Abstract
Microorganisms and plants produce a large diversity of secondary metabolites, whereas analyses of metazoan metabolomes have yielded comparatively few types of small molecules. We show that the nematode Pristionchus pacificus constructs elaborate molecular architectures from modified building blocks of primary metabolism, including an unusual xylopyranose-based nucleoside. These compounds act as signaling molecules controlling adult phenotypic plasticity and development and provide striking examples for modular generation of structural diversity in metazoans.
Keywords: metabolomics, model organisms, natural products, pheromones, small-molecule signaling
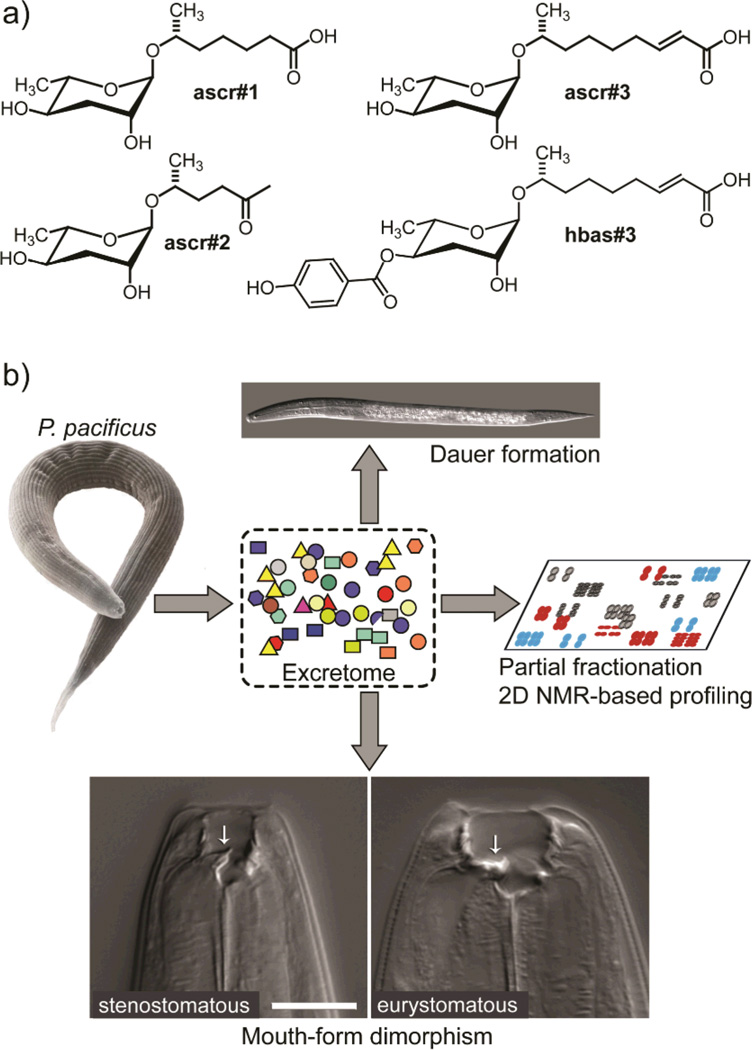
Small molecules serve a wide variety of essential biological functions.[1] They facilitate inter- and intra-specific chemical communication, are used for chemical defense and predation, function as hormones and second messengers in animals and plants, and serve as building blocks for biological macromolecules. In contrast to many groups of microorganisms and plants, whose genomes have revealed a great variety of "secondary" small-molecule biosynthetic pathways, e.g. for polyketides and non-ribosomal peptides,[2] most animals are not known to have dedicated biosynthetic machinery to generate structurally complex small molecules. Correspondingly, many unusual metabolites isolated from basal animals (e.g. sponges, bryozoans, etc.) and arthropods have turned out to be of microbial origin or diet-derived.[3] Recent studies of the model organism Caenorhabditis elegans showed that this nematode produces a family of small-molecule signals, the ascarosides, e.g. ascr#1–3, and hbas#3 (Figure 1a), which control multiple aspects of C. elegans life history, including larval development, mating, and social behaviors.[4] The structures of these signaling molecules derive from combination of the dideoxysugar ascarylose with a variety of lipid- and amino acid-metabolism derived moieties,[4e] suggesting that nematodes, and perhaps other animals, harbor yet unrecognized biosynthetic capabilities.
