Abstract
The unlimited differentiation and proliferation capacity of embryonic stem cells represents a great resource for regenerative medicine. Here, we describe a method for differentiating, isolating, and expanding endothelial cells (ECs) from mouse embryonic stem cells (mESCs). First, mESCs are expanded on a mouse embryonic fibroblast (mEF) feeder layer and partially differentiated into embryoid bodies (EBs) by growing the cells in an ultra-low attachment plate for up to 5 days. The EBs are then differentiated along the endothelial lineage using endothelial growth medium supplemented with 40 ng/mL vascular endothelial growth factor (VEGF). The differentiated endothelial population expresses both Fetal Liver Kinase 1 (Flk-1) and VE-Cadherin on the cell surface which can be further purified using a fluorescence-activated cell sorting (FACS) system and subsequently expanded on 0.1 % gelatin-coated plates. The differentiated cells can be analyzed by real-time PCR and flow cytometry to confirm enrichment of EC-specific genes and proteins.
Keywords: Embryonic stem cells, Differentiation, FACS, Endothelial cells
1. Introduction
Embryonic stem cells (ESCs), which are derived from the inner cell mass of blastocysts, are capable of self-renewal and differentiation along different cell lineages, a property known as pluripotence (1). Because ESC differentiation can be directed into particular lineages with specialized functional properties for tissue repair and replacement, they are considered to be an excellent resource for regenerative medicine (2). Three major obstacles associated with using ESCs for regenerative medicine are: (1) precise and controlled differentiation of ESCs toward a well-defined lineage; (2) isolation of homogeneous populations of stably differentiated cells that are fully functional; and (3) retaining the expansion potential.
A common approach is to pre-differentiate ESCs into three-dimensional cell aggregates known as embryoid bodies (EBs). These bodies contain cells from all three germ layers, mesoderm, ectoderm, and endoderm, and can self-renew and differentiate into different cell types. Embryoid bodies can be produced in three main ways: (1) hanging drop; (2) methylcellulose hydrogel; and (3) suspension culture. The most convenient and efficient of these is suspension culture of ESCs in non-adherent plates (3, 4). This approach produces large amounts of EBs in a short time, which is advantageous for high-throughput drug screening and tissue engineering. However, an important consideration is embryoid body size, which may affect the outcome and properties of differentiated cells. For example, uncontrolled overgrowth of EBs may result in cavity formation due to apoptosis, after which EBs eventually become cystic and contain fluid.
Methods for developing differentiated endothelial cells from EBs are well established and the corresponding gene expression patterns are well characterized (4– 6). Embryonic stem cells have been directly differentiated toward specific lineages without first making EBs by using conditioned medium for endothelial differentiation of ESC in collagen IV plates (7, 8).
Large-scale changes in gene expression accompany the initial differentiation of ESCs into EBs, and subsequent large-scale changes in gene expression are linked to lineage-specific differentiation of EBs along mesenchymal, epithelial, neural, or hematopoietic lineages (9, 10, 11). Embryoid bodies can be directed to give rise to hemangioblasts, which subsequently undergo further differentiation into either hematopoietic or endothelial cells. Hemangioblasts have been widely used to study the expression of transcription factors that control EC lineage and recapitulate many aspects of vascular development in vivo (12). The expression of vascular endothelial growth factor receptor-2 (VEGF-R2), also known as Flk-1, in mice, is a mesodermal indicator and the earliest functional marker for hemangioblasts (13).
After EBs are formed and exposed to endothelial differentiation medium containing VEGF, a heterogeneous cell population, including mesenchymal, hematopoietic, and epithelial cells, emerges. Only a subset of these cells will differentiate toward endothelial cells (less than 2 % with our current method). Therefore, once differentiation has been induced the endothelial cell population must be isolated and purified for further expansion and analysis.
Herein, we describe our methods for endothelial differentiation of mouse ESCs (mESCs) and FACS-based isolation and expansion of the resulting cell populations that express the endothelial-specific markers Flk-1 and VE-Cadherin on their surface.
2. Materials
2.1. Cells
mEF cells: Untreated mEF cells-C57BL/6 Cat. No. SCRC-1008, (ATTC Manassas, VA) OR primary mouse embryo fibroblasts, hygro-resistant strain C57BL/6, passage 3 (Millipore, Billerica, MA).
mESCs: mESC were gifts from the laboratory of Robert Blelloch, Eli and Edythe Broad Center of Regeneration Medicine and Stem Cell Research (University of California at San Francisco). The cells were made by targeted insertion of EGFP into the Rosa26 locus in C57BL/6 embryonic stem cells.
2.2. Cell Culture Media
mESCs and mEF media: Dulbecco’s Modified Eagle’s Serum (DMEM) supplemented with 15 % fetal bovine serum (FBS) (HyClone), 2 mM l-glutamine, 0.1 mM MEM nonessential amino acids (Invitrogen), 2.5 × 10−5 M 2-Mercaptoethanol (Sigma 99 %), 1 mM MEM sodium pyruvate (Invitrogen), 100 U/mL penicillin, 100 µg/mL streptomycin, and 1,000 U/mL Esgro LIF (Chemicon, Billerica, MA) (Only for mESCs).
mEF inactivation medium: Mitomycin C (Sigma): Mix 2 mg Mitomycin C powder with 4 mL Dulbecco’s Phosphate-Buffered Saline (D-PBS) (see Note 1).
Freezing medium: DMEM 60 %—FBS 20 %—Dimethyl sulfoxide (DMSO) 20 %: Add 6 mL DMSO (Sigma), 6 mL Hyclone FBS, 18 mL DMEM into the 50 mL falcon tube and mix properly (see Note 2).
