Abstract
The aim of this approach was to identify the major determinants, located at the 5′ end of the stop codon, that modulate translational read-through in Saccharomyces cerevisiae. We developed a library of oligonucleotides degenerate at the six positions immediately upstream of the termination codon, cloned in the ADE2 reporter gene. Variations at these positions modulated translational read-through efficiency ∼16-fold. The major effect was imposed by the two nucleotides immediately upstream of the stop codon. We showed that this effect was neither mediated by the last amino acid residues present in the polypeptide chain nor by the tRNA present in the ribosomal P site. We propose that the mRNA structure, depending on the nucleotides in the P site, is the main 5′ determinant of read-through efficiency.
INTRODUCTION
Translation termination initiates when a stop codon enters the ribosomal A site. However, under some conditions release of the polypeptide chain at the stop codon is suppressed by the misincorporation of an amino acid at this position (1). Continued translation in the original reading frame then results in synthesis of a longer protein. Numerous viruses, most of them infecting plants, are known to use such a programmed stop codon read-through to control their expression (2). In addition, read-through can be stimulated by aminoglycoside antibiotics such as paromomycin or gentamicin, allowing expression of a full-length protein from genes carrying a nonsense mutation (3–5). Deciphering the rules dictating read-through efficiency is thus of primary importance if one wants to modulate its efficiency for antiviral therapy or for pharmacological treatment of diseases due to premature termination codons (6,7).
Translation termination involves the interplay of several trans- and cis-acting factors (8,9). In eukaryotes, the class I release factor (eRF1) recognizes all three types of nonsense codons and promotes hydrolysis of the linkage between the nascent polypeptide and the P site tRNA (10). The class II eukaryotic release factor (eRF3) is a GTPase whose activity, upon binding to eRF1, is induced by eRF1 interaction with both the mRNA and the peptidyl transfer center of the ribosome (11,12). eRF3 is not involved directly in stop codon recognition and hydrolysis of the polypeptide ester bond, but it is important for inducing efficient termination, probably through recycling eRF1 to the ribosome (8). Other factors also participate directly or indirectly in the overall termination efficiency. The best known are components of the nonsense-mediated mRNA decay pathway (13). The three Upf proteins interact either with eRF1 or eRF3 and deletion of each of the genes encoding Upf1p, Upf2p and Upf3p results in enhanced nonsense codon read-through (13,14). The poly(A) binding protein (PABP), which plays a role in the stabilization of RNA messages, was also shown to interact with eRF3 in yeast, Xenopus and human cells (15–17). Its overexpression has an antisuppressor effect in Saccharomyces cerevisiae cells bearing sup35 mutations and was proposed to stimulate eRF3 in the post-termination recycling step. Similarly, overexpression of Mtt1p, a helicase that interacts with eRF3 and possibly with eRF1 stimulates translational read-through (18). Also, Itt1p, a TRIAD zinc finger protein, interacts with eRF1 and inhibits translation termination (19). Finally, several chaperone components interact directly with specific ribosomal domains, in particular the ribosomal exit tunnel, and may modulate translation accuracy (20). In addition to soluble factors, the ribosomal decoding site is involved in termination. Several experiments indicate that eRF1 interacts directly with 18S and 28S rRNA and that this interaction plays a role in the termination process (21).
cis-acting factors include the termination codon itself and the surrounding nucleotide context. Both upstream and downstream contexts are involved in efficient read-through in S.cerevisiae (22). The CA(A/G)N(U/C/G)A sequence downstream of the termination codon induces efficient read-through (23). Statistical and experimental analyses of some plant and animal viral RNAs carrying leaky termination codons revealed that the nucleotide context following the stop codon is a major determinant of translational read-through (24,25). The 5′ nucleotide context is also not random and has been shown to be involved in translation termination efficiency (26). The chemical properties of the penultimate amino acid in the nascent polypeptide chain were described as modulating translational read-through in eukaryotes, while in prokaryotes the ultimate amino acid was involved. The 5′ context may also mediate read-through by interaction between the P site tRNA directly with eRF1 or indirectly with the ribosome (27,28). Finally, a stacking of nucleotides in the vicinity of the stop codon might also be implicated (29).
We previously developed a combinatorial approach based on utilization of the ADE2 gene to dissect the cis-acting elements involved in termination read-through. This simple and efficient system allowed identification of 3′ contexts stimulating high read-through (23). Here, the same strategy was used to analyze the contribution of the 5′ context to the read-through phenomenon, and revealed that the presence of two adenines immediately upstream of the termination codon plays a major role in translational read-through. Neither tRNA identity nor amino acid chemical properties were found to be responsible for the 5′ context effect. We favor the hypothesis of a role of the mRNA structure, through interaction with the ribosome, in modulating the competition between the release factors and natural suppressor tRNAs.