Figure 1.
a) Structures of previously identified small-molecule signals regulating development and behavior in C. elegans. b) P. pacificus exo-metabolome samples induce dauer arrest and affect mouth-form dimorphism, promoting eurystomatous mouth development. We here identify the active components via activity guided fractionation and 2D NMR-spectroscopic profiling.
As part of a broad 2D NMR-spectroscopic screen of nematode metabolomes, we analyzed the exo-metabolome (the entirety of all secreted and excreted metabolites) of the necromenic roundworm Pristionchus pacificus. Like C. elegans, P. pacificus is a free-living nematode that has been established as a model organism for the study of developmental and evolutionary biology.[5] P. pacificus forms a necromenic association with beetles, which may represent a pre-adaptation to the evolution of true parasitism.[6] P. pacificus exhibits two types of phenotypic plasticity that are key to its survival in the wild. Like in many other nematode species, harsh environmental conditions, for example food shortage, trigger developmental arrest as dauer worms, a highly stress-resistant alternate larval stage.[7] P. pacificus further exhibits a unique dimorphism in mouth development, representing an example for phenotypic plasticity of morphology in an adult metazoan (Figure 1b). Adult worms can have either a narrow (“stenostomatous”) or a wide and more complex (“eurystomatous”) mouth opening, the latter developing in response to conditions of low food availability. The two different mouth forms are associated with different feeding habits: stenostomatous worms are considered to feed primarily on bacteria, whereas the eurystomatous form is suited for predatory behavior toward other nematodes. Previous studies of mixed-stage P. pacificus cultures showed that both dauer formation and mouth dimorphism are regulated by population density,[8] suggesting that these two phenotypes are driven by inter-organismal small-molecule signaling as part of a population density monitoring system (Figure 1b).[9]
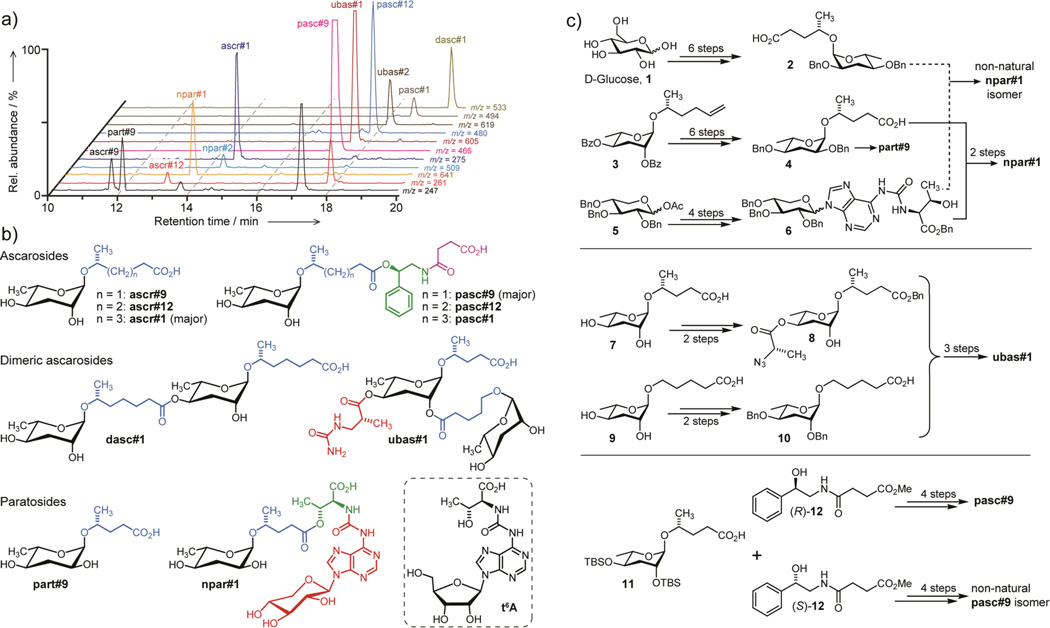
2D NMR-based metabolomics of P. pacificus exo-metabolome fractions with dauer-inducing activity (Supporting Information, Section 1.4) revealed a striking diversity of signals suggestive of primary metabolites, for example xylose, threonine, adenosine, short-chain fatty acids, succinate, phenylethanolamine, and several dideoxysugar derivatives (Supporting Information, Figure S1); however, their NMR data indicated that most of these moieties are part of larger assemblies. The dideoxysugar derivatives fell into two chemically different subgroups. One group was based on ascarylose, a sugar that is widely produced by nematodes,[10] whereas a second group of compounds appeared to include a related sugar, paratose, which previously had been reported only from bacteria (Figure S1).[11] Analyzing the P. pacificus exo-metabolome by HPLC-MS/MS for the presence of any of the ~150 ascarosides recently identified from C. elegans,[4e] we detected ascr#1, ascr#9, and ascr#12, as previously reported (Figure 2a).[10] However, these three compounds accounted only for a small portion of the structures detected by 2D NMR spectroscopy.