Endothelial differentiation medium: Endothelial Cell Growth Medium-2 (EGM® -2, Lonza) supplemented with 40 ng/mL recombinant human vascular endothelial growth factor (rhVEGF-R&D Systems): Add 100 µL of D-PBS to 10 µg of VEGF powder. Mix the solution by pipetting up and down. Add 40 µL of VEGF solution to 100 mL of EGM-2 (see Note 3).
2.3. Solution and Culture Plates
Gelatin coating solution and gelatinized plate: 0.1 % Gelatin Solution ES Cell Qualified (Chemicon/Millipore, Embryomax): Add 3 mL of gelatin per 10 cm dish or 1 mL per 6 cm dish, and coat the entire plate by swirling the plate. Allow the plate to sit for 30 min; then aspirate excess gelatin. Culture the cells immediately before the dish becomes dry.
mEF cell lysate solution: 0.25 % trypsin with Ethylenediaminete traacetic acid (EDTA) (1×) (Invitrogen).
mESCs cell lysate solution: Dissolve 0.5 g collagenase IV in 500 mL of Knockout D-MEM (Invitrogen) (see Note 4).
Poly (2-hydroxyethyl methacrylate) (Poly-HEMA) solution: Add 1 L of 100 % ethanol and a magnetic stirrer to an autoclaved 1 L bottle. Put the bottle on the stirrer and slowly add 25 g of Poly-HEMA (Sigma) preferably over a course of 20–30 min to avoid clumping. Stir the Poly-HEMA solution overnight.
Ultra-low attachment plates: Add 3 mL of Poly-HEMA solution per 10 cm dish or 1 mL per 6 cm dish. Coat the entire plate by gently swirling the Poly-HEMA solution over the surface. Heat the coated plates in a 45 °C oven for a minimum of 24 h. Place the plates under a UV light in a tissue culture hood with lids on. Wash the plates three times with D-PBS before use (see Note 5).
2.4. Fluorescence-Activated Cell Sorting
Harvesting solution: Use cell dissociation medium (Sigma) or 10 mM (EDTA) in D-PBS.
Washing buffer: dissolve 0.2 g sodium azide (NaN3, Sigma) in 200 mL D-PBS and prepare 0.09 % NaN3 -PBS solution and supplement it with 5 % D-FBS.
Blocking buffer: supplement the 0.09 % NaN3 -PBS solution with 10 % FBS.
Phycoerythrin (PE) conjugated anti-mouse Flk-1 (VEGF-R2, eBioscience, Inc., San Diego, CA).
Allophycocyanin (APC) conjugated anti-mouse VE-Cadherin (CD144, eBioscience).
PE conjugated rat IgG2a isotype control (eBioscience).
APC conjugated rat IgG1 isotype control (eBioscience).
3. Methods
One method to expand mESCs is to culture them on mEF feeder layers that have been inactivated with Mitomycin C or via irradiation and with the differentiation inhibitory factor, Leukemia inhibitory factor (LIF) (14). When mEF cells are inactivated, the mESCs can attach, form clumps, and use the growth factors produced from feeder layers, while mEF cell proliferation is inhibited. Once expanded LIF and stromal contact are both withdrawn, mESCs can be grown in ultra-low attachment plates and partially differentiated to form EBs containing all germ layers. The EBs are subsequently differentiated toward the endothelial cell lineage by growing in gelatinized plates and exposing the cells to differentiation medium containing VEGF. The differentiated ECs can be selected via cell sorting with endothelial-specific cell surface antigens.
3.1. mEF Feeder Layer
3.1.1. Plating mEF Cells
Thaw a frozen vial of mEF cells at 37 °C until all ice is completely melted.
Transfer the contents to 10 mL of mEF medium in a 15 mL falcon tube and spin down for 5 min at 125 × g, at room temperature.
Aspirate the supernatant and resuspend the cells in 1 mL mEF medium.
Determine cell viability using trypan blue stain and count the cells using a hemocytometer.
Plate 1 × 106 cells with >85 % viability on a 10 cm 0.1 % gelatinized plate and add 10 mL mEF medium.
Incubate mEF cultures at 37 °C in 5 % CO2 and change medium every 2–3 days.
3.1.2. Splitting mEF Cells
Wash confluent mEF cultures twice with D-PBS.
Add 2 mL of 0.25 % trypsin per 10 cm plate and incubate for 4–5 min at 37 °C in a 5 % CO2 incubator.
Tap the side of the plate to loosen the cells; then wash cells using 5 mL mEF medium.
Transfer the plate contents (cells + medium) to a 15 mL tube and spin down for 5 min at 125 × g at room temperature.
Aspirate the medium and resuspend the cells in the pellet in 3 mL mEF medium.
Split cells at a 1:3 ratio by plating 1 mL volume of the above suspension to each of 3 × 10 cm gelatinized plates and add an additional 10 mL of the mEF medium to each plate.
Feed the mEF plate with mEF medium every 2–3 days.
3.1.3. Inactivation of mEF Cells
Grow the mEF cells until they become 75–85 % confluent.
Aspirate the mEF medium and wash the cells with D-PBS.
Mix 200 µL of 0.5 mg/mL Mitomycin C in 10 mL MEF medium (10 µg/mL) and transfer it to each 10 cm mEF plate.
Incubate the mEF plates at 37 °C and 5 % CO2 for 2–3 h.
Wash the plates three times with 7–10 mL of D-PBS.
Add 1 mL of 0.25 % trypsin per 10 cm plate and incubate for 4–5 min at 37 °C in a 5 % CO2 incubator.
Tap the side of the plates to loosen the cells; then wash cells in each plate using 3 mL mEF medium.
Transfer the plate contents (cells + medium) to a 15 mL tube and spin down for 5 min at 125 × g at room temperature.
