Abstract
We recently reported an antibody-free targeted protein quantification strategy, termed high-pressure, high-resolution separations with intelligent selection and multiplexing (PRISM) for achieving significantly enhanced sensitivity using selected reaction monitoring (SRM) mass spectrometry. Integrating PRISM with front-end IgY14 immunoaffinity depletion, sensitive detection of targeted proteins at 50–100 pg/mL levels in human blood plasma/serum was demonstrated. However, immunoaffinity depletion is often associated with undesired losses of target proteins of interest. Herein we report further evaluation of PRISM-SRM quantification of low-abundance serum proteins without immunoaffinity depletion. Limits of quantification (LOQ) at low ng/mL levels with a median coefficient of variation (CV) of ~12% were achieved for proteins spiked into human female serum. PRISM-SRM provided >100-fold improvement in the LOQ when compared to conventional LC-SRM measurements. PRISM-SRM was then applied to measure several low-abundance endogenous serum proteins, including prostate-specific antigen (PSA), in clinical prostate cancer patient sera. PRISM-SRM enabled confident detection of all target endogenous serum proteins except the low pg/mL-level cardiac troponin T. A correlation coefficient >0.99 was observed for PSA between the results from PRISM-SRM and immunoassays. Our results demonstrate that PRISM-SRM can successful quantify low ng/mL proteins in human plasma or serum without depletion. We anticipate broad applications for PRISM-SRM quantification of low-abundance proteins in candidate biomarker verification and systems biology studies.
Keywords: SRM, PRISM, targeted quantification, low-abundance protein, human serum, sensitivity, reproducibility
Introduction
Highly sensitive, multiplexed quantitative assays hold a great promise for broad quantification of low-abundance target proteins in biomarker verification and systems biology studies.1–3 While immunoassays are still the commonly used approaches for targeted protein quantification, the technique presently suffers from issues related to reagent specificity or cross-reactivity,4, 5 as well as the lack of reagents for novel proteins. Selected reaction monitoring (SRM, or multiple reaction monitoring, MRM) has recently emerged as a promising alternative to immunoassays for targeted protein quantification given its relatively good reproducibility, sensitivity, and superior multiplexing capability.1, 2, 6–21 SRM coupled with liquid chromatography (LC) and stable isotope dilution permits the detection and quantification of hundreds of target proteins simultaneously in highly complex biological samples.9, 13, 20 However, a main limitation of SRM-based targeted quantification is the lack of sufficient sensitivity for quantification of low-abundance proteins or protein modifications.19, 22 For example, LC-SRM can typically detect moderately abundant proteins in human blood plasma/serum with concentrations at the low µg/mL or high ng/mL levels without the application of front-end fractionation and/or enrichment.6, 10, 11, 19 Although the detection of selected plasma proteins below 100 ng/mL levels has been reported in several recent multiplexed targeted quantification efforts in plasma without depletion,23–25 the majority of plasma proteins in the low ng/mL range are still not detectable in non-depleted plasma by direct LC-SRM.24 Many candidate protein biomarkers of proven clinical importance are present at the pg/mL to low ng/mL levels in human plasma/serum; thus, well below the limit of detection (LOD) for conventional LC-SRM.
Recent advances in sample prefractionation and/or enrichment strategies8, 26, 27 along with MS instrumentation19, 28 have proven useful for enhancing SRM sensitivity for the detection of low-abundance proteins. For example, following immunoaffinity depletion of high-abundance proteins and peptide fractionation by strong cation exchange chromatography, LC-SRM enabled quantification of plasma proteins at low ng/mL levels.8, 26 Peptide enrichment by anti-peptide antibodies, termed stable isotope standards and capture by anti-peptide antibodies (SISCAPA), coupled with SRM was demonstrated to be effective for quantifying target proteins at the low ng/mL range using as little as 10 µL of human plasma11, 29, 30 and even at the sub-ng/mL range if larger volumes (e.g., 1 mL) of human plasma are used.11 SISCAPA assays share with immunoassays in that antibodies are immobilized on various platforms to capture surrogate peptides or target proteins, but differ in that SRM measurements serve as the “secondary antibodies” for detection and quantification. However, specific antibody reagents for the target peptides are generally not available, and development of such reagents is relatively expensive ($4,000/anti-peptide antibody) and requires a long lead time (~6 months).17, 21 Furthermore, there are still technical limitations in multiplexing SISCAPA assays for large numbers of target proteins; studies published to date indicate that the highest multiplex level for SISCAPA assays is 50 peptides using a common set of reagents.31
More recently, we developed an antibody-free strategy, termed as high-pressure, high-resolution separations with intelligent selection and multiplexing (PRISM), that performs target peptide enrichment through high resolution reversed phase LC separation/fractionation and effective selection and multiplexing of targeted fractions for downstream LC-SRM measurements.32 We demonstrated that the integration of IgY14 immunoaffinity depletion (removing the 14 high-abundance plasma proteins) and PRISM-SRM can reliably quantify low-abundance proteins at 50–100 pg/mL levels in human plasma/serum .32 However, it is recognized that immunoaffinity depletion of high-abundance proteins is often associated with potential loss of target proteins of interest through either nonspecific binding to the depletion column or forming complexes with the bound high-abundance proteins.8, 27, 33, 34 For example, prostate-specific antigen (PSA) is known to complex with alpha-1-antichymotrysin and alpha-2-macroglobulin, and be partially removed by depletion.33
In this study, we assessed the sensitivity and reproducibility of PRISM-SRM assay for quantifying human serum proteins without immunoaffinity depletion. Accurate quantification with limit of quantification (LOQ) at low ng/mL levels was achieved for all standard proteins spiked into human female serum. The sensitivity of PRISM-SRM was also demonstrated by the successful detection of several endogenous serum proteins reported to be present at low ng/mL levels, including PSA, in clinical patient sera.
