Abstract
MUDENG (mu-2-related death-inducing gene, MuD) encodes a predicted ∼54-kDa protein in humans, considered to be involved in trafficking proteins from endosomes toward other membranous compartments as well as in inducing cell death. Here we report on the generation of a mouse monoclonal antibody (MAb) against the middle domain of human (h) MuD. This IgG sub 1 MAb, named M3H9, recognizes residues 244–326 in the middle domain of the MuD protein. Thus, the MuD proteins expressed in an astroglioma cell line and primary astrocytes can be detected by the M3H9 MAb. We showed that M3H9 MAb can be useful in enzyme-linked immunosorbent assay (ELISA) and immunoblot experiments. In addition, M3H9 MAb can detect the expression of the MuD protein in formalin-fixed, paraffin-embedded mouse ovary and uterus tissues. These results indicate that the MuD MAb M3H9 could be useful as a new biomarker of hereditary spastic paraplegia and other related diseases.
Introduction
Anovel gene, MUDENG was first identified by a screening method using hammerhead ribozymes to identify genes associated with Fas-mediated apoptosis.(1) Since then it has been revealed that the protein encoded by the MuD gene is composed of 490 amino acids, with a predicted size of 54.7 kDa.(2,3) It was first named MUDENG (mu-2-related death-inducing gene [MuD]) because ectopic expression of this gene was found to induce cell death.(4) The MuD sequence has been suggested to contain a μ homology domain (MHD) that is found in adaptor proteins (APs) that play an important role in intracellular trafficking pathways.(3,4) Hirst and associates reported that C14orf108 may be a component of AP-5, a newly identified AP complex involved in endosomal trafficking.(3) Finally, it was found that MuD is, in fact, the same gene as C14orf108.
We have generated mouse monoclonal antibodies (MAbs) against the MuD proteins using purified MuD peptides. These antibodies detect proteins approximately 54 kDa in size in the brain cell lysates, and can be used in studies on MuD in normal and cancer cells of human origin, for example, identification of MuD in various human cells or tissues by using enzyme-linked immunosorbent assay (ELISA), Western blot, and immunohistochemical analyses.
Materials and Methods
Primary rat astrocyte cultures
Primary glial cell cultures were established from neonatal rat cerebra.(5) Cells were cultured in high-glucose DMEM (supplemented with glucose to a final concentration of 6 g/L, 2 mM glutamine, 0.1 mM nonessential amino acid mixture, 0.1% gentamicin, and 10% FBS [HyClone, Logan, UT]). After 2 weeks in primary culture, oligodendrocytes and microglia were removed by mechanical dislodgement. Astrocytes were harvested by trypsinization (0.25% trypsin and 0.02% EDTA) and monitored for purity by immunofluorescence. Astrocyte cultures were >97% routinely positive for glial fibrillary acidic protein, an intracellular antigen unique to astrocytes.
Recombinant MuD polypeptide expression and purification
The human (h) MuD polypeptide (490 amino acids [aa]) and its fragments were expressed as histidine (His)-tagged proteins.(6) The recombinant proteins were expressed in BL21 cells induced with IPTG (Sigma-Aldrich, St. Louis, MO). The bacterial pellets were resuspended in binding buffer (7 M urea, 0.5 M NaCl, 20 mM Tris-HCl, 5 mM imidazole [pH 7.9]) and disrupted by sonication. The clarified sonicates were mixed with nickel beads (Qiagen, Valencia, CA). The bound proteins were eluted, refolded by dialysis for urea removal, and analyzed using SDS-PAGE.
Generation of anti-hMuD MAbs
BALB/c mice (Daehanbiolink, Umsung, Korea) were intraperitoneally immunized with 100 μg of purified His-tagged MuD protein emulsified in Freund's complete adjuvant (Sigma-Aldrich). Booster immunizations were carried out twice, every 14 days, with the same dosage of purified MuD proteins with Freund's incomplete adjuvant (Sigma-Aldrich). Three days before cellular fusion, mice were finally boosted through an intravenous administration of 30 μg of purified MuD proteins.
