SUMMARY
Molecular recognition between cognate SNAREs leads to formation of a four-helix bundle, which facilitates vesicle docking and membrane fusion. For a SNARE system involved in trafficking in yeast, target membrane (t-) SNARE Sso1p and vesicle associated (v-) SNARE Snc2p contribute one SNARE motif each, while another t-SNARE Sec9 donates two N-terminal and C-terminal SNARE motifs SN1 and SN2 to the helical bundle. Using EPR it is found that SN2 has a tendency to be uncoiled, leaving a significant population of the SNARE complexes to be partially unstructured on the membrane. In sharp contrast, SN2 is fully engaged in the four-helix bundle when removed from the membrane, showing that the membrane is the main destabilizing factor. Helix-breaking proline mutations in SN2 did not affect the rate of docking but reduced the rate of lipid mixing significantly, indicating that SN2 plays an essential role in activating the transition from docking to fusion.
INSTRUCTION
A wide variety of cellular processes, including trafficking of newly synthesized proteins, release of neurotransmitters and hormones, and fertilization, require membrane fusion (Jahn and Südhof, 1999; Rothman, 1994). A highly conserved family of proteins termed SNAREs (soluble N-ethyl maleimide sensitive factor attachment protein receptors) is known to mediate exocytotic membrane fusion (Brunger, 2005; Jahn et al., 2003; Jahn and Scheller, 2006; Lin and Scheller, 2000; McNew et al., 2000; Sollner et al., 1993; Ungar and Hughson, 2003). All SNARE proteins have the SNARE motifs, which are about 65 – 70 amino acid-long coiled coil sequences (Fasshauer et al., 1998; Poirier et al., 1998a; Weimbs et al., 1998). The SNARE motif of the vesicle-associated (v-) SNARE engages with those of the target membrane (t-) SNAREs to form a helical bundle structure (Hanson et al., 1997; Katz et al., 1998; Lin and Scheller, 1997). The structures of SNARE complexes involved in the neurotransmitter release, endocytosis in mammalian cells, and trafficking in yeast have been determined and it was found that they all have similar four-stranded coiled coil structures (Antonin et al., 2002; Furst et al., 2003; Poirier et al., 1998b; Strop et al., 2007; Sutton et al., 1998).
Although the coiled coil structure may be shared by nearly all SNARE systems, the structures are determined by employing the soluble parts of SNAREs lacking the transmembrane domains. Interestingly, however, a previous EPR study on yeast SNAREs raised the possibility that the membrane could influence the integrity of the coiled coil structure (Zhang et al., 2005). The results showed that the C-terminal part of the SNARE motif of membrane-anchored t-SNARE has a tendency to be uncoiled in the membrane bound state.
Formation of the coiled coil may proceed in multiple steps. The ‘zipper model’ predicts that association of t- and v-SNAREs starts from the N-terminal region and propagates towards the membrane-proximal C-terminal region, facilitating the stepwise apposition of two membranes. There is indirect biochemical and biophysical evidence that supports the zipper model (Chen et al., 2001; Fiebig et al., 1999; Hua and Charlton, 1999; Melia et al., 2002; Pobbati et al., 2006; Sørensen et al., 2006; Xu et al., 1999). However, the structure of the partially zipped complex has been illusive.
A SNARE family involved in golgi-to-plasma membrane trafficking in yeast is one of the best characterized SNARE systems biochemically and biophysically (Aalto et al., 1993; Brennwald et al., 1994; Chen et al., 2004; Ferro-Novick and Jahn, 1994; Protopopov et al., 1993; Xu et al., 2005; Yoon, 2006). Sso1p and Sec9 are the heavy and light chains of t-SNARE, respectively, while Snc2p is vesicle-anchored v-SNARE. In this work, we investigated the structure of the membrane-anchored SNARE complex using EPR (Hubbell et al., 1998). We were particularly concerned with the conformation of two N- and C-terminal SNARE motifs of Sec9 SN1 and SN2. The EPR analysis revealed that SN2 has tendency to be fully uncoiled, leaving a significant fraction of SNARE complexes partially unstructured. Combining the previously published data (Zhang et al., 2005) and the data from the present work we built a model for the partially formed complex on the membrane, which may represent a folding intermediate in the pathway of SNARE assembly.
Further, to dissect the functions of SN1 and SN2 of Sec9 in SNARE assembly and membrane fusion, two helix-breaking proline mutations were introduced to SN2. The effect of proline mutations on docking and fusion was examined with fluorescence resonance energy transfer (FRET) that employs fluorescence labeled SNAREs, fluorescence lipid mixing assay, and the newly developed single vesicle fusion assay (Yoon, 2006). The combined analyses revealed that the proline mutations did not affect the rate of docking but they reduced the rate of fusion significantly, indicating that SN2 controls the activation of fusion.
In summery, the EPR analyses show that part of the SNARE complex involved in trafficking in yeast can unfold to a significant extent on the membrane, suggesting that the stability of the SNARE complex is marginally low. The results are contradictory to the notion that the SNARE complex is the fusion machine that provides the energy required to overcome the high fusion energy barrier.