MATERIALS AND METHODS
Strains and cultivation conditions
The S.cerevisiae strains Y349 MATα lys2Δ201 leu2-3,112 his3Δ200 ura3-52 and its derivative FS1 MATα ade2-592 (frameshift point mutation) lys2Δ201 leu2-3,112 his3Δ200 ura3-52 were used. Yeast cells were transformed using the lithium acetate method (30). Transformants were grown in media selective for plasmid maintenance. To eliminate plasmids bearing URA3, SC medium containing 1 mg/ml 5-fluoroorotic acid (5-FOA) was used (31). Plasmid DNA preparation was done using the Escherichia coli DH5α strain.
Library construction
A first library corresponding to two different syntheses of double-stranded degenerated oligonucleotides, centered on a stop codon and carrying a 5′ degenerate context (CGGA NNN NNN TAG CAG TTA CAGG), was introduced in a frameshift-containing allele of the reporter ADE2 gene, cloned in the centromeric URA3 pFL38 vector (named pADE2 vector). The resulting constructs were transformed into the FS1 strain, as previously described (23). Cloning of the degenerate oligonucleotides was made in order to restore an in-frame ADE2 allele. Because both the frameshift and the premature stop codon containing alleles cause adenine auxotrophy and accumulation of a red pigment, the recoding efficiency of each cloned sequence was estimated by color screening on plates containing a drop-out medium CSM™ with all amino acids and 10 mg/l adenine after incubation for 5 days at 30°C. The interesting fragments were sequenced from plasmids producing white colored cells.
For the second library double-stranded degenerate oligonucleotides (GGGA NNN NNN TAG CAA GAA TAT TTA CAGC) carrying a 3′ context less efficient for read-through were used (32).
Quantification of read-through efficiency
The fragments of interest were cloned in the unique MscI site of the pAC99 plasmid, which bears a dual reporter gene system (14), and the resulting plasmids were transformed into the Y349 strain. The first gene encodes the β-galactosidase enzyme and, depending on the read-through efficiency of the cloned sequence, the second gene encodes the luciferase enzyme. Both enzyme activities were assayed in the same crude extract in at least three independent experiments, as previously described (33). The recoding efficiency was measured as the ratio of luciferase and β-galactosidase activities, compared to an in-frame control which contained a CAG codon instead of the TAG stop codon.
Experiments with the mutagenized tRNA were done with the target sequence ACA CAG TGA CAC TTA cloned into pAC99.
Site-directed mutagenesis of tRNA
tRNAUUGGln1 and tRNACUGGln2 genes were PCR amplified from the Y349 genomic DNA with promoter and terminator to yield PCR fragments encompassing ∼200 bp upstream and downstream of the genes, using Taq DNA polymerase (Amersham). The BamHI-ended oligonucleotide couples, Gln1w (5′-TATGGATCCTACTAAGTGGTGGAAGCGCG-3′) and Gln1c (5′-AATGGATCCAAGTTCAATAATTT CACTGG-3′) for tRNAUUGGln1 and Gln2w (5′-TGAG GATCCCTTCTACTATAAACCTCACTC-3′) and Gln2c (5′-TTGGGATCCTTCTTCGATATCTCTGGTATG-3′) for tRNACUGGln2, were used respectively. The BamHI-digested PCR fragments were then cloned into the BamHI site of the pFL44L (2µ, URA3) vector. The site-directed mutageneses of tRNAUUGGln1 in tRNACUGGln1* and tRNACUGGln2 in tRNAUUGGln2* were performed by PCR joining. The 5′ and 3′ parts of tRNAUUGGln1 were amplified using the oligonucleotide Gln1c with Gln1*w (5′-GTTATCACTTTCGGTT CTGATCCGGACAACC-3′) and Gln1*c (5′-GGTTGT CCGGATCAGAACCGAAAGTGATAAC-3′) with Gln1w, respectively. Similarly, tRNAUUGGln2* was generated using Gln2c with Gln2*w (5-GTTATCACTTTCGGTT TTGATCCGAACAACC-3′) and Gln2*c (5′-GGTTGTTC GGATCAAAACCGAAAGTGATAAC-3′) with Gln2w. Both overlapping fragments were then joined by PCR as described (34). The resulting PCR fragment was digested with BamHI and cloned into the BamHI-pFL44L vector.
Oligonucleotides and sequencing
Oligonucleotides were synthetized by MWG Biotech AG. All constructs were verified using Big Dye Terminator Sequencing Kit, followed by migration on an ABI sequencer (Applied Biosystems).
Statistics
Data were compared using the Mann–Whitney non-parametric test which investigates the difference between two sets of values from individual samples (Richard Lowry, 2000, Inferential Statistics, available at http://faculty.vassar.edu/lowry/ch11a.html).
