Abstract
Recognition of microbial products by members of the Toll-like receptor (TLR) family initiates intracellular signaling cascades that result in NF-κB activation and subsequent production of inflammatory cytokines. We explored the potential roles of microRNAs (miRNAs) in regulating TLR pathways. A target analysis approach to the TLR4 pathway adaptor molecules identified several putative targets of miR-200a, miR-200b and miR-200c. miRNA mimics were co-transfected with a NF-κB activity reporter plasmid into HEK293 cells stably expressing TLR4 (HEK293-TLR4). Mimics of both miR-200b and miR-200c, but not miR-200a, decreased NF-κB reporter activity in either untreated cells or in cells treated with endotoxin:MD2 as a TLR4 agonist. Transfection of HEK293-TLR4 cells with miR-200b or miR-200c significantly decreased expression of MyD88, whereas TLR4, IRAK-1 and TRAF-6 mRNAs were unaffected. When miR-200b or miR-200c mimics were transfected into the differentiated monocytic THP-1 cell line, the abundance of MyD88 transcripts, as well as LPS-induced expression of the pro-inflammatory molecules IL-6, CXCL9 and TNF-α were diminished. These data define miRNAs miR-200b and miR-200c as factors that modify the efficiency of TLR4 signaling through the MyD88-dependent pathway and can thus affect host innate defenses against microbial pathogens.
Keywords: Macrophage, microRNAs, Toll-like receptor, gene expression, pro-inflammatory response
Introduction
MicroRNAs (miRNAs) are evolutionarily conserved, small, non-coding RNAs that repress gene expression by hybridization to targeted transcripts. More than 1500 miRNAs have been discovered in the human genome (www.mirbase.org) and each miRNA has the potential to repress the expression of hundreds of mRNAs.1–11 Upon transcription, processing and transport out of the nucleus, active miRNAs regulate mRNAs through base-pairing interactions between the 8-base pair (bp) seed region of the miRNA with a complementary sequence of the targeted mRNA.11 Base-pairing usually occurs with sequences in the 3′untranslated region (UTR) although there are reports of miRNA targets in 5′UTRs or open reading frames (ORFs).3,12–14 In humans, miRNA-mediated suppression of gene expression was previously thought to occur by inhibiting translation, but recent studies suggest that miRNAs frequently function by promoting mRNA degradation.15,16 Regardless of the method of mRNA repression, mRNA/miRNA duplexes ultimately undergo degradation. Many studies have documented the role of miRNAs in cancer and development.17 The involvement of miRNAs in the regulation of innate and/or adaptive immune responses has recently begun to be explored.18
TLRs play important roles in initiating host defenses against invading pathogens. TLRs recognize a variety of molecules shared by pathogens, collectively referred to as pathogen-associated molecular patterns (PAMPs).19–21 PAMP recognition by TLRs initiates a cascade of intracellular signals leading to NF-κB activation and, consequently, pro-inflammatory molecule expression.22 Although there are alternative pathways for the activation of NF-κB, such as TNF-α-induced signaling,23 TLRs often mediate inflammatory responses to microbial products. The ability to negatively regulate the pro-inflammatory responses initiated by TLR ligation is important to avoid pathology. Indeed, dysregulation of TLR-mediated responses can lead to the development of chronic inflammatory diseases.24,25 Based on changes in miRNA abundance observed after TLR ligation, several miRNAs have been found to down-modulate TLR-mediated responses, notably miR-155, miR-146 and miR-21.26,27 We reasoned that a constellation of miRNAs may contribute toward fine tuning strong inflammatory responses, including some with changes in abundance that were not detected in profiling studies. We therefore approached the question through the use of prediction programs to identify miRNAs that might target TLR pathway proteins. The outcome yielded several predicted targets for miR-200a, miR-200b and miR-200c, leading us to hypothesize that these miRNAs might function as intracellular modulators of responses initiated by TLR signaling.28,29
TLRs function through two discrete downstream signaling pathways, dependent on the adaptor molecules TRIF or MyD88, leading to activation of the transcription factors IRF3 or NFκB. MyD88 is essential for signaling through all TLRs with the exception of TLR3 and the MyD88-independent, TRIF-dependent pathway downstream of TLR4.20–22,30 Using mimics of miRNAs and reporter assays to study function, we found evidence suggesting that, (i) miR-200b and miR-200c suppress signaling pathways leading to NF-κB activation after TLR4 ligation and (ii) the effect is at least partially accounted for by diminished MyD88 transcript levels. Further studies in the THP-1 monocytic cell line, differentiated to resemble macrophages, showed that miR-200b or miR-200c mimics caused a decrease in the levels of transcripts encoding MyD88 and induced the expression of inflammatory molecules in response to LPS. Overall, these findings suggest that miR-200b and miR-200c are novel regulators of TLR4 signaling.
Materials and methods
microRNA target analysis
Potential miRNA target interactions with mRNAs in the TLR4 pathway were identified using TargetScan 6 and miRanda (August 2010 release) prediction programs to systematically query each of the proteins along the pathway (see Figure 1). TargetScan predicts biological targets of miRNAs according to matches with the 7–8 bp seed region, surrounding context and the degree of conservation across species (www.targetscan.org).31 miRanda incorporates the likelihood of mRNA down-regulation using a regression model trained on sequence context features of the miRNA::mRNA duplex (microRNA.org).