Aspirate the medium and resuspend the pellet in 1 mL mEF medium.
Culture the cells in a gelatinized plate at a density of 2–3 × 106 cells per 10 cm plate.
Add 10 mL mEF medium to each plate; then incubate them at 37 °C and 5 % CO2.
Feed the mEF plates with mEF medium every 2–3 days (see Notes 6 and 7).
3.1.4. Freezing the mEF Cells
Wash cells twice with D-PBS.
Add 2 mL of 0.25 % trypsin per 10 cm plate and incubate for 4–5 min at 37 °C.
Tap the plate sides to loosen cells and wash them using 10 mL mEF medium, then transfer cells to a 15 mL tube, and spin them for 5 min at 125 × g.
Aspirate the trypsin and medium, and resuspend the cells using 0.5 mL mEF medium.
Transfer cells to cryo-vial/freezing vials and add 0.5 mL cold freezing medium per vial; then put the vials on ice.
Place vials in −80 °C freezer overnight; after 24 h, place them in liquid nitrogen.
3.2. Culturing mESCs
3.2.1. Plating mESCs on Feeder Layer
Thaw a frozen vial of mESC at 37 °C until ice is completely melted.
Transfer contents to a 15 mL falcon tube with 10 mL of mESC medium and spin down for 5 min at 125 × g at room temperature.
Aspirate the medium and resuspend the cells in 1 mL mESC medium.
Count the cell numbers and determine cell viability using trypan blue stain and a hemocytometer.
Plate 1 × 106 cells on a 10 cm plate of inactivated mEF feeder layer and add 10 mL of mESC medium.
Incubate in 37 °C and 5 % CO2 and feed mESC every day with mESC medium (see Note 8).
3.2.2. Splitting mESCs
This method is for splitting the mESCs once they have been cultured on the mEF feeder layer. The mESCs will begin to form colonies on the mEF feeder layer that can eventually merge. Because mESCs only maintain their undifferentiated state when colonies are not merged, cells must be passaged before colonies come in contact with each other.
Wash the mESC plates twice with D-PBS.
Add 5 mL 0.1 % collagenase IV per 10 cm plate and incubate for 5–10 min at 37 °C at 5 % CO2.
Tap plate sides to loosen cells and wash the cells with 10 mL of mESC medium (collagenase IV primarily detaches the mESCs colonies; however some mEFs may also become detached during this treatment).
Transfer contents to a 15 mL tube and spin down for 5 min at 125 × g at room temperature.
Aspirate the supernatant; then resuspend the cells in 4 mL mESC medium.
Split the cells at a 1:4 ratio by plating 1 mL volume of the above suspension to each of 4 × 10 cm inactivated new mEF feeder layers and add 10 mL of mESC medium into each plate.
Feed the plates with mESC medium every day. Colonies merge together, usually between 3 and 4 days.
3.2.3. Freezing the mESCs
Wash cells twice with D-PBS.
Add 5 mL 0.1 % collagenase IV per10 cm plate and incubate for 5–10 min at 37 °C and 5 % CO2.
Tap the plate sides to loosen cells and wash using 10 mL mESC medium, then transfer cells to a 15 mL tube, and spin for 5 min at 125 × g.
Aspirate the supernatant and resuspend the cells with 0.5 mL mESC medium.
Transfer cells to cryo-vial/freezing tubes and add 0.5 mL cold freezing medium per vial; then put them on ice.
Place vials in −80 °C freezer overnight; after 24 h place them in liquid nitrogen.
3.3. Preparing the Cells for Differentiation
After treating the mESCs and mEFs cells with 0.1 % collagenase IV (see Subheading 3.2.2), mESCs will primarily detach; however, some mEF cells may also detach from the plate. As only mESCs will be used for EB formation, mESCs can be separated from detached mEF cells by allowing the mEF cells to differentially adhere onto new 10 cm culture dishes.
Subculture all cells (mESC + mEF) onto a new 10 cm plate using mESC medium and incubate at 37 °C and 5 % CO2 for 0.5–1 h. The mEF cells will attach during this time and mESCs will stay in suspension. After 0.5–1 h, transfer the non-adherent mESCs into a 15 mL falcon tube and triturate the cells using a 5 mL pipette to break down the colonies. These cells will be used to prepare EBs.
3.4. EB Formation in Suspension Culture
Transfer the mESC suspensions isolated by differential adhesion (see Subheading 3.3) to a 15 mL tube and spin down for 5 min at 125 × g.
Aspirate the mESC medium and gently resuspend the cells in 1 mL mEF medium.
Add 10 mL of mEF medium to a Poly-HEMA coated plate (ultra-low attachment plate) and transfer the suspended cells to this plate. Let the cells sit for 2 days before feeding with fresh medium.
Use a 10 mL tissue culture pipette to transfer suspended EBs from each 10 cm plate to a 15 mL falcon tube and let the EBs settle to bottom of tube by gravity for 10–15 min at 37 °C and 5 % CO2.
Aspirate used medium down to the 2 mL mark; then add 8–10 mL fresh mEF medium.
Use a 10 mL tissue culture pipette to transfer contents back into the original 10 cm plates and incubate them at 37 °C and 5 % CO2.
Change the medium every other day and grow the EBs for up to 5 days.
Switch the medium from mEF medium to EC differentiation medium a day before transferring the EBs to gelatinized plate (see Subheading 3.4) and treat the EBs with EC differentiation medium in Poly-HEMA plate overnight. The following day, transfer the EBs to gelatinized plates for use in EC differentiation (see Subheading 3.5).