RESULTS
Primary screening
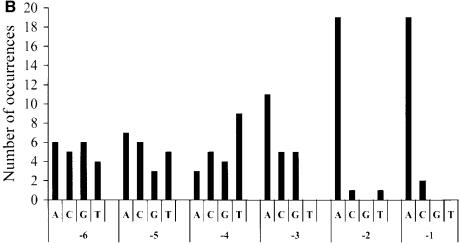
We used a combinatorial approach to identify 5′ nucleotidic contexts of stop codon giving high translational read-through efficiency. A degenerate target sequence carrying variations on the six nucleotides upstream of the stop codon was used to construct a library of recombinant ADE2 gene mutants (pADE2 vector). We chose a 3′ stop codon nucleotide context (CAG TTA) that allows a moderate read-through efficiency (23) in order to select for sequences driving a higher level of read-through. The degenerate library was transformed into the FS1 yeast strain [Ade–]. Under these conditions, the screen gave rise mostly to red colored colonies (low read-through level) and a few white/pink colonies (high read-through level). Among 104 transformants, 91 white clones (1%) were analyzed. Each clone was plated on 5-FOA medium to counterselect the presence of the pADE2 vector. All growing cells produced red colonies, confirming that the white phenotype had occured following a read-through event during translation on the pADE2 vector. Plasmids isolated from these cells were then sequenced and nucleotides surrounding the stop codon identified. Among the 61 different sequences identified (Fig. 1A), we observed a high frequency of adenine at the two positions immediately upstream of the termination codon. No significant bias was seen at positions –3 and –4. In contrast, thymine was not observed at position –5 and seemed also counterselected at position –6.
Figure 1.
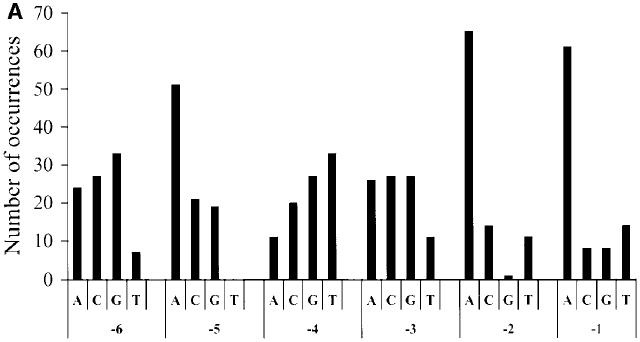
Nucleotide repartition by position among all identified sequences. The y-axis represents the number of occurrences for each of the four nucleotides found immediately upstream of the stop codon from position –1 to –6. (A) First synthesis and (B) second synthesis of the double-stranded degenerated oligonucleotides.
To check whether these biases were meaningful, a second round of screening was done. Two new libraries were constructed. One derived from a new synthesis of the same degenerate oligonucleotide and the other from an oligonucleotide carrying a different 3′ stop codon nucleotide context (CAA GAA), allowing a lower level of read-through (32). Analysis of plasmids from 21 white/pink clones obtained from the first library (17 different sequences) showed that thymine was neither counterselected at position –5 nor at position –6 (Fig. 1B). Moreover, a designed target sequence with T at positions –5 and –6 (TTA codon at position –2 and CAA codon at position –1) gave a read-through efficiency of 13%. These results clearly demonstrate that a bias in the first synthesis of the degenerate oligonucleotides rather than a real biological role was responsible for the under-representation of thymine at positions –5 and –6 in Figure 1A. The strong bias for two adenines just upstream of the stop codon was confirmed, just as with the analysis of some white clones from the second library (data not shown).
Quantification of read-through efficiency
Twenty-one different read-through target sequences, corresponding to 45 clones, were sub-cloned at the junction of a lacZ–luc fusion gene in the pAC99 dual reporter vector (see Materials and Methods). The estimated read-through efficiencies are shown in Table 1, ranging from 1 to 16%. The mean read-through efficiency computed from the 45 clones is 8%, which illustrates the stringency of the screening procedure, with a majority of isolated clones having a read-through level higher than 5%. Results show that variations of the ribosomal P site codon imposed a 16-fold effect on translational read-through. The presence of CAA, GAA or AAA codons just upstream of the stop codon allows a high level of read-through ranging from 6 to 16%. Two separate groups with read-through motifs carrying or not carrying adenine at the –1 and –2 positions were statistically analyzed using the Mann–Whitney non-parametric test. The test demonstrated that the difference in read-through efficiencies observed between the two groups is highly significant [P value (α) = 0.00047]. In contrast, variation of the ribosomal E site codon had only a 1.5-fold effect on translational read-through (12% for GAT GAA compared to 8% for GGG GAA). This demonstrates the nucleotide sequence just upstream of the stop codon plays a critical role in translational read-through, whereas the further upstream sequence has no major influence.