Experimental procedures
Reagents
All 3 target proteins (bovine carbonic anhydrase, bovine beta-lactoglobulin, and human PSA) were purchased from Sigma-Aldrich (St. Louis, MO). Urea, dithiothreitol (DTT), iodoacetamide, ammonium formate, trifluoroacetic acid (TFA) and formic acid were obtained from Sigma (St. Louis, MO). Synthetic peptides labeled with 13C/15N on C-terminal lysine and arginine for all targeted proteins were from Thermo Scientific (San Jose, CA).
Human Specimens
A human female serum sample was from BioChemed Services (Winchester, VA). Clinical serum samples from prostate cancer patients undergoing PSA screening were provided by the Johns Hopkins Medical Institutions. The use of human serum samples was approved by the Institutional Review Boards of the University of Washington, Pacific Northwest National Laboratory, and Johns Hopkins University in accordance with federal regulations.
Surrogate peptide selection
Selection of highly responsive surrogate peptides was critical for successful quantification of low abundance proteins in complex biosamples. For endogenous serum proteins, 10 tryptic peptides without miscleavages (except those peptides containing inhibitory motifs for trypsin)35 were initially chosen for representing each target protein based upon in silico digestion data. Existing LC-MS/MS results from the Global Proteome Machine (GPM) and data from our own laboratory, and then evaluated by ESP predictor36 and CONSeQuence software.37 All peptides were further blasted for their uniqueness to target proteins. All selected peptides were unique to the given proteins except that the peptide IVGGWECEK, a commonly used surrogate peptide for PSA SRM assay8, 10, is shared by PSA and kallikrein-2 (KLK2). Four peptides per protein with moderate hydrophobicity and high score from the prediction tools were selected for peptide synthesis. The synthesized crude heavy isotopic peptides were further evaluated for peptide response and fragmentation pattern. Two final surrogate peptides were selected for the detection and quantification of the corresponding target protein. For each peptide, 3 transitions were selected based on their abundances and the best transition (i.e., the one with the most intense SRM signal and without clear evidence of co-eluting interference) was used to generate calibration curves, and to quantify the target protein. The potential interference for given transitions were assessed based on the relative intensity ratios between the 3 transitions for both light and heavy peptides using an approach similar as previously reported.38 Optimal collision energy (CE) values were achieved by direct infusion of the individual peptides. For standard proteins spiked into serum, their surrogate peptides were selected based on the results from a previous study.32 High purity heavy peptides (>95%) were used for these standard proteins.
Target protein spike-in and human serum protein digestion
Three target proteins (carbonic anhydrase, beta-lactoglobulin, and PSA) were spiked into female serum at 0, 0.1, 0.3, 0.5, 1, 2.5, 5, 10, and 100 ng/mL levels to generate individual samples. The concentrations of target protein stock solutions were determined by the BCA protein assay (Pierce). Each 12.5 µL aliquot of the serum (~1 mg) was diluted 10-fold with 50 mM NH4HCO3 (pH 8.0). The diluted serum samples were denatured and reduced with 8 M urea and 10 mM DTT in 50 mM NH4HCO3 buffer for 1 h at 37° C. Protein cysteine residues were alkylated with 40 mM iodoacetamide for 1 h at room temperature. The resulting sample was diluted 6-fold with 50 mM NH4HCO3, and sequencing grade modified porcine trypsin (Promega, Madison, WI) was added at a trypsin:protein ratio of 1:50 (w/w) for digestion at 37° C for 3 h. The protein digest was then loaded onto a 1 mL SPE C18 column (Supelco, Bellefonte, PA) and washed with 4 mL of 0.1% TFA, 5% acetonitrile. Peptides were eluted from the SPE column with 1 mL of 0.1% TFA, 80% acetonitrile and lyophilized. The final peptide concentration was determined by the BCA assay (Pierce). Peptide samples were stored at −80° C until time for use. The peptide stock was then diluted to 1 µg/µL with 0.1% formic acid in water and isotope-labeled synthetic peptides were spiked at 0.5 fmol/µL.