3.5. EC Differentiation of mESCs
The size of EBs is an important factor that can affect properties of differentiated cells. The mean size of EBs after 5–6 days of culture in ultra-low attachment plates described above should be around 100–250 µm (approximately to 10–25 cells wide). Smaller EBs might not contain all three germ layers, whereas overgrown EBs will start undergoing apoptosis (Fig. 1).
Transfer the optimally sized (100–250 µm) EBs from each 10 cm Poly-HEMA plate to a 15 mL falcon tube and let the EBs settle to the bottom of the tube for 10–15 min at 37 °C and 5 % CO2.
Aspirate used medium down to the 2 mL mark.
Plate the EBs onto 0.1 % gelatinized plates and add 10 mL fresh EC differentiation medium.
Replace the medium every other day and culture for 7 days.
EBs will attach to 0.1 % gelatinized plates usually overnight and adherent cells will grow out of the EBs after 1–2 days.
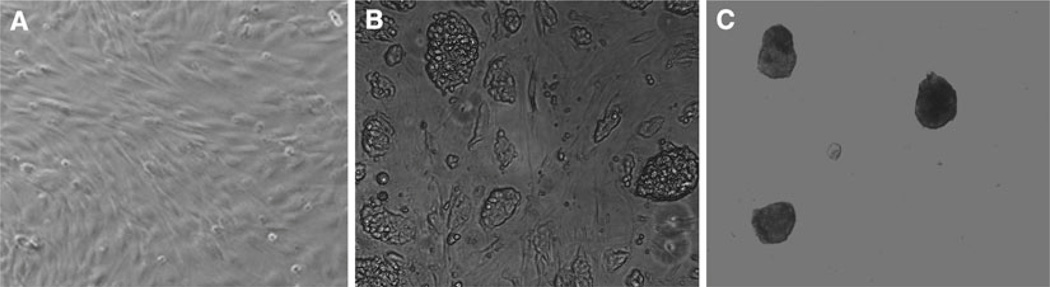
Fig. 1.
Photomicrographs of (A) mouse embryonic fibroblasts (mEF) as feeder layer, (B) mouse embryonic stem cells (mESCs) cultured on the feeder layer, (C) embryoid bodies cultured on ultra-low attachment plates.
Differentiation starts when EBs form (while in suspension culture, see Subheading 3.4) and will continue after the EBs adhere to the 0.1 % gelatinized plates. The cells adhering and growing out from the attached EBs have an extended morphology at early time points (day 1–2) in gelatinized plate (Fig. 2A) and become more rounded at later time points (day 7) (Fig. 2D).
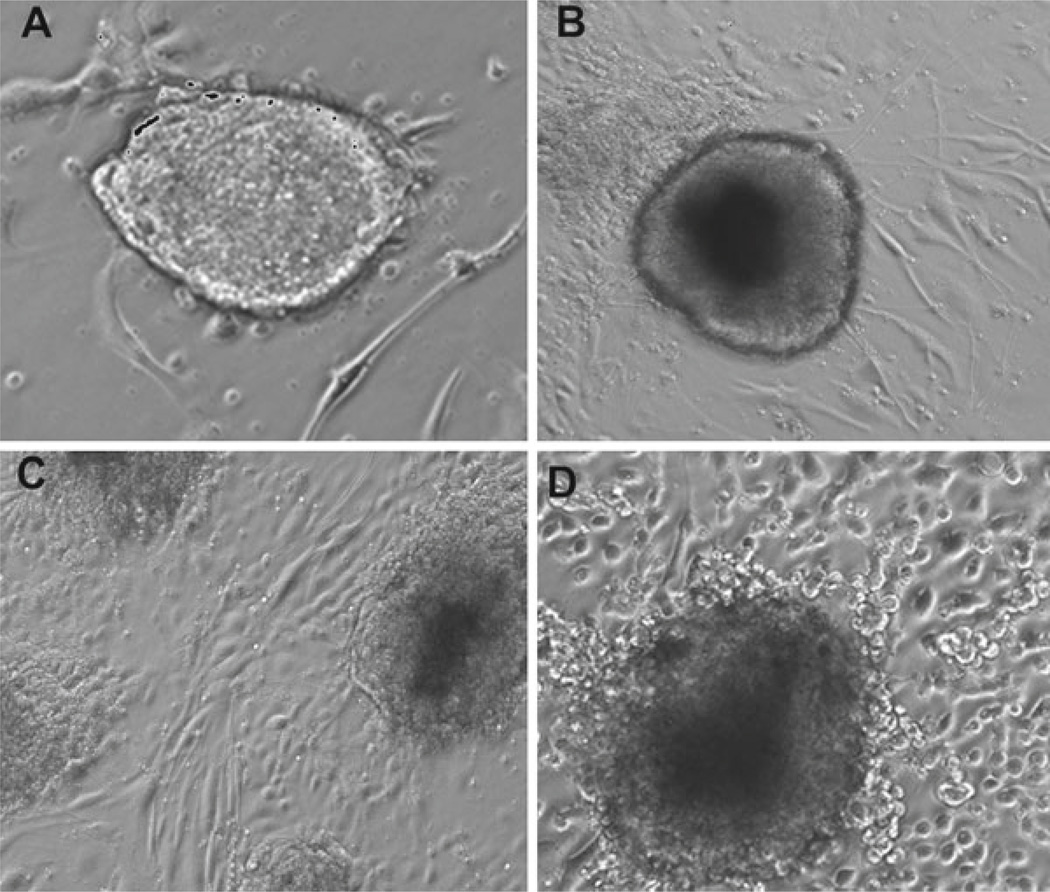
Fig. 2.
Emergence of differentiating endothelial cells from mESCs cultured on gelatin-coated plates at various time points: (A) day 1, (B) day 3, (C) day 5, and (D) day 7.
