDRG1 have also been identified as 5’ partners with ERG [19, 20]. It is becoming evident that despite a growing number of 5’ partners fusing to ERG, they commonly appear to be androgen responsive and ultimately activate ERG expression.
In addition to ERG, various 5’ fusion partners have been found to fuse with ETV1, which has been observed in approximately 5% to 10% of all prostate cancer patients and represents the second largest category of ETS fusions. From the initial discovery using outlier expression analysis two prostate-specific genes were identified, androgen-induced SLC45A3 and androgen-repressed C150rf21, as well as the retroviral element HERV-K_22q11.23, and the housekeeping gene HNRPA2B1 [21]. Other 5’ partners found to fuse with ETV1 are FLJ35294, EST14, and ACSL3 [17, 22, 23]. Continued exploration of ETS family member expression in prostate cancer using an outlier analysis, COPA, revealed additional ETS family members involved in gene fusions including ETV4 and ETV5. ETV4 was found to have multiple 5’ partners including TMPRSS2 [24], KLK2, CANT1 [25] and DDX5 [17]. ETV5, an ETS family member, was also found to be activated by androgen responsive 5’ partners TMPRSS and SLC45A3 the gene is TMPRSS2 [26]. Together, ETV4 and ETV5 gene fusions account for approximately 2% of all gene fusions found in prostate cancer.
Recent transcriptome sequencing analysis revealed the recurrent RNA chimera involving SLC45A3 and ELK4, which are adjacent genes, in the same orientation, located on chromosome 1. Unlike other fusion events, this represents a transcriptional-induced chimera, or read-through event, that lacks any disruption at the genomic level [6, 27, 28]. Recent work has focused on the mechanism of the SLC45A3-ELK4 RNA chimera formation and determined that there is androgen-dependent cis-splicing involving exon 1 of SLC45A3 and exon 2 of ELK4 due to a decrease in chromatin association of CCTC-binding factor (CTCF) [29]. “Insulator” sequences bind CTCF to inhibit transcription at the boundaries of genes [30] and are known to exist in the intergenic region between SLC45A3 and ELK4 [31]. With continued technological advances, such as RNA-Seq, we expect to continue unraveling more of these RNA chimera events in prostate and other cancers [32].
Non-ETS subtype gene fusions
Given the prevalence of gene fusions in prostate cancer, many groups have focused on identifying novel driver gene fusions in ETS rearrangement negative patients that may serve as biomarkers and therapeutic targets. Analysis of transcriptome sequencing data led to the discovery of gene fusions involving different RAF kinases (SLC45A3-BRAF and ESRP1-CRAF1) that ultimately activate the signaling pathway [33]. Screening across larger patient cohorts revealed that RAF kinase gene fusions occurred in approximately 1 to 2% of patients. Using FISH, the prevalence of BRAF and CRAF1 has been independently validated across a cohort comprised of Chinese patients occurring in 2.5% and 1.5% of the 218-naïve cases, respectively. RAF positive patients were found to have a higher Gleason score and a trend toward higher clinical stage cancer [34]. The discovery of RAF fusions has been especially appealing due to the potential of targeting the RAS pathway with existing FDA approved drugs.
In addition to RAF kinase gene fusions, a recent study analyzing RNA-Seq data generated from the tumor and adjacent normal tissue from 14 prostate cancer patients in China revealed additional recurrent non-ETS gene fusions. The first novel gene fusion occurred between USP9Y-TTTY15 which are adjacent genes, in the same orientation, located approximately 9-kilobases apart on chromosome Y. USP9Y-TTTY15 was reported to occur at a frequency of 21.4%, and subsequently confirmed in a larger cohort of 54 tumor samples (frequency 35.2%) by RT-PCR and Sanger sequencing [14]. The lincRNA (long intergenic noncoding RNA) TTTY15 has not been studied, but USP9Y mutations and deletions are currently being studied in the context of male infertility [35] and as of yet have not been connected to cancer. Three additional novel gene fusions were also discovered and validated in a cohort of 54 tumor cases, CTAGE5-KHDRBS3, SDK1-AMACR, and RAD50-PDLIM4 [14]. CTAGE5 (cutaneous T-cell lymphoma–associated antigen 5), previously shown to be overexpressed in many cancer cell lines [36], was shown to fuse with KHDRBS3 (KH domain containing, RNA binding, signal transduction associated 3), a gene also associated with cancer [37], at a frequency of 37% [14]. SDK1 (located within a common fragile site of chromosome 7p) [38] was also found fused to AMACR (an androgen responsive gene) [39] at a frequency of 24%. Lastly, RAD50-PDLIM4 was seen in the Chinese population at a frequency of 27.8%. The RAD50 protein is known to be important in localizing the cohesion complex of newly replicated chromatids in yeast [40]. Overall, this study highlighted the lower prevalence of the TMPRSS2-ERG fusion across different ethnicities while reporting novel recurrent gene fusions that, upon further characterization, may provide insights into prostate cancer pathogenesis of Chinese men.
SPINK1 subtype
The last molecular subtype distinction accounts for 10 to 15% of prostate cancer and is classified by the overexpression of SPINK1, or serine protease inhibitor Kazal-type 1. SPINK1 was found through outlier analysis but has not been shown to be involved in a genomic rearrangement. SPINK1 has been associated with a more aggressive form of prostate cancer and interestingly appears to be mutually exclusive from ETS and RAF rearrangements [41].