RESULTS
Site-Directed Spin Labeling of the SNARE Complex on the Membrane
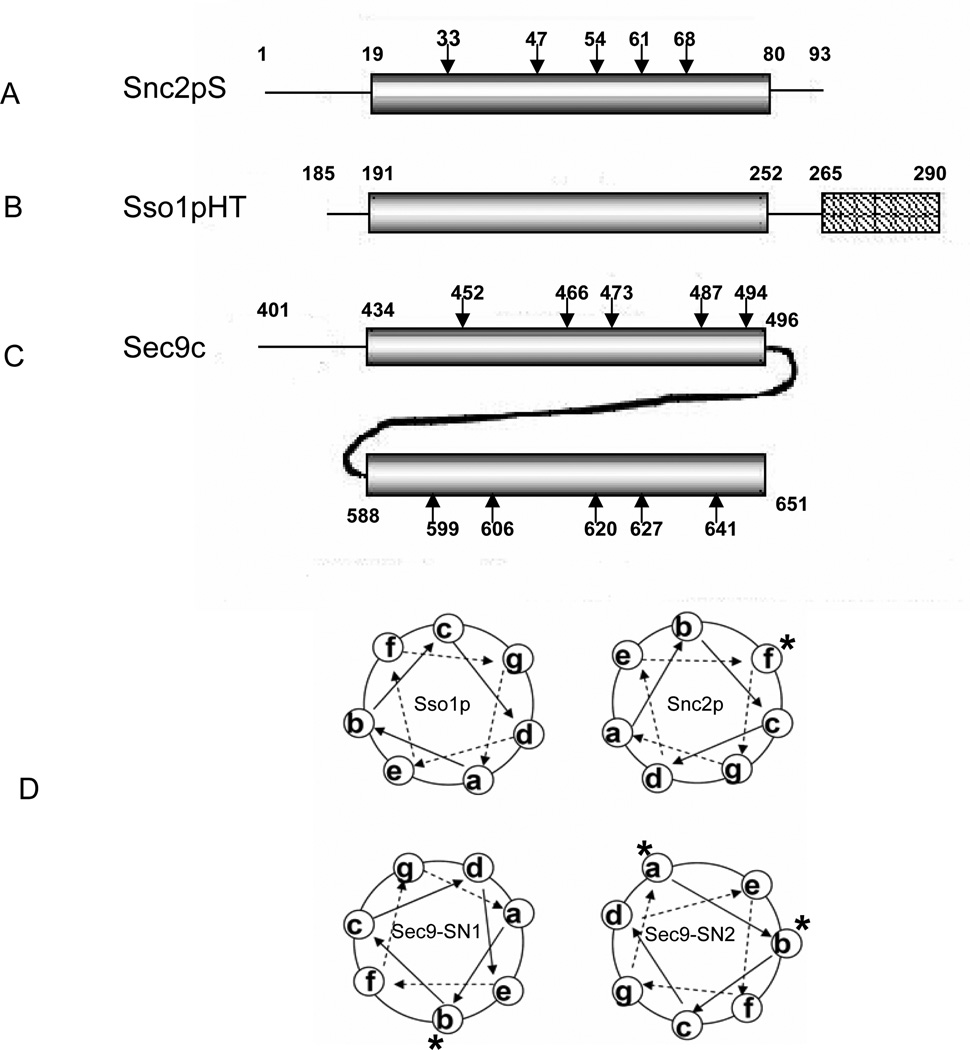
To investigate the global conformation of SN1 and SN2 of Sec9c (amino acids 401–651 of Sec9) by itself in solution as well as in the SNARE complex, we prepared ten cysteine mutants, five each in SN1 (Q452C, G466C, N473C, K487C, and A494C) and SN2 (D599C, N606, D620C, N627C and H641C) (Figure 1). The selected residues are all at the predicted ‘b’ position in the heptad repeats of the Sec9 SNARE motifs (Figure 1D). The fully exposed b positions are chosen to minimize the potential perturbations to the integrity of the SNARE complex due to the nitroxide side chain. The cysteine mutants were derivatized with methanethiosulfonate spin label (MTSSL).
Figure 1. Primary Structures of Recombinant Yeast SNAREs.
(A) Soluble Snc2pS contains amino acids 1–93 of Snc2p. Positions selected for site-directed spin labeling (SDSL) are marked by arrows.
(B) Sso1pHT contains the amino acids 185–290 of Sso1p lacking the N-terminal regulatory Habc domain. This polypeptide includes both the SNARE motif and the transmembrane domain, which are represented by the cylinder and the rectangle, respectively.
(C) Sec9c represents amino acids 401–651 of Sec9 and contains two SNARE motif region, SN1 (amino acids 434–496) and SN2 (amino acids 588–651). The amino acids selected for SDSL are marked by arrows. SNARE motif regions are represented by the cylinders.
(D) Schematic helical wheel model for the SNARE four-helix bundle. The heptad repeats a-g are shown. The labeled a, b and f positions are marked with *.
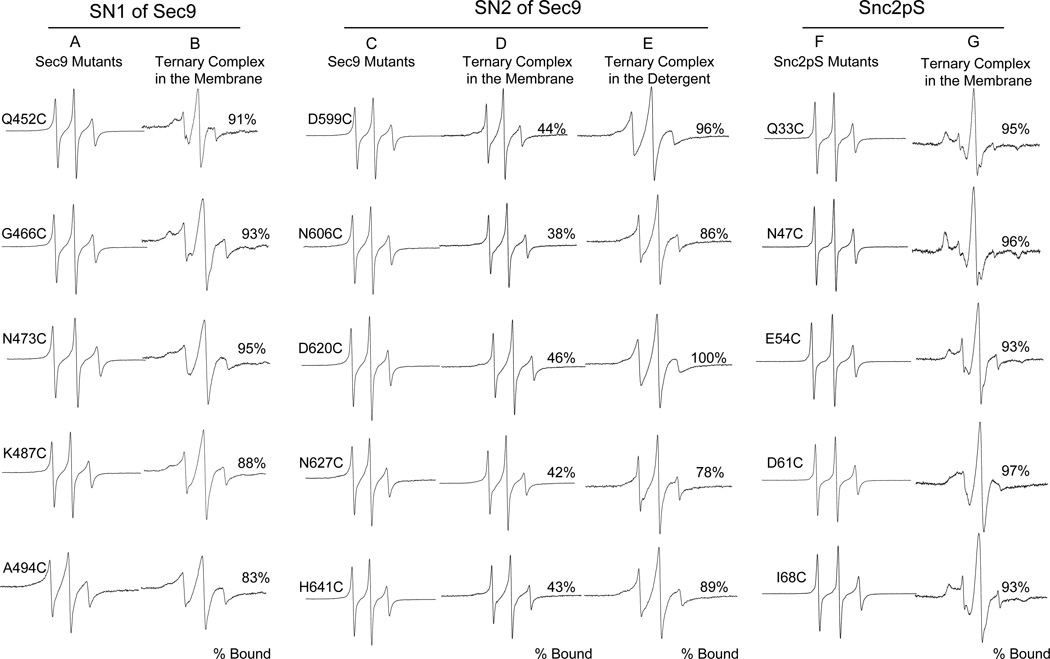
The EPR line shape is sensitive to the motional rate of the nitroxide (Kweon et al., 2003; Schneider, 1989). The sharp EPR spectra typical for the nitroxide attached to a freely diffusing random coil were observed for all ten positions for Sec9c alone in solution (Figures 2A and 2C). The results indicate that both SN1 and SN2 motifs are unstructured prior to complex formation.
Figure 2. EPR Spectra of the Spin Labeled Sec9c and Snc2pS Mutants.
The membrane-bound ternary SNARE complex is anchored to the membrane through the transmembrane domain of Sso1pHT. Soluble version of Snc2p Snc2pS was used as v-SNARE. For each mutants, the percentage of complex formation was calculated from the equation [fcomplex/(fcomplex + ffree) × 100%]. The standard spectral decomposition analysis (Thorgeirsson et al., 1996) provides the fraction of the mutant in the complex (fcomplex) and that in an unstructured form (ffree).
(A and B) EPR spectra for spin labeled SN1 of Sec9c in solution (A) and those in the membrane-bound ternary SNARE complex (B).
(C, D and E) EPR spectra for spin labeled SN2 of Sec9c in solution (C), those in the membrane-bound ternary SNARE complex (D), and in detergent-solubilized ternary SNARE complex (E).
(F and G) EPR spectra for spin labeled Snc2pS in solution (F) and those in the membrane-bound ternary SNARE complex (G).