3.6. Cell Sorting
3.6.1. FACS
In the early stages of differentiation, the culture conditions to promote endothelial differentiation described above give rise to a heterogeneous population with less than 2 % of the cells being pure endothelial lineage. Thus, to study the characteristics and fate of purified endothelial progenitors, this small subpopulation of cells must be isolated from the mixed cultures. FACS is a specialized type of flow cytometry for separating heterogeneous mixtures of biological cells into multiple fractions, one cell at a time, based on light scattering and fluorescent characteristics of each cell. Lasers are used to excite intrinsic or extrinsic fluorescence of cells and the fluorescence intensity is measured from cells or particles through sensitive photomultiplier tubes (15, 16).
The combination of flow cytometry and single-cell sorting is a powerful way to identify and isolate cells with particular characteristics, for instance, based upon markers expressed on the cell surface during differentiation. Control setup and data validation can sometimes be complex, especially when cells have a high fluorescence background or are transfected with a fluorescence reporter before being labeled. Nevertheless, compared to other techniques, FACS facilitates rapid data acquisition, specific multiparameter analysis, and functional separation with high accuracy (17, 18).
During the cell sorting process, a tunable transducer permits the fluid sheath to be broken into individual droplets such that each droplet encapsulates single cells. An electric circuitry places an electrical charge on the fluid stream and the individual droplets. The point at which a cell passes through the laser focus and enters into a droplet corresponds to a specific delay. Because the droplets carry a charge on their surface, a de flecting plate can redirect these charged droplets to collection tubes. Sorting criteria, region designation, multiparameter acquisition, and/or analysis are defined by a software system that includes display platforms.
An alternative to FACS system for cell sorting is magnetic cell separation. This technique is based on magnetic labeling of cells with very small microbeads that do not alter cell structure and function of cells. Separation of labeled cells takes place within a column that provides a magnetic field for cell sorting.
3.6.2. Sorting Cells with Endothelial Phenotype
As mentioned above, conditions that support endothelial differentiation of mESCs yield a heterogeneous population of cells with less than 2 % pure endothelial cells. Although hemangioblasts, which express Flk-1, can differentiate into both hematopoietic and endothelial cells, endothelial progenitor cells also express VE-Cadherin. Thus, to distinguish endothelial precursor cells from hemangioblasts, mesenchymal, epithelial, and other mixed cell types within the differentiating cultures, selecting for cells expressing both Flk-1 and VE-Cadherin is advisable (see Note 9). By labeling putative ECs with two fluorescent-tagged antibodies against Flk-1 and VE-Cadherin, ECs can be selected via FACS. We have used mESCs that were transfected with green fluorescent protein (GFP), which required initial compensation (18–20 %), to de fine negative populations before sorting by Flk-1 and VE-Cadherin surface markers.
Samples can then be analyzed on a Becton Dickinson FACS Vantage TM/DIVA, with a nozzle size of 80 µm, in which forward light scatter (FSC) is collected through a neutral density filter in the forward light scatter path, and side scatter (SSC) is collected through a neutral density filter at a 90° angle. The 488 nm lasers excite fluorescein isothiocyanate (FITC)/GFP and PE, while the 633 nm laser excites APC. Fluorescence emissions can be collected through the FITC (533/30 BP), PE (585/42 BP), and APC (660/20 BP) filters in fluorescence channels FL1, FL2, and FL4, respectively. Below are the steps for sample preparation for FACS sorting and expansion of sorted cells:
Remove the differentiation medium from 10 cm plates and wash the cells twice with D-PBS (5 plates × 10 cm should yield sufficient cell numbers for sorting and subsequent gene expression analysis of sorted cells).
Harvest the cells with 3–5 mL D-PBS–10 mM EDTA or cell dissociation medium and incubate for 5–10 min at 37 °C and 5 % CO2.
Pipette the cells up and down with a 5 mL cell culture pipette and wash the plate to detach all the cells (use a cell scraper if needed).
Pass the cell suspension through a 22 g needle five times to dissociate the EBs into single cells, verified by an inverted microscope.
Centrifuge the cells at 4 °C, at 125 × g for 5 min.
Wash the cells with FACS washing buffer.
Centrifuge the cells at 4 °C and 125 × g for 5 min and remove the supernatant.
Resuspend the cells in 5 mL FACS blocking buffer and incubate them for 30 min on ice.
Centrifuge the cells at 4 °C, 125 × g for 5 min, and remove the supernatant.
Resuspend the cells in 500 µL blocking medium and count the cells using trypan blue stain and a hemocytometer.
Prepare six round-bottomed polystyrene 5-mL FACS tubes for unstained control, FLK-1-PE single color control, VE-Cadherin-APC single color control, double-stain FLK-1-PE + VE-Cadherin-APC, IgG-PE, and IgG-APC isotype controls to confirm specific antibody binding and blocking of nonspecific receptor binding (see Note 10).
Transfer 1–2 × 105 cells in the above-defined control tubes and transfer the rest of the cells to the tube for double-stained FLK-1-PE + VE-Cadherin-APC.
Add APC conjugated anti-mouse CD144 (VE-Cadherin) to the VE-Cadherin-APC single-control tube and add PE conjugated anti-mouse FLK-1(VEGF-R2) to the FLK-1-PE single-control tube. Add a mixture of both antibodies (1:1) to the double-stain FLK-1-PE + VE-Cadherin-APC tube. The final concentration of antibody should be 1 µg of antibody/106 cells in 100 µL block solution. Adjust the final concentration with blocking solution.
Use the unstained sample, which was not exposed to antibody, as a negative control. Use the EOMA cell line (ATTC # CRL-2586) that expresses both VE-Cadherin and Flk-1 as a positive control. Label EOMA cells with both APC conjugated anti-mouse VE-Cadherin and PE conjugated anti-mouse Flk-1 under the same (see Note 11) labeling conditions.