For clinical patient sera, serum sample containing ~1 mg proteins (8.65–11.65 µL) was diluted 10-fold with 50 mM NH4HCO3 (pH 8.0). The diluted samples were processed as described above. All stocks of peptide samples from patient sera were individually diluted to 1 µg/µL with 0.1% formic acid in water and heavy synthetic peptide standards were spiked at 0.5 fmol/µL for PSA (high purity peptides) and 5 fmol/µL (crude peptides) for the other endogenous proteins being monitored. The targeted endogenous serum proteins include epidermal growth factor receptor (EGFR), kallikrein 6 (KLK6), matrix metalloproteinase 9 (MMP9), periostin (POSTN), and cardiac troponin T (cTnT).
High-pH reversed phase LC fractionation
The high-pH reversed phase LC fractionation is one of the main components of the PRISM workflow (Fig. S1).32 A nanoACQUITY UPLC® system (Waters Corporation, Milford, MA) equipped with a reversed phase capillary LC columns and an autosampler was used for fractionation. Capillary reversed phase column, 200 Im inner diameter (i.d.) × 50 cm long, were packed in-house with 3 µm Jupiter C18 bonded particles (Phenomenex, Torrence, CA). Separations were performed at mobile phase flow rates of 3.3 µL/min on the binary pump systems using 10 mM ammonium formate (pH 10) in water as mobile phase A and 10 mM ammonium formate (pH 10) in 90% acetonitrile as mobile phase B. A 45 µL of sample with a peptide concentration of 1 µg/µL was typically loaded onto the reversed phase capillary column and separated using a binary gradient of 5–15% B in 15 min, 15–25% B in 25 min, 25%–45% B in 25 min, 45–90% B in 38 min. Following the LC separation, the eluent from the capillary column was split into two flowing streams (1:10 split) via a Tee union. A small fraction of the eluent at a flow rate of ~300 nL/min was directed to a TSQ Quantum Ultra triple quadrupole mass spectrometer (Thermo Scientific, San Jose, CA) for on-line SRM monitoring of heavy peptide standards. The TSQ Quantum Ultra instrument operating parameters were optimized for all SRM transitions by infusion of each heavy peptide. Typically, the TSQ Quantum Ultra mass spectrometer was operated with ion spray voltages of 2,400 ± 100 V, a capillary offset voltage of 35 V, a skimmer offset voltage of −5 V and a capillary inlet temperature of 220° C. The tube lens voltages were obtained from automatic tuning and calibration without further optimization. A single scan event was used to monitor a total of 24 SRM transitions, 4 SRM transitions per heavy peptide, using the following parameters: Q1 and Q3 unit resolution of 0.7 FWHM, Q2 gas pressure of 1.5 mTorr, scan width of 0.002 m/z and a scan time of 25 ms. The large fraction of the eluent, at a flow rate of ~3 µL/min, was automatically dispensed every minute into a 96-well plate during ~100 min LC run using the Triversa NanoMate® system (Advion BioSciences, Ithaca, NY). Prior to peptide fraction collection, 17 µL of water was added to each well of the 96-well plate to avoid the loss of peptides and dilute the peptide fraction (~1:7 dilution) for LC-SRM analysis.
LC-SRM analysis
Following the intelligent selection (ISelection) of target peptide fractions, the peptide fraction of interest was subjected to LC-SRM measurement (Fig. S1). All peptide fractions were analyzed by using nanoACQUITY UPLC® system coupled on-line to a TSQ Vantage triple quadrupole mass spectrometer (Thermo Scientific, San Jose, CA). Solvents used were 0.1% formic acid in water (mobile phase A) and 0.1% formic acid in 90% acetonitrile (mobile phase B). Peptide fraction samples were loaded onto an ACQUITY UPLC 5 µm C18 trap column (180 µm i.d. × 20 mm) for 5 min at 10 µL/min with 3% B. Peptide separations were performed at a flow rate of 400 nL/min using an ACQUITY UPLC BEH 1.7 µm C18 column (75 µm i.d. × 25 cm), which was connected to a chemically etched 20 µm i.d. fused-silica emitter via a Valco stainless steel union. Either 1 µL of unfractionated serum digests or 4 µL of individual peptide fractions (total volume 20 µL; ~0.11 µg was loaded onto the analytical column for LC-SRM analysis) following PRISM was injected for LC separations using a binary gradient of 10–20% B in 7 min, 20–25% B in 17 min, 25–40% B in 1.5 min, 40–95% B in 2.5 min and at 95% B for 6 min for a total of ~35 min and the analytical column was re-equilibrated at 98% A for 15 min. The TSQ Vantage was operated in the same manner as the TSQ Quantum Ultra. A dwell time of 40 ms was used for all SRM transitions.