Annotation of gene fusions
As our ability to comprehensively detect gene fusions in human cancers has improved, we have been able to observe distinct patterns amongst the different fusions that can help in their classification and provide potential insights into their mechanism of formation. Maher et al. proposed five different groupings to classify gene fusions [6]. The first category (Class I) is caused by an inter-chromosomal translocation resulting in the juxtaposition of two genes thereby creating a gene fusion such as BCR-ABL. Class II are complex inter-chromosomal rearrangements that may result in the regulatory region of a nearby gene being altered following a rearrangement as exemplified by the gene fusion of MIPOL1-DGKB activating ETV1 in LNCaP cells. Class III fusions are classified by an intra-chromosomal deletion resulting in the fusion of the two genes flanking the deleted region as exemplified by TMPRSS2-ERG. Class IV fusions are complex intra-chromosomal rearrangements in which a single gene may harbor multiple rearrangements as exemplified by HJURP harboring multiple breakpoints and thereby generated the HJURP-EIF4E2 and INPP4-HJURP fusion transcripts. The last category is comprised of read-through events (Class V) that occur when two consecutive genes on a chromosome, in the same orientation, are fused together. This class of fusions could either be the by-product of a focal deletion, a splicing event, or is a commonly occurring transcript that to date has been annotated as two independent transcripts. Overall, such a classification system may distinguish gene fusions that may arise from different mechanisms.
MECHANISMS OF GENE FUSION EVENTS
Genomic rearrangements are necessary and essential to normal development producing biological diversity within the immune system to combat pathogens. In lymphocytes V(D)J recombination and class switching recombination (CRS) involve protein complexes that help to coordinate DNA breaking and rejoining [42, 43]. It is now understood that many of the genomic rearrangements in cancer are also well-orchestrated events and not only spontaneous DNA damage caused by environmental agents or normal cellular metabolism. Within prostate cancer disease, there is significant evidence to support gene rearrangements or gene fusions as driving events in the progression of prostate cancer similar to the discovery of BCR-ABL in CML 50 years ago.
The majority of androgen receptor binding sites are transcriptional enhancers in distal regions and not within promoter regions [44]. Therefore, the looping model proposes a mechanism to explain the well-established interaction of the distal enhancers with promoter regions of AR targets genes, many of which are fusion partners such as TMPRSS2. This model suggests chromatin rearrangement, or “looping”, results in distal enhancers in close proximity with promoter regions [45–47]. It was hypothesized that AR may also play a role in the generation of gene fusions. Interestingly, treatment with the AR ligand, dihydrotestostrone (DHT), caused gene fusion-free LNCaP cells to induce colocalization of TMPRSS2 and ERG [48, 49]. Furthermore, androgen induced chromatin looping also appeared to be regulated by AR, which was found to be recruited to TMPRSS2 and ERG breakpoints when LNCaP cells were treated with DHT [49, 50].
Just as AR was found near breakpoints, it is also believed that breakpoints are associated with regions of active histone modifications. New technology has enabled monitoring of the elaborate protein/chromatin interactions by combining chromatin immunoprecipitation with Conformation Capture Chromatin technology (ChIP-3C) to capture the protein interaction with distal and proximal chromatin via crosslinking [51]. If close association of distal and proximal chromatin exists upon crosslinking then digestion, re-ligation and PCR amplification will result in the presence of both fragment ends in sequence alignment. The Hi-C method advances the previous technology by utilizing the unbiased approach of massively parallel sequencing for a high-resolution mapping of chromatin [52]. With this method, the 3D structure of DNA was further eluted and predicted to form highly compact chromatin into a “fractal globule”, a configuration that allows easy folding and unfolding for gene activation and repression. Moreover, these studies also assessed a compartmentalization of the chromatin within the nucleus segre- gated by open and closed chromatin domains. This technology has improved the understanding of chromatin organization and dynamic movement. Moreover, the Hi-C method was applied in prostate caner to determine ERG overexpression in a benign prostate cell line (RWPE1) as sufficient to cause global changes in chromatin structure [53]. Potential evidence of chromosome looping was indicated with the sequencing and analysis of the whole genome of seven primary tumors and matched controls of prostate patients. Berger et al. describe a ‘closed chain’ pattern of chromatin rearrangements, an equilibrium between breaking and rejoining of chromatin producing chimeric chromosomes, without the loss of genetic material, thus implying simultaneous disruption of many genes driving disease progression [54]. Inspection of breakpoints from TMPRSS2-ERG positive tumors using publicly available ChIP-sequencing (ChIP-Seq) data determined some correlation to open chromatin marks (RNA polymerase II, H3K3me3, H3K36me, H3ace) in VCaP cells as well as AR and ERG binding sites [55]. Moreover, ETS fusion negative tumors correlated more closely with marks of inactive closed chromatin (H3K27me3) in VCaP cells.