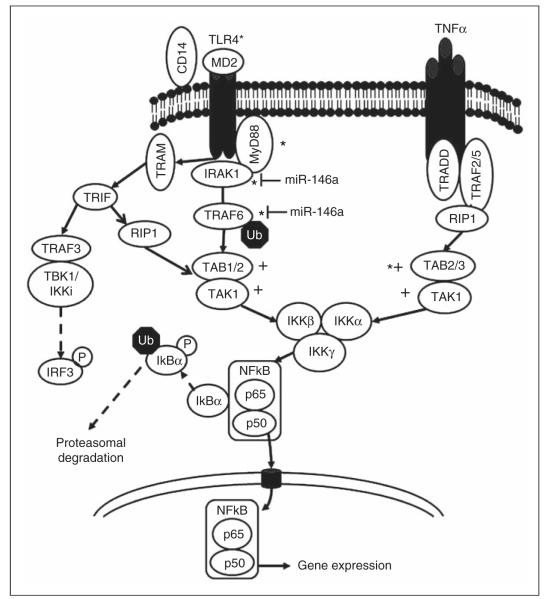
Figure 1.
Putative miRNA target proteins in the signaling pathways activated after TLR4 ligation or TNF-α exposure. Components of the signaling pathways activated by TLR4 ligation or TNF-α exposure, which ultimately lead to NF-κB activation, are shown. Please note the MyD88-dependent and -independent pathways resulting in NF-κB activation after TLR4 ligation. Potential targets for miR-200a (+) or for miR-200b and miR-200c (*) within the TLR4 and TNF-α pathways, identified through a miRNA query of 3′UTRs, are indicated. Analysis of the IRAK1 3′UTR revealed a 7 bp exact complementary sequence match to the miR-200b and miR-200c seed sequence, even though IRAK1 was not a predicted miRNA200b target in TargetScan version 6.0. Experimentally validated miR-146a targets are indicated.28
Cell lines
HEK293 cells stably expressing episomal TLR4 by transfection with pcDNA3.0 conferring resistance to G418 (HEK293-TLR4) were a gift from Dr Jesse Chow (Eisai Research Institute Andover, MA, USA).32,33 The cells maintained the plasmid in culture even without G418 addition. Prior to experiments, cells were cultured in DMEM supplemented with 10% FBS and 10 μg/ml of the antibacterial ciprofloxacin (DM-10) at 37° C and 5% CO2. The THP-1 human monocytic cell line (ATCC, Manassas, VA) was cultured in RPMI with 10% FBS (RP-10) at 37° C and 5% CO2. Media and FBS were from GIBCO (Life Technologies, Grand Island, NY).
Cloning of putative 3′UTR targets
Full-length 3′UTRs of human MyD88, TLR4, IRAK1, TRAF6 or TAB2 were amplified from human liver cDNA (Clontech, Mountain View, CA) by PCR and subsequently cloned downstream of the Renilla reniformis luciferase gene in the XhoI/EagI sites of the psiCHECK-2 vector (Promega, Madison, WI, USA). Cycling conditions were: 94° C for 2 h followed by 30 cycles at 94° C for 30 min, 60° C for 30 min, then 72° C for 4 min followed by 72° C for 10 min. The predicted miR-200b/miR-200c seed sequence binding site of the MyD88 3′UTR was mutated using the QuikChange II XL Site-Directed Mutagenesis kit (Stratagene, Santa Clara, CA). Wild-type and mutant 3′UTRs were confirmed by sequencing. See Table 1 for primer sequences. Primers were made by Integrated DNA Technologies (Iowa City, IA).
Table 1.
Primers for cloning 3′ UTRs. Sequences for 3′UTRs of the indicated human genes were obtained from the National Center for Biotechnology Information website. Primers were designed to include the entire UTR beginning just downstream of the stop codon and extending to the site of poly(A) addition. With the exception of the MyD88 mutated clone, a second set of primers was used on PCR products to generate an XhoI site or an EagI site at the 5′ end of the sense or antisense primers, respectively.
| Gene (accession #) | Sense primer | Antisense primer |
|---|---|---|
| MyD88 (NM_002468) | 5′-CCTGCACCAAATCTTGGTTCTGG ACTCG-3′ |
5′-CAAGGTAGAATATTATTTATTATT ATAAACTCAGGATGCAAGATATATT CCAGG-3′ |
| MyD88 mutated | 5′-CCCAATGTACCAGCCCTTATAC CTCTAATGAAGCACAGAGAGAGG-3′ |
5′-CCTCTCTCTGTGCTTCATTAGAGG TATAAGGGCTGGTACATTGGG-3′ |
| TLR4 (NM_138554) | 5′-GGTAAATCATGGAATCCAGAA GGAACAGTGGG-3′ |
5′-CATAACTTTTATTACAATATA TTATTATAACTATTCAATTTTATTGTTAGT- TATTGTTAATCTCT-3′ |
| IRAK1 (NM_008363) | 5′-AACCTGAAAAGAGCCAGGGACC TGAAGAAA-3′ |
5′-TCAGAGTAATCACCCCCAAAACA AGTGGGG-3′ |
| TRAF6 (NM_004620.3) | 5′-TTTTGAAGACTTTCGACTTGCC CTCACTTGCTC-3′ |
5′-TTTTCGGCCGAACACTTAAA CAAGTATTATTCAACAG-3′ |
| TAB2 (NM_015093.3) | 5′-GCCAAATGGCCCTGTATCTTC TCTAAAACCA-3′ |
5′-CCACATTAGAATATAAGTTTTTTA ATTTTTATAAATAACTTATCTGTTACAAA- GATAGTT-3′ |
MicroRNA target luciferase assays
HEK293-TLR4 cells were plated in 48-well dishes at 2 × 105 cells per well for luciferase assays. miRNA mimics were Pre-miR™ miRNA precursor molecules and the negative control Pre-miR™ from Applied Biosystems (ABI; Forest City, CA). The negative control is a random sequence miRNA mimic that has not been found to suppress any human target genes. The psiCHECK-2 Renilla luciferase-MyD88 3′UTR reporter plasmid (50 ng) was co-transfected with either a negative control or a functional miRNA mimic (50 nM final concentration) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Twenty-four hours post-transfection, lysates were collected and analyzed with the Dual Luciferase Reporter Assay System (Promega) using a FLUOstar luminometer (BMG Labtech).