Data analysis
SRM data acquired on the TSQ Vantage were analyzed using Xcalibur 2.0.7 (Thermo Scientific). The relative light to heavy peptide (L/H) SRM signal ratios of the four transitions selected and optimized for the SRM assay were predefined by internal standard heavy peptides in buffer. Matrix inferences from co-eluting peptides with a transition that falls within the mass width of Q1 and Q3 were detected by deviation from the expected L/H SRM signal ratios. The best transition for each peptide was used for quantification. Peak detection and integration were determined based on two criteria: 1) same retention time; 2) approximately same relative SRM peak intensity ratios across multiple transitions between light peptides and heavy peptide standards. All data were manually inspected to ensure correct peak detection and accurate integration. Signal-to-noise ratio (S/N) was calculated by peak intensity at the apex over the highest background noise in a retention time window of ± 15 s for the target peptides. The background noise levels were conservatively estimated by visual inspection of the chromatographic peak regions.32 The LOD and LOQ were defined as the lowest concentration point of each target protein at which the S/N of surrogate peptides was at least 3 and 10, respectively. For conservatively determining the LOQ values, in addition to the requirements of the S/N to equal or be above 10, two other criteria were applied: the coefficient of variation (CV) at the concentration point be less than 20%; surrogate peptide response over the protein concentration be within the linear dynamic range. The L/H SRM peak area ratio was used to generate calibration curves and evaluate reproducibility. All calibration and correlation curves were plotted using Microsoft Excel 2007. The RAW data from TSQ Vantage were loaded into Skyline software39 to display graphs of extracted ion chromatograms (XICs) of multiple transitions of target proteins monitored.
Results
PRISM-SRM Protein Quantification in Non-depleted Serum
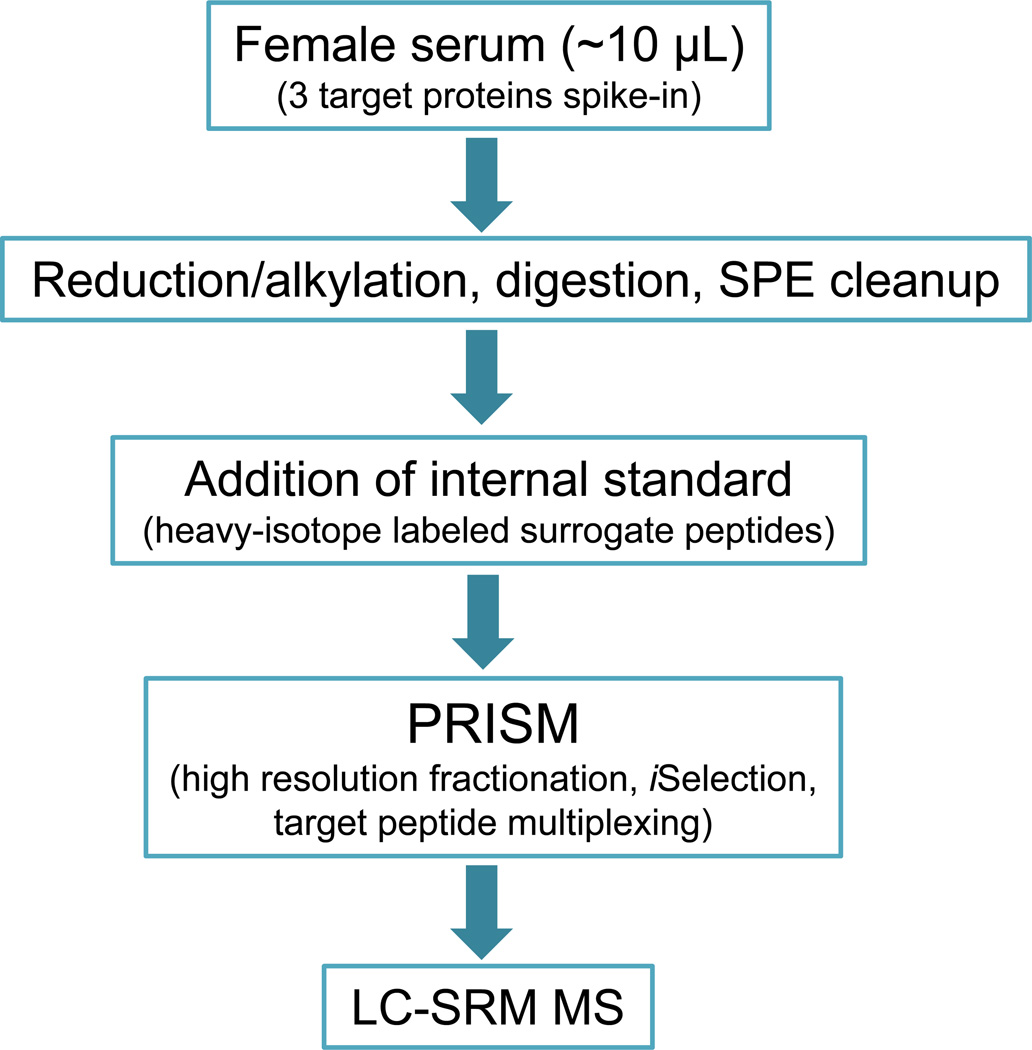
To assess the effectiveness of PRISM-SRM quantification in non-depleted serum in terms of sensitivity and reproducibility, we utilized a workflow as illustrated in Fig. 1. Bovine carbonic anhydrase, bovine beta-lactoglobulin, and PSA were spiked into female serum at different levels for this assessment. Following sample processing and protein digestion, the peptide samples were spiked with heavy peptide internal standards prior to PRISM fractionation.32 After sample processing including SPE clean-up, the heavy peptides were spiked into peptide mixtures prior to PRISM to ensure the identical levels of heavy peptides present in different samples so that a robust on-line SRM monitoring for PRISM can be achieved.32 The 3 proteins and their surrogate peptides are shown in Table 