Two recent studies have proposed models incorporating the well-coordinated and precise molecular complexes involved in double strand breaks (DBS) as key mechanisms to forming gene fusions such as TMPRSS2-ERG. These groups independently determined that DSB are not random events happening throughout the genome, but in fact a sequential process initiated by activated androgen receptor at determin-able genomic locations. This rationale stems from recent work demonstrating that estrogen receptor (ER) can initiate reorganization of chromatin by utilizing actin/myosine-1 machinery in the nucleus and other factors within nuclear speckles [56] in breast cancer. Lin et al. demonstrated in a prostate cancer model, using the LNCaP cell line, that two pathways, ligand-dependent and genotoxic stress, act in synergy to induce TMPRSS2-ERG fusion events [49]. In the presence of genotoxic stress such as irradiation, DHT treatment has been shown to increase both mRNA and protein levels of activation-induced cytidine deaminase (AID) which can lead to DSB. Taken together, AR induces spatial proximity of chromosomes through the myosin/actin motor system while concurrently recruiting AID, through the coactivator Gadd45, and the LINE-1 repeat-encoded ORF2 endonuclease to site specific locations to facilitate DBS [49]. Another enzyme recruited to activated AR and necessary to facility DBS is topoisomerase II. Topoisomerase II (TOP2) is an enzyme responsible for resolving DNA topological constraints during transcription by catalyzing transient double strand breaks; TOP2 activity was implicated in estrogen receptor-mediated double strand breaks [57]. Expanding upon this observation, Haffner et al. determined that TOP2B is recruited during normal regulation of androgen receptor-mediated genes; moreover, TOP2B-AR-liganded complexes colocalize specifically at previously identified TMPRSS2 and ERG break points [50]. As proof of principle, studies in various leukemias have investigated the increase in gene rearrangements in conjunction with DNA-topoisomerase inhibitors and determined development of therapy-related rearrangements of the AML1 and MLL genes [58].
Last, studies have also demonstrated that androgen stimulation may play a role in DSB repair. DSB repair occurs via homologous recombination (HR) or non-homologous end joining (NHEJ). The error-prone NHEJ is the major pathway for the repair of DSB in eukaryotes [59]. Recent work has shown that androgen stimulation increases the recruitment of several proteins involved in the error-prone NHEJ pathway to the TMPRSS2-ERG fusion breakpoints including Ku70-Ku80 and ataxia telangiectasia-mutated protein (ATM) [49, 50]. Furthermore, inhibiting various components of the NHEJ pathway decreased the generation of the TMPRSS2–ERG gene fusions. Overall, androgen stimulation may be involved in the spatial association of gene fusion partners, inducing DSB, and involved in the recruitment of proteins involved in NHEJ.
CLINICAL RELEVANCE OF GENE FUSIONS AND POTENTIAL THERAPIES
Clinical Markers of Disease
Various studies have shown that samples exhibiting TMPRSS2-ERG as well as intronic deletion (2+Edel) between the genes correspond to a more aggressive prostate cancer [60–63]. One group, looking for an alternative to PSA testing, used in situ hybridization assays to assess TMPRSS2-ERG status and found 25% survival (after 8 years) with cancers harboring the fusion and 2+Edel compared to 90% survival of cancers without the fusion after the same period of time [60]. Moreover, the frequency of ‘2+Edel’ fusion status was found to be the main mechanism of TMPRSS2-ERG fusion in one study [61] and associated more closely with androgen-independent metastatic cancer in two other independent studies [62, 63].
TMPRSS2-ERG expression has also been associated with moderate to poorly differentiated tumors [64] and indicative of disease progression [65]. Nam and colleagues studied two different patient groups (26 and 165 patients) after radical prostatectomy and found expression of TMPRSS2-ERG between 42% and 49% in both studies as well as increased incidents of relapse in cancers harboring tumors as determined by PSA elevation (mean follow up 12 months) [66, 67]. The larger study allowed for a longer follow up (mean 28 months) and more conclusive evaluation of TMPRSS2-ERG as a diagnostic marker of disease independent of Gleason score, stage, or PSA testing with 58.4% recurrence after surgery in fusion positive patients compared to 8.1% recurrence in patients lacking the gene fusion [67]. Watchful-waiting cohort studies using prostate cancer-related death as end point analysis have confirmed the findings that prostate cancer patients harboring the TMPRSS2-ERG fusion are clinically more aggressive. One hundred and eleven early stage tumors collected from Orebro University Hospital in Sweden were assessed for TMPRSS2-ERG expression and determined a 12 year cancer-free survival in 26% of patients who were fusion negative compared to only 6% for patients who harbored the fusion [68]. Another watchful-waiting cohort of 445 prostate cancer samples determined an even higher survival rate of 90% after 8 years in TMPRSS2-ERG negative samples [60].
However, there are other studies that did not reach the same conclusion. Petrovics et al. concluded higher expression of ERG in prostate tumor tissue captured by laser microdissection and correlation to longer PSA recurrence-free survival in a cohort of 114 paired matched tumor and normal tissues [69]. PSA recurrence is defined by a rising PSA whereas clinical recurrence is associated with the presence of a metastatic site. Winnes et al. also used PSA levels to access prostate cancer status in 50 prostate samples. Based on PSA levels, this study determined a correlation, but not a significant difference, between TMPRSS2-ERG positive and negative cancers leading them to conclude that TMPRSS2-ERG positive cancers led to lower Gleason scores and a better cancer prognosis [70]. Saramaki et al. also correlated TMPRSS2-ERG status with a good disease prognosis and did not observe a correlation with fusion expression and disease progression after studying a wide array of prostate models and tissue [71]. Moreover, other studies have found no association with TMPRSS2-ERG status and disease prognosis [72–74].
Many studies have shown detection of TMPRSS2-ERG in urine as a new noninvasive assay for prostate cancer in combination and instead of PSA detection [75–79]. Hessels et al. used quantitative RT-PCR to detect TMPRSS2-ERG in combination with prostate cancer antigen 3 (PCA3), a noncoding RNA specifically expressed in prostate and overexpressed in cancer, and concluded a combined sensitivity of 73% for prostate cancer determination [76]. The Chinnaiyan group expanded upon these findings to develop a quantitative RT-PCR based multiplex urine-based diagnostic test including the biomarkers: PCA3, AMACR, GOLPH2, TMPRSS2-ERG [77]. On a much larger scale, including samples from multiple centers, this group also refined the Prostate Cancer Prevention Trial (PCPT) risk calculator, which incorporated PSA serum levels and clinical factors to include urine-based detection of PCA3 and TMPRSS2-ERG, thus better stratifying prostate cancer risk [79].