Figure 2.
a) High-resolution HPLC-MS analysis reveals species whose molecular formulae provide additional constraints for structures proposed based on NMR-spectroscopic analysis of the P. pacificus exo-metabolome. b) Major components of the P. pacificus exo-metabolome derived from assembly of building blocks from carbohydrate (black), amino acid (green), and nucleoside (red) metabolism, as well as likely TCA cycle-derived succinate (magenta). Also shown is the related tRNA nucleoside, N6-threonylcarbamoyladenosine (t6A). c) Abbreviated synthesis scheme for identified P. pacificus metabolites and several non-natural stereoisomers (see Supporting Information for details)
Combining results from NMR and high resolution HPLC-MS/MS analyses (Figures 2a, S2, Table S1), we proposed structures for the major unknown components of the P. pacificus exo-metabolome, in some cases after additional fractionation to increase the content of minor components (Figure 2b). The most abundant ascaroside derivative, named pasc#9 (Supporting Information, Section 1.1 and www.smid-db.org for nomenclature) was proposed as an N-succinyl 1-phenylethanolamide linked to ascarylose via a 4-hydroxypentanoic acid chain (Figure 2b). This compound was accompanied by two dimeric ascaroside derivatives, a dimer (dasc#1) of the known ascr#1 in which one ascr#1-unit is attached to carbon 4 of the other ascr#1 unit, and second dimer (ubas#1) consisting of ascr#9 to which the (ω)-oxygenated ascaroside oscr#9 is attached in position 2 (Figure 2b). This second dimer, ubas#1, additionally bears a 3-ureido isobutyrate moiety attached to carbon 4. To our knowledge, neither dimeric ascarosides nor ureido isobutyrate-substituted metabolites have previously been reported from nature.
These ascarosides were accompanied by two abundant metabolites that included paratose instead of ascarylose, npar#1 and part#9 (Figure 2b). In npar#1, the paratose moiety was linked to a short lipid side chain, which in turn was connected to threonine. This amino acid was connected further via a carbamoyl group to a derivative of the nucleoside adenosine (Figure 2b). Strikingly, this adenosine was found to include a xylopyranose, and not ribofuranose or deoxyribofuranose as in DNA, RNA, and known nucleoside-based signaling molecules. The accompanying part#9 represents the paratose and side-chain portions of npar#1 (Figure 2b).
To confirm these structural assignments, for determining stereochemistry, and to explore biological functions we developed total syntheses for the proposed structures, taking advantage of their modular nature (Figure 2c, Supporting Information). Comparison of NMR spectra of the synthetic and natural samples established the configuration of the N-succinyl 1-phenylethanolamide moiety in natural pasc#9 as R (Figure S3). Similarly, comparison of NMR spectroscopic data and HPLC retention times enabled assigning the stereochemistry of the dimeric ascarosides dasc#1 and ubas#1 (Figures S4, S5). The R-configuration of the 3-ureido isobutyrate moiety in ubas#1 is consistent with its likely origin from thymine catabolism.[12] Lastly we synthesized samples of the paratoside part#9 and the unusual xyloadenosine derivative, npar#1. Comparing HPLC-retention times of natural part#9 and several synthetic part#9 diastereomers we found that natural part#9 must be either d-paratosyl-4S-hydroxypentanoic acid or l-paratosyl-4R-hydroxypentanoic acid (Figure S6). We initially assumed that part#9 and npar#1 contained d-paratose, which had previously been described from bacteria whereas l-paratose had not been found in nature. Furthermore, d-paratose is a putative intermediate in the biosynthesis of l-ascarylose,[11] on which all ascarosides in nematodes are based.[4e,10,13] However, the HPLC retention time and NMR spectra of a synthetic npar#1 diastereomer including d-paratosyl-4S-hydroxypentanoic acid did not match the data obtained for natural npar#1 (Figure S7a). Therefore, we concluded that npar#1 must be based on l-paratosyl-4R-hydroxypentanoic acid, which was confirmed via synthesis of this diastereomer and comparison of its spectroscopic data with those of natural npar#1 (Figure S7b,c). Using chiral derivatization agents (Mosher's acid chlorides),[14] we further showed that part#9 is also based on l-paratose (Figure S8). The sugar l-paratose has not previously been found in nature; however, its occurrence in nematodes could plausibly result from epimerization of l-ascarylose at position 2.