Table 1. Identified 5′ sequences and number of occurrences.
| Nucleotide sequence | Number of occurrencesa |
Read-through efficiency (%) |
|
|---|---|---|---|
| Codon –2 | Codon –1 | ||
| ACC |
CAA |
2 |
16 |
| AAG |
CAA |
3 |
15 |
| GAT |
CAA |
6 |
14 |
| GAT |
GAA |
3 |
12 |
| CAT |
ATA |
1 |
11 |
| ACT |
GAA |
2 |
11 |
| CAG |
CAA |
5 |
10 |
| CAT |
GAA |
2 |
9 |
| GGT |
GAA |
3 |
8 |
| GGG |
GAA |
3 |
8 |
| CGG |
AAA |
1 |
7 |
| ACC |
AAA |
1 |
6 |
| CCG |
ATC |
1 |
6 |
| GAT |
AAG |
1 |
5 |
| TAC |
TCT |
2 |
5 |
| GAC |
ACG |
2 |
5 |
| CCT |
TCA |
1 |
4 |
| ACT |
GAT |
1 |
4 |
| GAG |
ATT |
2 |
3 |
| CCG |
TAC |
2 |
2 |
| GAG | TTT | 1 | 1 |
The experiment was repeated at least three times for each sequence, with a variation of ∼30% in relative read-through efficiencies.
aNumber of recombinant plasmids carrying the same target sequence.
Penultimate and ultimate amino acid charges and chemical properties do not modulate translational read-through
We then looked at whether the 5′ nucleotide context may mediate read-through by the charge or chemical properties of the penultimate and ultimate amino acid residues in the nascent peptide chain. As shown in Table 2, ultimate glutamine, glutamic acid, lysine or isoleucine amino acids (corresponding respectively to CAA, GAA, AAA and ATA codons), which are associated with a high level of read-through, have different charges and chemical properties. Conversely, amino acids with the same charge and chemical properties are associated with different levels of translational read-through. Similar results were observed with various E site codons. Finally, if amino acid identity is the main determinant, one would expect that various P site codons, encoding the same amino acid but decoded by different tRNAs, direct the same read-through efficiency. We constructed and analyzed several such target sequences (i.e. codons CAA and CAG for glutamine, GAA and GAG for glutamic acid and AAA and AAG for lysine). As previously observed (28), the read-through efficiencies revealed a 4-fold difference between the glutamine isocodons and an ∼2-fold difference between either the glutamic acid or lysine isocodons (data not shown). Thus, at least for the four tested and probably for all codons, the amino acid identity does not seem to be a major determinant of efficient read-through.
Table 2. Charge and chemical properties of the ultimate and the penultimate amino acid residues in the nascent polypeptide chain with the observed read-through efficiencies.
| Position –2 | Position –1 | Read-through efficiency (%) | ||
|---|---|---|---|---|
| Nature | Amino acid | Nature | Amino acid | |
| Polar |
Thr |
Polar |
Gln |
16 |
| Basic |
Lys |
Polar |
Gln |
15 |
| Acidic |
Asp |
Polar |
Gln |
14 |
| Polar |
Gln |
Polar |
Gln |
10 |
| Acidic |
Asp |
Polar |
Thr |
5 |
| Polar |
Tyr |
Polar |
Ser |
5 |
| Non-polar |
Pro |
Polar |
Ser |
4 |
| Non-polar |
Pro |
Polar |
Tyr |
2 |
| Acidic |
Asp |
Acidic |
Glu |
12 |
| Polar |
Thr |
Acidic |
Glu |
11 |
| Basic |
His |
Acidic |
Glu |
9 |
| Non-polar |
Gly |
Acidic |
Glu |
8 |
| Polar |
Thr |
Acidic |
Asp |
4 |
| Basic |
Arg |
Basic |
Lys |
7 |
| Polar |
Thr |
Basic |
Lys |
6 |
| Acidic |
Asp |
Basic |
Lys |
5 |
| Basic |
His |
Non-polar |
Ile |
11 |
| Non-polar |
Pro |
Non-polar |
Ile |
6 |
| Acidic |
Glu |
Non-polar |
Ile |
3 |
| Acidic | Glu | Non-polar | Phe | 1 |
Influence of ribosomal P site tRNAs on translational read-through
This last result suggested that the P site tRNA might determine the read-through efficiency. This hypothesis has already been proposed by Mottagui-Tabar and co-workers (28). To investigate the role of the ribosomal P site tRNA in conjunction with the corresponding glutamine codon, the UUG anticodon of tRNAGln1, which decodes the CAA codon and promotes a high level of read-through, was mutated to CUG. Similarly, the CUG anticodon of tRNAGln2, which decodes the CAG codon and allows a low level of read-through, was replaced by UUG. Nucleotide sequence differences between both tRNAs are shown in Figure 2. Glutaminyl tRNA species have 94.4% similarity and GLnRS can aminoacylate glutaminyl tRNAs carrying anticodons with U or G at the 5′ end with similar kinetic parameters (35,36). To test the effect of the mutations, an appropriate target sequence must be chosen, taking into consideration that (i) the glutaminyl tRNAs are known to act as natural suppressors of amber (UAG) and ochre (UAA) stop codons and (ii) in our constructs the CAG codon following the stop codon, which is known to be involved in read-through efficiency, might also be affected by the mutated tRNAGln. To eliminate these potential side-effects and substantiate the influence of the tRNA structure on read-through specifically through the P site, we used constructs carrying a UGA termination codon instead of a UAG, followed by a CAC codon, which is decoded by a tRNAHis, instead of CAG. With this construct, we observed a similar effect of the 5′ codon (CAA versus CAG) compared to the previous one (UAG stop codon followed by CAG) (Table 3).