1, where 2 surrogate peptides per protein were selected. For each target peptide, 4 transitions were monitored to achieve maximum selectivity and sensitivity in the SRM assays, and the best SRM transition for each peptide was used to generate the calibration curve and estimate the reproducibility of PRISM-SRM assay (Table 1). In female serum, it was reported that both the free and total PSA were below the detection limits of the immunoassays (≤0.01 ng/mL);8 thus, the contribution of endogenous PSA concentration was deemed negligible.
Figure 1.
Workflow for PRISM-SRM detection and quantification of target proteins in non-depleted serum.
Table 1.
Target proteins and their surrogate peptides. All internal standards were synthesized with 13C and 15N heavy-isotope labeled at the C-terminal arginine or lysine.
| Protein | Accession number | Molecular weight | Surrogate peptide | SRM transitions | ||||
|---|---|---|---|---|---|---|---|---|
| (kDa) | Q1 | Q3b | ||||||
| Bovine carbonic anhydrase | P00921 | 29 | DFPIANGER | 509.82+ | 378.72+ | 546.3+ | 756.4+ | 658.3+ |
| DGPLTGTYR | 490.22+ | 597.3+ | 496.3+ | 404.22+ | 710.4+ | |||
| Bovine beta-lactoglobulin | P02754 | 20 | VLVLDTDYKK | 597.32+ | 981.5+ | 882.5+ | 769.4+ | 491.3+ |
| VYVEELKPTPEGDLEILLQK | 771.83+ | 1026.12+ | 976.52+ | 790.92+ | 847.52+ | |||
| Prostate-specific antigen | P07288 | 30 | IVGGWECcamEKa | 539.22+ | 964.4+ | 865.3+ | 436.2+ | 213.1+ |
| LSEPAELTDAVK | 636.82+ | 943.5+ | 472.32+ | 846.5+ | 775.4+ | |||
Cysteine was synthesized as carbamidomethyl cysteine.
All 4 transitions were monitored and the best transitions in bold were used for plotting calibration curves for target proteins and correlation curves between calculated and expected target protein concentrations in female serum.
PRISM incorporated a first dimensional high resolution reversed-phase capillary LC (cLC) separation and fractionation as a core component for effective enrichment of target peptides using pH 10 mobile phases.32 This approach addressed the general drawback of fractionation strategies on throughput (i.e., the need to analyze many fractions per sample limiting overall throughput) by introducing the concept of iSelection of target fractions, and only the selected target fractions were analyzed by the second dimensional LC-SRM. A limited number of target fractions could also be multiplexed prior to nanoLC-SRM to enhance throughput by taking advantage of the partial orthogonality between high and low pH reversed-phase separations. To assess the reproducibility, 3 injection replicates for LC-SRM were performed for each individual target peptide fraction, and 3 process replicates of PRISM (i.e., three aliquots of the same biological sample being processed independently by PRISM fractionation) followed by 3 injection replicates were performed only at the concentration level of 2.5 ng/mL.