To exclude the possibility that the identified compounds are bacterial metabolites, we additionally analyzed the metabolome of the E. coli OP50 bacteria used as food for P. pacificus. None of the identified compounds were found to be present in the bacterial metabolome (Figure S9). Furthermore, we showed that all identified compounds are also produced in P. pacificus cultures fed with Pseudomonas sp. instead of E. coli as well as in axenic (bacteria-free) cultures[4d, 15] (Figure S10).
Next we asked whether the identified combinations of sugar, amino acid, lipid, and nucleoside-derived building blocks are specific. To address this, we carefully re-analyzed the entire P. pacificus exo-metabolome by high-resolution HPLC-MS/MS, quantified the identified compounds using synthetic standards (Table S1), and screened for homologues or alternative combinations of the primary metabolism-derived building blocks in our structures. We found that pasc#9 is accompanied by trace amounts of two homologues including six- and seven-carbon side chains, which were also detected by NMR spectroscopy (Figure S11). In addition, we detected a small amount of a homologue of ubas#1 as well as a derivative of npar#1 whose MS data indicated loss of the xylose (ubas#2 and npar#2 in Figure 2a, Table S1). Importantly, we did not observe any non-specific or seemingly random combinations of building blocks that would suggest non-enzymatic genesis of the identified compounds. These results show that assembly of the identified compounds is highly specific.
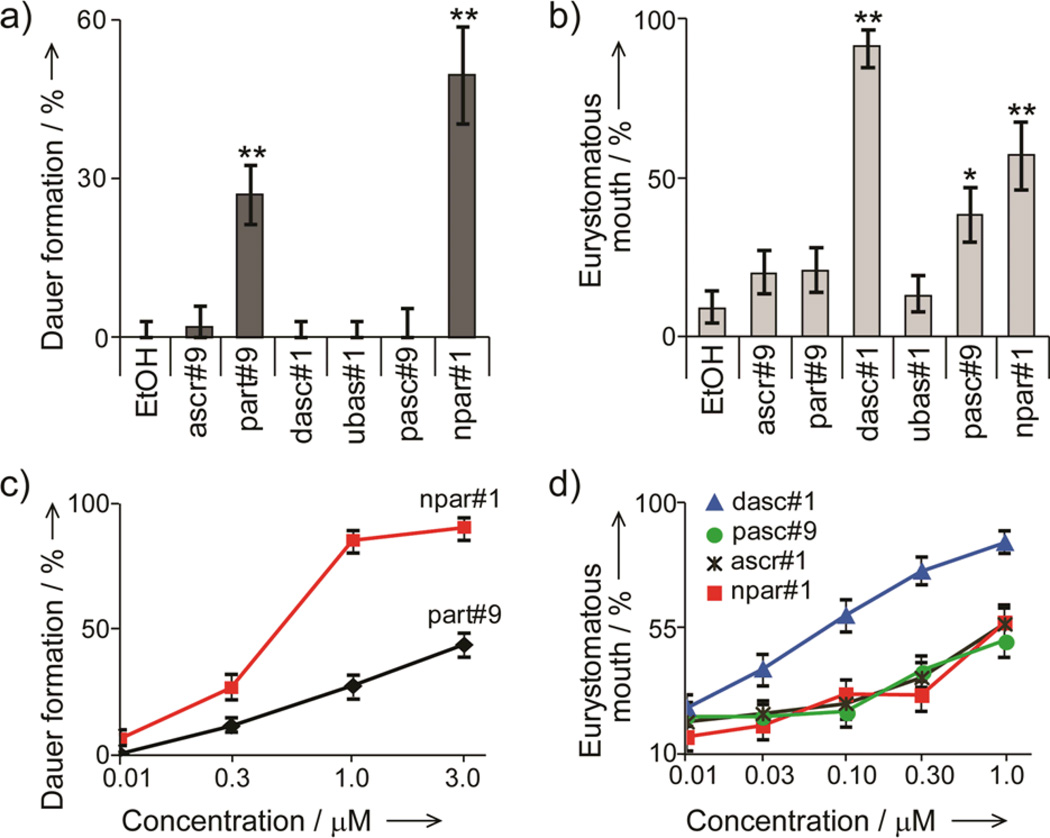
We then tested synthetic samples of the identified compounds for their activity in the P. pacificus dauer and mouth-form dimorphism assays. As expected from previous studies that showed that C. elegans exo-metabolome samples are not active in the P. pacificus mouth-form dimorphism and dauer assays,[16] we found that ascr#1, a compound abundantly excreted by C. elegans,[4e,17] has no dauer-inducing activity in P. pacificus, even at unphysiologically high concentrations (20 µM, Figure S12). In contrast, physiological concentrations of the nucleoside derivative npar#1 strongly induced dauer formation and appears to account for most of the reported dauer-inducing activity of unfractionated exo-metabolome (Figure 3a, c).[9] Additionally, we observed weaker dauer induction with part#9, whereas all other compounds tested were inactive in this assay. Testing our synthetic compounds in the mouth dimorphism assay, we found that the dimeric compound dasc#1, which was inactive in the dauer formation assay, strongly induces the eurystomatous mouth form (Figure 3b, d). In addition, weaker induction of the eurystomatous mouth form was observed for high concentrations of pasc#9, ascr#1, and npar#1, whereas ascr#9 and part#9 as well as the dimeric ubas#1 were inactive at physiological concentrations in the tested wild-type strain (Figure 3b Table S1).