The C-terminal SNARE Motif (SN2) of Sec9 Has Tendency to be Unstructured
In order to prepare the ternary SNARE complex, the labeled Sec9c mutants were mixed with the two-fold molar excess of soluble Snc2p (Snc2pS: amino acids 1–93) and membrane-bound Sso1pHT (amino acids 185–290 of Sso1p lacking the regulatory Habc domain) (Figure 1). Sso1pHT was reconstituted into the vesicles made of 1-palmitoyl-2-dioleoyl-sn-glycero-3-phosphatidylcholine (POPC) and 1,2-dioleoyl-sn-glycero-3-phosphatidylserine (DOPS) in molar ratio of 65:35. The EPR spectra of the SN1-labeled mutants became much broader when compared with those taken before complex formation, reflecting the retarded motion of the nitroxides due to helix formation (Figure 2B). The EPR spectra still had small sharp components, which most likely represent the uncomplexed species. The spectral subtraction analysis (Thorgeirsson et al., 1996) revealed that about 10% of SN1 are not engaged in complex formation.
For the SN2-labeled mutants, however, the sharp component appears to be much bigger, indicating that a large fraction of SN2 do not participate in complex formation (Figure 2D). The spectral subtraction analysis showed that approximately 60% of SN2 was in an unstructured conformation. The fraction unstructured for SN2 did not change at various molar ratios of Sec9c to Sso1pHT and Snc2p, suggesting that unstructured SN2 is neither due to the insufficient numbers of the binding partners, nor due to intermolecular equilibrium (Figure 1S). Therefore, the results suggest that as much as one out of two SNARE complexes has unstructured SN2, even though SN1 is fully engaged with other SNARE motifs. An alternative possibility, although less likely, is that SN2 is bound to the coiled coil full time but partially, and different parts alternates its attachment to the coiled coil.
In contrast, previous NMR and crystallographic studies from Brunger’s group has shown that all SNARE motifs from t- and v-SNAREs are fully engaged each other to form a complete four helical bundle when the recombinant SNARE motifs were used (Fiebig et al., 1999; Strop et al., 2007). To verify if destabilization of SN2 is mainly due to the presence of the membrane, we collected the EPR spectra of SNARE complexes with the SN2-labed mutants in detergent solutions (Figure 2E). As expected, we observed full participation of SN2 in four-helix bundle formation within experimental uncertainties.
To further investigate the structure of the ternary SNARE complex from yeast, we prepared five cysteine mutants (Q33C, N47C, E54C, D61C, and I68C) of soluble v-SNARE Snc2pS and the mutants were labeled with MTSSL. This time, the predicted ‘f’ positions in the SNARE motif were chosen as the labeling sites (Figures 1A and 1D). The EPR lineshapes of the mutants are all sharp, reflecting the freely diffusing random coil prior to complex formation (Figure 2F). When the spin labeled mutants were combined with membrane-bound Sso1pHT and Sec9c, the EPR lineshapes for all mutants experienced dramatic broadening reflecting the helical conformation (Figure 2G). Therefore, the results show that the SNARE motif of Snc2p is fully involved in complex formation.
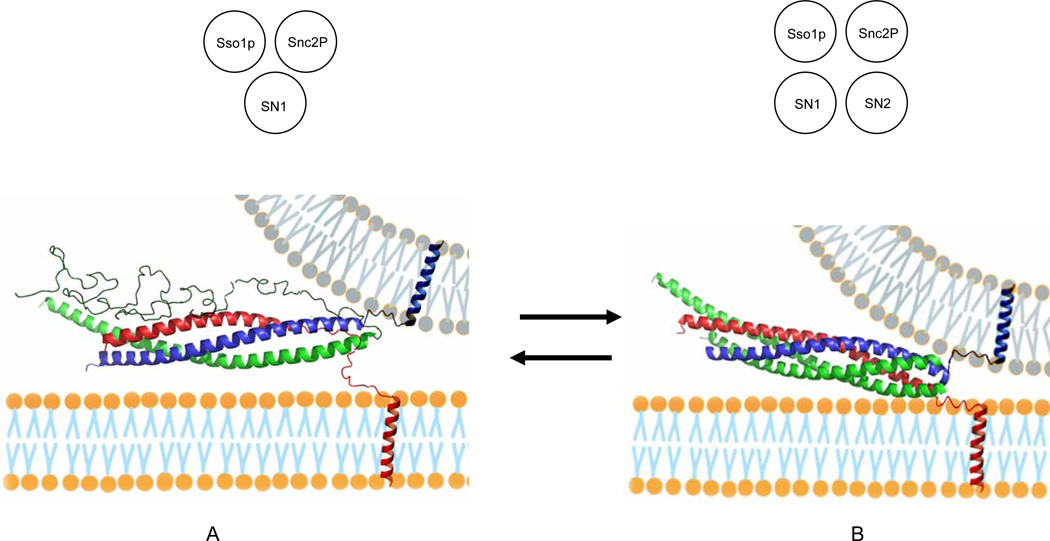
A Structural Model for the Partially Assembled SNARE Complex
The EPR results show that about 50 % of SNARE complexes have a partially frayed conformation in which SN2 of Sec9c is completely uncoiled while SN1 and Snc2p are fully structured in the coiled coil. As for the secondary structure of transmembrane t-SNARE Sso1p is concerned, a previous EPR study revealed that the C-terminal half of the SNARE motif has strong tendency to be uncoiled (Zhang et al., 2005). In fact, it was shown that about 50 % of SNARE complexes carry the partially unstructured Sso1p SNARE motif.
A structural model for the partially assembled complex is shown in Figure 3A. In this model, we speculate that Sso1p, SN1, and Snc2p form a three-stranded coiled coil in the membrane-distal N-terminal region (see the inset in Figure 3A). In the membrane-proximal C-terminal region, however, only SN1 and Snc2p intertwine to form a two-helix bundle. The extensive interaction between SN1 and Snc2p throughout the entire length appears to be quite different from that in the fully assembled four-helix bundle. In the crystal structure of the SARE core, SN1 and Snc2p are located at the opposite sides and make only limited contacts (Figure 3B). Therefore, binding of SN2 to this partial complex would involve weakening of the interaction between SN1 and Snc2p, which opens up a crevice for SN2 to fit in between. In addition, we expect that the partially formed SNARE complex is in equilibrium with the four-helix bundle with little free energy difference between two conformations.
Figure 3. Models for the Partially Assembled SNARE Complex and the Fully Assembled Four–Helix Bundle.
(A) Partially assembled complex with the C-terminal region of Sso1pHT and the entire length of SN2 of Sec9c unstructured. (Inset) a diagram for a three-stranded coiled coil composed of Sso1p, SN1 and Snc2p.
(B) Fully assembled four-helix bundle. The EPR analysis show that A and B are nearly equally populated. The SNARE proteins are color-coded: red, Sso1p; green, Sec9c; blue, Snc2p. The apposing membrane in gray is drawn arbitrarily to help speculate the trans SNARE complexes. (Inset) a diagram for a four-stranded coiled coil composed of Sso1p, SN1, SN2, and Snc2p. Both partially and fully assembled complexes were constructed based on the crystal structure (Sutton et al., 1998) with PyMOL.
