Abstract
Allelic variation at the Cu–Zn superoxide dismutase (SOD1) locus has been shown to be associated with resistance of the snail, Biomphalaria glabrata, to infection by the trematode parasite, Schistosoma mansoni. SOD1 catalyses the production of hydrogen peroxide, a known cytotoxic component of the oxidative burst used in defence against pathogens. In our laboratory population of B. glabrata, the most resistant allele at SOD1 is over-expressed relative to the other two alleles. Because hydrogen peroxide also causes oxidative stress on host tissues, we hypothesised that over-expression of SOD1 might be compensated by epistatic interactions with other loci involved in oxidation–reduction (redox) pathways. Catalase, peroxiredoxins and glutathione peroxidases all degrade hydrogen peroxide. We tested whether alleles at each of these loci were in linkage disequilibrium with SOD1 in our population, as might be expected given strong epistatic selection. We found that SOD1, catalase (CAT) and a peroxiredoxin locus (PRX4) are in strong linkage disequilibrium in our population. We also found that these loci are tightly linked, within 1–2 cM of each other, which explains the high linkage disequilibrium. This result raises the possibility that there is a linked cluster of redox genes, and perhaps other defence-relevant genes, in the B. glabrata genome. Whether epistatic interactions for fitness actually exist among these loci still needs to be tested. However the close physical linkage among SOD1, PRX4 and CAT, and subsequent high disequilibrium, makes such interactions a plausible hypothesis.
Keywords: Snail, Schistosoma mansoni, Genetics, Superoxide dismutase, Catalase, Peroxiredoxin
1. Introduction
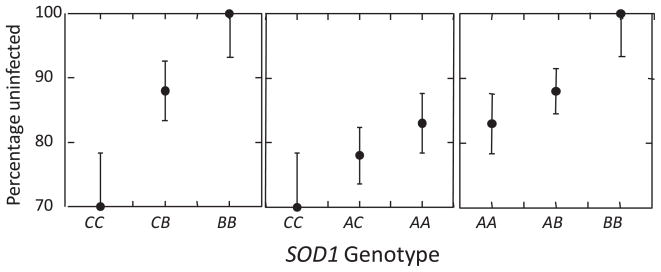
The snail, Biomphalaria glabrata, is an intermediate host for the human pathogen, Schistosoma mansoni. Resistance in B. glabrata to infection with S. mansoni is highly heritable in many laboratory and field populations (Richards and Shade, 1987; Shoukry et al., 1997; Webster and Woolhouse, 1998; Webster et al., 2004; Theron et al., 2008). One locus at which allelic variation associates with resistance to the parasite is copper–zinc superoxide dismutase (SOD1) (Goodall et al., 2004, 2006; Bonner et al., 2012). Increased hydrogen peroxide (H2O2) production and increased expression of SOD1 were correlated with the difference in resistance between snails from the M-line strain and the more resistant 13-16-R1 strain (Bender et al., 2005; Bayne, 2009). Within strain 13-16-R1, there are three common alleles (intron sequence variants A, B and C) that associate additively with resistance (Fig. 1). Furthermore, the most protective allele, B, is constitutively over-expressed relative to alleles A and C (Bender et al., 2007).
Fig. 1.
Association between resistance to infection and copper–zinc superoxide dismutase (SOD1) genotype in the 13-16-R1 population of Biomphalaria glabrata at Oregon State University, USA (percentage of snails not infected after challenge by the PR1 strain of Schistosoma mansoni, ±1 S.E.). There are three alleles, (A, B and C), so relationships among the genotypes are shown for each pair of alleles. Data are from Table 2 in Goodall et al. (2006). Note that the B allele confers the highest resistance and the C allele the lowest, and that allelic effects appear additive.
SOD1 is a ubiquitous protein involved in several cellular functions including signalling and immune response (Nappi and Ottaviani, 2000; Fink and Scandalios, 2002; Ramasarma, 2007; Abreu and Cabelli, 2010). SOD1 catalyses the reduction of highly reactive superoxide (O2−) to H2O2. H2O2 is a known cytotoxic component of the oxidative burst, which is one of the defence mechanisms for parasite clearance in molluscs (Bayne, 2009; Hahn et al., 2001; Loker, 2010). When PR-1 strain schistosomes invade a 13-16-R1-strain snail, hemocytes surround the invading parasite and are thought to generate H2O2 as part of the killing mechanism (Hahn et al., 2001; Bender et al., 2007; Bayne, 2009).