Immune serum was first tested using immunoblotting and ELISA, and then spleens from the selected mice were used for fusion to generate hybridomas. Fusion was performed by mixing splenocytes with mouse sp2/0-Ag14 myeloma cells at 10:1 ratio with polyethylene glycol according to the method previously described.(7) Fused cells were selected in HAT medium (Sigma-Aldrich). Hybridomas, which were selected by screening supernatants with immunoblotting and ELISA, were generated by a standard limiting dilution protocol.
Purification of anti-hMuD MAbs
Hybridomas were grown in DMEM containing 10% FBS, pooled, and resuspended in PBS at a concentration of 5×106/mL. These cell suspensions were then injected intraperitoneally into groups of pristane-primed BALB/C mice (1 mL pristine/mouse; mice were primed 7 days earlier). Ascitic fluid was collected 10–14 days later. The collected ascitic fluid or hybridoma cell culture supernatant was pooled, and the γ-globulin (IgG) from this fluid was passed through a Protein G Sepharose 4 Fast Flow column (GE Biosciences, Piscataway, NJ). The bound antibody was eluted with 0.1 M glycine buffer (pH 2.7) and immediately neutralized with 1 M Tris-base (pH 8.0). Then, the purified IgG fraction was dialyzed against PBS (0.01 M, pH 7.0), concentrated, and stored at −80°C until use.(8)
Enzyme-linked immunosorbent assay
Polystyrene plates (96-well; Immunomodules, Nunc, Roskilde, Denmark) were coated with hMuD protein (10 μg/mL) in coating buffer at 4°C overnight. The plates were washed and then blocked by PBST containing 3% BSA. Hybridoma culture supernatants containing anti-MuD M3H9 MAbs were added and incubated for 1.5 h at 37°C. The plates were washed three times with PBST, then horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (H+L) (1:5000; Bio-Rad, Hercules, CA) for detecting IgG binding was added, and incubation was continued for 60 min at 37°C. Wells were washed three times with PBST, and bound HRP was detected using a substrate solution containing 2.2′-azino-bis(3-etylbenzo-thiazoline-6-sulfonic acid) diammonium salt (ABTS; Sigma-Aldrich) and H2O2 for 10 min at room temperature. The reaction was stopped with 2 M H2SO4, and the plates were read at 630 nm with an automatic microplate reader (Bio-Tek, Winooski, VT).
Immunoblot analysis
Cells were grown and lysed in lysis buffer (M-PER® mammalian protein extraction reagent [Pierce, Rockford, IL], supplemented with 1 mM phenylmethylsulfonylfluoride [PMSF], 2 μg/mL aprotinin, 1 μg/mL leupeptin, 1 μg/mL pepstatin A, 2 mM sodium fluoride, and 1 mM sodium orthovanadate). Cell lysates (30 μg) were subjected to SDS-PAGE and transferred onto a polyvinyl difluoride (PVDF) membrane, which was then blocked with 5% nonfat milk. The membrane was incubated with hybridoma culture supernatant or purified M3H9 MAb overnight at 4°C, followed by incubation with HRP-labeled goat anti-mouse IgG antibody for 1 h. The bands were detected by enhanced chemiluminescence (GE Healthcare Biosciences, Pittsburgh, PA).
Binding domain mapping
Wild-type MuD and its fragments (pEGFPc1-hMuD/C14orf108; residues: 1–172, 1–243, 1–391, 1–460, 173–490, 244–490, 392–490, and 197–443) were tagged with green fluorescent protein (GFP) and expressed in human colon cancer cell line, HCT116, by transient transfection using Lipofectamine2000® reagent (Invitrogen, Gaithersburg, MD). Immunoblotting was performed using the M3H9 MAbs.