Other molecular subtypes of gene fusion events have also been correlated to disease prognosis. SPINK1 overexpression is associated with aggressive prostate cancer. Two different cohorts of 79 cases (37 cases of recurrence) and 817 cases (200 cases of recurrence) of prostate cancer showed SPINK1 outlier expression to associate with disease recurrence independent of other markers of disease [41]. Furthermore, confirmation of these findings was determined with knockdown studies in a cell line overexpressing SPINK1, 22RV1, and the resultant inhibition of cellular invasiveness [41]. Unlike the findings from Tomlins et al., Leinonen and colleagues did not find SPINK1 expression to be mutually exclusive from ETS-positive fusions in their cohort of 196 hormonally-treated needle biopsies; but this group did determine a strong association (lacking the power for significance) between SPINK1 expression and shorter progression-free survival [74]. Recently, using a tissue microarray of 640 samples, it was shown that the protein expression of SLC45A3 is down-regulated through SLC45A3-ERG in metastatic disease and correlates with a shorter survival time as measured by PSA [80]. Similar down-regulation of SLC45A3 protein expression was also noted in a study by Yin et al. in their analysis for another marker independent of PSA levels [81]. The read-through chimera, SLC45A3-ELK4 is also implemented in cancer progression and as a potential diagnostic marker. Knockdown studies of SLC45A3-ELK4 in LNCaP and PC3 cells showed decreased cell proliferation and cell cycle arrest in an androgen-dependent and independent pathway whereas wild-type SLC45A3 and ELK4 transcripts had no effect [29]. Moreover, in quantitative RT-PCR analysis of SLC45A3-ELK4 expression in 10 clinical samples and 29 controls indicated a correlation with higher transcript levels and a Gleason score of 7 or more with the highest transcript expression seen in metastatic caner samples [29]. More studies with larger sample sizes are needed to determine if SLC45A3-ELK4 is a biomarker for advanced prostate cancer.
Molecular Targeted Therapies
Androgen is an important hormone that binds and thus activates the androgen receptor (AR) and initiates downstream signaling essential and necessary for proper prostate development [82]. This cascade of signaling events leads to the activation of such genes as PSA and TMPRSS2 [83, 84]. Following initial diagnosis of metastatic prostate cancer, the standard of care is androgen deprivation therapy. However, most prostate cancers will invariably develop resistance to initial androgen deprivation therapy and the disease will evolve into castration recurrent prostate cancer (CRCP). While recent phase III trials have demonstrated survival benefits to next-generation anti-androgens such as abiraterone and MDV3100, or other systemic therapies such as cabazitaxel or alpharadin, each of these agents results in median improvements in survival of only a few months. Given the preclinical oncogenic phenotypes conferred by alterations such as ETS fusions, RAF fusions, and SPINK1, targeting these alterations represents a potentially promising treatment strategy for patients with aggressive metastatic disease.
The impact and AR-mediated reactivation of gene fusions post androgen deprivation therapy (ADT) is not well documented. Therefore, to determine the regulation of TMPRSS2-ERG by AR in CRCP, various models of prostate cancer were analyzed. Cai and colleagues reported gene expression of AR and AR-regulated genes using quantitative RT-PCR in CRPC samples as well as primary tumors distinguished by TMPRSS2-ERG status to determine an increase in gene expression in TMPRSS2-ERG positive CRPC samples compared to TMPRSS2-ERG negative samples [85]. This group showed the greatest expression of ERG, and by extension TMPRSS2-ERG expression, in fusion positive CRPC samples indicating reactivation of the gene fusion by AR-mediated signaling [85]. A direct link between AR-regulated increase in TMPRSS2-ERG expression was determined by examination of the VCaP cell line, which express the TMPRSS2-ERG gene fusion, and xenografts pre- and post-AR deprivation through monitoring of ERG mRNA and protein levels. Evidence of androgen axis-based regulation of TMPRSS2-ERG was observed in a phase I clinical trial of abiraterone acetate, a selective inhibitor of cytochrome P450 17 (CYP17). CYP17 is a necessary enzyme in the synthesis of androgen and estrogen. Five out of six patients harboring the TMPRSS2-ERG fusion showed decline greater than 50% in PSA serum levels, indicative of diminished AR signaling, for greater than six months [86]. However, another study did not find similar results using PCR analysis to detect TMPRSS2-ERG in 71 naïve lymph node-positive prostate tumors. The authors postulated a quicker and longer response to hormone therapy as measured by PSA levels and survival in fusion positive tumors; however, there was no correlation to fusion status and response to ADT and therefore it was concluded that TMPRSS2-ERG is not a good predictive marker of hormone response [87]. Thus, based on these conflicting studies, it is uncertain whether TMPRSS2-ERG gene status can be used to predict response to next-generation anti-androgen therapy.