H2O2 is one of several reactive oxygen species that are used by cells for various biological processes in addition to defence against infection (e.g. signalling, growth and development, programmed cell death; Dowling and Simmons, 2009). However, reactive oxygen species are a double-edged sword. Their effects are dose-dependent and so mismatch between oxidative and anti-oxidative pathways can exert oxidative stress on cells, including damage to DNA, RNA and proteins (Beckman and Ames, 1998). Therefore, if over-expression of the B allele at SOD1 is responsible for its protective effect against parasites, then that raises the question of whether there are fitness costs to having that allele in the absence of parasites (Dowling and Simmons, 2009; Monaghan et al., 2009).
We recently showed that under laboratory conditions there was no fitness cost to having the B allele versus the C allele in terms of survival and fecundity, and there was a slight advantage in terms of growth (Bonner et al., 2012). One explanation for this unexpected result is that over-expression of the B allele is somehow compensated by epistatic interactions with other loci involved in oxidation–reduction (redox) pathways. Candidate loci that might compensate epistatically for over-expression of SOD1 include those coding for catalase, peroxiredoxins and glutathione peroxidases because these enzymes catalyse the conversion of H2O2 into water and other less harmful byproducts. However, such a compensatory mechanism would require SOD1 to be in substantial linkage disequilibrium (LD) with the interacting loci, which could occur via close physical linkage or strong, recurring epistatic selection (Sinervo and Svensson, 2002; Phillips, 2008).
Here we tested whether loci coding for Cu–Zn superoxide dismutase (SOD1, GenBank accession number AY505496.2), catalase (CAT, FC856064), peroxiredoxin 4 (PRX4; Knight et al., 2009) and glutathione peroxidase (GPX; DW474737) were in LD in the 13-16-R1 snail population and if so, whether that could be explained by physical linkage in the genome. We show that SOD1, PRX4 and CAT are in substantial LD above background levels and that they occur in a <2 cM cluster on the same chromosome. GPX was not linked to, nor was it in LD with, the other three loci.
2. Materials and methods
2.1. Study population
We used a population of the 13-16-R1 strain of B. glabrata that has been maintained as a large population (hundreds) in the Bayne laboratory at Oregon State University (OSU), USA since the mid-1970s. The 13-16-R1 snail strain was reportedly created from one or more crosses between strains of snails isolated from Brazil and Puerto Rico. Biomphalaria glabrata is a facultative self-fertilising hermaphrodite such that snails will preferentially outcross when given access to a mate, but when isolated will usually reproduce through self-fertilisation (e.g. Vianey-Liaud and Dussart, 2002; and personal observations). For example, our laboratory population is in Hardy–Weinberg equilibrium for all the loci used in this study. This population has been maintained in the absence of selection pressure by S. mansoni.
Previous work on SOD1 in this population looked at sequence variation in an approximately 540 bp piece of intron 4 (Goodall et al., 2006). Three main sequence variants (alleles) are segregating at this locus (alleles A, B and C) in addition to a rare variant of allele C (allele D at ~1% allele frequency), which in this study was scored as a C allele (only two copies were observed out of 80 individuals and the results were almost identical to when the D allele was included in the analyses).
2.2. Loci used
We used eight previously published microsatellite loci (Table 1; Jones et al., 1999; Mavarez et al., 2000) to assess the background level of LD in the population. We assumed that none of these eight loci are closely linked. Size variation at microsatellites was scored on an ABI 3100 capillary sequencer. Allelic variation at intron 4 of SOD1, intron 2 of GPX and intron 2 of PRX4 was assessed via Sanger sequencing or by high-resolution melt analysis on an ABI Fast Real-Time PCR system. The three alleles from intron 6 of CAT differed substantially in size (490, 566 and 592 bp), and therefore were scored by size on an ABI capillary sequencer. Sequences for the SOD1 alleles are available in GenBank under accession numbers DQ239577–DQ239579 (from Goodall et al., 2006), and for the alleles at CAT and PRX4 under JX989079–JX989081 and JX989076– JX989078. Allele sequences for GPX are shown in Supplementary Fig. S1. Primers and PCR conditions for the microsatellite and redox loci used in this study can be found in Supplementary Tables S1 and S2.
Table 1.