Figure 2.
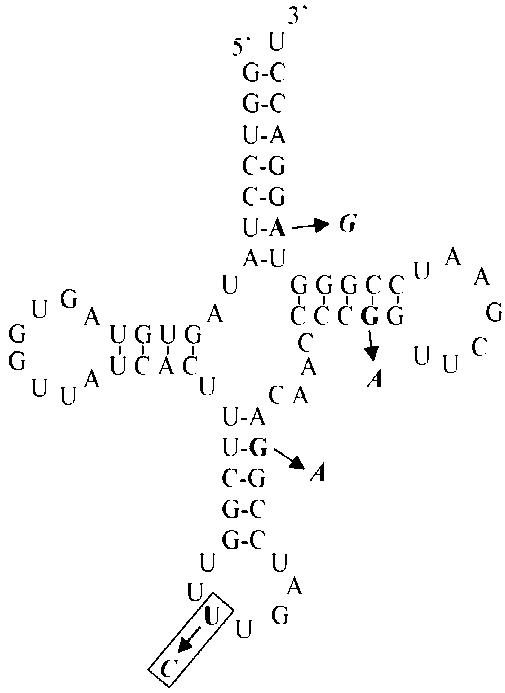
Secondary structure of the yeast glutaminyl tRNA species (from 42). Representation of tRNAUUGGln1 with the four tRNACUGGln2 sequence changes indicated in bold, with anticodon one boxed.
Table 3. Influence of ribosomal P site tRNAs on translational read-through.
| Nucleotide sequence | Read-through efficiency (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| –2 | –1 | Stop | +1 | +2 | pFL44La | tRNAUUGGln1 | tRNACUGGln1* | tRNACUGGln2 | tRNAUUGGln2* |
| ACA |
CAA |
TGA |
CAC |
TTA |
8 |
9 |
8 |
8 |
6 |
| ACA | CAG | TGA | CAC | TTA | 2 | 3 | 3 | 2 | 3 |
Each experiment was done at least five times with a variability of ∼20%.
aThe empty pFL44L vector.
The effect of wild-type or mutant glutaminyl tRNAs was estimated using a target sequence carrying either a CAA or a CAG P site codon. The results are shown in Table 3. The analysis revealed only a slight effect of the tRNA structure on the read-through efficiency (9% for tRNAUUGGln1 versus 6% for tRNAUUGGln2* with the CAA target; 2% for tRNACUGGln2 versus 3% for tRNACUGGln1* with the CAG target). This suggested that the major effect of the 5′ context was not mediated by the tRNA structure itself.
DISCUSSION
In the present work we attempted to better understand the effect of the nucleotide context located 5′ of the stop codon on translational read-through. Up to six nucleotides upstream of the stop codon have been previously shown to be involved in read-through efficiency (27), however, no definitive mechanistic model has emerged for the role of these nucleotides. A systematic study is thus needed to precisely identify the elements involved (nucleotide, codon, amino acid, etc.). A classical site-directed mutagenesis approach would not permit generation and analysis of the 4 × 103 sequences that are needed to examine all the possibilities. To circumvent this problem, we recently set up a combinatorial approach, allowing testing of thousands of constructs in a single experiment by using a yeast-based selective system (23). Here, we used a similar strategy to analyze the six positions immediately upstream of the termination codon. It should be kept in mind that this selective procedure allows the identification of the major determinants involved in read-through, but does not rule out minor effects mediated by other factors.
The obtained results demonstrate that varying the 5′ nucleotide context modulates translational read-through from 1 to 16%. The major effect was mediated by the codon immediately upstream of the stop codon.