Quantitative PCR
THP-1 cells were plated in 12-well dishes in RP-10 supplemented with 5 ng/ml PMA at 1 × 106 cells/well to cause their differentiation toward macrophage-like characteristics. HEK293-TLR4 cells were plated in 12-well dishes in DM-10 at 1 × 106 cells/well. Cells were transfected in the presence of 50 nM negative control or experimental miRNA mimics (Pre-miRs™, ABI) using Lipofectamine RNAiMAX Transfection Reagent (Invitrogen). Fortyeight hours post-transfection, differentiated THP-1 cells were stimulated with 100 ng/ml of Escherichia coli 055:B5 LPS (Sigma, St Louis, MO, USA) for 3 h. Total RNA was extracted with TRIzol (Invitrogen) and reverse transcribed with the SuperScript III First-Strand Synthesis System using random hexamer primers (Invitrogen). Quantitative PCR was performed using TaqMan Gene Expression Assays (Applied Biosystems). Cycling conditions were: 95° C for 20 min followed by 40 cycles at 95° C for 1 min then 60° C for 20 min as per the manufacturer’s instructions. Changes in the abundance of mRNAs encoding MyD88, TNF-α, IL-6, CXCL9 or β-actin cDNA were quantified by qPCR. Ct values were normalized to the ACTB values. ΔCt values for cytokine/chemokine transcripts in miRNA mimic-transfected cells were compared with negative control-transfected cells using the ΔΔCt method.33 In separate experiments, MyD88 expression was determined in HEK293 and THP-1 cells at 24 or 48 h post-transfection with miRNA mimics. Changes in MyD88 levels were calculated using the ΔΔCt method with β-actin as an internal control.
For determining miR-200b and miR-200c basal expression levels, THP-1 cells were plated in 12-well dishes at 1 × 106 cells/well in RP-10 supplemented with 5 ng/ml PMA. Twenty-four hours post-plating, total RNA was extracted with TRIzol (Invitrogen) and reverse transcribed using the TaqMan miRNA reverse transcription kit (Applied Biosystems). Reverse transcriptase-qPCR (RT-qPCR) was performed using TaqMan Gene Expression Assays (Applied Biosystems) for miR-200b or miR-200c with RNU48 serving as an internal control. Fold change was calculated by the ΔΔCt method.
NF-κB reporter assays
HEK293-TLR4 (1 × 106 cells/well) were seeded in 12-well plates overnight and then co-transfected with 150 ng pNF-κB-luc (Clontech), 15 ng phRL-SV40 Renilla (Promega) and 50 nM functional or negative control miRNA mimics (Pre-miRs™, ABI) with Lipofectamine 2000 (Invitrogen). Forty-eight hours post-transfection, cells were stimulated with 0.5–5.0 ng/ml of 1:1 lipooligosaccharide (LOS; Neisseria meningitidis serotype B):MD2 for 6 h to induce signaling through TLR4, then harvested and assayed using the Dual Luciferase Reporter Assay System.35 Firefly luciferase values were normalized to the Renilla luciferase values in triplicate wells of each condition. The mean normalized luciferase values in the experimental conditions were expressed as a ratio with the negative control condition. LOS:MD-2 was generated by treatment of LOS:CD14 (30 min at 37° C) with conditioned insect cell medium (Expression Systems ESF921) containing soluble human His6-MD-2 generated as described previously.33
Statistics
Analyses were done using Graph Pad Prism version 5 software.
Results
Analysis of miRNAs active on TLR pathway genes
The MyD88-dependent and -independent TLR4 pathways are depicted in Figure 1. miRNAs have been shown previously to target key members of the TLR signaling pathway.29 To identify other candidate miRNAs that might target this pathway, we used TargetScan and miRanda to assemble extensive lists of miRNAs that are predicted to target each member of this pathway. We noted that several members of the TLR4 signaling pathway were predicted targets of miR-200a, miR-200b and miR-200c (see Supplementary Table 1). Targets for miR-200a differed from genes targeted by miR-200b or miR-200c, but none have been reported to target immune regulatory transcripts. Given the importance of the TLR pathways in immunity, we investigated potential roles for miR-200a, miR-200b and miR-200c on TLR4 activity.
MicroRNAs regulate NF-κB activation in resting cells
To determine the effects of miRNAs on NF-κB activity in resting cells, HEK293-TLR4 cells were co-transfected with miRNA mimics, a reporter construct containing the gene encoding firefly luciferase under control of an NF-κB-responsive promoter, and phRL-SV40 Renilla that constitutively expresses Renilla luciferase. One of the following miRNA mimics was included in each condition: miR-200a, miR-200b, miR-200c, miR-29b, miR-146a or a negative control (non-targeting) mimic. The miR-29b mimic was included as a negative control and the miR-146a mimic was included as a positive control for NF-κB repression as it was previously shown to repress IRAK1 and TRAF6 expression, two key proteins in TLR4 signaling.28 Firefly and Renilla luciferase activity data were normalized as described in the Materials and methods section. As illustrated in Figure 2, cells transfected with miR-200a exhibited no decrease in NF-κB activity compared with cells containing a negative control miRNA. Conversely, luciferase activity values in cells containing miR-146a, miR-200b or miR-200c mimics were significantly decreased in comparison with control miRNA transfected cells, suggesting each of these mimics caused a decrease in NF-κB activation. As expected, the miR-29b mimic showed no effect on NF-κB activation (Figure 2). These results suggest that miR-200b and miR-200c suppress the basal activity of the NF-κB reporter in resting cells, whereas miR-200a does not.