Incubate all samples on ice for 30 min.
Centrifuge samples at 4 °C, 125 × g for 5 min, and remove the supernatant.
Wash the cells three times with washing solution and centrifuge to remove supernatant.
If cell clumps are observed, filter the cell suspension through a 40 µm cell strainer into a 5-mL round bottom cap tube to prevent blockages in the flow cytometer before sorting.
Keep all the samples on ice and in the dark until FACS analysis is carried out.
Vortex all FACS tubes for a short period of time before analysis.
Run unstained cells through the sorter as the first control.
Analyze 30,000 events for each sample.
Adjust the FSC and SSC settings until the cells appear in the middle of the FSC versus SSC dot plot, and exclude cell aggregate and cell debris. Then adjust the PMT voltage of the FITC, APC, and PE detectors until the cells appear within the lower quadrant of the different dot plots, thus setting the background or unstained fluorescence levels according to these parameters.
Run the single color sample and correct the spectral overlap using digital compensation by subtracting the spectral overlap signal of single colors from the overall signals detected in each channel.
Apply the correct amount of compensation to confirm that the positive population was directly horizontal or vertical to the negative population and not detectable in the other detectors. FITC compensation is not required for the APC detector, but should be checked.
Run the positive control and the isotype-matched antibody to verify antibody specificity.
After multiparameter acquisition setup for de fining background/negative signal and compensation values, run double-stain FLK-1-PE + VE-Cadherin-APC sample.
Draw a PE versus APC two-color quadrant graph that displays the fluorescence pro file of double-stained cells.
Select the cells residing in the double-positive FLK-1-PE/VE-Cadherin-APC quadrant (Q2) for sorting and collection (Fig. 3).
Using the previously defined setup, run the double-positive sample for sorting with low speed and collect the Q2 subpopulations in a FACS tube containing EC differentiation medium and 10 mg/mL gentamicin.
Centrifuge the collected subpopulations at 4 °C, 125 × g for 5 min, and replace the medium with fresh EC differentiation medium and 10 mg/mL gentamicin.
Plate the cells in a 0.1 % gelatinized plate and incubate the cells at 37 °C in 5 % CO2. Change the medium every other day (see Note 12).
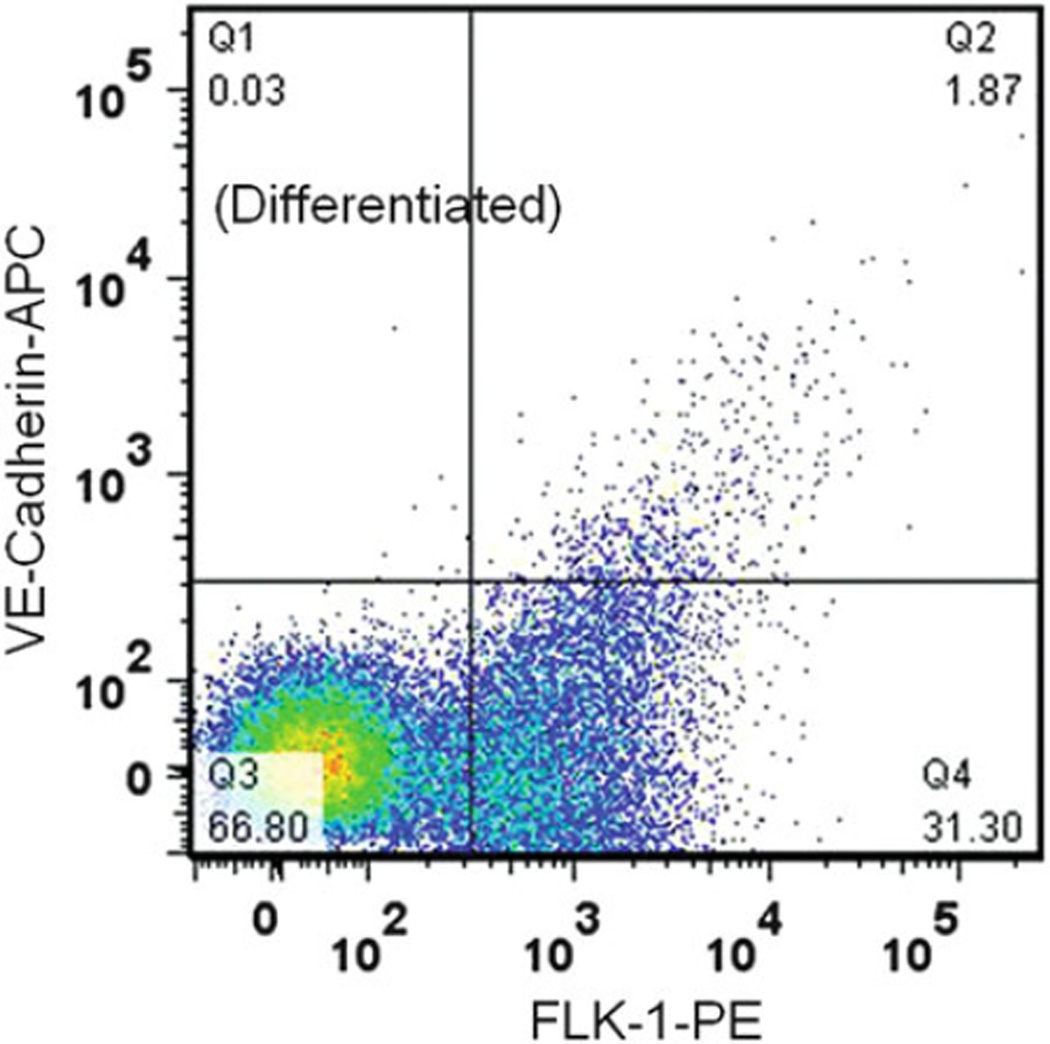
Fig. 3.