SN2 of Sec9 may not be Essential for Docking
Previously, An and Almers proposed that in PC 12 cells SN2 of SNAP-25 might play a role in docking of vesicles (An and Almers, 2004). However, the structural model for the partially formed complex (Figure 3A) suggests that docking can be achieved without the involvement of SN2. To further test if SN2 is involved in docking for yeast SNAREs, we prepared two proline mutants, L626P and L647P in SN2. These are the internal ‘a’ positions of the heptad repeats and are chosen to maximally disrupt the interaction of SN2 with other SNARE motifs (Figure 1D).
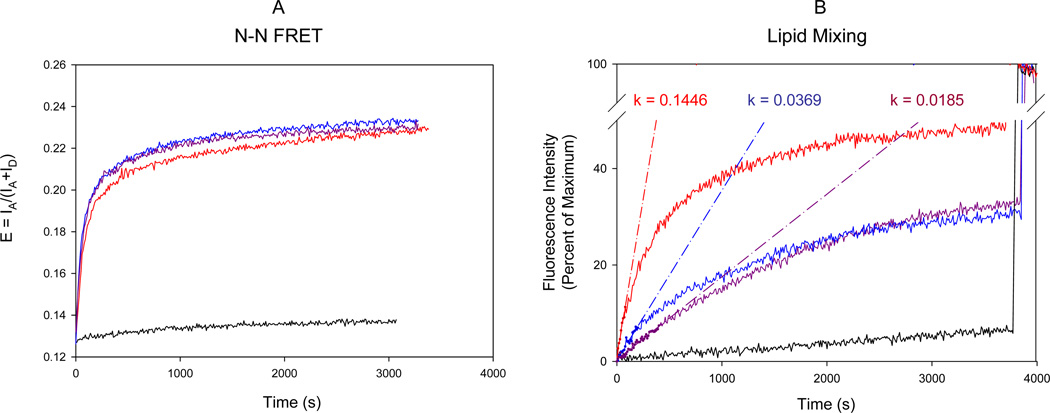
Docking was measured with fluorescence resonance energy transfer (FRET) employing dye-labeled SNAREs. The N-terminal residues, amino acid 185 of t-SNARE Sso1pHT and amino acid 13 of full length v-SNARE Snc2p (amino acids 1–115), were changed to cysteines and the cysteine mutants were labeled with fluorescence donor Cy3 and acceptor Cy5, respectively. Labeled Sso1pHT and labeled Snc2p were reconstituted into two separate populations of vesicles made of POPC and DOPS in a molar ratio of 65:35. Docking was conveniently induced by adding wild-type Sec9c or the proline mutants to the mixture of t-SNARE vesicles and v-SNARE vesicles. When docking happens, t-SNARE captures the v-SNARE and the distance between the Cy3 on Sso1pHT and Cy5 on Snc2p decrease greatly, which results in the increase of FRET. Without Sec9c, the FRET efficiency defined as IA/(IA+ID), where IA and ID are the acceptor and the donor fluorescence intensities respectively, was negligible (Figure 4A, black line). Addition of wild-type Sec9c increased E rapidly in time (Figure 4A, red line), reporting docking of v- and t-SNARE vesicles. Interestingly, when the proline mutants were used, the efficiency and the kinetics of docking were nearly identical to those of the wild-type Sec9c (Figure 4A, blue and purple lines). Therefore, the results suggest that SN2 of Sec9c is not involved in docking. We however note that this tentative conclusion must be verified further with the SN2-deletion mutant.
Figure 4. FRET Assays of SNARE Assembly and Membrane Fusion.
(A) FRET assay of trans SNARE assembly and vesicle docking. The fluorescence change was monitored in two channels with the excitation wavelength of 545 nm and the emission wavelengths of 570 and 668 nm, respectively. FRET efficiency E was defined as E = IA/(IA+ID). Little fluorescence change was observed without Sec9c (black line). The reaction was initiated by adding wild type Sec9c (red line) or its proline mutants (L626C, blue line; L647C, purple line) to the mixture of t-SNARE vesicles containing Cy3-labeled Sso1pHT and the v-SNARE vesicles carrying Cy5-labeled Snc2p. Labels were attached near the N-terminal tips.
(B) Fluorescence lipid mixing assay of SNARE-mediated membrane fusion. Fluorescence changes for lipid mixing were normalized with respect to the maximum fluorescence intensity (MFI) obtained by adding 0.1% reduced triton x-100. The black line represents the negative control in the absence of Sec9c. The red, blue and purple lines represent the fluorescence changes after adding wild-type Sec9c, L626P and L647P, respectively. The k values represent the initial rate of the fusion kinetics in units of percent per sec.
SN2 of Sec9c Plays a Role in Activating Membrane Fusion
For neuronal SNAREs involved in transmitter release, SN2 of SNAP-25, the neuronal counterpart of Sec9, is the target of botulinum neurotoxins (BoNTs) which are the major naturally-occurring inhibitors of the transmitter release (Binz et al., 1994; Blasi et al., 1993; Chen and Barbieri, 2006; Niemann et al., 1994). BoNT E cuts the N-terminal region of SN2 between residues 180 and 181, which completely abrogates exocytosis, while BoNT A removes the last 9 residues from the C-terminal end of SN2, partially inhibiting the release. For yeast SNAREs, we ask now if SN2 of Sec9 plays a similar role in activating membrane fusion. If so, we expect that the aforementioned proline mutants of Sec9c will impair membrane fusion significantly. To test this hypothesis, the fluorescence lipid mixing assay was used to measure the fusion activity of the proline mutants (McNew et al., 2000; Weber et al., 1998; Xu et al., 2005). This time, we incorporated 1.5 mole % each of fluorescent lipids, NBD-PS and rhodamine-PE in the v-SNARE vesicles. In the lipid mixing assay, membrane fusion results in the recovery of NBD fluorescence due to the increase of the average distance between the fluorescence donor and the acceptor.
Little lipid mixing was observed in the absence of Sec9c, confirming that Sec9 is absolutely required for membrane fusion (Figure 4B, black line). In the presence of the wild type Sec9c, lipid mixing reached 40% of the maximum fluorescence intensity after one hour of the fusion reaction. However, with the two proline mutants, the fusion activity was much reduced. Quantitatively, the initial rates of lipid mixing for L626P and L647P were only one fourth and one eighth of that for wild-type Sec9c, respectively (Figure 4B, blue and purple lines). Therefore, the results support that the SN2 of Sec9 play a key role in activating membrane fusion.