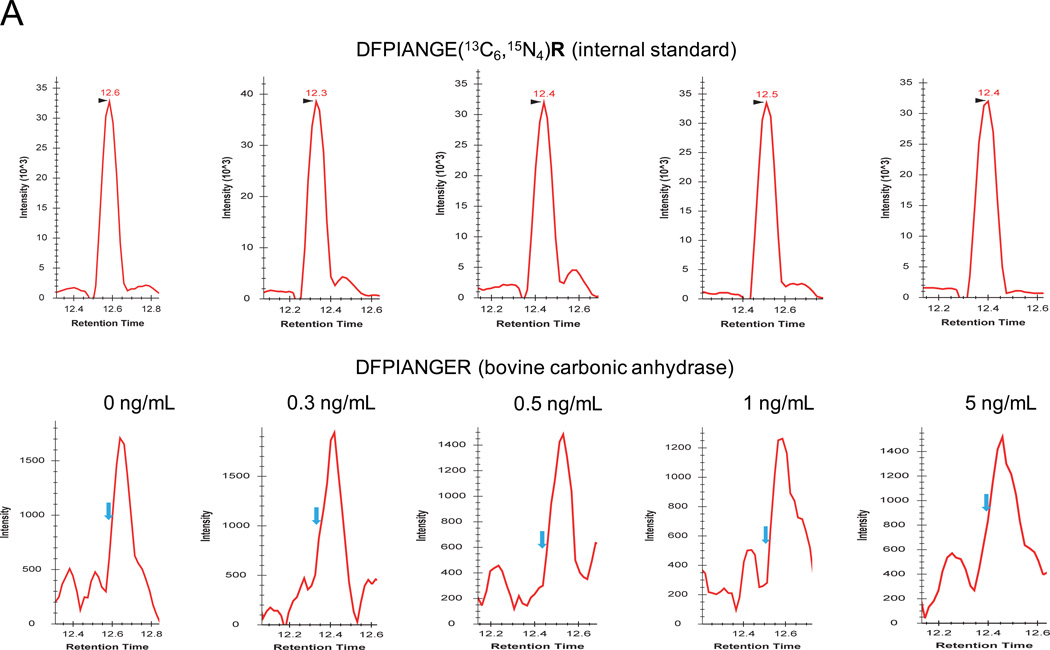
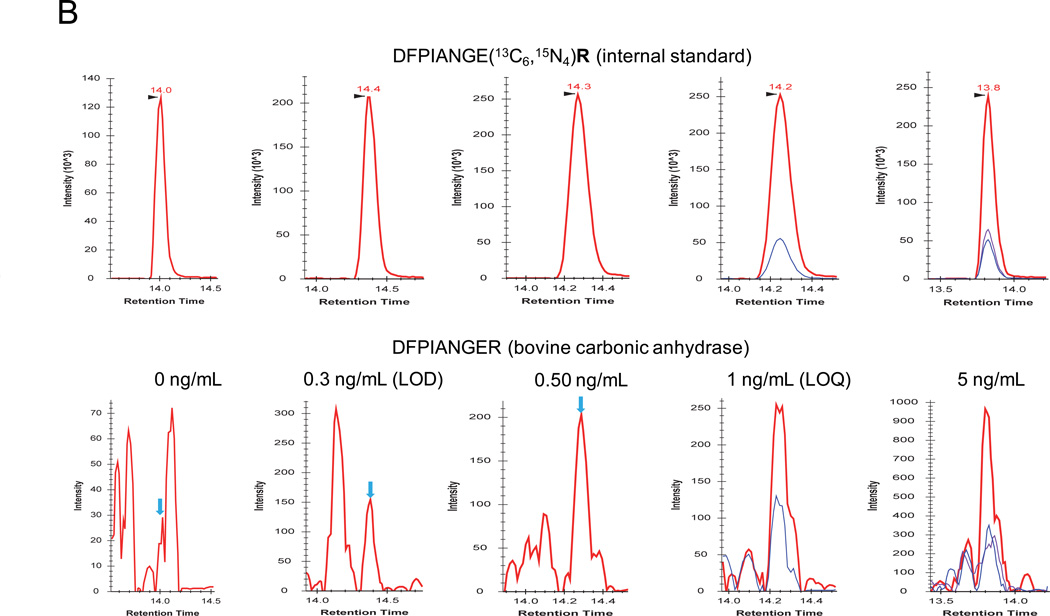
The linear dynamic range, LOD, LOQ, and reproducibility for each surrogate peptide were assessed with target protein concentrations ranging from 100 pg/mL to 100 ng/mL. A side-by-side comparison of the SRM signals of surrogate peptides with and without the application of PRISM were also performed to assess the improvement on the SRM sensitivity based on the LOD and LOQ values. Fig. 2A and 2B shows XICs of transitions monitored for peptide DFPIANGER derived from bovine carbonic anhydrase at 5 concentration points using conventional LC-SRM and PRISM-SRM, respectively. The lower-abundance transitions were observed with heavy matrix interferences at low concentration points and they are not presented here. Without PRISM, the signals for light peptide transitions were dominated by co-eluting interferences, and little changes in overall signal levels were observed from 0 to 5 ng/mL (Fig. 2A). PRISM significantly reduced background interference levels and enhanced S/N when compared to LC-SRM measurements (Fig. 2B, Fig. S2.1–S7.1 and Tables S1–S2), which lowered the LOD and LOQ to 300 pg/mL and 1 ng/mL, respectively. The LOQ values obtained from the best transition for each surrogate peptide were in the range of 500 pg/mL to 5 ng/mL (Table 2). The PRISM strategy improved the overall SRM sensitivity by more than 100-fold, and up to 1,000-fold depending on the targeted peptide, when compared to LC-SRM analyses without PRSIM (Table 2). Two peptides, VLVLDTDYKK and DGPLTGTYR, showed only 50- and 100-fold improvements in LOQ, respectively, primarily due to co-eluting interferences. In our previous study, with prior IgY14 immunoaffinity depletion the PRISM-SRM assay was able to quantify proteins in human plasma/serum at a concentration range of 50–100 pg/mL.32 This suggested that the IgY14 depletion contributed to the improvement of SRM sensitivity by a nearly 10-fold, which was consistent with ~90% removal of the protein mass by IgY14 depletion.40 Without depletion, PRISM-SRM allowed quantification of all 3 target proteins at low ng/mL levels in human serum with at least 1 surrogate peptide (Table 2).