More recently, discovery of novel ERG interactions have yielded a promising therapeutic approach in ETS positive cancers such as Ewing’s sarcoma and prostate cancer [88, 89]. Specifically, Brenner and colleagues used mass spec-trometry in tandem with liquid chromatography to identify proteins interacting with ERG. They found that ERG physically interacted with a series of DNA repair proteins, including poly(ADP-ribose) polymerase (PARP1). PARP1 is best known for its role in initiating the base excision DNA repair pathway and is needed to maintain DNA stability. The absence of PARP1 leads to accumulation of single stranded DNA breaks, which can lead to accumulation of double stand breaks at replication forks, normally repaired via the homologous recombination (HR) DNA repair pathway. PARP inhibition alone is not lethal to the cell, but inhibition in combination with a defective HR repair, as seen in BRCA1/BRCA2 defective cells, leads to collapse of the replication fork and eventually cell death; a process termed “synthetic lethality” [90, 91]. Based on this principle, ongoing trials are assessing PARP1 inhibition as a targeted therapy in BRCA-mutant cancers. Brenner and colleagues demonstrated that pharmacologic inhibition or genetic knockdown of PARP1 preferentially potentiated DNA damage in preclinical models of ETS-positive cancers, and also inhibited ERG-mediated transcription and invasion. In a series of xenograft models, PARP1 inhibition was preferentially effective in inhibiting tumor growth of ETS-positive versus ETS-negative malignancies (both prostate cancer and Ewing’s sarcoma). Based on this preclinical research, investigators at the University of Michigan have initiated a randomized multi-institutional phase II clinical trial (NCT01576172), which stratifies patients with metastatic CRPC based on biopsy-proven ETS fusion status and then randomizes these patients to treatment with the next-generation anti-androgen abiraterone, alone or in combination with the PARP1 inhibitor veliparib. This effort represents one of the first biomarker-driven therapeutic trials in metastatic prostate cancer.
Another developing area of therapeutic potential is targeting SPINK1 overexpression. Ateeq et al. exploited the secretory properties of the SPINK1 protein by investigating monoclonal antibody therapy as a potential target to inhibit its actions in prostate cancer [92]. The similar structure and sequence homology of SPINK1 to epidermal growth factor (EGF) led to studies determining interaction and crosstalk between EGFR and SPINK1. Given the FDA approved drugs for EGFR cancer treatment, this was an exciting finding and a promising potential for future drug therapies. Indeed, nude mice implanted with 22RV1 cells and treated for 4 weeks with anti-SPINK1 antibody showed a 65% reduction in tumor size whereas mice treated with anti-EGFR antibody (C225) showed a 41% reduction. Furthermore, combination therapy of anti-SPINK1 and anti-EGFR antibodies showed an additive effect of 74% reduction in tumor size [92]. These findings provide a mechanism for the limited results seen in clinical trials for prostate cancer with the anti-EGFR antibody, cetuximab [93], in which only one patient (2.7%) had an 80% PSA decline from baseline and two patients (5.6%) had a PSA decline between 50% to 80%. It also provides an incentive for establishing a molecule subtype classification of prostate cancer.
Due to the heterogeneity of epithelial cancers drug treatment regiments are often too broad and not as beneficial from patient to patient. Therefore, increasing the hunt for diagnostic markers to stratify prostate cancer subtypes would provide a significant clinical impact as exemplified by molecular profiles of disease heterogeneity in breast cancer [94]. To date, it is established that prostate cancer can be distinctly delineated into at least three subtypes via gene fusion status with the largest compromising ETS family members and encompassing 50 to 75% of all prostate cancer. ETS negative prostate cancer can be separated into smaller subsets based on RAF expression or SPINK1 overexpression. Studies being conducted in Asia are confirming these stratifications found in western countries and also delineating other potential genomic markers. The unraveling of the genetic complexity of prostate cancer with the finding of various recurrent gene fusions has been attributed to the advances in technology specifically next-generation whole genome and RNA sequencing. Furthermore, these findings can now be validated and confirmed with such technologies as 3C and Hi-C that have the power to inspect the 3D conformation of chromatin at distinct locations. Utilizing all of these techniques has led to the beginning of our understanding of the mechanism of gene fusion formation. The looping model proposed very precise and coordinated double strand breaks at specific locations with AR-dependent or independent formation of protein complexes. This knowledge could have significant impact on how prostate cancer is diagnosed and treated in the future as we learn about the mechanisms and pathways that are altered and regulated by these gene fusion events.
In summary, the discovery of gene fusions in prostate cancer has led to a molecular stratification of prostate cancer that has improved our understanding of prostate cancer initiation and progression. Moreover, gene fusions have also proven useful as biomarkers detectable in urine. As the complexity of the disease is unraveled, it is now understood that relying solely on PSA oversimplifies and results in an overestimation of instances of prostate cancer. As “active surveillance” is getting more of a foothold as the preferred standard of care [95], it is clear that other biomarkers, such as gene fusions, must be considered and implemented in clinical diagnostic assays to improve patient care. Gene fusions and other genomic alterations, such as lincRNAs, which were recently discovered to play a role in prostate cancer [96], are unequivocally usurping PSA as better diagnostic marker. More recently, significant advances have been made to understanding the mechanism of AR in gene fusion formation. The finding that inhibition of PARP1 reduces tumor formation in ETS positive patients offers a rationale for ongoing, and future, clinical trials combining PARP inhibitors with drugs targeting AR (abiraterone). Overall, androgen-driven gene fusions have led to improved early detection markers, stratification of prostate cancer patients, and new therapies that will hopefully reduce the incidence of prostate cancer as well as alleviate, if not eliminate, the pitfalls of current treatment regiments.
ACKNOWLEDGEMENTS
CAM is a Prostate Cancer Foundation Young Investigator and is funded by an NIH Pathway to Independence Award (R00 CA149182).
Footnotes
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
DISCLAIMER: The above article has been published in Epub (ahead of print) on the basis of the materials provided by the author. The Editorial Department reserves the right to make minor modifications for further improvement of the manuscript.