Single Vesicle Fusion Assay Supports that SN2 of Sec9 is Essential for the Transition from Docking to Fusion
The recently developed single vesicle fusion assay was used to further investigate the function of Sec9 SN2 (Yoon, 2006). The v-SNARE vesicles were made by reconstituting Snc2p into the vesicles that was doped with the lipid fluorescence acceptor 1,1’-dioctadecyl-3,3,3’,3’,-tertramethyl-indodicarbo-cyanine perchlorate (DiD, 2 mol%) and the biotinylated lipid (0.1 mol%). The v-SNARE vesicles were immobilized on a PEG-coated quartz surface in a flow cell through the biotin-neutravidin conjugation. On the other hand, t-SNARE Sso1pHT was reconstituted into a separate population of vesicles that contained the lipid fluorescence donor 1,1’-dioctadecyl-3,3,3’,3’,-tertramethylindocarbocyanine perchlorate (DiI, 2 mol%). Sec9c was added separately to the Sso1pHT vesicles to make the t-SNARE vesicles. The t-SNARE vesicles were flown into the flow cell at 25 pM concentration at 37 °C. The low concentration of t-SNARE vesicles was intended to minimize the interaction of multiple t-SNARE vesicles with single v-SNARE vesicles.
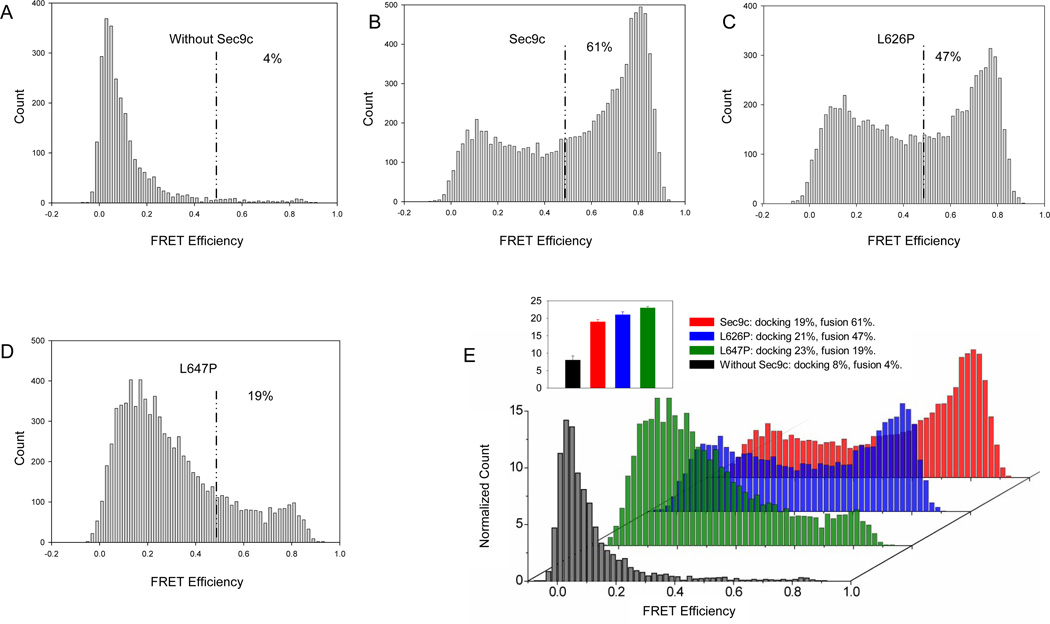
Docking of a t-SNARE vesicle to a surface tethered v-SNARE vesicle can be clearly distinguished from subsequent fusion and other intermediate steps by observing an abrupt increase of the donor fluorescence intensity, ID (Yoon, 2006). After 20 min incubation, unbound t-SNARE vesicles were removed by flow washing. The docking probability was obtained by dividing the total number of the t-SNARE vesicles by the average number of the v-SNARE vesicles in an imaging area. The docking probability for the wild-type Sec9c was 19%, while those of two proline mutants are 21% and 23 %, respectively (Figure 5E). Therefore, the results again show that SN2 of Sec9 may not be essential in vesicle docking.
Figure 5. Single Vesicle Assay of SNARE –Mediated Docking and Membrane Fusion.
Single vesicle assay was performed between surface tethered v-SNARE vesicle (acceptor dye) and non-tethered t-SNARE vesicle (donor dye) with Sec9c and its proline mutants. Histograms represent the distribution of FRET signals from v- and t-SNARE vesicle mixing (after 20 min incubation). The vesicle populations showing FRET efficiency below 0.5 and above 0.5 were assigned to be unfused and fused population, respectively (Yoon et al., 2006). The ratio between Sec9c (or praline mutant) to Sso1pHT was 1:1. These histograms were built by combining data from 26–44 imaging locations and by analyzing 2507–9390 vesicles.
(A to D) The distribution of the FRET efficiency in the absence of Sec9c (A), in the presence of Sec9c (B), L626P (C) and L647P (D).
(E) 3D overlay of histograms normalized to count per imaging area allows direct comparison and clearly shows the decrease of fused population as a result of helix disruption of SN2.
(Inset) the bar graph representing the docking probabilities for the wild-type and the proline mutants of Sec9c measured at t = 20 min.
Lipid mixing due to fusion leads to an increase in FRET efficiency, E. For our convenience, we analyzed the FRET distribution at 20 min after the start of the fusion reaction. In the absence of the Sec9c, we observed an E distribution which was centered at ~ 0.1 and mostly less than 0.25. The low E value suggested that some v-and t-SNARE vesicle pairs make close contact without lipid mixing (Figure 5A). In contrast, when Sec9c was included (Sec9c/Sso1pHT = 1:1), approximately 61% of vesicle pairs showed the E values greater than 0.5 (Figure 5B), indicating that extensive membrane fusion occurred. For the proline mutant L647P, the population of E above 0.5 was reduced to 19% (Figure 5D), showing that the proline mutation made significant impact on the fusion activity of SNAREs. For L626P (Figure 5D), the disturbing effect of the proline mutation was somewhat milder than that of L647P. The results might suggest that C-terminal region of SN2 is more critical for fusion than the N-terminal region, which is consistent with the results of bulk fusion assay in which L647P was twice more disruptive than L627P (Figure 4B). Therefore, again, single vesicle fusion assay suggests that SN2 of Sec9 may not be essential for docking but important for fusion.
DISCUSSION
For yeast SNAREs, the EPR analysis revealed that a partially assembled complex coexists with the complete four helix bundle on the membrane. A striking feature of the partial complex is that entire SN2 is unstructured and moves freely in solution. The EPR analysis revealed that the destabilization of SN2 is mainly due to the presence of the membrane. Moreover, it is likely that the C-terminal half of the Sso1p SNARE motif is unstructured, too (Zhang et al., 2005). Our previous EPR results showed that the membrane destabilizes the complex and causes the C-terminal region of Sso1p to dissociate from the rest of the helical bundle (Zhang et al., 2005).
How can the membrane disrupt the SNARE complex structurally? The membrane proximal region of t-SNAREs is rich in positively charged amino acids. It was previously shown that the basic region can insert into the membrane near the head group/acyl chain interface as a random coil (Kim et al., 2002; Kweon et al., 2002). In this conformation, the positive charges could snorkel out and the hydrophobic residues could insert into the acyl chain region to maximize the binding energy. We expect that membrane binding would compete with coiled coil formation and such competitive membrane binding would force the four-helix bundle to partially unfold and refold (Figure 3). The stability of the yeast SNARE complex is intrinsically low with a water filled cavity in the core region (Munson et al., 2000; Strop et al., 2007). Therefore, it would not take much energy to disturb such a marginally stable structure.