Immunohistochemical staining
Paraffin-embedded mouse ovary and uterus tissue sections were deparaffinized, rehydrated, and incubated overnight with anti-MuD MAb M3H9 at 4°C. The slides were then washed and incubated with HRP-polymer conjugate (Reagent A, SuperPicture Polymer Detection Kit, Invitrogen, Carlsbad, CA) for 10 min at room temperature, followed by an additional 10-min incubation with AEC-chromogen solution (Reagent B, Invitrogen). Reactivity was detected using AEC substrate solution according to the manufacturer's protocol. Slides were fixed in 2% paraformaldehyde, counterstained with hematoxylin (Sigma-Aldrich) for 5 min, and visualized using a light microscopy (Olympus, Tokyo, Japan).
Statistical analysis
Student's t-test was used to assess significant differences among treatment groups. The criterion for statistical significance was set at p<0.05.
Results and Discussion
Purification of MuD protein
The recombinant MuD proteins were expressed in BL21 cells that were induced with and without IPTG (Fig. 1A, lanes 1, 2). The recombinant MuD proteins purified with nickel resins (Fig. 1A, lane 3) were analyzed by Coomassie Blue staining. The recombinant MuD bands on SDS-PAGE were seen to have a molecular weight of ∼54 kDa (Fig. 1A). To verify whether the bands represented the recombinant MuD protein, we performed immunoblot analysis with anti-histidine antibody (Santa Cruz Biotechnology, Dallas, TX). As shown in Figure 1B (lanes 2, 3), anti-histidine antibody recognized the same band of recombinant MuD protein as did the M3H9 MAb.
FIG. 1.
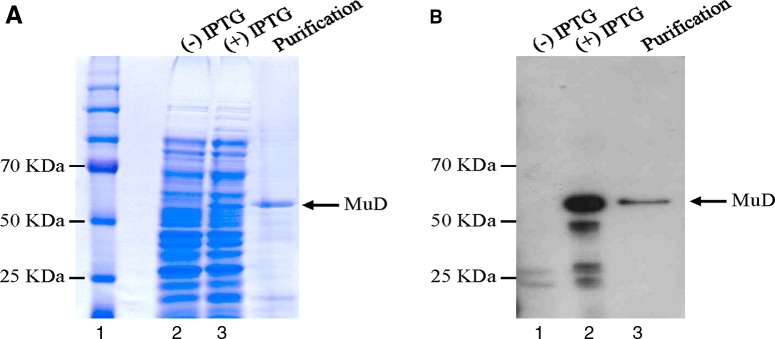
SDS-PAGE and Western blot analyses of recombinant MuD protein. (A) SDS-PAGE for the expression of histidine-tagged MuD protein in BL21 cells. Lane 1, total proteins without IPTG in BL21 cells; lane 2, total proteins with IPTG in BL21 cells; lane 3, purified MuD on nickel beads. (B) Immunoblot analysis with anti-histidine antibody. Lane 1, total proteins without IPTG in BL21 cells; lane 2, total proteins with IPTG in BL21 cells; lane 3, purified MuD with nickel beads. Results are representative of three experiments.