The two-color fluorescence pro file of differentiating ECs in ESC cultures stained for VE-Cadherin and FLK-1. The Q3 quadrant contained a negative population as determined by unstained and isotype controls. The Q1 and Q4 quadrants contained cells that only expressed VE-Cadherin and FLK-1 detected by APC conjugated anti-mouse CD144 (VE-Cadherin) and PE conjugated anti-mouse Flk-1 (VEGF-R2).
All the steps related to endothelial differentiation of mouse embryonic stem cells have been summarized in Fig. 4.
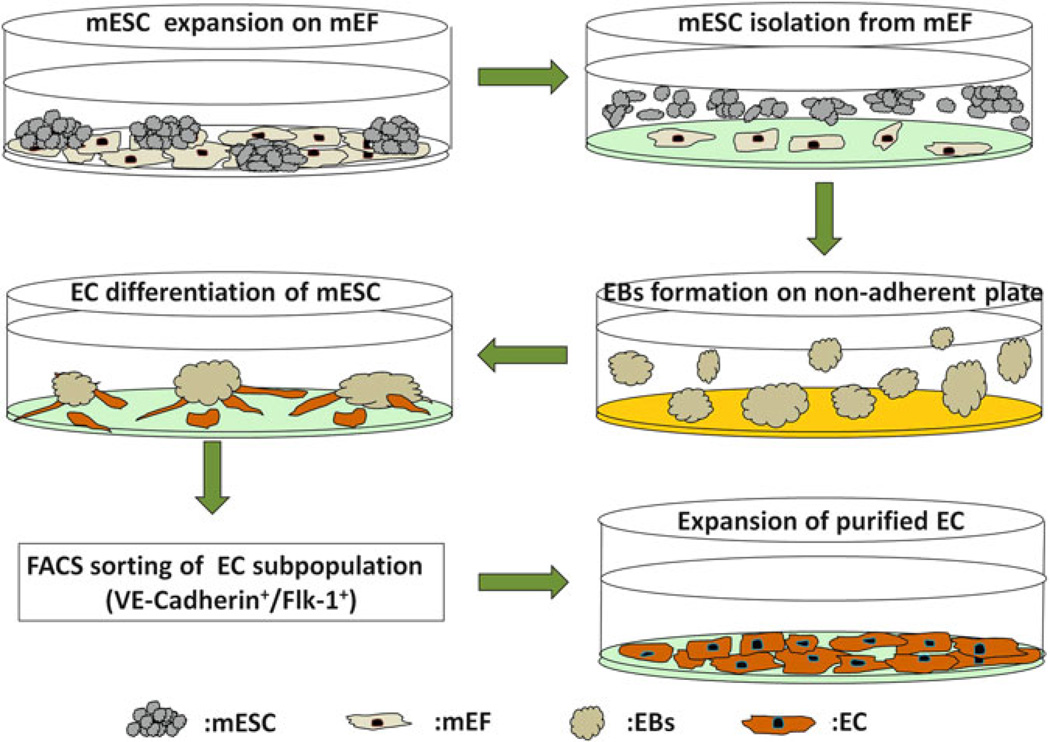
Fig. 4.
Schematic diagram demonstrating all steps during EC differentiation of ESCs and expansion of purified cells. The mESCs expanded on the mEF feeder layer are first subcultured on 0.1 % gelatinized plates to separate mEFs from the mESCs before EB formation. The non-adherent mESCs are then transferred to non-adherent plates coated with Poly-HEMA; the loss of stromal contact and withdrawal of Leukemia inhibitory factor (LIF) induce formation of 3D EBs. After 5 days of culture, the optimally sized EBs are switched to EC differentiation medium and are transferred to gelatinized plates the next day. EBs are cultured on the gelatinized plates in EC differentiation medium for up to 7 additional days. FACS sorting is then used to purify the emerging endothelial subpopulation expressing the endothelial markers (Flk-1+ and VE-Cadherin+). Purified sorted ECs can then be further expanded by culturing on gelatinized coated plates.
Acknowledgment
This work is supported by grants from NIH/NCI TMEN grant (U54CA126552.) to Nancy Boudreau and Mina J Bissell and U.S. Department of Energy, Office of Biological and Environmental Research (DE-AC02-05CH1123), a Distinguished Fellow Award and Low Dose Radiation Program (03-76SF00098) to Mina J. Bissell. Mandana Veiseh was supported by a postdoctoral fellowship from the NCI of the NIH (F32 CA132491A). We thank Pamela Derish in the Department of Surgery at UCSF for editorial review of the manuscript.
Footnotes
Mitomycin C solution is light sensitive. Store the mixture at 4 °C in a dark container for up to 6 weeks or at −20 °C for up to 4 months.
Freezing medium is light sensitive. Turn off the tissue culture hood lights during sample preparation. Aliquot the freezing medium into smaller batches (3–5 mL) and keep at 4 °C in a foil-covered container for up to 2 weeks, or store at −20 °C for up to 4 months.
Differentiation medium is light sensitive. Cover the medium container with aluminum foil. Also prepare fresh medium in smaller batches for each experiment and store at 4 °C for up to 4 weeks.
After dissolving the Collagenase IV in Knockout D-MEM medium, filter the solution using a 0.2 µm filter, then aliquot the solution into smaller batches (3–5 mL), and store at −20 °C. Samples can be kept up to 4 months.
To store poly-HEMA coated plates, wrap them in parafilm after exposing them to UV light and keep them at room temperature. Wash the plates three times with sterile D-PBS in a tissue culture hood before use.