Figure 3.
Regulation of mouth dimorphism and dauer induction by synthetic samples of identified P. pacificus metabolites. All experiments were performed in triplicate for each treatment. a, b) Compounds were assayed at 1 µM concentration (*P < 0.01, **P < 0.001). c, d) Compounds with significant activity at 1 µM (P < 0.01) were subsequently tested at a range of concentrations.
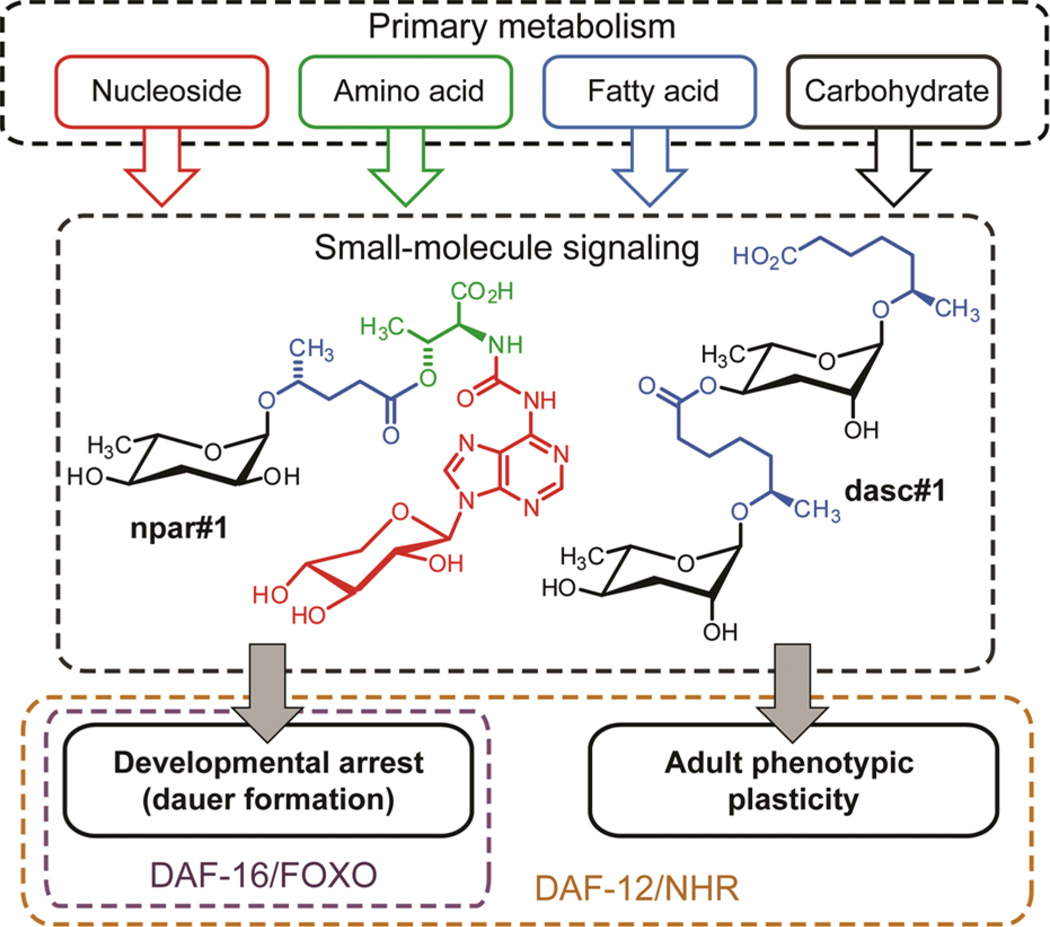
These results show that adult phenotypic plasticity and larval development in P. pacificus are controlled by distinct yet partially overlapping sets of signaling molecules. Whereas mouth-form dimorphism is primarily regulated by dasc#1, the product of highly specific ascaroside dimerization, dauer formation is controlled by a molecule combining a paratoside with an unusual nucleoside. Previous work showed that the signaling molecules controlling phenotypic plasticity in P. pacificus act upstream of evolutionarily conserved transcription factors including DAF-16/FOXO and the nuclear hormone receptor DAF-12 (Figure 4),[18] whereby daf-12 is required for both dauer induction and mouth-form dimorphism, whereas daf-16, is required for dauer induction but dispensable for regulation of mouth-form dimorphism.[18a] Therefore, the different subsets of small molecules regulating dauer formation and mouth-form dimorphism appear to target different downstream effectors. Based on the recent identification of several G-protein coupled receptors (GPCRs) of the C. elegans dauer pheromone,[19] it is probable that GPCRs expressed in specific chemosensory neurons connect npar#1 and dasc#1 with their respective downstream pathways in P. pacificus. Our results further demonstrate species-specific co-option of small molecule biosynthetic pathways for regulating different aspects of development, as ascr#1 contributes to dauer in C. elegans,[17] whereas its dimer dasc#1 regulates adult morphology in P. pacificus. Similar to the multifunctional signaling properties of ascarosides in C. elegans,[4] it is possible that dasc#1, npar#1, and the other identified compounds serve additional signaling functions in P. pacificus, for example, in mediating social behaviors and sex-specific attraction.
Figure 4.