Screening for MAbs against recombinant MuD protein
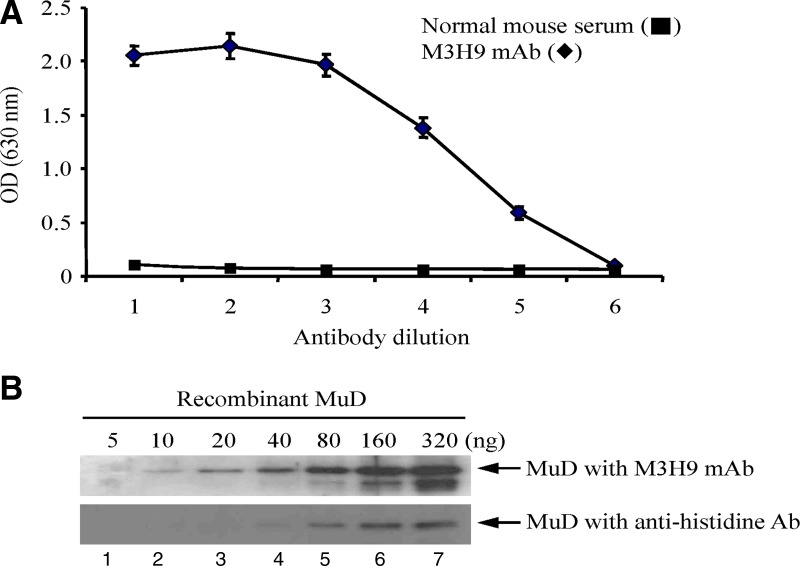
Sera from a selected mouse immunized with MuD protein were screened using ELISA for reactivity with MuD protein. Immune serum showed a significant reactivity with the recombinant MuD protein, indicating the induction of a response to the recombinant MuD protein (data not shown). These results led us to fuse splenocytes from immunized mouse with mouse sp2/0-Ag14 myeloma cells. ELISA screening of the supernatants was performed to identify several parental hybridomas that were similarly reactive with hMuD. After subcloning, further screening led us to select a clone that specifically reacted strongly with the MuD protein; the clone was designated M3H9 MAb, an anti-MuD antibody. Isotyping of M3H9 MAb showed it to be of the IgG sub 1. The M3H9 MAb from ascitic fluid was purified on a Protein G column and screened by indirect ELISA with recombinant MuD protein. At the antigen concentration of 10 μg/mL, the M3H9 MAb had a titer higher than 1:125,000 (0.01 μg/mL), as determined by indirect ELISA (Fig. 2A). Pre-immune serum had a low signal with MuD (Fig. 2A). Immunoblot analysis of the M3H9 MAb confirmed its reactivity against various amounts of recombinant MuD protein (Fig. 2B) and showed the detection limit of M3H9 MAb to be ∼10 ng (lane 2). Compared with anti-histidine antibody, M3H9 MAb showed stronger reactivity (detection limit: 10 ng:40 ng [Fig. 2B, lane 2 vs. lane 4]).
FIG. 2.
Reactivity of anti-MuD M3H9 MAb and immunoblot. (A) Wells were coated with purified MuD protein (10 μg/mL) and then incubated with purified, 5-fold serially diluted M3H9 MAb (♦) [1, 1:200 (8.15 μg/mL); 2, 1:1000 (1.63 μg/mL); 3, 1:5000 (0.32 μg/mL); 4, 1:25,000 (0.06 μg/mL); 5, 1:125,000 (0.01 μg/mL); 6, 1:625,000 (0.002 μg/mL)] and with normal mouse sera (■). Each point represents the mean±SD of three replicates (significant versus the control, p<0.05). (B) Immunoblot with the M3H9 MAb. Indicated amounts of MuD proteins (5–320 ng) were loaded on 10% SDS-PAGE, transferred onto a PVDF membrane, and hybridized with M3H9 MAb. The membrane was then incubated with HRP-conjugated mouse IgG antibody, stripped, and reprobed for histidine detection.