Diversity statistics at the redox and microsatellite loci in the 13-16-R1 population of Biomphalaria glabrata at Oregon State University.
| Sample set 1
|
Sample set 2d
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hexp | Na | Fisa | Pb | Nc | Hexp | Na | Fis | P | N | |
| Redox loci | ||||||||||
| SOD1 | 0.68 | 3 | −0.1 | 0.71 | 40 | 0.65 | 3 | −0.02 | 0.59 | 38 |
| PRX4 | 0.66 | 3 | −0.08 | 0.93 | 40 | 0.65 | 3 | 0.03 | 0.46 | 32 |
| CAT | 0.4 | 3 | 0.009 | 0.19 | 40 | 0.48 | 3 | 0.07 | 0.57 | 40 |
| GPX | – | – | – | – | – | 0.68 | 3 | −0.18 | 0.45 | 29 |
| Microsatellite loci | ||||||||||
| bgc8 | 0.45 | 2 | −0.05 | 1 | 40 | |||||
| bgc7 | 0.6 | 3 | −0.08 | 0.47 | 40 | |||||
| bge4 | 0.11 | 2 | −0.05 | 1 | 40 | |||||
| bge5 | 0.66 | 6 | −0.14 | 0.48 | 40 | |||||
| bgu8 | 0.29 | 2 | −0.03 | 1 | 40 | |||||
| bgu10 | 0.55 | 4 | 0.09 | 0.8 | 40 | |||||
| bgu15 | 0.38 | 2 | −0.19 | 0.8 | 40 | |||||
| ubg1 | 0.74 | 4 | 0.07 | 0.23 | 40 | |||||
Hexp, expected heterozygosity; Na, Number of alleles; SOD1, Cu–Zn superoxide dismutase; PRX4, peroxiredoxin 4; CAT, catalase; GPX, glutathione peroxidase.
Deviation from Hardy–Weinberg genotype proportions (Fis estimated using Weir and Cockerham’s method, and tested for significance using the exact test, in GENEPOP).
P value for test of deviation from Hardy–Weinberg equilibrium.
Sample size (number of individuals successfully genotyped at each locus).
GPX was scored only in Sample set 2, while the microsatellite loci were scored only in Sample set 1.
2.3. Linkage disequilibrium
Allelic variation was scored at SOD1, PRX4, CAT and the eight microsatellites on a sample of 40 snails taken from the laboratory population in 2006 (Sample set 1). We obtained complete genotypes for all 11 loci in all 40 snails in that sample. There was not enough DNA remaining in many of those samples to also score GPX. Therefore, we scored the four redox loci (but not the microsatellites) in an additional sample of 40 snails collected in 2008 (Sample set 2). LD among loci was analysed separately for each sample set. Multi-allelic LD was estimated between each pair of loci in each sample using Weir (1979) correlation coefficient (R) as implemented in GENETIX 4.05.
2.4. Linkage mapping of SOD1, PRX4, CAT and GPX
There were three alleles at each locus. These were designated as follows: SOD1 (A, B and C), PRX4 (1, 2 and 3), CAT (490, 566 and 592) and GPX (100, 200 and 300) (Tables 1 and 2). Two crosses were performed. Family 1 was informative for mapping all four loci (CC 11 592592 100300 × AC 21 490592 200300). Family 2 (AB 22 566592 200300 × AC 12 592592 200200) was informative for mapping all pairs of loci except GPX-PRX4 and CAT-PRX4. Two hundred off-spring from each family were genotyped at all four loci, with the exception that we genotyped only 22 individuals from Family 1 for GPX. This was because after first genotyping Family 2, it was obvious that GPX was unlinked to the other loci. Therefore we used a subset of individuals to verify the pattern in Family 1.
Table 2.
Allele frequencies at the redox loci in each sample seta from the 13-16-R1 snail population.
| Locus | Allele | Frequency
|
|
|---|---|---|---|
| Sample set 1 | Sample set 2 | ||
| SOD1 | A | 0.375 | 0.421 |
| B | 0.3 | 0.395 | |
| C | 0.325 | 0.184 | |
| 40 | 38 | ||
| PRX4 | 1 | 0.275 | 0.203 |
| 2 | 0.375 | 0.359 | |
| 3 | 0.35 | 0.438 | |
| 40 | 32 | ||
| CAT | 490 | 0.062 | 0.025 |
| 566 | 0.188 | 0.263 | |
| 592 | 0.75 | 0.712 | |
| 40 | 40 | ||
| GPX | 100 | – | 0.345 |
| 200 | – | 0.345 | |
| 300 | – | 0.31 | |
| 29 | |||
SOD1, Cu–Zn superoxide dismutase; PRX4, peroxiredoxin 4; CAT, catalase; GPX, glutathione peroxidase.