In view of the accumulating evidence for nascent peptide-mediated regulation of translation (37), one can imagine that distortion of the decoding center might be triggered by the nascent polypeptide, ending in an altered interaction with the release factor. However, from the presented results, various amino acids exhibiting different chemical properties were associated with similar levels of read-through, either at the –1 or –2 positions. Similarly, synonymous –1 codons decoded by different tRNAs induced various levels of translational read-through. We therefore concluded that the chemical properties of the ultimate amino acid residues in the nascent polypeptide chain do not have a major influence on read-through in yeast.
Possibly the ribosomal P site tRNA directs the competition between eRF1 and natural suppressor tRNA(s) for stop codon recognition (28). Wild-type and mutant glutaminyl tRNAs were used to study the influence of ribosomal P site tRNA on translational read-through over TGA leaky termination codon constructs. tRNAGln1 and mutant tRNAGln2*, which have the same UUG anticodon but a different backbone, direct very similar translational read-through efficiencies as do tRNAGln2 and mutant tRNAGln1* for the anticodon CUG. Although overexpression of a tRNA may lead to under-modification, these results suggest that the P site tRNA has no major influence on the competition between eRF1 and potential suppressor tRNA for termination codon recognition.
Finally, since the common determinant of most sequences driving a high read-through level is the presence of adenine at the two positions 5′ of the stop codon, the effect could be mediated directly by the nucleotides themselves, possibly through mRNA structure and interaction with ribosomal components. Previous reports ended with conflicting observations concerning the role of adenine 5′ of the stop codon. Mottagui-Tabar and co-workers concluded from their study that the 5′ context influence is independent of a C or an A nucleotide switch at the –1 position. In contrast, an A at this position was associated with a high read-through level in mouse cells (38) and in plant cells (39). Furthermore, Beier and Grimm analyzed 53 plant and animal viral RNAs containing leaky termination codons and observed that 39 viral sequences have an adenine in position –1 of the stop codon, 37 have an adenine in position –2 and 29 have adenines in both positions (24). Similarly, Harrell and co-workers showed that among 91 plant and animal viral RNAs using translational read-through, 65 sequences have an adenine in position –1, 69 have an adenine in position –2 and 50 carry two adenines in positions –1 and –2 of the stop codon (25). So the presence of adenines just upstream of a stop codon in sequences regulated by read-through appears to have been evolutionarily selected. Here, using a combinatorial approach, we showed that 67% of the identified sequences have an adenine in position –1, 70% in position –2 and 62% in both positions. Furthermore, 80% (11/13) of the sequences driving a read-through efficiency of >5% were shown to have two adenines immediately upstream of the stop codon. Thus it seems that the presence of adenines in positions –1 and –2 of the UAG termination codon is the main 5′ determinant to obtain high efficiency translational read-through.
It is highly probable that the presence of two adenines also affects read-through on UGA and UAA stop codons. Indeed, from the study of Harrell and co-workers a similar bias can be identified for all three stop codons (25). More generally, the stop codon context seems to act independently of the identity of the stop. This has been observed in several studies where the effect of a given context was tested on all three stop codons (14,22,39) and is illustrated here by the results shown in Table 3.
On the whole, 958 of 5836 (16%) yeast natural termination signals carry an AA_STOP motif (GenBank release 13 November 2003), which suggests that there is no strong counter-selection against 5′ read-through stimulating contexts. This is consistent with previous studies demonstrating that the effect of the 5′ context is highly dependent on the 3′ context (4,5,22,23,38,39).
We hypothesize that these adenines modify the mRNA structure at the P site, which in turn alters decoding at the A site through distortion of the ribosome structure. Indeed, a recent cryo-electron microscopy reconstruction of the yeast 80S ribosome demonstrates that the functional centers and the majority of bridges binding the two subunits are mainly composed of rRNA (40). Remarkably, most of the ribosomal bridges are conserved between eukaryotes and prokaryotes, in particular the B2a bridge that connects the decoding and the peptidyl transferase centers and locates at the subunit interface. B2a is formed by interaction between helix 44 of the 18S rRNA and helix 69 of domain IV of the 25S rRNA. Similarly, the ribosomal A site is composed mainly of helix 69 and of the upper part of helix 44. The latter region contains an invariant adenosine dinucleotide (A1823, A1824), which corresponds to the A1492–A1493 dinucleotide of prokaryotic 16S rRNA interacting directly with the termination signal. On the body/platform side of the small ribosomal subunit, the P site codon also appears to directly contact the top of helix 44, whereas the tRNA interacts with helix 24 of 18S rRNA. Thus, the upper part of helix 44 is implicated in the formation of both the A and P sites and of the B2a bridge, which acts as a signal transmitter between the two active sites of the ribosome (41). We propose that the presence of two adenines upstream of the stop codon, possibly through their high stacking potential, induces structural modifications in the ribosomal P site, which are then transmitted via the B2a bridge to the ribosomal A site. Subsequently, competition between the eRF1 and/or eRF3 release factor(s) and a potential natural suppressor tRNA would be displaced in favor of read-through.