Identification of MuD binding domain recognized by M3H9 MAb
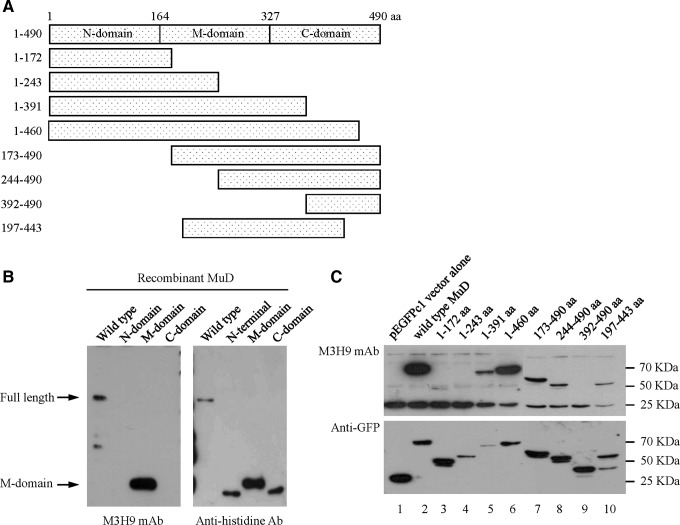
The MuD protein consists of 490 aa(2) (Fig. 3A). To define the binding domain recognized by the mouse anti-MuD M3H9 MAb, we tested the binding reactivity of M3H9 MAb with wild-type and mutant MuD proteins (1–490 aa, 1–164 aa, 165–327 aa, and 328–490 aa) expressed in BL21 cells and then performed immunoblot analysis of the M3H9 MAb. The middle (M)-domain of MuD protein was only recognized by M3H9 MAb (Fig. 3B, left), suggesting that the reactive site is located in the M-domain (residues 164–326) of the protein. Anti-histidine antibody was also used to detect the presence of MuD (Fig. 3B, right). To further characterize the binding site, we transfected HCT116 cells with wild-type and various mutant plasmids (Fig. 3A) and then performed immunoblot analysis of the M3H9 MAb. The M3H9 MAb showed binding to the MuD mutants 1–391, 1–460, 173–490, 244–490, and 197–443 (Fig. 3C, top). However, M3H9 MAb did not bind to the MuD mutants 1–172, 1–243, and 392–490, indicating that the reactive site is located between residues 244 and 326 of the M-domain region. The GFP antibody detections of various MuD polypeptides were seen (Fig. 3C, bottom).
FIG. 3.
M3H9 MAb reacts with residues 244–326 of the MuD protein. (A) Wild-type and three kinds of mutant constructs (MuD 1–490 aa, 1–163 aa, 164–326 aa, and 327–490 aa) are fused in the His-tagged plasmids. The rest of the mutant constructs (1–172 aa, 1–243 aa, 1–391 aa, 1–460 aa, 173–490 aa, 244–490 aa, 392–490 aa, and 197–443 aa) are fused in the pEGFP plasmids. (B) Immunoblot analysis of wild-type MuD protein and its fragments. Full-length (1 μg) and purified MuD (100 ng) polypeptides belonging to each N-/M-/C region were loaded on 10% SDS-PAGE, transferred onto a PVDF membrane, and probed with M3H9 MAb. The membrane was incubated with HRP-conjugated mouse IgG antibody, then stripped and reprobed for histidine detection. (C) The GFP-MuD proteins (1–490 aa, 1–172 aa, 1–243 aa, 1–392 aa, 1–460 aa, 173–490 aa, 244–490 aa, 392–490 aa, and 197–443 aa) expressed in the HCT116 cells by transient transfection were lysed, loaded on 10% SDS-PAGE, and probed with M3H9 MAb. Then the membrane was incubated with HRP-conjugated mouse IgG and reprobed for GFP detection.