Allele frequency differences between Sample sets 1 and 2 are not statistically significant (tested for each locus using the exact test for population differentiation in GENEPOP).
3. Results
3.1. Allelic variation
No loci showed significant deviations from Hardy–Weinberg equilibrium in either sample set (Table 1). The eight microsatellites showed levels of variation similar to those of the redox loci, with an average of 3.1 alleles per locus (range 2–6) and average heterozygosity = 0.47 (range 0.11–0.74), versus three alleles per locus and average heterozygosity of 0.62 (range 0.40–0.68) at the redox loci (Table 1). Thus, the microsatellites were good comparators for the background level of LD in the population. Table 2 shows allele frequencies at each of the redox loci.
3.2. Linkage disequilibrium in the 13-16-R1 population
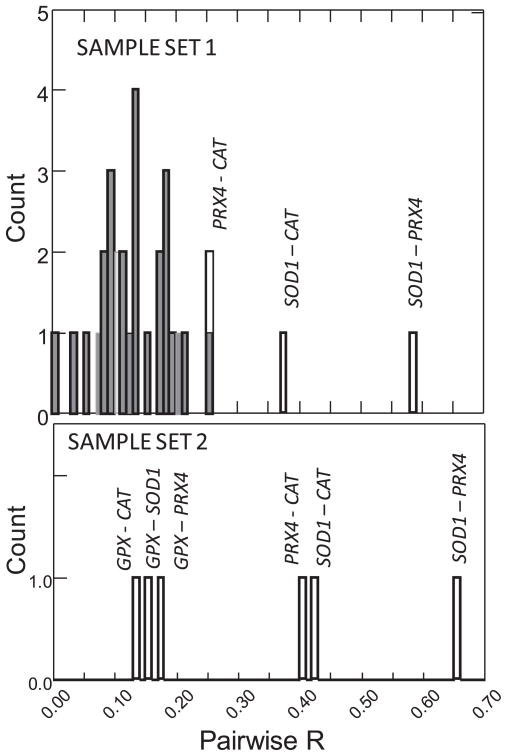
LD among the eight putatively unlinked microsatellite loci ranged from 0 to 0.25, averaging 0.13 (Fig. 2). Two pairs of microsatellites (bge5 with ubg1, and bgu8 with bgu10) showed significant LD but not after correction for multiple testing (28 pairwise tests). In contrast, LD was very high and statistically significant between SOD1 and PRX4 (R = 0.58 and 0.65 in Sample sets 1 and 2, respectively), as well as high and significant between CAT and SOD1 (R = 0.37 and R = 0.40) and between CAT and PRX4 (R = 0.25 and R = 0.42) (Fig. 2; Table 3). LD between GPX and each of the three other redox loci ranged from 0.13 to 0.15 (Fig. 2; Table 3). Relative to the background level of LD in the population (inferred from the microsatellites), there is very high LD between SOD1 and PRX4, and high LD between those two loci and CAT (Fig. 2). LD between GPX and the other three redox loci was in line with the background level among putatively unlinked microsatellite loci in this population.
Fig. 2.
Frequency distribution of pairwise R values (multi-allelic measure of linkage disequilibrium) between loci in Sample sets 1 and 2 (histogram with 0.01 unit bins). Pairwise values between microsatellite loci are shown in black bars, and between redox loci in white bars. Values for each pair of redox loci are labelled. Note that values among Cu–Zn superoxide dismutase (SOD1), peroxiredoxin 4 (PRX4) and catalase (CAT) are much larger than the background level in this population described by the eight putatively-unlinked microsatellites.
Table 3.