Figure 2.
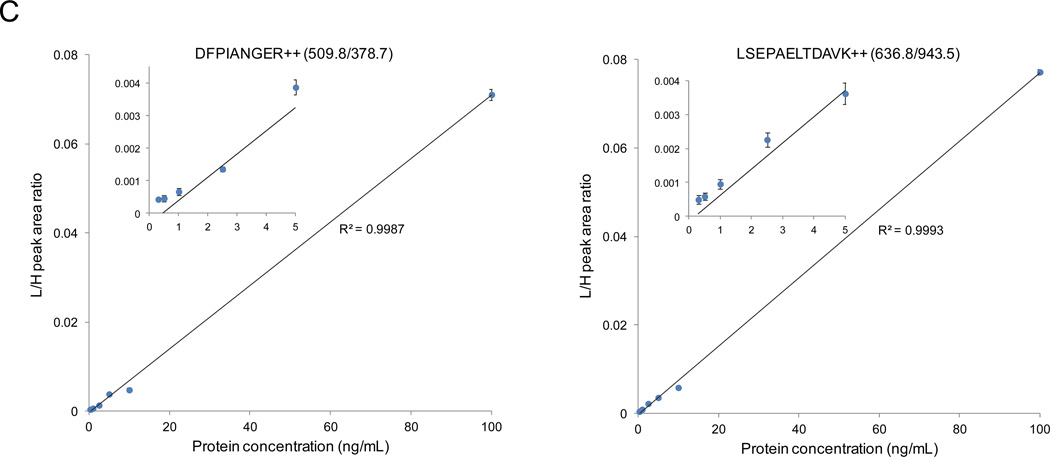
Sensitivity, reproducibility, and accuracy of PRISM-SRM assay. XICs of transitions monitored for DFPIANGER derived from bovine carbonic anhydrase at various concentrations: (A) without PRISM fractionation; (B) with PRISM fractionation. DFPIANGER: 509.8/378.7 (red), 509.8/756.5 (blue), 509.8/546.3 (purple). Internal standards were spiked in at 0.5 fmol/µL. The blue arrows indicate the locations of expected SRM peak of light peptides based on the retention time of heavy internal standards. (C) Calibration curves for quantifying bovine carbonic anhydrase and PSA.
Table 2.
Summary of LOD and LOQ of 3 standard proteins spiked into female serum by PRISM-SRM and conventional LC-SRM (Direct LC) assays.
| Protein | Accession number | Surrogate peptide | SRM assay | LOD | LOQ |
|---|---|---|---|---|---|
| (ng/mL) | (ng/mL) | ||||
| Bovine carbonic anhydrase | P00921 | DGPLTGTYR | Direct LC | 100 | 500 |
| PRISM | 1 | 5 | |||
| DFPIANGER | Direct LC | 1000b | 1000 | ||
| PRISM | 0.3 | 1 | |||
| Bovine beta-lactoglobulin | P02754 | VLVLDTDYKK | Direct LC | 100 | 250 |
| PRISM | <0.1c | 5c | |||
| VYVEELKPTPEGDLEILLQK | Direct LC | 100 | 2000d | ||
| PRISM | <0.1c | 5c | |||
| Prostate-specific antigen | P07288 | IVGGWECcamEKa | Direct LC | 500b | 1000 |
| PRISM | 0.1 | 1 | |||
| LSEPAELTDAVK | Direct LC | 250 | 2000e | ||
| PRISM | 0.3 | 0.5 | |||
Cysteine was synthesized as carbamidomethyl cysteine.
Large co-eluting interference.
Strong SRM signal of both light peptides at low pg/mL levels due to interferences of co-eluting species.
Low SRM response for heavy internal standard.
Co-eluting interference for heavy internal standard.