As the partial complex is in equilibrium with the full four-helix bundle, it is reasonable to assume that the partial complex observed in this work (Figure 3A) may represent a folding intermediate in the SNARE assembly pathway. The structural model for the folding intermediate based on EPR is somewhat different from the hypothetical “partially zipped complex” that was previously envisioned with the limited experimental data (Hua and Charlton, 1999; Melia et al., 2002; Sørensen et al., 2006). The zipper model predicted that this intermediate state has the intact four helix bundle in the N-terminal half, but has the fully unstructured C-terminal half.
Two principal interactions hold the partial complex together. Firstly, the N-terminal half of the Sso1p SNARE motif associates with SN1 of Sec9 and the Snc2p SNARE motif to form the three-helix bundle in the N-terminal region. Secondly, SN1 of Sec9 engages with Snc2p through the entire length of the SNARE motifs to maintain the two-stranded coiled coil in the C-terminal half. Therefore, it appears that SN1 of Sec9 plays a principal role in binding to v-SNARE Snc2p. For neuronal SNAREs, An and Almers showed that in PC12 cells SN2 of SNAP-25 in the binary complex of t-SNAREs Syntaxin 1A and SNAP-25 remains unstructured (An and Almers, 2004). They hypothesized that unstructured SN2 play a role in capturing v-SNARE VAMP2. In contrast, however, our results suggest that SN2 may not be essential for binding to v-SNARE, although this hypothesis must be tested further with the SN2-deletion mutant.
The results show SN2 of Sec9 plays essential role in activating membrane fusion. The helix-breaking proline mutants in this domain greatly reduced the fusion activity, indicating that SN2 must be incorporated into the SNARE core to drive membrane fusion. Why is the SN2 helix required for fusion, even though it is not membrane anchored? The key structural element that draws our attention is the C-terminal region of Sso1p that is predicted to be unstructured as well in the intermediate complex (Figure 3A). If this region remains to be unstructured, it will confer too loose an attachment of one membrane to another for fusion to occur. Therefore, we speculate that helix formation of SN2 must be coupled to the conformational change of the C-terminal region of Sso1p. We envision the incorporation of SN2 into the SNARE core induces helix formation of the C-terminal region of Sso1p, which allows formation of complete four helix bundle. Formation of the complete four-helix bundle would drive close apposition of two membranes, facilitating the fusion. In cells, binding of a SM protein Sec1p to the SNARE core would stabilize the four-helix bundle (Togneri et al., 2006), which would facilitate membrane fusion. This new mechanistic model for SNARE assembly and membrane fusion is not in conflict of the previous zipper model (Sørensen et al., 2006) in its essence, but provides a molecular picture detailing the conformational changes of the SNARE complex in the process.
The most striking feature of the SNARE complex is the four stranded coiled coil that may be conserved throughout all SNARE systems (Fasshauer et al., 1998; Skehel and Wiley, 1998; Weimbs et al., 1997). The SNARE coiled coil is thought to be the central part of the fusion machinery that provides the necessary energy to overcome the fusion energy barrier (Jahn and Scheller, 2006; Sutton et al., 1998; Ungar and Hughson, 2003). Our EPR analysis however shows that the four helix bundle is not stable and can be easily converted to the partially structured complex. Therefore, the results cast doubts on the generality of the idea that the SNARE core is the fusion machine releasing the energy required to overcome the fusion energy barrier. Alternatively, it is possible that the SNARE complex play a setup role in arranging the lipids in the right geometry for hemifusion and pore formation to occur (Jun et al., 2007). Although speculative, the fusion energy barrier might not be as high as what has been predicted from the lipid-stalk model in which the protein components are not considered. We expect that the presence of several SNARE transmembrane domains (TMDs) in the active zone will reduce the fusion energy significantly. The SNARE TMDs are enriched with β-branching amino acids, which will help frustrate lipid molecules to be susceptible for remodeling.
In summary, our structural EPR and the fluorescence results on SNARE-mediated membrane fusion shed a light on the mechanism by which Sec9 controls vesicle docking and fusion. Vesicle docking could occur without the intimate involvement of SN2 of Sec9. However, SN2 is found to be essential in activating membrane fusion. Perhaps, the conformational change of SN2 is required to complete the zipping of the C-terminal region of Sso1p SNARE motif. Therefore, the SN1 and SN2 of Sec9 appear to work sequentially to activate the vesicle docking and fusion.
EXPERIMENTAL PROCEDURES
Protein Sample Preparation
Plasmid construction, mutagenesis, protein expression, purification and spin labeling for yeast SNAREs were described in detail elsewhere (Chen et al., 2004). In brief, Sso1pHT (amino acids 185–290), for which the N-terminal α-helical Habc domain was deleted, soluble v-SNARE Snc2pS (amino acids 1–93 of Snc2p), and full-length Snc2p (amino acids 1–115) were expressed as the N-terminal glutathione S-transferase (GST) fusion proteins. Sec9c (amino acids 401–651 of Sec9) was expressed as a C-terminal His6-tagged protein. The QuickChange site-directed mutagenesis kit (Stratagene) was used to produce the cysteine as well as the proline mutants. The DNA sequences were confirmed by the Iowa State University DNA Sequencing Facility.
Recombinant proteins were expressed in Escherichia coli.Rosetta (DE3) pLysS (Novagen). His6-tagged Sec9c was purified using the Ni-NTA agarose beads (Qiagen). The beads were washed with washing buffer (Hepes buffer with 20 mM immidazole, pH 7.4), then the protein was eluted out by elution buffer (Hepes buffer with 150 mM immidazole, pH 7.4). The glutathione-agarose beads (Sigma) was used to purify Sso1pHT , full-length Snc2p and Snc2pS. The protein-bound GST beads was washed excessively with washing buffer (PBS, pH7.4) and then protein was cleaved by thrombin in cleavage buffer (50 mM Tris-HCL, 150mM NaCl, pH 8.0). We added 0.8% n-Octylglucoside (OG) in the cleavage buffer for Sso1pHT and full length Snc2p. The purified proteins were examined by SDS-PAGE, and the purity was at least 90% for all SNARE proteins.
Cysteine mutants for Sec9c and Snc2pS were spin labeled while the protein was still bound to the beads. After the protein-bound beads were washed with excessive washing buffer, approximately 20 fold excess of (1-oxyl-2,2,5,5 tetramethylpyrrolinyl-3 -methyl) methanethiosulfonate spin label (MTSSL) was added to the column, and the reaction mixture was left overnight at 4 °C. Labeling Sso1pHT and Snc2p cysteine mutants with the fluorescence labels was carried out after the thrombin cleavage. Sso1pHT E185C and Snc2p P13C were labeled with Cy3 and Cy5 maleimide (Amersham), respectively. The free dye was removed using the PD-10 desalting columns (Amersham).