Immunoblotting and immunohistochemical staining using M3H9 MAb
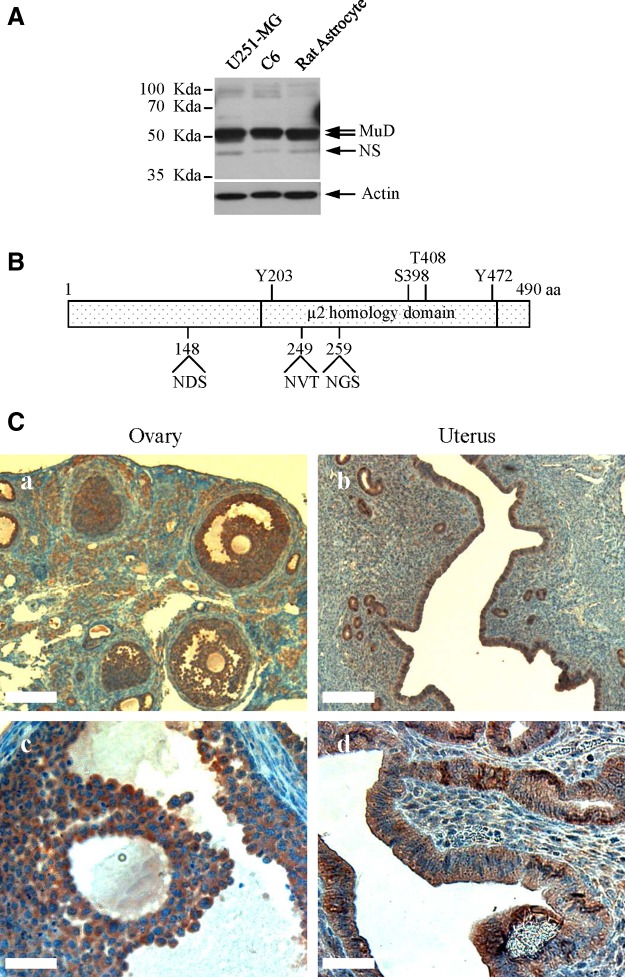
We investigated whether M3H9 MAb could detect endogenous MuD protein inside cells. Therefore, we examined the reactivity of M3H9 MAb with cells of astroglial origin such as U251-MG, C6, and rat primary astrocytes. Interestingly, as shown in Figure 4A, two specific bands were detected between 50 and 55 kDa, suggesting the existence of two types of endogenous MuD in the cells because of the size differences caused by either transcriptional alternative splicing or post-translational modifications, including glycosylation and phosphorylation. According to the structural prediction from UniProt (www.uniprot.org/uniprot/Q9H0R1) and Omin (www.omin.org/614368), there are three putative N-glycosylation sites at asparagines residues and four putative phosphorylation sites in hMuD(9) (Fig. 4B). Interestingly, these post-translational modification sites are found in the μ2 homology domain. Moreover, this antibody recognized all the MuD proteins from human, rat, and mouse cells.
FIG. 4.
Expression of MuD in astroglial cells and immunohistochemical staining of normal mouse ovary and uterus tissues with M3H9 MAb. (A) M3H9 MAb recognition of MuD protein in the primary astrocyte and astrocytic cell lines. Total protein extracts (30 μg) from human U251-MG, rat C6, and rat primary astrocytes were loaded on 10% SDS-PAGE, transferred onto a PVDF membrane, and probed with M3H9 MAb. The membrane was incubated with HRP-conjugated mouse IgG antibody, then stripped and reprobed for actin detection. NS, non-specific. (B) Prediction of putative glycosylation and phosphorylation sites in the MuD protein. (C) Formalin-fixed, paraffin-embedded sections of normal mouse ovary (a and c) and uterus tissues (b and d). Scale bar (a and b, 2 mm; c and d, 0.5 mm).
To determine whether the M3H9 MAb can be used for detecting mouse (m) MuD proteins in fixed tissues, we analyzed formalin-fixed sections of normal mouse ovary and uterus. Staining of mMuD in the fixed tissues was evident in the cumulus oophorus, follicular epithelial cells, and columnar epithelial cells (Fig. 4C). In addition, under a higher power microscope, staining was seen to be restricted to epithelial cells and not the stromal cells in lamina propria, supporting the specific reactivity of M3H9 MAb with MuD-expressing cells.