Figure 2.
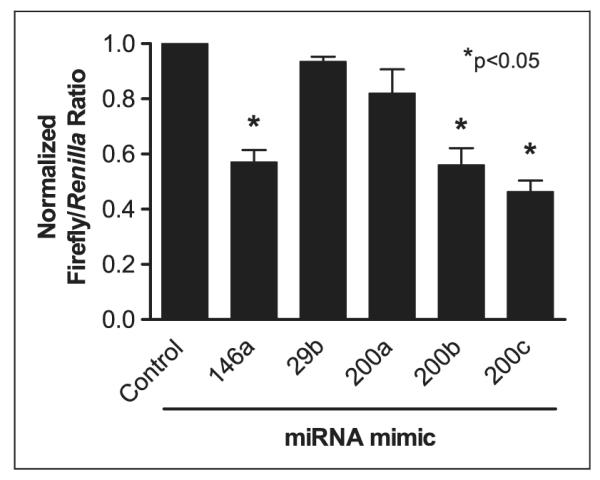
Effects of miRNA mimics on basal NF-κB activity. HEK293-TLR4 cells were co-transfected with an NF-κB reporter plasmid and Renilla luciferase expression plasmid, and incubated for 48 h in the presence of 50 nM of the indicated miRNA mimics. Data indicate the mean ± SEM ratio of firefly/Renilla luciferase activity in transfected cells, normalized to negative control miRNA mimic (first bar). miR-146a, which has been reported to be a repressor of NF-κB activation, served as a positive control.28 miR-29b, which has no reported effect on NF-κB activation, served as a negative control. The bars represent the ratios of firefly/Renilla luciferase activity in the absence of activating stimulus. Data were generated from three independent experiments, each with three replicates per condition. *P < 0.05 compared with negative control miRNA, unpaired t-test.
Dose-dependent suppression of TLR4-mediated NF-κB activation by miR-146a, miR-200b and miR-200c
No matter what stimulus maintains basal NF-κB activity, our predictions suggest that miR-200b and miR-200c will inhibit NF-κB activation in response to a TLR4 agonist. Therefore we studied miRNA effects on NF-κB activation in HEK293-TLR4 cells transfected with miRNA mimics prior to addition of a TLR4 ligand. LOS:MD2 was chosen over LPS as a TLR4 ligand because HEK293-TLR4 cells do not produce MD2, a necessary co-factor for TLR4 activation.32 TNF-α was added as a TLR4-independent means of NF-κB activation.
Similar to Figure 2, HEK293-TLR4 cells were co-transfected with the NF-κB reporter, the phRL-SV40 Renilla expression plasmid and either negative control or functional miRNA mimics. Forty-eight hours post-transfection, cells were stimulated with varying doses of LOS:MD2 (0–5.0 ng/ml) for 6 h and lysates were analyzed by the dual luciferase assay. The normalized firefly/Renilla luciferase ratios demonstrated that, as expected, the positive control miR-146a caused a decrease in NF-κB activation by LOS:MD2 (Figure 3A). miR-200b or miR-200c also significantly suppressed NF-κB reporter expression in response to some doses of LOS:MD2 (Figure 3B and Figure 3C respectively). These observations show that miR-200b and miR-200c are capable of suppressing the activation of NF-κB when stimulated with a TLR4 agonist.
Figure 3.
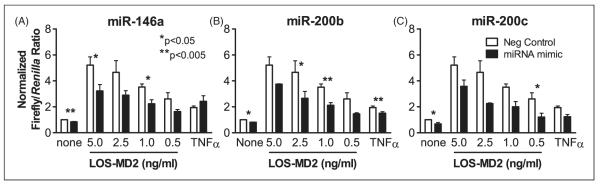
Effect of miRNA mimics on LPS-stimulated NF-κB activation. HEK293-TLR4 cells were co-transfected with an NF-κB reporter plasmid, a transfection control Renilla luciferase expression plasmid, and the indicated miRNA mimic at 50 nM final concentration. The latter included a negative control miRNA mimic, or miRNA mimics for miR-146a (A), miR-200b (B) or miR-200c (C). After 48 h cultivation, cells were stimulated with buffer alone (none), the indicated doses of LOS:MD2 or TNF-α and cultured for an additional 6 h. Data indicate the mean±SEM firefly/Renilla luciferase activity compared with the negative control of uninduced miRNA from three independent experiments, each with three replicates per condition. *P < 0.05, **P < 0.005 comparing the test with the negative control miRNA unpaired t-test. LOS, lipooligosaccharide.