Linkage disequilibrium among redox loci in each sample set from the 13-16-R1 snail population.
| Locus pair | Sample set 1
|
Sample set 2
|
||
|---|---|---|---|---|
| Ra | Pb | R | P | |
| SOD1–PRX4 | 0.58 | 0.0001 | 0.65 | 0.0001 |
| SOD1–CAT | 0.37 | 0.0003 | 0.40 | 0.0004 |
| SOD1–GPX | – | – | 0.15 | 0.62 |
| PRX4–CAT | 0.25 | 0.03 | 0.42 | 0.0001 |
| PRX4–GPX | – | – | 0.17 | 0.60 |
| CAT–GPX | – | – | 0.13 | 0.72 |
SOD1, Cu–Zn superoxide dismutase; PRX4, peroxiredoxin 4; CAT, catalase; GPX, glutathione peroxidase.
Multi-allelic correlation coefficient of Weir (1979) as implemented in the GENETIX software.
P value from chi-square test of R = 0.
3.3. Estimated map distances among the four redox loci
No loci showed deviation from expected Mendelian inheritance in either family. In Family 1 we observed no recombinants out of 200 individuals among SOD1, PRX1 and CAT, giving an exact one-sided 95% confidence interval (CI) for the pairwise recombination rate, r, among those three loci of ≤0.018. In Family 2 we observed three recombinants out of 200 between SOD1 and CAT (r = 0.015; 95% CI 0.003–0.043), and one recombinant between SOD1 and PRX4 (r = 0.005; 95% CI 0.0001–0.028). Combining results from both families shows four recombinants out of 400, giving an estimate of 1 cM over the entire haplotype block (95% CI of 0.003– 0.025). Therefore, we conclude that SOD1, PRX4 and CAT are closely linked to each other in a chromosomal block that probably spans less than 2 cM. Both families showed free recombination between GPX and the other loci.
3.4. Inference of gene order
Because we observed no recombinants in the fully-informative cross (Family 1), we cannot infer which locus is in the middle. However, from the strong positive or negative pairwise correlations between alleles across loci observed in the LD analysis, it was obvious that alleles at the three linked loci fall into three haplotype blocks (Supplementary data S1 and Fig. S2). The strongest associations among alleles distinguish haplotype 3, B, 566 from haplotypes 1, C, 592/490 and 2, A, 592/490 (for alleles at PRX4, SOD1 and CAT from right to left). Here 592/490 means either allele 490 or 592 at CAT (allele 490 is rare (Table 2) so there are effectively only two alleles at that locus). Given the strong signal of three ancestral haplotypes, we can count the number of putative recombinant chromosomes currently segregating in our population (e.g. a CB 13 566566 genotype would imply a recombination event occurred between CAT and the other two loci). In sample sets 1 and 2 we observed a total of 11 putative recombinants between PRX4 and the other two loci, and 15 putative recombinants between CAT and the other two loci. There were no putative recombinants between SOD1 and the other two loci. Therefore, we conclude that SOD1 is in the middle.
4. Discussion
Here we observed strong LD among SOD1, PRX4 and CAT that can be explained by their close physical linkage in a 1–2 cM block of chromosome. The three clear haplotype blocks present in the 13-16-R1 snail population probably reflect its hybrid origin between Caribbean and South American populations.
We currently do not know what physical distance corresponds to 1–2 cM in the B. glabrata genome. Average recombination rates of 1–5 cM per megabase of DNA are typical in other organisms, albeit with substantial variation across chromosomal regions (e.g. Backström et al., 2010; Stevison and Noor, 2010; Hohenlohe et al., 2012). We recently completed a single nucleotide polymorphism (SNP)-based linkage map from an outcrossed pair of 13-16-R1 snails, from which we estimated a very rough average of 4–10 cM/Mb (unpublished data). Thus, given no other information, it seems reasonable to tentatively conclude that SOD1, PRX4 and CAT are within several hundred thousand bp of each other. These three loci are found on three different, relatively small contigs in the current draft of the B. glabrata genome (which is still quite fragmented). An improved assembly will be required to determine the actual positions of these loci relative to each other.
We also have 20 inbred lines from the 13-16-R1 population in which we typed the four redox loci and several thousand SNPs (unpublished data). Relative to LD between SNPs that are known to be within 1–2 cM of each other, the LD observed among SOD1, PRX4 and CAT was not significantly higher. Thus, tight linkage alone is a sufficient explanation for the high LD among the redox loci (i.e. retarded decay of LD that was generated by genetic drift and/or mixture disequilibrium). Therefore, it is not necessary to invoke epistatic selection to explain the high LD.