Small molecule signaling in nematodes occupies a central position connecting primary metabolism to evolutionarily conserved transcription factors, including DAF-16/FOXO and the nuclear hormone receptor (NHR) DAF-12, a vitamin D and liver X receptor homolog.
The structures of the identified small-molecule signals appear to integrate specific inputs from the major primary metabolic pathways: fatty acid, carbohydrate, amino acid, and nucleoside metabolism (Figure 4). Specific assembly of building blocks from these pathways via ester and amide linkages generates the identified modular architectures, which are further distinguished from previously known animal metabolites by the inclusion of l-paratose and a xylopyranose-based adenosine. This unprecedented nucleoside is likely derived from metabolism of canonical (ribo)-threonylcarbamoyl adenosine (t6A, Figure 2b), a highly conserved nucleoside found directly adjacent to the anticodon triplet of a subset of tRNAs.[20] t6A plays an important role for maintaining translational fidelity; however, it usually accounts for only a very small fraction of tRNA, and the production of large quantities of a xylopyranose derivative in P. pacificus is surprising.
Our results provide direct evidence for small-molecule control of adult phenotypic plasticity in a metazoan that relies on conserved endocrine signaling pathways. Whether the biosynthesis of the identified signaling molecules also involves conserved pathways or depends on dedicated enzymes specific to P. pacificus (and perhaps other nematodes) is not known. However, the finding that the P. pacificus compounds are derived from assembly of modified primary metabolites suggests that their biosynthesis is largely based on conserved biochemical pathways (Figure 4). Notably, the P. pacificus genome contains more than 25,000 gene predictions with many specific gene duplication events among genes encoding primary metabolic enzymes.[21] Supporting the involvement of primary metabolism in the biosynthesis of nematode signaling molecules, recent investigations of ascaroside biogenesis in C. elegans showed that the lipid-like ascaroside side chains are derived from conserved peroxisomal-β-oxidation.[4e,22] Known signaling molecules and co-factors in higher animals, e.g. S-adenosyl methionine or phosphatidylinositols, often rely on combination of building blocks derived from one or two different primary metabolic pathways. Our results demonstrate that metazoans may extend such strategies to produce signaling molecules of much greater structural complexity, suggesting that detailed spectroscopic re-analysis of metabolomes from higher animals, including mammals, may also reveal novel types of modular small-molecule signals.