Membrane Reconstitution and Complex Formation
The mixture of POPC (1-palmitoyl-2-dioleoyl-sn-glycero -3-phosphatidylcholine) and DOPS (1,2-dioleoyl-sn-glycero-3-phosphatidylserine) (molar ratio of 65:35) in chloroform was dried in a vacuum and was resuspended in a buffer (25mM HEPES/KOH, 100 mM KCl, pH 7.4) to make the total lipid concentration about 50 mM. Protein-free large unilamellar vesicles (~100 nm in diameter) were prepared by extrusion through polycarbonate filters (Avanti Polar Lipids). For the lipid mixing assay, the vesicles containing POPC, DOPS, NBD-PS (1,2-dioleoyl-sn-glycero-3-phosphoserine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl)), and rhodamine-PE (1,2-dioleoyl-snglycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl)) in the molar ratio of 62:35:1.5:1.5 were prepared following the procedure described above, and the final lipid concentration was ~10 mM. For the single-lipid mixing assay, the POPC/DOPS (65/35) vesicles were made by the same extrusion method except that the t-SNARE vesicles contain 2 mol% DiI while the v-SNARE vesicles contain 2 mol% DiD and 0.1 mol% biotinylated lipid.
SsoHT were mixed with vesicles at a lipid/protein molar ratio of 100 at 4 °C for 20 min. The protein/lipid mixture was diluted two times to make the concentration of OG below the critical micelle concentration. The mixture was then dialyzed overnight against dialysis buffer (25 mM HEPES/KOH, 100 KCl, 5% (w/v) glycerin, pH 7.4) at 4 °C.
For complex formation, Sec9c was mixed with reconstituted SsoHT and Snc2pS in a molar ratio of 1:2:2. The mixture was incubated at room temperature for 60 min. The vesicle solution was concentrated using a 100-kDa cutoff centrifugal filter (Millipore) before the EPR measurement. The final protein concentration for EPR was approximately 50µM.
EPR Data Collection
The EPR spectra were obtained using a Bruker ESP 300 spectrometer (Bruker, Germany) equipped with a low noise microwave amplifier (Miteq, Hauppauge, New York) and a loop-gap resonator (Medical Advances, Milwaukee, Wisconsin). The modulation amplitude was set to be no greater than one-fourth of the line width. The spectra were collected at room temperature in first-derivative mode with 1 mM microwave power.
FRET Assay of SNARE Assembly
Fluorescence labeled Sso1pHT and Snc2p were reconstituted into two separate populations of POPC/DOPS vesicles in a lipid/protein ratio 100:1, respectively. Reconstituted SsoHT, Sec9c and reconstituted Snc2p were mixed at a molar ratio 1:1:1. The total lipid concentration in the reaction is 0.4 mM. Fluorescence intensity was monitored in two channels with the excitation wavelength of 545 nm and emission wavelengths of 570 and 668 nm respectively. Fluorescence changes were recorded with a Varian Cary Eclipse model fluorescence spectrophotometer using a quartz cell of 100 µL with a 2 mm path length. All measurements were carried out at 35°C.
Fluorescence Lipid Mixing Assay
Sso1pHT was reconstituted into the POPC/DOPS vesicles while Snc2p was reconstituted into a separate population of POPC/DOPS vesicles doped with 1.5 mol% each NBD-PE and rhodamin-PS. The lipid to protein molar ratio was 100:1. The fusion reaction was conveniently initiated by adding Sec9c to the mixture of two vesicle populations. The final reaction solution contained ~0.5 mM lipids. Fluorescence was measured at the excitation and emission wavelengths of 465 and 530 nm, respectively. The fluorescence changes were recorded with the same instrument under the same conditions described above. The maximum fluorescence intensity (MFI) was obtained by adding 1% (v/v) reduced triton x-100 (Sigma).
Single Vesicle Fusion Assay
The single vesicle fusion assay was performed as previously described(Yoon, 2006). Briefly, the Snc2p–reconstituted vesicles were immobilized on a PEG-coated quartz surface through the biotin-neutravidin conjugation. The Sso1pHT vesicles were mixed with Sec9c in a molar ratio of 1:1, diluted to a final vesicle concentration of 25 pM to make the t-SNARE vesicles. The t-SNARE vesicles were flown into the flow cell to induce fusion on the surface. The flow cell was incubated at 37 °C for 20 min prior to data acquisition. The single fusion events were monitored in a prism-type total-internal reflection fluorescence microscope (IX70, Olympus) by using an electron multiplying charge-coupled device camera (iXon DV 887-BI, Andor Technology). Fluorescence data was acquired with custom Visual C++ (Microsoft) routines and donor and corresponding acceptor intensities were obtained through running IDL (Research Systems, Boulder, CO) programs.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the grants from National Institute of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aalto MK, Ronne H, Keranen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. The EMBO journal. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An SJ, Almers W. Tracking SNARE complex formation in live endocrine cells. Science. 2004;306:1042–1046. doi: 10.1126/science.1102559. [DOI] [PubMed] [Google Scholar]
- Antonin W, Fasshauer D, Becker S, Jahn R, Schneider TR. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nature structural biology. 2002;9:107–111. doi: 10.1038/nsb746. [DOI] [PubMed] [Google Scholar]
- Binz T, Blasi J, Yamasaki S, Baumeister A, Link E, Südhof TC, Jahn R, Niemann H. Proteolysis of SNAP-25 by types E and A botulinal neurotoxins. The Journal of biological chemistry. 1994;269:1617–1620. [PubMed] [Google Scholar]
- Blasi J, Chapman ER, Link E, Binz T, Yamasaki S, De Camilli P, Südhof TC, Niemann H, Jahn R. Botulinum neurotoxin A selectively cleaves the synaptic protein SNAP-25. Nature. 1993;365:160–163. doi: 10.1038/365160a0. [DOI] [PubMed] [Google Scholar]
- Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- Brunger AT. Structure and function of SNARE and SNARE-interacting proteins. Quarterly reviews of biophysics. 2005;38:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- Chen S, Barbieri JT. Unique substrate recognition by botulinum neurotoxins serotypes A and E. The Journal of biological chemistry. 2006;281:10906–10911. doi: 10.1074/jbc.M513032200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Xu Y, Zhang F, Shin YK. Constitutive versus regulated SNARE assembly: a structural basis. The EMBO journal. 2004;23:681–689. doi: 10.1038/sj.emboj.7600083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Scales SJ, Scheller RH. Sequential SNARE assembly underlies priming and triggering of exocytosis. Neuron. 2001;30:161–170. doi: 10.1016/s0896-6273(01)00270-7. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro-Novick S, Jahn R. Vesicle fusion from yeast to man. Nature. 1994;370:191–193. doi: 10.1038/370191a0. [DOI] [PubMed] [Google Scholar]
- Fiebig KM, Rice LM, Pollock E, Brunger AT. Folding intermediates of SNARE complex assembly. Nature structural biology. 1999;6:117–123. doi: 10.1038/5803. [DOI] [PubMed] [Google Scholar]