On the other hand, given we chose, a priori, to examine LD among CAT, PRX4, SOD1 and GPX based entirely on their proteins’ enzymatic function, it does seem very coincidental that three turn out to be so closely linked. After obtaining our results, we checked whether loci for catalase, superoxide dismutase and peroxiredoxins are closely linked in some other well-annotated genomes for which good information on recombination rates is available. No evidence was found for linkage in vertebrates (human, chicken, cattle, zebra fish) or two insects (Apis mellifera, Anopheles gambiae). However, the three loci are linked in the same order (SOD1 in the middle) in Drosophila melanogaster and Caenorhabditis elegans, although not as tightly linked as they appear to be in B. glabrata (39 cM across the entire interval in D. melanogaster and 24 cM in C. elegans; Hillier et al., 2007; Fiston-Lavier et al., 2010). This similarity could simply reflect the arrangement in the common ancestor of insects, nematodes and molluscs. It is also possible that there is some selective advantage to having these loci physically linked.
Mounting evidence suggests that the oxidative burst is an important component of defence in molluscs (Bayne, 2009; Green et al., 2009; Mourao et al., 2009; Loker, 2010; Mone et al., 2011). Thus it is worth considering the possibility that there is a linked cluster of redox genes, and perhaps other defence-relevant genes, in the B. glabrata genome (e.g. similar to the Plasmodium resistance island in Anopheles; Riehle et al., 2006). Frequency-dependent biotic interactions (e.g. host-parasite) are considered among the most likely to increase LD among epistatically interacting loci (Sinervo and Svensson, 2002). Selection for reduced recombination among such loci would reduce the recombinational load, as in traditional models of the evolution of co-adapted gene complexes (Dobzhan-sky, 1970; Ford, 1975). Whether SOD1, CAT and PRX4 really occur in a defence-related genomic island in B. glabrata is, of course, speculation at this point. However it is an intriguing hypothesis that deserves further study.
Finally, our results also raise the question of whether the statistical association between SOD1 alleles and resistance to S. mansoni that was reported by Goodall et al. (2006) is actually caused by SOD1 alone. It is possible that SOD1 is simply in strong LD with a closely-linked locus that actually controls resistance. More likely, SOD1 may interact epistatically with other linked loci and it is the entire haplotype block that is driving the association with resistance. Fine mapping of recombinants in this region will go a long way towards teasing apart those causal relationships.
Our original motivation for testing whether the loci that control degradation of H2O2 were in LD with SOD1 was to explore the possibility that any costs of carrying the B allele at SOD1 are compensated by other loci (see Bonner et al., 2012). Whether such epistatic interactions for fitness actually exist still needs to be tested. However the close physical linkage and high LD among SOD1, PRX4 and CAT makes such interactions a plausible hypothesis.
Supplementary Material
Acknowledgments
Thank you to J. Tennessen for assistance with the bioinformatics and SNP analysis, and to J. Tennessen and M. Steinauer for comments on an earlier draft. C. Goodall and R. Bender identified the PRX4 alleles and developed the high resolution melt assay. The Center for Genome Research and Biocomputing at Oregon State University (USA) assisted with genotyping. This work was partially funded by the US National Institutes of Health Grant AI016137.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ijpara.2012.10.020.
Footnotes
Note: Nucleotide sequence data reported in this paper are available in GenBank under accession numbers JX989076–JX989081.