Supplementary Material
Footnotes
We thank Parag Mahanti and Alex Artyukhin for helpful discussions and Maciej Kukula for assistance with High Resolution Mass Spectrometry. This work was supported in part by the National Institutes of Health (GM088290 and GM085285), the Cornell/Rockefeller/Sloan-Kettering Training Program in Chemical Biology and the Max-Planck Society.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Neelanjan Bose, Boyce Thompson Institute and Department of Chemistry and Department of Chemical Biology, Cornell University Ithaca, New York, USA.
Akira Ogawa, Department for Evolutionary Biology, Max-Planck-Institute for Developmental Biology, Tübingen, Germany.
Stephan H. von Reuss, Boyce Thompson Institute and Department of Chemistry and Department of Chemical Biology, Cornell University Ithaca, New York, USA
Joshua J. Yim, Boyce Thompson Institute and Department of Chemistry and Department of Chemical Biology, Cornell University Ithaca, New York, USA
Erik J. Ragsdale, Department for Evolutionary Biology, Max-Planck-Institute for Developmental Biology, Tübingen, Germany
Ralf J. Sommer, Department for Evolutionary Biology, Max-Planck-Institute for Developmental Biology, Tübingen, Germany.
Frank C. Schroeder, Boyce Thompson Institute and Department of Chemistry and Department of Chemical Biology, Cornell University Ithaca, New York, USA.
References
- 1.Meinwald J. J. Nat. Prod. 2011;74:305–309. doi: 10.1021/np100754j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Walsh CT. Acc. Chem. Res. 2008;41:4–10. doi: 10.1021/ar7000414. [DOI] [PubMed] [Google Scholar]; b) Meier JL, Burkart MD. Chem. Soc. Rev. 2009;38:2012–2045. doi: 10.1039/b805115c. [DOI] [PubMed] [Google Scholar]
- 3.a) Piel J. Nat. Prod. Rep. 2004;21:519–538. doi: 10.1039/b310175b. [DOI] [PubMed] [Google Scholar]; b) Piel J. Nat. Prod. Rep. 2009;26:338–362. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]; c) Dossey AT. Nat. Prod. Rep. 2010;27:1737–1757. doi: 10.1039/c005319h. [DOI] [PubMed] [Google Scholar]; d) Gronquist M, Schroeder FC, Lew M, Hung-Wen L. Comprehensive Natural Products II. Elsevier; 2010. [Google Scholar]
- 4.a) Butcher RA, Fujita M, Schroeder FC, Clardy J. Nat. Chem. Biol. 2007;3:420–422. doi: 10.1038/nchembio.2007.3. [DOI] [PubMed] [Google Scholar]; b) Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn HT, Teal PEA, Malik RU, Edison AS, Sternberg PW, Schroeder FC. Nature. 2008;454:1115–1118. doi: 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Pungaliya C, Srinivasan J, Fox BW, Malik RU, Ludewig AH, Sternberg PW, Schroeder FC. Proc. Natl. Acad. Sci. USA. 2009;106:7708–7713. doi: 10.1073/pnas.0811918106. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Srinivasan J, von Reuss SH, Bose N, Zaslaver A, Mahanti P, Ho MC, O'Doherty OG, Edison AS, Sternberg PW, Schroeder FC. PLoS Biol. 2012;10:e1001237. doi: 10.1371/journal.pbio.1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) von Reuss SH, Bose N, Srinivasan J, Yim JJ, Judkins JC, Sternberg PW, Schroeder FC. J. Am. Chem. Soc. 2012;134:1817–1824. doi: 10.1021/ja210202y. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Izrayelit Y, Srinivasan J, Campbell SL, Jo Y, von Reuss SH, Genoff MC, Sternberg PW, Schroeder FC. ACS Chem. Biol. 2012;7:1321–1325. doi: 10.1021/cb300169c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong RL, Sommer RJ. Bioessays. 2006;28:651–659. doi: 10.1002/bies.20404. [DOI] [PubMed] [Google Scholar]