Acknowledgments
ACKNOWLEDGEMENTS
We warmly thank Maryse Godon for her technical assistance during this work. We thank Michaël Bekaert and David Abergel for help with statistic analyses and for stimulating discussions. We are especially grateful to Robert Dickson and Yih-Ling Tzeng for critical reading of the manuscript. This work was supported, in part, by grants from the Association Française contre les Myopathies (contract no. 7757) and from the Association pour la Recherche sur le Cancer (contract no. 4699).
REFERENCES
- 1.Baranov P.V., Gesteland,R.F. and Atkins,J.F. (2002) Recoding: translational bifurcations in gene expression. Gene, 286, 187–201. [DOI] [PubMed] [Google Scholar]
- 2.Bertram G., Innes,S., Minella,O., Richardson,J. and Stansfield,I. (2001) Endless possibilities: translation termination and stop codon recognition. Microbiology, 147, 255–269. [DOI] [PubMed] [Google Scholar]
- 3.Burke J.F. and Mogg,A.E. (1985) Suppression of a nonsense mutation in mammalian cells in vivo by the aminoglycoside antibiotics G-418 and paromomycin. Nucleic Acids Res., 13, 6265–6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howard M., Frizzell,R.A. and Bedwell,D.M. (1996) Aminoglycoside antibiotics restore CFTR function by overcoming premature stop mutations [see comments]. Nature Med., 2, 467–469. [DOI] [PubMed] [Google Scholar]
- 5.Howard M.T., Shirts,B.H., Petros,L.M., Flanigan,K.M., Gesteland,R.F. and Atkins,J.F. (2000) Sequence specificity of aminoglycoside-induced stop codon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann. Neurol., 48, 164–169. [PubMed] [Google Scholar]
- 6.Barton-Davis E.R., Cordier,L., Shoturma,D.I., Leland,S.E. and Sweeney,H.L. (1999) Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice [see comments]. J. Clin. Invest., 104, 375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Politano L., Nigro,G., Nigro,V., Piluso,G., Papparella,S., Paciello,O. and Comi,L.I. (2003) Gentamicin administration in Duchenne patients with premature stop codon. Preliminary results. Acta Myol., 22, 15–21. [PubMed] [Google Scholar]
- 8.Kisselev L., Ehrenberg,M. and Frolova,L. (2003) Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J., 22, 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura Y. and Ito,K. (2003) Making sense of mimic in translation termination. Trends Biochem. Sci., 28, 99–105. [DOI] [PubMed] [Google Scholar]
- 10.Kisselev L.L. and Buckingham,R.H. (2000) Translational termination comes of age. Trends Biochem. Sci., 25, 561–566. [DOI] [PubMed] [Google Scholar]
- 11.Frolova L., Le Goff,X., Zhouravleva,G., Davydova,E., Philippe,M. and Kisselev,L. (1996) Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA, 2, 334–341. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhouravleva G., Frolova,L., Le Goff,X., Le Guellec,R., Inge-Vechtomov,S., Kisselev,L. and Philippe,M. (1995) Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J., 14, 4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W., Czaplinski,K., Rao,Y. and Peltz,S.W. (2001) The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J., 20, 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bidou L., Stahl,G., Hatin,I., Namy,O., Rousset,J.P. and Farabaugh,P.J. (2000) Nonsense-mediated decay mutants do not affect programmed –1 frameshifting. RNA, 6, 952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uchida N., Hoshino,S., Imataka,H., Sonenberg,N. and Katada,T. (2002) A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J. Biol. Chem., 277, 50286–50292. [DOI] [PubMed] [Google Scholar]
- 16.Cosson B., Couturier,A., Chabelskaya,S., Kiktev,D., Inge-Vechtomov,S., Philippe,M. and Zhouravleva,G. (2002) Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol. Cell. Biol., 22, 3301–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosson B., Berkova,N., Couturier,A., Chabelskaya,S., Philippe,M. and Zhouravleva,G. (2002) Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol. Cell, 94, 205–216. [DOI] [PubMed] [Google Scholar]