References
- Abreu IA, Cabelli DE. Superoxide dismutases – a review of the metal-associated mechanistic variations. BBA Proteins Proteom. 2010;1804:263–274. doi: 10.1016/j.bbapap.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Backström N, Forstmeier W, Schielzeth H, Mellenius H, Nam K, Bolund E, Webster MT, Öst T, Schneider M, Kempenaers B, Ellegren H. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res. 2010;20:485–495. doi: 10.1101/gr.101410.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne CJ. Successful parasitism of vector snail Biomphalaria glabrata by the human blood fluke (trematode) Schistosoma mansoni: a 2009 assessment. Mol Biochem Parasitol. 2009;165:8–18. doi: 10.1016/j.molbiopara.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- Bender RC, Broderick EJ, Goodall CP, Bayne CJ. Respiratory burst of Biomphalaria glabrata hemocytes: Schistosoma mansoni-resistant snails produce more extracellular H2O2 than susceptible snails. J Parasitol. 2005;91:275–279. doi: 10.1645/GE-415R. [DOI] [PubMed] [Google Scholar]
- Bender RC, Goodall CP, Blouin MS, Bayne CJ. Variation in expression of Biomphalaria glabrata SOD1: a potential controlling factor in susceptibility/resistance to Schistosoma mansoni. Dev Comp Immunol. 2007;31:874–878. doi: 10.1016/j.dci.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Bonner KM, Bayne CJ, Larson MK, Blouin MS. Effects of Cu/Zn superoxide dismutase (sod1) genotype and genetic background on growth, reproduction and defense in Biomphalaria glabrata. PLoS Negl Trop Dis. 2012;6:e1701. doi: 10.1371/journal.pntd.0001701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics of the Evolutionary Process. Columbia University Press; Columbia, NY: 1970. [Google Scholar]
- Dowling DK, Simmons LW. Reactive oxygen species as universal constraints in life-history evolution. Proc Biol Sci. 2009;276:1737–1745. doi: 10.1098/rspb.2008.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink RC, Scandalios JG. Molecular evolution and structure-function relationships of the superoxide dismutase gene families in angiosperms and their relationship to other eukaryotic and prokaryotic superoxide dismutases. Arch Biochem Biophys. 2002;399:19–36. doi: 10.1006/abbi.2001.2739. [DOI] [PubMed] [Google Scholar]
- Fiston-Lavier AS, Singh ND, Lipatov M, Petrov DA. Drosophila melanogaster recombination rate calculator. Gene. 2010;463:18–20. doi: 10.1016/j.gene.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Ford EB. Ecological Genetics. 4. Chapman and Hall; London: 1975. [Google Scholar]
- Goodall CP, Bender RC, Broderick EJ, Bayne CJ. Constitutive differences in Cu/Zn superoxide dismutase mRNA levels and activity in hernocytes of Biomphalaria glabrata (Mollusca) that are either susceptible or resistant to Schistosoma mansoni (Trematoda) Mol Biochem Parasitol. 2004;137:321–328. doi: 10.1016/j.molbiopara.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Goodall CP, Bender RC, Brooks JK, Bayne CJ. Biomphalaria glabrata cytosolic copper/zinc superoxide dismutase (SOD1) gene: association of SOD1 alleles with resistance/susceptibility to Schistosoma mansoni. Mol Biochem Parasitol. 2006;147:207–210. doi: 10.1016/j.molbiopara.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Green TJ, Dixon TJ, Devic E, Adlard RD, Barnes AC. Differential expression of genes encoding anti-oxidant enzymes in Sydney rock oysters, Saccostrea glomerata (Gould) selected for disease resistance. Fish Shellfish Immunol. 2009;26:799–810. doi: 10.1016/j.fsi.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Hahn UK, Bender RC, Bayne CJ. Killing of Schistosoma mansoni sporocysts by hemocytes from resistant Biomphalaria glabrata: role of reactive oxygen species. J Parasitol. 2001;87:292–299. doi: 10.1645/0022-3395(2001)087[0292:KOSMSB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Hillier LW, Miller RD, Baird SE, Chinwalla A, Fulton LA, Koboldt DC, Waterston RH. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 2007;5:e167. doi: 10.1371/journal.pbio.0050167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Currey M, Cresko WA. Extensive linkage disequilibrium and parallel adaptive divergence across threespine stickleback genomes. Philos Trans R Soc Lond B Biol Sci. 2012;367:395–408. doi: 10.1098/rstb.2011.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CS, Lockyer AE, Rollinson D, Piertney SB, Noble LR. Isolation and characterization of microsatellite loci in the freshwater gastropod, Biomphalaria glabrata, an intermediate host for Schistosoma mansoni. Mol Ecol. 1999;8:2149–2151. doi: 10.1046/j.1365-294x.1999.00802-5.x. [DOI] [PubMed] [Google Scholar]