REFERENCES
- 1.Nowell PC, Hungerford DA. Chromosome Studies on Normal and Leukemic Human Leukocytes. J Natl Cancer Inst. 1960;25:85–109. [PubMed] [Google Scholar]
- 2.Nowell PC, Hungerford DA. Chromosome studies in human leukemia. II. Chronic granulocytic leukemia. J Natl Cancer Inst. 1961;27:1013–1035. [PubMed] [Google Scholar]
- 3.Druker BJ, Sawyers CL, Kantarjian H, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344(14):1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 4.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 5.MacDonald JW, Ghosh D. COPA--cancer outlier profile analysis. Bioinformatics. 2006;22(23):2950–2951. doi: 10.1093/bioinformatics/btl433. [DOI] [PubMed] [Google Scholar]
- 6.Maher CA, Kumar-Sinha C, Cao X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458(7234):97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer MK, Chinnaiyan AM, Maher CA. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics. 2011;27(20):2903–2904. doi: 10.1093/bioinformatics/btr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pflueger D, Terry S, Sboner A, et al. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21(1):56–67. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bass AJ, Lawrence MS, Brace LE, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43(10):964–968. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Annunziata CM, Davis RE, Demchenko Y, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12(2):115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. JJ CLin Oncol. 2011;29(27):3659–3668. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 13.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8(7):497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren S, Peng Z, Mao JH, et al. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res. 2012;22(5):806–821. doi: 10.1038/cr.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magi-Galluzzi C, Tsusuki T, Elson P, et al. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate. 2011;71(5):489–497. doi: 10.1002/pros.21265. [DOI] [PubMed] [Google Scholar]
- 16.Lee K, Chae JY, Kwak C, Ku JH, Moon KC. TMPRSS2-ERG gene fusion and clinicopathologic characteristics of Korean prostate cancer patients. Urology. 2010;76(5):1268.e7–1268.e13. doi: 10.1016/j.urology.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Han B, Mehra R, Dhanasekaran SM, et al. A fluorescence in situ hybridization screen for E26 transformation-specific aberrations: identification of DDX5-ETV4 fusion protein in prostate cancer. Cancer Res. 2008;68(18):7629–7637. doi: 10.1158/0008-5472.CAN-08-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esgueva R, Perner S, J LaFargue C, et al. Prevalence of TMPRSS2-ERG and SLC45A3-ERG gene fusions in a large prostatectomy cohort. Mod Pathol. 2010;23(4):539–546. doi: 10.1038/modpathol.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maher CA, Palanisamy N, Brenner JC, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. PNAS. 2009;106(30):12353–12358. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pflueger D, Rickman DS, Sboner A, et al. N-myc downstream regulated Gene 1 ( NDRG1 ) is fused to ERG in prostate cancer. Neoplasia. 2009;11(8):804–811. doi: 10.1593/neo.09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448(7153):595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 22.Gasi D, Van der Korput HA, Douben HC, et al. Overexpression of full-length ETV1 transcripts in clinical prostate cancer due to gene translocation. PloS One. 2011;6(1):e16332. doi: 10.1371/journal.pone.0016332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Attard G, Clark J, Ambroisine L, et al. Heterogeneity and clinical significance of ETV1 translocations in human prostate cancer. Br J Cancer. 2008;99(2):314–320. doi: 10.1038/sj.bjc.6604472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomlins SA, Mehra R, Rhodes DR, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66(7):3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 25.Hermans KG, Bressers AA, van der Korput HA, Dits NF, Jenster G, Trapman J. Two unique novel prostate-specific and androgen-regulated fusion partners of ETV4 in prostate cancer. Cancer Res. 2008;68(9):3094–3098. doi: 10.1158/0008-5472.CAN-08-0198. [DOI] [PubMed] [Google Scholar]
- 26.Helgeson BE, Tomlins SA, Shah N, et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68(1):73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- 27.Rickman DS, Pflueger D, Moss B, et al. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res. 2009;69(7):2734–2738. doi: 10.1158/0008-5472.CAN-08-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nacu S, Yuan W, Kan Z, et al. Deep RNA sequencing analysis of readthrough gene fusions in human prostate adenocarcinoma and reference samples. BMC Med Genomics. 2011;4(1):11. doi: 10.1186/1755-8794-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Gong M, Yuan H, Park HG, Frierson HF, Li H. Chimeric transcript generated by cis-splicing of adjacent genes regulates prostate cancer cell proliferation. Cancer Discov. 2012;2(7):598–607. doi: 10.1158/2159-8290.CD-12-0042. [DOI] [PubMed] [Google Scholar]
- 30.Williams A, Flavell RA. The role of CTCF in regulating nuclear organization. J Exp Med. 2008;205(4):747–750. doi: 10.1084/jem.20080066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Kannan K, Wang L, Wang J, Ittmann MM, Li W, Yen L. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. PNAS. 2011;108(22):9172–9177. doi: 10.1073/pnas.1100489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palanisamy N, Ateeq B, Kalyana-Sundaram S, et al. Rearrangements of the RAF kinase pathway in prostate cancer. gastric cancer and melanoma Nat Med. 2010;16(7):793–798. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren G, Liu X, Mao X, Zhang Y, Stankiewicz E, Hylands L. Identification of frequent BRAF copy number gain and alterations of RAF genes in chinese prostate cancer. Genes Chromosomes Cancer. 2012;51(11):1014–1023. doi: 10.1002/gcc.21984. [DOI] [PubMed] [Google Scholar]