- Furst J, Sutton RB, Chen J, Brunger AT, Grigorieff N. Electron cryomicroscopy structure of N-ethyl maleimide sensitive factor at 11 A resolution. The EMBO journal. 2003;22:4365–4374. doi: 10.1093/emboj/cdg420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- Hua SY, Charlton MP. Activity-dependent changes in partial VAMP complexes during neurotransmitter release. Nature neuroscience. 1999;2:1078–1083. doi: 10.1038/16005. [DOI] [PubMed] [Google Scholar]
- Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- Jahn R, Lang T, Südhof TC. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- Jahn R, Scheller RH. SNAREs - engines for membrane fusion. Nature reviews. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Jahn R, Südhof TC. Membrane fusion and exocytosis. Annual review of biochemistry. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- Jun Y, Xu H, Thorngren N, Wickner W. Sec18p and Vam7p remodel trans-SNARE complexes to permit a lipid-anchored R-SNARE to support yeast vacuole fusion. The EMBO journal. 2007;26:4935–4945. doi: 10.1038/sj.emboj.7601915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L, Hanson PI, Heuser JE, Brennwald P. Genetic and morphological analyses reveal a critical interaction between the C-termini of two SNARE proteins and a parallel four helical arrangement for the exocytic SNARE complex. The EMBO journal. 1998;17:6200–6209. doi: 10.1093/emboj/17.21.6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CS, Kweon DH, Shin YK. Membrane topologies of neuronal SNARE folding intermediates. Biochemistry. 2002;41:10928–10933. doi: 10.1021/bi026266v. [DOI] [PubMed] [Google Scholar]
- Kweon DH, Kim CS, Shin YK. The membrane-dipped neuronal SNARE complex: a site-directed spin labeling electron paramagnetic resonance study. Biochemistry. 2002;41:9264–9268. doi: 10.1021/bi025934+. [DOI] [PubMed] [Google Scholar]
- Kweon DH, Kim CS, Shin YK. Regulation of neuronal SNARE assembly by the membrane. Nature structural biology. 2003;10:440–447. doi: 10.1038/nsb928. [DOI] [PubMed] [Google Scholar]
- Lin RC, Scheller RH. Structural organization of the synaptic exocytosis core complex. Neuron. 1997;19:1087–1094. doi: 10.1016/s0896-6273(00)80399-2. [DOI] [PubMed] [Google Scholar]
- Lin RC, Scheller RH. Mechanisms of synaptic vesicle exocytosis. Annual review of cell and developmental biology. 2000;16:19–49. doi: 10.1146/annurev.cellbio.16.1.19. [DOI] [PubMed] [Google Scholar]
- McNew JA, Parlati F, Fukuda R, Johnston RJ, Paz K, Paumet F, Sollner TH, Rothman JE. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407:153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- Melia TJ, Weber T, McNew JA, Fisher LE, Johnston RJ, Parlati F, Mahal LK, Sollner TH, Rothman JE. Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. The Journal of cell biology. 2002;158:929–940. doi: 10.1083/jcb.200112081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson M, Chen X, Cocina AE, Schultz SM, Hughson FM. Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly. Nature structural biology. 2000;7:894–902. doi: 10.1038/79659. [DOI] [PubMed] [Google Scholar]
- Niemann H, Blasi J, Jahn R. Clostridial neurotoxins: new tools for dissecting exocytosis. Trends in cell biology. 1994;4:179–185. doi: 10.1016/0962-8924(94)90203-8. [DOI] [PubMed] [Google Scholar]
- Pobbati AV, Stein A, Fasshauer D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313:673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- Poirier MA, Hao JC, Malkus PN, Chan C, Moore MF, King DS, Bennett MK. Protease resistance of syntaxin.SNAP-25.VAMP complexes. Implications for assembly and structure. The Journal of biological chemistry. 1998a;273:11370–11377. doi: 10.1074/jbc.273.18.11370. [DOI] [PubMed] [Google Scholar]
- Poirier MA, Xiao W, Macosko JC, Chan C, Shin YK, Bennett MK. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nature structural biology. 1998b;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- Protopopov V, Govindan B, Novick P, Gerst JE. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell. 1993;74:855–861. doi: 10.1016/0092-8674(93)90465-3. [DOI] [PubMed] [Google Scholar]
- Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Schneider DJ, Freed JH. Lasers, Molecules and Methods. 1989 [Google Scholar]
- Skehel JJ, Wiley DC. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Sørensen JB, Wiederhold K, Müller EM, Milosevic I, Nagy G, de Groot BL, Grubmuller H, Fasshauer D. Sequential N- to C-terminal SNARE complex assembly drives priming and fusion of secretory vesicles. The EMBO journal. 2006;25:955–966. doi: 10.1038/sj.emboj.7601003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strop P, Kaiser SE, Vrljic M, Brunger AT. The structure of the yeast plasma membrane SNARE complex reveals destabilizing water filled cavities. The Journal of biological chemistry. 2007 doi: 10.1074/jbc.M707912200. [DOI] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson TE, Russell CJ, King DS, Shin YK. Direct determination of the membrane affinities of individual amino acids. Biochemistry. 1996;35:1803–1809. doi: 10.1021/bi952300c. [DOI] [PubMed] [Google Scholar]
- Togneri J, Cheng YS, Munson M, Hughson FM, Carr CM. Specific SNARE complex binding mode of the Sec1/Munc-18 protein, Sec1p. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17730–17735. doi: 10.1073/pnas.0605448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar D, Hughson FM. SNARE protein structure and function. Annu Rev Cell Dev Biol. 2003;19:493–517. doi: 10.1146/annurev.cellbio.19.110701.155609. [DOI] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Weimbs T, Low SH, Chapin SJ, Mostov KE, Bucher P, Hofmann K. A conserved domain is present in different families of vesicular fusion proteins: a new superfamily. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimbs T, Mostov K, Low SH, Hofmann K. A model for structural similarity between different SNARE complexes based on sequence relationships. Trends in cell biology. 1998;8:260–262. doi: 10.1016/s0962-8924(98)01285-9. [DOI] [PubMed] [Google Scholar]
- Xu T, Rammner B, Margittai M, Artalejo AR, Neher E, Jahn R. Inhibition of SNARE complex assembly differentially affects kinetic components of exocytosis. Cell. 1999;99:713–722. doi: 10.1016/s0092-8674(00)81669-4. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhang F, Su Z, McNew JA, Shin YK. Hemifusion in SNARE-mediated membrane fusion. Nature structural & molecular biology. 2005;12:417–422. doi: 10.1038/nsmb921. [DOI] [PubMed] [Google Scholar]
- Yoon TY, Okumus B, Zhang F, Shin YK, Ha T. Multiple intermediates in single SNARE-induced membrane fusion. Proceedings of the National Academy of Sciences of the United States of America. 2006 doi: 10.1073/pnas.0606032103. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su Z, Zhang F, Chen Y, Shin YK. A partially zipped SNARE complex stabilized by the membrane. The Journal of biological chemistry. 2005;280:15595–15600. doi: 10.1074/jbc.M500736200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.