- Knight M, Raghavan N, Goodall C, Cousin C, Ittiprasert W, Sayed A, Miller A, Williams DL, Bayne CJ. Biomphalaria glabrata peroxiredoxin: effect of Schistosoma mansoni infection on differential gene regulation. Mol Biochem Parasitol. 2009;167:20–31. doi: 10.1016/j.molbiopara.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loker ES. Gastropod immunobiology. Adv Exp Med Biol. 2010;708:17–43. doi: 10.1007/978-1-4419-8059-5_2. [DOI] [PubMed] [Google Scholar]
- Mavarez J, Amarista M, Pointier JP, Jarne P. Microsatellite variation in the freshwater schistosome-transmitting snail Biomphalaria glabrata. Mol Ecol. 2000;9:1009–1011. doi: 10.1046/j.1365-294x.2000.00939-11.x. [DOI] [PubMed] [Google Scholar]
- Monaghan P, Metcalfe NB, Torres R. Oxidative stress as a mediator of life history trade-offs: mechanisms, measurements and interpretation. Ecol Lett. 2009;12:75–92. doi: 10.1111/j.1461-0248.2008.01258.x. [DOI] [PubMed] [Google Scholar]
- Mone Y, Ribou AC, Cosseau C, Duval D, Theron A, Mitta G, Gourbal B. An example of molecular co-evolution: reactive oxygen species (ROS) and ROS scavenger levels in Schistosoma mansoni/Biomphalaria glabrata interactions. Int J Parasitol. 2011;41:721–730. doi: 10.1016/j.ijpara.2011.01.007. [DOI] [PubMed] [Google Scholar]
- de Mourao MM, Dinguirard N, Franco GR, Yoshino TP. Role of the endogenous antioxidant system in the protection of Schistosoma mansoni primary sporocysts against exogenous oxidative stress. PLoS Negl Trop Dis. 2009;3:e550. doi: 10.1371/journal.pntd.0000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nappi AJ, Ottaviani E. Cytotoxicity and cytotoxic molecules in invertebrates. BioEssays. 2000;22:469–480. doi: 10.1002/(SICI)1521-1878(200005)22:5<469::AID-BIES9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Phillips PC. Epistasis – the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasarma T. Many faces of superoxide dismutase, originally known as erythrocuprein. Curr Sci. 2007;92:184–191. [Google Scholar]
- Richards CS, Shade PC. The genetic variation of compatibility in Biomphalaria glabrata and Schistosoma mansoni. J Parasitol. 1987;73:1146–1151. [PubMed] [Google Scholar]
- Riehle MM, Markianos K, Niaré O, Xu J, Li J, Touré AM, Podiougou B, Oduol F, Diawara S, Diallo M, Coulibaly B, Ouatara A, Kruglyak L, Traoré SF, Vernick KD. Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science. 2006;312:577–579. doi: 10.1126/science.1124153. [DOI] [PubMed] [Google Scholar]
- Shoukry N, El-Assal FM, Soliman GN, Mansour NS. Susceptibility of three successive snail generations from positive and negative laboratory bred Biomphalaria alexandrina from different localities in Egypt to infection with Schistosoma mansoni from Giza. J Egypt Soc Parasitol. 1997;27:317–329. [PubMed] [Google Scholar]
- Sinervo B, Svensson E. Correlational selection and the evolution of genomic architecture. Heredity. 2002;89:329–338. doi: 10.1038/sj.hdy.6800148. [DOI] [PubMed] [Google Scholar]
- Stevison L, Noor M. Genetic and evolutionary correlates of fine-scale recombination rate variation in Drosophila persimilis. J Mol Evol. 2010;71:332–345. doi: 10.1007/s00239-010-9388-1. [DOI] [PubMed] [Google Scholar]
- Theron A, Coustau C, Rognon A, Gourbiere S, Blouin MS. Effects of laboratory culture on compatibility between snails and schistosomes. Parasitology. 2008;135:1179–1188. doi: 10.1017/S0031182008004745. [DOI] [PubMed] [Google Scholar]
- Vianey-Liaud M, Dussart G. Aspects of pairing and reproduction in the hermaphrodite freshwater snail Biomphalaria glabrata (Gastropoda: Pulmonata) J Mollus Stud. 2002;68:243–248. [Google Scholar]
- Webster JP, Gower CM, Blair L. Do hosts and parasites coevolve? Empirical support from the Schistosoma system. Am Nat. 2004;164 (Suppl 5):S33–S51. doi: 10.1086/424607. [DOI] [PubMed] [Google Scholar]
- Webster JP, Woolhouse MEJ. Selection and strain specificity of compatibility between snail intermediate hosts and their parasitic schistosomes. Evolution. 1998;52:1627–1634. doi: 10.1111/j.1558-5646.1998.tb02243.x. [DOI] [PubMed] [Google Scholar]
- Weir BS. Inferences about linkage disequilibrium. Biometrics. 1979;35:235–254. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.