In addition to TLR4 ligation, miR-200b significantly suppressed NF-κB activation caused by 50 ng/ml TNF-α, and miR-200c exhibited a trend toward suppression (P = 0.0520). miR-146a, in contrast, had no effect on TNF-α-induced NF-kB activity.
miR-200b and miR-200c directly target mRNA encoding the adaptor molecule MyD88
TLR4, MyD88, IRAK1 and TRAF6 are TLR4 pathway intermediates identified as putative targets of miR-200a, miR-200b or miR-200c in our initial screen (Figure 1). Analysis of the IRAK1 3′UTR revealed a 7 bp exact complementary sequence match to the miR-200b and miR-200c seed sequence, even though IRAK1 was not a predicted target in TargetScan version 6.0. To assess whether any of these were actually suppressed by miR-200b and miR-200c, we cloned the full-length 3′UTR of their respective genes downstream of a Renilla luciferase gene in the psiCHECK-2 vector. The vector contains a firefly luciferase gene as an internal control to normalize transfection effciency. HEK293-TLR4 cells were co-transfected with 3′-UTR reporter constructs along with miRNA mimics of miR-200b, miR-200c or a negative control for 24 h prior to cell harvest and luciferase assay. Overall repression was calculated by the ratio of Renilla/firefly luciferase activity in samples treated with negative control or the miR-200b or miR-200c miRNA mimics.
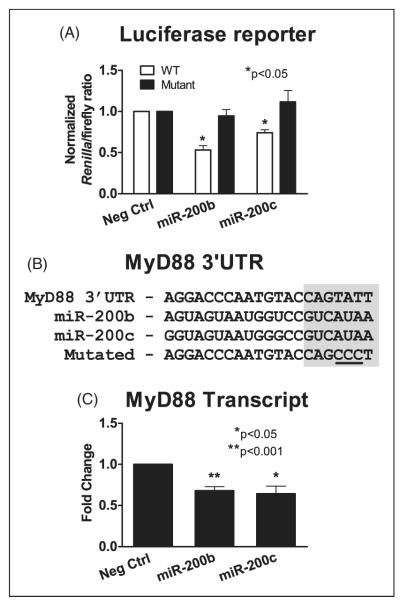
Initial screens were done on HEK293-TLR4 cells containing 3′UTR luciferase reporters of TLR4, IRAK1, TAB2 or TRAF6. These assays showed there was no difference between Renilla luciferase activity in cells that were transfected with the negative control mimic or miR-200a, miR-200b or miR-200c mimics (data not shown). In contrast, studies of cells containing Renilla luciferase followed by the 3′UTR of MyD88 showed a significant and repeatable decrease in activity in cells containing either the miR-200b or miR-200c mimic compared with the negative control (Figure 4A). This suggested that MyD88 may be a miR-200b or miR-200c target. To validate this interaction, we generated another psiCHECK-2 plasmid with a mutated MyD88 3′UTR reporter containing mismatches in the common miR-200b/miR-200c seed region (Figure 4B). Mutations in this region should block miR-200b and miR-200c binding and abrogate the miR-200b- or miR-200c-mediated repression of luciferase expression. The mutated reporter vector was co-transfected into HEK293-TLR4 cells along with miRNA mimics for miR-200b, miR-200c or a negative control. The dual luciferase assays (Figure 4A) showed Renilla luciferase activity in HEK293-TLR4 cells containing the mutant MyD88 reporter did not change significantly upon treatment with mimics of miR-200b or miR-200c. In contrast, cells containing the WT MyD88 3′UTR reporter showed a significant reduction in Renilla luciferase activity in the presence of mimics of miR-200b or miR-200c. These results demonstrate a direct interaction between miR-200b or miR-200c and the MyD88 3′UTR.
Figure 4.
MyD88 luciferase assays validating a direct interaction between miR-200b or miR-200c and MyD88. HEK293-TLR4 cells were transfected with constructs containing the luciferase reporter gene followed by either the 3′UTR of MyD88 or a mutated version of the MyD88 3′UTR containing mutations within the predicted miR-200b and miR-200c seed region. (A) Cells were co-transfected with 25 nM of miRNA mimics for miR-200b, miR-200c or a negative control. Data represent the mean ± SEM Renilla/firefly luciferase ratios. (B) TargetScan predicted interaction between miR-200b and miR-200c and the putative MyD88 3′UTR. Shaded bases represent miRNA seed regions. Mutated bases are underlined. (C) HEK293-TLR4 cells were transfected with miRNA mimics for miR 200b, miR-200c or a negative control. RNA harvested at 48 h post-transfection was used as template for RT-qPCR to measure MyD88 transcript levels. Data are mean ± SEM of three independent experiments with three replicates per condition. *P < 0.05, **P < 0.001.
The ability of miR-200b or miR-200c to regulate wild-type MyD88 expression in cultured HEK293-TLR4 cells was further verified by RT-qPCR. The data showed statistically significant knock down of MyD88 transcript levels by miR-200b and miR-200c compared with cells treated with the negative control (25% and 30% respectively) (Figure 4C).
miR-200b and miR-200c represses MyD88 and the TLR4-induced expression of cytokines/chemokines in macrophages
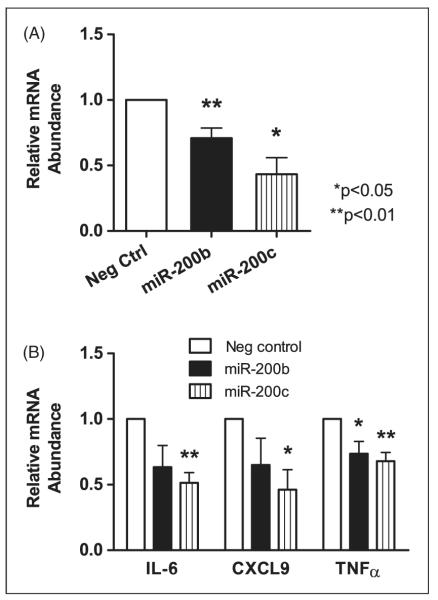
Ligation of TLR4 in macrophages can lead to expression of classical activation markers including IL-6, CXCL9 and TNF-α.36 To determine whether macrophages express miR-200b and miR-200c, we performed RT-qPCR using RNA from the THP-1 monocytic cell line, differentiated toward a macrophage phenotype with PMA. The data revealed that both miR-200b and miR-200c were expressed in this cell type, although a high Ct for miR-200b suggested this miRNA may be expressed at a lower basal concentration than miR-200c (data not shown). The effects of miR-200b or miR-200c on MyD88 transcript levels were analyzed in differentiated THP-1 cells transfected with mimics of each of these miRNAs or a negative control mimic. Twenty-four hours post-transfection, RT-qPCR revealed a statistically significant decrease in MyD88 transcripts in THP-1 cells transfected with either miR-200b or miR-200c mimics compared with a negative control mimic thus further verifying MyD88 as a target of miR-200b and miR-200c (Figure 5A).