Targeting of MuD in disease
The MuD gene was first identified during screening of novel genes associated with Fas-mediated apoptosis.(1) Later, it was reported that overexpression of MuD in cells induces cell death,(3,4) supporting the theory that MuD may have an important role in cell death or survival. In addition, a recent study has reported that MuD may interact with KIAA0415/SPG48 and play an important role in intracellular trafficking pathways.(3) MuD and KIAA0415/SPG48 are the two subunits of AP-5 complexes. Slabicki and colleagues suggested SPG48 as a new gene associated with hereditary spastic paraplegia, a group of genetic disorders characterized by progressive weakness and stiffness of the legs.(3,10,11) Thus, studies of MuD expression using the anti-MuD antibody may be useful in the prognosis of patients with hereditary spastic paraplegia and other related diseases.
Our results show that M3H9 MAb recognizes MuD by binding with the protein between the residues 244 and 326 in the M-domain. The recognition site of the antibody varies according to the protein structure.(12) Therefore, further information on the specific recognition sites of M3H9 MAb is necessary to design future therapeutic approaches. Already under way is the construction of new MuD fragments for determining the narrow epitope specificity of M3H9 MAb. Information on the epitope structure of MuD would be vital for antibody drug development.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2011-0010920), and by a grant from the Korea Healthcare Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A080485).
Author Disclosure Statement
The authors have no financial interests to disclose.
References
- 1.Kawasaki H. Taira K. A functional gene discovery in the Fas-mediated pathway to apoptosis by analysis of transiently expressed randomized hybrid-ribozyme libraries. Nucleic Acids Res. 2002;30:3609–3614. doi: 10.1093/nar/gkf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee MR. Shin JN. Moon AR. Park SY. Hong G. Lee MJ. Yun CW. Seol DW. Piya S. Bae J. Oh JW. Kim TH. A novel protein, MUDENG, induces cell death in cytotoxic T cells. Biochem Biophys Res Commun. 2008;370:504–508. doi: 10.1016/j.bbrc.2008.03.139. [DOI] [PubMed] [Google Scholar]
- 3.Hirst J. Barlow LD. Francisco GC. Sahlender DA. Seaman MN. Dacks JB. Robinson MS. The fifth adaptor protein complex. PLoS Biol. 2011;9:e1001170. doi: 10.1371/journal.pbio.1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee K. Kim TH. Bae J. MUDENG is a mediator of Fas-induced apoptosis signaling and induces BAX-dependent cell death. ASCB Late Abstract Contents. 2009;2433:S-L29. [Google Scholar]
- 5.Qin H. Niyongere SA. Lee SJ. Baker BJ. Benveniste EN. Expression and functional significance of SOCS-1 and SOCS-3 in astrocytes. J Immunol. 2008;181:3167–3176. doi: 10.4049/jimmunol.181.5.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green MR. Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 7.Eshhar Z. Monoclonal antibody strategy, techniques. In: Springer TA, editor. Hybridoma Technology in the Biosciences and Medicine. Plenum; New York: 1985. pp. 3–41. [Google Scholar]
- 8.Harlow E. Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 9.Blom N. Sicheritz-Ponten T. Gupta R. Gammeltoft S. Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 10.Slabicki M. Theis M. Krastev DB. Samsonov S. Mundwiller E. Junqueira M. Paszkowski-Rogacz M. Teyra J. Heninger AK. Poser I. Prieur F. Truchetto J. Confavreux C. Marelli C. Durr A. Camdessanche JP. Brice A. Shevchenko A. Pisabarro MT. Stevanin G. Buchholz F. A genome-scale DNA repair RNAi screen identifies SPG48 as a novel gene associated with hereditary spastic paraplegia. PLoS Biol. 2010;8:e1000408. doi: 10.1371/journal.pbio.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang S. And now there are five: a new player in intracellular trafficking pathways. PLoS Biol. 2011;9:e1001173. doi: 10.1371/journal.pbio.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kortagere S. Krasowski MD. Ekins S. The importance of discerning shape in molecular pharmacology. Trends Pharmacol Sci. 2009;30:138–147. doi: 10.1016/j.tips.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]