- 6.Rae R, Riebesell M, Dinkelacker I, Wang Q, Herrmann M, Weller AM, Dieterich C, Sommer RJ. J. Exp. Biol. 2008;211:1927–1936. doi: 10.1242/jeb.014944. [DOI] [PubMed] [Google Scholar]
- 7.Weller AM, Mayer WE, Rae R, Sommer RJ. J. Parasitol. 2010;96:525–531. doi: 10.1645/GE-2319.1. [DOI] [PubMed] [Google Scholar]
- 8.Bento G, Ogawa A, Sommer RJ. Nature. 2010;466:494–497. doi: 10.1038/nature09164. [DOI] [PubMed] [Google Scholar]
- 9.Mayer MG, Sommer RJ. Proc. Roy. Soc. B. 2011;278:2784–2790. doi: 10.1098/rspb.2010.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choe A, von Reuss SH, Kogan D, Gasser RB, Platzer EG, Schroeder FC, Sternberg PW. Curr. Biol. 2012;22:772–780. doi: 10.1016/j.cub.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorson JS, Lo SF, Ploux O, He XM, Liu HW. J. Bacteriol. 1994;176:5483–5493. doi: 10.1128/jb.176.17.5483-5493.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Kuilenburg ABP, Stroomer AEM, van Lenthe H, Abeling NGGM, van Gennip AH. Biochem. J. 2004;379:119–124. doi: 10.1042/BJ20031463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartley JP, Bennett EA, Darben PA. J. Nat. Prod. 1996;59:921–926. doi: 10.1021/np960236+. [DOI] [PubMed] [Google Scholar]
- 14.Hoye TR, Jeffrey CS, Shao F. Nat. Protoc. 2007;2:2451–2458. doi: 10.1038/nprot.2007.354. [DOI] [PubMed] [Google Scholar]
- 15.Lu NC, Goetsch KM. Nematologica. 1993;39:303–311. [Google Scholar]
- 16.Ogawa A, Streit A, Antebi A, Sommer RJ. Curr. Biol. 2009;19:67–71. doi: 10.1016/j.cub.2008.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong PY, Jung M, Yim YH, Kim H, Park M, Hong E, Lee W, Kim YH, Kim K, Paik YK. Nature. 2005;433:541–545. doi: 10.1038/nature03201. [DOI] [PubMed] [Google Scholar]
- 18.a) Ogawa A, Bento G, Bartelmes G, Dieterich C, Sommer RJ. Development. 2011;138:1281–1284. doi: 10.1242/dev.058909. [DOI] [PubMed] [Google Scholar]; b) Sommer RJ, Ogawa A. Curr. Biol. 2011;21:R758–R766. doi: 10.1016/j.cub.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 19.a) Kim K, Sato K, Shibuya M, Zeiger DM, Butcher RA, Ragains JR, Clardy J, Touhara K, Sengupta P. Science. 2009;326:994–998. doi: 10.1126/science.1176331. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) McGrath PT, Xu YF, Ailion M, Garrison JL, Butcher RA, Bargmann CI. Nature. 2011;477:321–325. doi: 10.1038/nature10378. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Park D, O'Doherty I, Somvanshi RK, Bethke A, Schroeder FC, Kumar U, Riddle DL. Proc. Natl. Acad. Sci. USA. 2012;109:9917–9922. doi: 10.1073/pnas.1202216109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deutsch C, El Yacoubi B, de Crecy-Lagard V, Iwata-Reuyl D. J. Biol. Chem. 2012;287:13666–13673. doi: 10.1074/jbc.M112.344028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.a) Dieterich C, Clifton SW, Schuster LN, Chinwalla A, Delehaunty K, Dinkelacker I, Fulton L, Fulton R, Godfrey J, Minx P, Mitreva M, Roeseler W, Tian H, Witte H, Yang SP, Wilson RK, Sommer RJ. Nat. Genet. 2008;40:1193–1198. doi: 10.1038/ng.227. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Borchert N, Dieterich C, Krug K, Schutz W, Jung S, Nordheim A, Sommer RJ, Macek B. Genome Res. 2010;20:837–846. doi: 10.1101/gr.103119.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butcher RA, Ragains JR, Li W, Ruvkun G, Clardy J, Mak HY. Proc. Natl. Acad. Sci. USA. 2009;106:1875–1879. doi: 10.1073/pnas.0810338106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.