Figure 5.
LPS-stimulated cytokine/chemokine induction is repressed by miR-200b or miR-200c mimics. Repression of MyD88 by miR 200b or miR-200c leads to a decrease in classical macrophage activation markers. miRNA mimics (50 nM) of miR-200b or miR-200c were transfected into differentiated THP-1 cells. (A) Twenty-four hours post-transfection, RNA was harvested and RT-qPCR was performed using TaqMan primers for β-actin and MyD88 with β-actin serving as an internal control. Data represent the mean ± SEM fold change in expression comparing experimental to negative control miRNA mimics from three independent experiments. (B) Forty-eight h post-transfection, cells were treated with 100 ng/ml LPS and RNA was harvested after an additional 3 h. RT-qPCR was performed using TaqMan primers for β-actin, IL-6, CXCL9, or TNF-α. Relative mRNA abundance was determined by normalizing to the b-actin cDNA, then comparing cDNA abundance in miR mimic-transfected compared to the negative control miRNA mimic transfected samples. Data represent the mean ± SEM relative expression from three independent experiments. *P < 0.05, **P < 0.01.
The above effect of miR-200b and miR-200c on MyD88 mRNA abundance led us to investigate whether augmenting the levels of miR-200b or miR-200c would suppress the downstream consequences of TLR4 ligation in this cell type. THP-1 cells over-expressing miR-200b, miR-200c or a negative control were treated with LPS for 3 h followed by RT-qPCR analysis for IL-6, CXCL9 and TNF-α. CXCL9 and TNF-α transcripts were significantly suppressed by miR-200b and miR-200c relative to LPS-treated cells transfected with the negative control miRNA mimic. The IL-6 transcript decreased significantly in cells containing the miR-200c mimic and there was a trend toward significance caused by miR-200b mimic (P = 0.09) (Figure 5B). These observations show that some of the transcripts that respond to TLR4 ligation are suppressed by either miR-200b or miR-200c in macrophages.
Discussion
Innate immune responses are critical for the host defense against invading pathogens. Key to initiating these defenses is the innate response of host cells to PAMPs. TLRs constitute a group of transmembrane pathogen recognition receptors (PRRs) that respond to a variety of PAMPs. Ligation of TLRs leads to activation of signaling pathways that culminates in activation of transcription factors, including NF-κB, AP-1, IRF3 and the MAP kinase family.37 These signals can lead to up-regulation of pathways involved in cell survival, proliferation, inflammation and immune regulation.22
Owing to their immediate and dramatic effects on host cellular responses, regulation of the TLRs must be precise and rapidly responsive. Control of TLRs occurs at multiple levels. Mechanisms to down-modulate the consequences of TLR activation include, but are not limited to, the following: compartmentalization of TLRs within the cell; negative regulation of protein abundance acting through ubiquitin-proteasome pathway such as Triad3A or SOCS-1; competition for binding to MyD88 or TLR4; and negative regulation of mRNA by miRNAs.38–40 TargetScan and miRanda predicted several potential miR-200a, miR-200b and miR-200c targets in the TLR4 pathway. This led us to examine a potential role for these three miRNAs in the regulation of TLR4 signaling and consequent NF-κB activation. Our data showed that exogenously introduced mimics of miR-200b or miR-200c resulted in diminished NF-κB reporter activity in a HEK293-TLR4 cell line. miR-200a had no effect on the NF-κB reporter. The latter has a distinct seed sequence even though it is clustered with miR-200b on chromosome 1p36.33. Examination of genes encoding additional proteins participating in the TLR4 signaling pathway indicated that the 3′UTRs of the adaptor protein MyD88 or the signaling pathway proteins IRAK1, TRAF6 or TAB2 could each constitute a miR-200b and miR-200c target. In contrast, transcripts encoding proteins in the alternate pathway, TRIF, TRAM and IRF3, were not predicted targets of either miR-200b or miR-200c. MyD88 is essential for mediating signals through all TLR pathways with the exception of TLR3 and the alternate TLR4 pathway.20,22 miR-200b and miR-200c were also able to suppress the expression of IL-6, CXCL9 and TNF-α induced by exposure of a macrophage-like human monocytic cell line to the TLR4 ligand LPS. Thus, miR-200b and miR-200c may be modifiers of the genes expressed during either classical or type 2 macrophage activation,36 leading to hypotheses that these miRNAs play an important role in antimicrobial responses.
miR-200b and miR-200c are well-recognized modulators of epithelial to mesenchymal transition (EMT) during embryogenesis—a process that facilitates tissue remodeling.41 The ZFH family zinc-finger E-box binding homeobox proteins ZEB1 and ZEB2 negatively regulate E-cadherin,42 activate EMT and thus promote tumor progression. It has been shown that ZEB1 directly suppresses transcription of miR-200c and miR-14.43 In turn, miR-200c is a direct suppressor of EMT and targets both ZEB1 and TGF-β2 transcripts for degradation. EMT is reportedly an essential early step in tumor metastasis, and loss of miR-200b and miR-200c expression in some breast cancers led to the suggestion that miR-200b and miR-200c may, in part, suppress tumor progression.44 miR-200b and miR-200c levels are decreased in response to TGF-β or the protein tyrosine phosphatase Pez.42,44 As such, there is a feedback loop between miR-200b and miR-200c and ZEB1, regulating proliferation. The current study constitutes the first report implicating miR-200b and miR-200c in regulating innate immune responses.