- 35.Sun C, Skaletsky H, Birren B, et al. An azoospermic man with a de novo point mutation in the Y-chromosomal gene USP9Y. Nature Gen. 1999;23(4):429–432. doi: 10.1038/70539. [DOI] [PubMed] [Google Scholar]
- 36.Usener D, Schadendorf D, Koch J, Dübel S, Eichmüller S. cTAGE: a cutaneous T cell lymphoma associated antigen family with tumor-specific splicing. J Invest Dermatol. 2003;121(1):198–206. doi: 10.1046/j.1523-1747.2003.12318.x. [DOI] [PubMed] [Google Scholar]
- 37.Lu Y, Ryan SL, Elliott DJ, et al. Amplification and overexpression of Hsa-miR-30b, Hsa-miR-30d and KHDRBS3 at 8q24.22-q24.23 in medulloblastoma. PloS One. 2009;4(7):e6159. doi: 10.1371/journal.pone.0006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosco N, Pelliccia F, Rocchi A. Characterization of FRA7B, a human common fragile site mapped at the 7p chromosome terminal region. Cancer Genet Cytogenet. 2010;202(1):47–52. doi: 10.1016/j.cancergencyto.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Vaarala MH, Hirvikoski P, Kauppila S, Paavonen TK. Identification of androgen-regulated genes in human prostate. Mol Med Report. 2012;6(3):466–472. doi: 10.3892/mmr.2012.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tittel-Elmer M, Lengronne A, Davidson MB, et al. Cohesin association to replication sites depends on Rad50 and promotes fork restart. Mol Cell. 2012;48(1):98–108. doi: 10.1016/j.molcel.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomlins SA, Rhodes DR, Yu J, et al. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell. 2008;13(6):519–528. doi: 10.1016/j.ccr.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Ann Rev Genet. 2011;45(D):167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 43.Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu D, Zhang C, Shen Y, Nephew KP, Wang Q. Androgen receptor-driven chromatin looping in prostate cancer. Trends Endocrinol Metab. 2011;22(12):474–480. doi: 10.1016/j.tem.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19(5):631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 46.Chen Z, Zhang C, Wu D, et al. Phospho-MED1-enhanced UBE2C locus looping drives castration-resistant prostate cancer growth. EMBO J. 2011;30(12):2405–2419. doi: 10.1038/emboj.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bastus NC, Boyd LK, Mao X, et al. Androgen-induced TMPRSS2:ERG fusion in nonmalignant prostate epithelial cells. Cancer Res. 2010;70(23):9544–9548. doi: 10.1158/0008-5472.CAN-10-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mani RS, Tomlins SA, Callahan K, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326(5957):1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin C, Yang L, Tanasa B, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139(6):1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haffner MC, Aryee MJ, Toubaji A, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nature Genet. 2010;42(8):668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 52.Lieberman-Aiden E, Van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rickman DS, Soong TD, Moss B, et al. Oncogene-mediated alterations in chromatin conformation. PNAS. 2012;109(23):9083–9088. doi: 10.1073/pnas.1112570109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berger MF, Lawrence MS, Demichelis F, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu J, Yu J, Mani RS, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17(5):443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu Q, Kwon Y-S, Nunez E, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. PNAS. 2008;105(49):19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ju BG, Lunyak VV, Perissi V, et al. A topoisomerase llbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312(5781):1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 58.Pedersen-Bjergaard J, Andersen MK, Johansson B. Balanced chromosome aberrations in leukemias following chemotherapy with DNA-topoisomerase II inhibitors. J Clin Oncol. 1998;16(5):1897–1898. doi: 10.1200/JCO.1998.16.5.1897. [DOI] [PubMed] [Google Scholar]
- 59.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 60.Attard G, Clark J, Ambroisine L, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27(3):253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tu JJ, Rohan S, Kao J, Kitabayashi N, Mathew S, Chen YT. Gene fusions between TMPRSS2 and ETS family genes in prostate cancer: frequency and transcript variant analysis by RT-PCR and FISH on paraffin-embedded tissues. Mol Pathol. 2007;20(9):921–928. doi: 10.1038/modpathol.3800903. [DOI] [PubMed] [Google Scholar]
- 62.Perner S, Demichelis F, Beroukhim R, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66(17):8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 63.Mehra R, Tomlins SA, Yu J, et al. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent metastatic prostate cancer. Cancer Res. 2008;68(10):3584–3590. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rajput AB, Miller MA, De Luca A, et al. Frequency of the TMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007;60(11):1238–1243. doi: 10.1136/jcp.2006.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66(17):8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 66.Nam RK, Sugar L, Wang Z, et al. Expression of TMPRSS2:ERG gene fusion in prostate cancer cells is an important prognostic factor for cancer progression. Cancer Biol Ther. 2007;6(1):40–45. doi: 10.4161/cbt.6.1.3489. [DOI] [PubMed] [Google Scholar]