- 18.Czaplinski K., Majlesi,N., Banerjee,T. and Peltz,S.W. (2000) Mtt1 is a Upf1-like helicase that interacts with the translation termination factors and whose overexpression can modulate termination efficiency. RNA, 6, 730–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urakov V.N., Valouev,I.A., Lewitin,E.I., Paushkin,S.V., Kosorukov,V.S., Kushnirov,V.V., Smirnov,V.N. and Ter-Avanesyan,M.D. (2001) Itt1p, a novel protein inhibiting translation termination in Saccharomyces cerevisiae. BMC Mol. Biol., 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rospert S., Dubaquie,Y. and Gautschi,M. (2002) Nascent-polypeptide-associated complex. Cell. Mol. Life Sci., 59, 1632–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velichutina I.V., Hong,J.Y., Mesecar,A.D., Chernoff,Y.O. and Liebman,S.W. (2001) Genetic interaction between yeast Saccharomyces cerevisiae release factors and the decoding region of 18 S rRNA. J. Mol. Biol., 305, 715–727. [DOI] [PubMed] [Google Scholar]
- 22.Bonetti B., Fu,L.W., Moon,J. and Bedwell,D.M. (1995) The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol., 251, 334–345. [DOI] [PubMed] [Google Scholar]
- 23.Namy O., Hatin,I. and Rousset,J.P. (2001) Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep., 2, 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beier H. and Grimm,M. (2001) Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res., 29, 4767–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrell L., Melcher,U. and Atkins,J.F. (2002) Predominance of six different hexanucleotide recoding signals 3′ of read-through stop codons. Nucleic Acids Res., 30, 2011–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arkov A.L., Korolev,S.V. and Kisselev,L.L. (1995) 5′ Contexts of Escherichia coli and human termination codons are similar. Nucleic Acids Res., 23, 4712–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mottagui-Tabar S. and Isaksson,L.A. (1997) Only the last amino acids in the nascent peptide influence translation termination in Escherichia coli genes. FEBS Lett., 414, 165–170. [DOI] [PubMed] [Google Scholar]
- 28.Mottagui-Tabar S., Tuite,M.F. and Isaksson,L.A. (1998) The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur. J. Biochem., 257, 249–254. [DOI] [PubMed] [Google Scholar]
- 29.Bossi L. (1983) Context effects: translation of UAG codon by suppressor tRNA is affected by the sequence following UAG in the message. J. Mol. Biol., 164, 73–87. [DOI] [PubMed] [Google Scholar]
- 30.Ito H., Fukuda,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boeke J.D., LaCroute,F. and Fink,G.R. (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet., 197, 345–346. [DOI] [PubMed] [Google Scholar]
- 32.Namy O., Duchateau-Nguyen,G. and Rousset,J.P. (2002) Translational readthrough of the PDE2 stop codon modulates cAMP levels in Saccharomyces cerevisiae. Mol. Microbiol., 43, 641–652. [DOI] [PubMed] [Google Scholar]
- 33.Stahl G., Bidou,L., Rousset,J.P. and Cassan,M. (1995) Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res., 23, 1557–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fabret C., Ehrlich,S.D. and Noirot,P. (2002) A new mutation delivery system for genome-scale approaches in Bacillus subtilis. Mol. Microbiol., 46, 25–36. [DOI] [PubMed] [Google Scholar]
- 35.Hong K.W., Ibba,M., Weygand-Durasevic,I., Rogers,M.J., Thomann,H.U. and Soll,D. (1996) Transfer RNA-dependent cognate amino acid recognition by an aminoacyl-tRNA synthetase. EMBO J., 15, 1983–1991. [PMC free article] [PubMed] [Google Scholar]
- 36.Weiss W.A. and Friedberg,E.C. (1986) Normal yeast tRNA(CAGGln) can suppress amber codons and is encoded by an essential gene. J. Mol. Biol., 192, 725–735. [DOI] [PubMed] [Google Scholar]
- 37.Tenson T. and Ehrenberg,M. (2002) Regulatory nascent peptides in the ribosomal tunnel. Cell, 108, 591–594. [DOI] [PubMed] [Google Scholar]
- 38.Cassan M. and Rousset,J.P. (2001) UAG readthrough in mammalian cells: effect of upstream and downstream stop codon contexts reveal different signals. BMC Mol. Biol., 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skuzeski J.M., Nichols,L.M., Gesteland,R.F. and Atkins,J.F. (1991) The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol., 218, 365–373. [DOI] [PubMed] [Google Scholar]
- 40.Ramakrishnan V. (2002) Ribosome structure and the mechanism of translation. Cell, 108, 557–572. [DOI] [PubMed] [Google Scholar]
- 41.Bashan A., Zarivach,R., Schluenzen,F., Agmon,I., Harms,J., Auerbach,T., Baram,D., Berisio,R., Bartels,H., Hansen,H.A. et al. (2003) Ribosomal crystallography: peptide bond formation and its inhibition. Biopolymers, 70, 19–41. [DOI] [PubMed] [Google Scholar]
- 42.Murray L.E., Rowley,N., Dawes,I.W., Johnston,G.C. and Singer,R.A. (1998) A yeast glutamine tRNA signals nitrogen status for regulation of dimorphic growth and sporulation. Proc. Natl Acad. Sci. USA, 95, 8619–8624. [DOI] [PMC free article] [PubMed] [Google Scholar]