Data reported herein are focused on the TLR4 pathway because of its importance in antibacterial innate immune responses.21,22,45 Nonetheless, amongst the 10 currently recognized human-encoded TLRs, 9 utilize MyD88 for downstream signaling and regulation of immune response genes.20–22,30 Relevant to this article, MyD88 is essential for expression of pro-inflammatory genes after TLR4 ligation, including TNF-α, CXCL9 and IL-6 (as studied here in Figure 5).36 We have not yet examined the effects miR-200b and miR-200c on signaling through other MyD88-dependent TLRs, although our data suggest that their functions would also be suppressed by miR-200b or miR-200c ligation. Ligation of TLR4 can lead to NF-κB signaling through a pathway that depends upon TRIF but not MyD88 (see Figure 1). The fact that the enhanced expression of NF-κB following TLR4 ligation was significantly suppressed by mimics of miR-200b or miR-200c, suggests that either a significant portion of signaling following TLR4 ligation must go through the MyD88 pathway or that these miRNAs have unpredicted effects suppressing proteins in this arm of the pathway. Indeed, NF-κB activation initiated by TNF-α was also significantly suppressed by the miR-200b mimic. These observations make it seem highly likely that miR-200b and miR-200c have additional targets leading to repressed NF-kB activity. Although our data suggest that TAB2 was not a target, it is likely that miR-200b (and possibly miR-200c) targets an alternate transcript. Further studies warrant investigation of TAB3, as well as other predicted targets of the common miR-200b/miR-200c seed region.
The first report suggesting that miRNAs are capable of regulating the innate immune system demonstrated the ability of miR-146a to suppress TLR4 signaling through targeting two key adaptor molecules, IRAK1 and TRAF6.28 Since this report, other studies have documented a role for miRNAs in the regulation of selected aspects of innate and adaptive immune responses.46,47 Similar to our findings, several miRNAs (miR-101, miR-155, miR-199 a, miR-105) have been shown to target intermediates in signaling pathways essential for TLRs and other innate immune responses.29,48–51 Furthermore, several miRNAs were shown to be differentially regulated in peripheral blood cell responses to the TLR4 ligand LPS, including miR-146b, miR-150, miR-342, let-7 g and miR-143.52
The goal of the current study was to discover miRNAs capable of modifying the cellular response after TLR ligation. Our approach was to screen for predicted targets, which contrasted with many published reports that relied upon significant changes in miRNA abundance to identify potentially regulatory miRNAs. Both miR-200b and miR-200c down-modulated the response to an NF-κB reporter after TLR4 ligation in a (HEK-293 TLR4) cell line expressing recombinant TLR4. Experimental manipulation of miR-200b and miR-200c levels in the same line showed they were able to down-modulate expression of the adaptor molecule MyD88, suggesting they might cause the effect. Furthermore, miR-200b and miR-200c were able to modulate the abundance of MyD88 mRNA, as well as LPS-mediated expression of chemokines downstream of NF-κB activation (IL-6, CXCL9, TNF-α) in a macrophage cell line. These observations suggest that miR-200b and miR-200c may function as modifiers of innate responses delivered through TLR4. Further studies should investigate the potential involvement of these miRNAs in down-modulating the response to LPS or other TLR ligands, possibly in concert with other miRNAs that regulate TLR signaling pathways such as miR-146a, miR-155 and miR-21.29 A cadre of miRNAs each controlling different intermediates in the TLR pathways could have far-reaching effects in controlling innate immune responses.
Supplementary Material
Acknowledgments
Funding This work was supported in part by NIH grants R01 AI045540, R01 AI067874, R01 AI076233, R21 AI080801 and Merit Review and OEF/OIF Merit grants from the Department of Veterans’ Affairs. EBW was supported by NIH training grant T32 AI007511. JWG was supported by a CDA-1 award from the Department of Veterans’ Affairs.