Cultured, mitotically-inactivated mEF cells can be kept for up to 2 weeks in mEF medium while changing the medium every 2–3 days.
Primary mouse embryo fibroblasts, Hygro-resistant strain C57BL/6 (Millipore, Billerica, MA) also can be used as an alternative to mEF cells. These primary cells have been treated with Mitomycin-C and can be directly plated onto gelatin-coated culture vessels. 2–3 × 106 cells will produce the appropriate density feeder layer on a 10 cm plate.
It is important to monitor the cells and change their medium every day. mESCs can easily become contaminated.
One potential way of improving the percentage of ECs differentiated from ESCs is to first sort the Flk-1+ cells and then continue differentiation of this subpopulation on collagen type IV coated plate prior to sorting for endothelial cells. The percentage of double-positive cells for VE-Cadherin and Flk-1 can be increased up to fivefold.
Perform the identical steps for staining and sample preparation on all control samples.
Control samples are used for de fining the background/negative signal, compensation values, and multiparameter sort criteria. Sort region is defined on the double-positive quadrant of a double-stained sample.
The EC differentiation medium can be switched to EGM-2 medium after 1 week.
References
- 1.Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler RJ. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J Embryol Exp Morph. 1985;87:27–45. [PubMed] [Google Scholar]
- 2.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 3.Vittet D, Prandini MH, Berthier R, Schweitzer A, Martin-Sisteron H, Uzan G, Dejana E. Embryonic stem cells differentiate in vitro to endothelial cells through successive maturation steps. Blood. 1996;88:3424–3431. [PubMed] [Google Scholar]
- 4.Feraud O, Cao Y, Vittet D. Embryonic stem cell-derived embryoid bodies development in collagen gels recapitulates sprouting angiogenesis. Lab Invest. 2001;81:1669–1681. doi: 10.1038/labinvest.3780380. [DOI] [PubMed] [Google Scholar]
- 5.Blancas AA, Lauer NE, McCloskey KE. Endothelial differentiation of embryonic stem cells. Curr Protoc Stem Cell Biol. 2008;Chapter 1(Unit 1) doi: 10.1002/9780470151808.sc01f05s6. F 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchetti S, Gimond C, Iljin K, Bourcier C, Alitalo K, Pouyssegur J, Pages G. Endothelial cells genetically selected from differentiating mouse embryonic stem cells incorporate at sites of neovascularization in vivo. J Cell. Sci. 2002;115:2075–2085. doi: 10.1242/jcs.115.10.2075. [DOI] [PubMed] [Google Scholar]
- 7.Nishikawa SI, Nishikawa S, Hirashima M, Matsuyoshi N, Kodama H. Progressive lineage analysis by cell sorting and culture identifies FLK1 + VE-cadherin + cells at a diverging point of endothelial and hemopoietic lineages. Development. 1998;125:1747–1757. doi: 10.1242/dev.125.9.1747. [DOI] [PubMed] [Google Scholar]
- 8.Hirashima M, Kataoka H, Nishikawa S, Matsuyoshi N. Maturation of embryonic stem cells into endothelial cells in an in vitro model of vasculogenesis. Blood. 1999;93:1253–1263. [PubMed] [Google Scholar]
- 9.Bahrami SB, Veiseh M, Dunn AA, Boundreau NJ. Temporal changes in Hox gene expression accompany endothelial cell differentiation of embryonic stem cells. Cell Adhesion & Migration. 2011;5(2):133–141. doi: 10.4161/cam.5.2.14373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Ceballos E, Chambon P, Gudas LJ. Differences in gene expression between wild type and Hoxa1 knockout embryonic stem cells after retinoic acid treatment or leukemia inhibitory factor (LIF) removal. J Biol Chem. 2005;280:16484–16498. doi: 10.1074/jbc.M414397200. [DOI] [PubMed] [Google Scholar]
- 11.Loring JF, Porter JG, Seilhammer J, Kaser MR, Wesselschmidt R. A gene expression pro file of embryonic stem cells and embryonic stem cell-derived neurons. Restor Neurol Neurosci. 2001;18:81–88. [PubMed] [Google Scholar]
- 12.Lugus JJ, Park C, Choi K. Developmental relationship between hematopoietic and endothelial cells. Immunol. Res. 2005;32:57–74. doi: 10.1385/IR:32:1-3:057. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi TP, Dumont DJ, Conlon RA, Breitman ML, Rossant J. flk-1, an flt-related receptor tyrosine kinase is an early marker for endothelial cell precursors. Development. 1993;118:489–498. doi: 10.1242/dev.118.2.489. [DOI] [PubMed] [Google Scholar]
- 14.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 15.Preffer F, Dombkowski D. Advances in complex multiparameter flow cytometry technology: applications in stem cell research. Cytometry B Clin Cytom. 2009;76:295–314. doi: 10.1002/cyto.b.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Hammes F, De Roy K, Verstraete W, Boon N. Past, present and future applications of flow cytometry in aquatic microbiology. Trends Biotechnol. 2010;28:416–424. doi: 10.1016/j.tibtech.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Hulspas R, O’Gorman MR, Wood BL, Gratama JW, Sutherland DR. Considerations for the control of background fluorescence in clinical flow cytometry. Cytometry B Clin Cytom. 2009;76:355–364. doi: 10.1002/cyto.b.20485. [DOI] [PubMed] [Google Scholar]
- 18.Lugli E, Roederer M, Cossarizza A. Data analysis in flow cytometry: the future just started. Cytometry A. 2010;77:705–713. doi: 10.1002/cyto.a.20901. [DOI] [PMC free article] [PubMed] [Google Scholar]