- 67.Nam RK, Sugar L, Yang W, et al. Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br J Cancer. 2007;97(12):1690–1695. doi: 10.1038/sj.bjc.6604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26(31):4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 69.Petrovics G, Liu A, Shaheduzzaman S, et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24(23):3847–3852. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- 70.Winnes M, Lissbrant E, Damber JE, Stenman G. Molecular genetic analyses of the TMPRSS2-ERG and TMPRSS2-ETV1 gene fusions in 50 cases of prostate cancer. Oncol Rep. 2007;17(5):1033–1036. [PubMed] [Google Scholar]
- 71.Saramäki OR, Harjula AE, Martikainen PM, Vessella RL, Tammela TLJ, Visakorpi T. TMPRSS2:ERG fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin Cancer Res. 2008;14(11):3395–3400. doi: 10.1158/1078-0432.CCR-07-2051. [DOI] [PubMed] [Google Scholar]
- 72.Yoshimoto M, Joshua AM, Chilton-Macneill S, et al. Three-color FISH analysis of TMPRSS2/ERG fusions in prostate cancer indicates that genomic microdeletion of chromosome 21 is associated with rearrangement. Neoplasia. 2006;8(6):465–469. doi: 10.1593/neo.06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lapointe J, Kim YH, Miller MA, et al. A variant TMPRSS2 isoform and ERG fusion product in prostate cancer with implications for molecular diagnosis. Mod Pathol. 2007;20(4):467–473. doi: 10.1038/modpathol.3800759. [DOI] [PubMed] [Google Scholar]
- 74.Leinonen KA, Tolonen TT, Bracken H, et al. Association of SPINK1 expression and TMPRSS2:ERG fusion with prognosis in endocrine-treated prostate cancer. Clinical Cancer Res. 2010;16(10):2845–2851. doi: 10.1158/1078-0432.CCR-09-2505. [DOI] [PubMed] [Google Scholar]
- 75.Laxman B, Tomlins SA, Mehra R, et al. Noninvasive detection of TMPRSS2:ERG fusion transcripts in the urine of men with prostate cancer. Neoplasia. 2006;8(10):885–888. doi: 10.1593/neo.06625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hessels D, Smit FP, Verhaegh GW, Witjes JA, Cornel EB, Schalken JA. Detection of TMPRSS2-ERG fusion transcripts and prostate cancer antigen 3 in urinary sediments may improve diagnosis of prostate cancer. Clinical Cancer Res. 2007;13(17):5103–5108. doi: 10.1158/1078-0432.CCR-07-0700. [DOI] [PubMed] [Google Scholar]
- 77.Laxman B, Morris DS, Yu J, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68(3):645–649. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nguyen PN, Violette P, Chan S, et al. A panel of TMPRSS2:ERG fusion transcript markers for urine-based prostate cancer detection with high specificity and sensitivity. Eur Urol. 2011;59(3):407–414. doi: 10.1016/j.eururo.2010.11.026. [DOI] [PubMed] [Google Scholar]
- 79.Tomlins SA, Aubin SMJ, Siddiqui J, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 3(94):94ra72. doi: 10.1126/scitranslmed.3001970. 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perner S, Rupp NJ, Braun M, et al. Loss of SLC45A3 protein (prostein) expression in prostate cancer is associated with SLC45A3-ERG gene rearrangement and an unfavorable clinical course. Int J Cancer. 2012;132(4):807–812. doi: 10.1002/ijc.27733. [DOI] [PubMed] [Google Scholar]
- 81.Yin M, Dhir R, Parwani AV. Diagnostic utility of p501s (prostein) in comparison to prostate specific antigen (PSA) for the detection of metastatic prostatic adenocarcinoma. Diagn Pathol. 2007;2:41. doi: 10.1186/1746-1596-2-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grönberg H. Prostate cancer epidemiology. Lancet. 2003;361(9360):859–864. doi: 10.1016/S0140-6736(03)12713-4. [DOI] [PubMed] [Google Scholar]
- 83.Afar DE, Vivanco I, Hubert RS, et al. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001;61(4):1686–1692. [PubMed] [Google Scholar]
- 84.Lin B, Ferguson C, White JT, et al. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59(17):4180–4184. [PubMed] [Google Scholar]
- 85.Cai C, Wang H, Xu Y, Chen S, Balk SP. Reactivation of androgen receptor-regulated TMPRSS2:ERG gene expression in castration-resistant prostate cancer. Cancer Res. 2009;69(15):6027–6032. doi: 10.1158/0008-5472.CAN-09-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Attard G, Reid AH, Yap TA, et al. Phase I clinical trial of a selective inhibitor of CYP17, abiraterone acetate, confirms that castration-resistant prostate cancer commonly remains hormone driven. J Clin Oncol. 2008;26(28):4563–4571. doi: 10.1200/JCO.2007.15.9749. [DOI] [PubMed] [Google Scholar]
- 87.Boormans JL, Hermans KG, Made AC, et al. Expression of the androgen-regulated fusion gene TMPRSS2-ERG does not predict response to endocrine treatment in hormone-naïve, node-positive prostate cancer. Eur Urol. 57(5):830–835. doi: 10.1016/j.eururo.2009.08.013. 201. [DOI] [PubMed] [Google Scholar]
- 88.Brenner JC, Feng FY, Han S, et al. PARP-1 inhibition as a targeted strategy to treat Ewing’s sarcoma. Cancer Res. 2012;72(7):1608–1613. doi: 10.1158/0008-5472.CAN-11-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brenner JC, Ateeq B, Li Y, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19(5):664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 91.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 92.Ateeq B, Tomlins SA, Laxman B, et al. Therapeutic targeting of SPINK1-positive prostate cancer. Sci Transl Med. 2011;3(72):1–18. doi: 10.1126/scitranslmed.3001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Slovin SF, Kelly WK, Wilton A, et al. Anti-epidermal growth factor receptor monoclonal antibody cetuximab plus Doxorubicin in the treatment of metastatic castration-resistant prostate cancer. Clin Genitourin Cancer. 2009;7(3):E77–E82. doi: 10.3816/CGC.2009.n.028. [DOI] [PubMed] [Google Scholar]
- 94.Vargo-Gogola T, Rosen JM. Modelling breast cancer: one size does not fit all. Nat Rev Cancer. 2007;7(9):659–672. doi: 10.1038/nrc2193. [DOI] [PubMed] [Google Scholar]
- 95.Wilt TJ, Brawer MK, Jones KM, et al. Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med. 2012;367(3):203–213. doi: 10.1056/NEJMoa1113162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]