Footnotes
Reprints and permissions: sagepub.co.uk/journalsPermissions.nav
References
- 1.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, et al. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 3.Grosshans H, Filipowicz W. Molecular biology: the expanding world of small RNAs. Nature. 2008;451:414–416. doi: 10.1038/451414a. [DOI] [PubMed] [Google Scholar]
- 4.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 5.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 9.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 10.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 11.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anglicheau D, Muthukumar T, Suthanthiran M. MicroRNAs: small RNAs with big effects. Transplantation. 2010;90:105–112. doi: 10.1097/TP.0b013e3181e913c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carissimi C, Fulci V, Macino G. MicroRNAs: novel regulators of immunity. Autoimmun Rev. 2009;8:520–524. doi: 10.1016/j.autrev.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Pedersen I, David M. MicroRNAs in the immune response. Cytokine. 2008;43:391–394. doi: 10.1016/j.cyto.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67:129–139. doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 18.Dai R, Ahmed SA. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res. 2011;157:163–179. doi: 10.1016/j.trsl.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Jenkins KA, Mansell A. TIR-containing adaptors in Toll-like receptor signalling. Cytokine. 2010;49:237–244. doi: 10.1016/j.cyto.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 21.Medzhitov R, Janeway CA., Jr Innate immune recognition and control of adaptive immune responses. Semin Immunol. 1998;10:351–353. doi: 10.1006/smim.1998.0136. [DOI] [PubMed] [Google Scholar]
- 22.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 23.Wajant H, Scheurich P. TNFR1-induced activation of the classical NF-kappaB pathway. FEBS J. 2011;278:862–876. doi: 10.1111/j.1742-4658.2011.08015.x. [DOI] [PubMed] [Google Scholar]
- 24.Cook DN, Pisetsky DS, Schwartz DA. Toll-like receptors in the pathogenesis of human disease. Nat Immunol. 2004;5:975–979. doi: 10.1038/ni1116. [DOI] [PubMed] [Google Scholar]
- 25.Lee JY, Hwang DH. The modulation of inflammatory gene expression by lipids: mediation through Toll-like receptors. Mol Cell. 2006;21:174–185. [PubMed] [Google Scholar]
- 26.Quinn SR, O’Neill LA. A trio of microRNAs that control Toll-like receptor signalling. Int Immunol. 2011;23:421–425. doi: 10.1093/intimm/dxr034. [DOI] [PubMed] [Google Scholar]
- 27.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O’Leary JJ, Ruan Q, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 28.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the finetuners of Toll-like receptor signalling. Nat Rev Immunol. 2011;11:163–175. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- 30.Armant MA, Fenton MJ. Toll-like receptors: a family of pattern-recognition receptors in mammals. Genome Biol. 2002;3:3011.1–3011.6. doi: 10.1186/gb-2002-3-8-reviews3011. REVIEWS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Young DW, Gusovsky F, Chow JC. Cellular events mediated by lipopolysaccharide-stimulated toll-like receptor 4. MD-2 is required for activation of mitogen-activated protein kinases and Elk-1. J Biol Chem. 2000;275:20861–20866. doi: 10.1074/jbc.M002896200. [DOI] [PubMed] [Google Scholar]
- 33.Gioannini TL, Teghanemt A, Zhang D, Coussens NP, Dockstader W, Ramaswamy S, et al. Isolation of an endotoxin-MD-2 complex that produces Toll-like receptor 4-dependent cell activation at picomolar concentrations. Proc Natl Acad Sci USA. 2004;101:4186–4191. doi: 10.1073/pnas.0306906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Marquez RT, Wendlandt E, Galle CS, Keck K, McCaffrey AP. MicroRNA-21 is upregulated during the proliferative phase of liver regeneration, targets Pellino-1, and inhibits NF-kappaB signaling. Am J Physiol Gastrointest Liver Physiol. 2010;298:G535–G541. doi: 10.1152/ajpgi.00338.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benoit M, Desnues B, Mege JL. Macrophage polarization in bacterial infections. J Immunol. 2008;181:3733–3739. doi: 10.4049/jimmunol.181.6.3733. [DOI] [PubMed] [Google Scholar]
- 37.Moresco EM, LaVine D, Beutler B. Toll-like receptors. Curr Biol. 2011;21:R488–R493. doi: 10.1016/j.cub.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 38.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheedy FJ, O’Neill LA. Adding fuel to fire: microRNAs as a new class of mediators of inflammation. Ann Rheum Dis. 2008;67(Suppl 3):iii50–iii55. doi: 10.1136/ard.2008.100289. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Hu Y, Deng WW, Sun B. Negative regulation of Toll-like receptor signaling pathway. Microbes Infect. 2009;11:321–327. doi: 10.1016/j.micinf.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 41.Mongroo PS, Rustgi AK. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol Ther. 2010;10:219–222. doi: 10.4161/cbt.10.6312548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katoh Y, Katoh M. Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA (review) Int J Mol Med. 2008;22:271–275. [PubMed] [Google Scholar]
- 43.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 45.Beutler B, Poltorak A. The sole gateway to endotoxin response: how LPS was identified as Tlr4, and its role in innate immunity. Drug Metab Dispos. 2001;29(4 Pt 2):474–478. [PubMed] [Google Scholar]
- 46.Taganov KD, Boldin MP, Baltimore D. MicroRNAs and immunity: tiny players in a big field. Immunity. 2007;26:133–137. doi: 10.1016/j.immuni.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Garmire LX, Shen Z, Briggs S, Yeo G, Subramaniam S, Glass C. Regulatory network of microRNAs in RAW 264.7 macrophage cells. Conf Proc IEEE Eng Med Biol Soc. 2010;2010:6198–6201. doi: 10.1109/IEMBS.2010.5627742. [DOI] [PubMed] [Google Scholar]
- 48.Yan D, Ng WL, Zhang X, Wang P, Zhang Z, Mo YY, et al. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS One. 2010;5:e11397. doi: 10.1371/journal.pone.0011397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang P, Hou J, Lin L, Wang C, Liu X, Li D, et al. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol. 2010;185:6226–6233. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- 50.Chen R, Alvero AB, Silasi DA, Kelly MG, Fest S, Visintin I, et al. Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene. 2008;27:4712–4723. doi: 10.1038/onc.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benakanakere MR, Li Q, Eskan MA, Singh AV, Zhao J, Galicia JC, et al. Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes. J Biol Chem. 2009;284:23107–23115. doi: 10.1074/jbc.M109.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidt WM, Spiel AO, Jilma B, Wolzt M, Muller M. In vivo profile of the human leukocyte microRNA response to endotoxemia. Biochem Biophys Res Commun. 2009;380:437–441. doi: 10.1016/j.bbrc.2008.12.190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.