The calibration curves of PRISM-SRM measurements (from the best transition for each protein) showed excellent linearity for all 3 target proteins for concentrations ranging from 1 ng/mL to 100 ng/mL with a median CV of ~10% for triplicates (Fig. 2C and Table S2). However, at concentrations lower than 1 ng/mL non-linearity was observed, which could be attributed to signal contribution from matrix interferences. The background signal presumably originated from co-eluting peptides with similar transition m/z values as the target peptides. The reproducibility of PRISM-SRM was further evaluated by 3 process replicates for each target protein at 2.5 ng/mL concentration. An average CV across the 3 process replicates for the 3 target proteins was ~10%, illustrating the quantification precision for low ng/mL levels of proteins in human plasma/serum (Table S2).
Quantification of Endogenous Proteins in Prostate Cancer Patient Sera
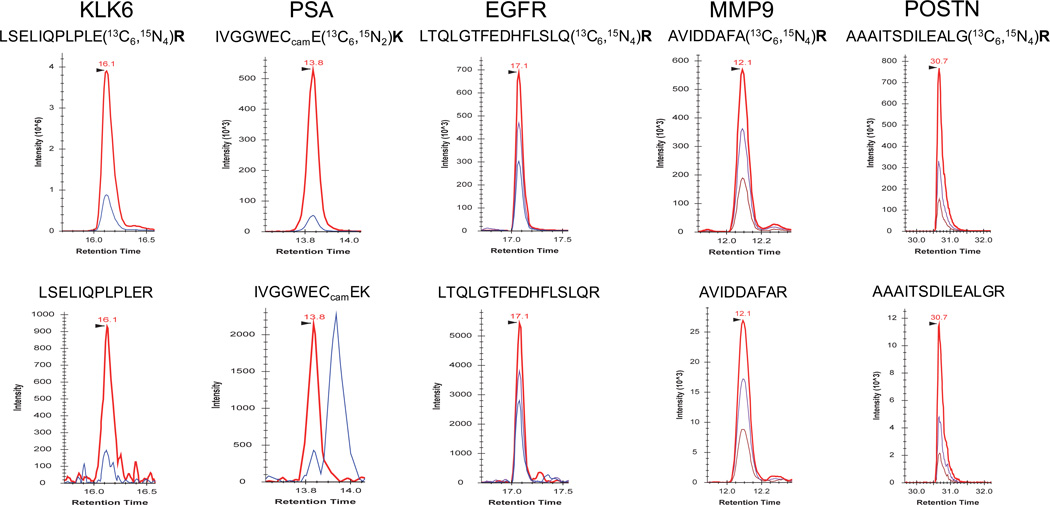
PRISM-SRM was next applied to detect 6 endogenous proteins in 2 prostate cancer patient serum samples without depletion. The 6 proteins were PSA, epidermal growth factor receptor (EGFR), kallikrein 6 (KLK6), matrix metalloproteinase 9 (MMP9), periostin (POSTN), and cardiac troponin T (cTnT), all of which were reported with plasma concentrations ranging from low pg/mL to ng/mL levels.41–46 The XICs in Fig. 3 demonstrated that PRISM-SRM was able to confidently detect and quantify 5/6 endogenous proteins (PSA, EGFR, KLK6, MMP9, and POSTN) in the 2 clinical serum samples (also see Fig. S8 and Table S3). The cTnT was detected by only one surrogate peptide with lower confidence due to the relatively low signal-to-noise ratios (Fig. S8), which is anticipated given the low pg/mL level reported in human serum/plasma (50–100 pg/mL),42 which was lower than the LOD of the PRISM-SRM assay for non-depleted serum. The concentrations of KLK6, EGFR, MMP9, and POSTN in human serum were reported to be at 2.9–6.8 ng/mL,41 43.2–114.2 ng/mL,43 261.6–305.2 ng/mL,44 and 120–513 ng/mL,45 respectively. These results further illustrated that PRISM-SRM provided sufficient sensitivity for quantifying endogenous proteins at low ng/mL levels in human plasma/serum without immunoaffinity depletion. We also compared the PRISM-SRM measurement results of PSA with immunoassay data for the same set of prostate cancer patient sera (Table S4). An excellent correlation, R2 > 0.99, was observed (Fig. S9–S10).
Figure 3.
XICs of transitions for 5 endogenous proteins in a prostate cancer patient serum from PRISM-SRM measurements. For PSA protein pure internal standards were spiked in at 0.5 fmol/µL; while for the other proteins, crude internal standards were used with the spiking level at 5 fmol/µL. LSELIQPLPLER: 704.4/724.4 (red), 704.4/965.6 (blue); IVGGWECcamEK: 539.2/865.4 (red), 539.2/964.4 (blue); LTQLGTFEDHFLSLQR: 635.7/725.4 (red), 635.7/781.9 (purple), 635.7/845.9 (blue); AVIDDAFAR: 489.3/807.4 (red), 489.3/694.3 (purple), 489.3/579.3 (chestnut); AAAITSDILEALGR: 700.9/1074.6 (red), 700.9/973.5 (purple); 700.9/771.5 (chestnut).
Discussion
As demonstrated in our previous study, the sensitivity of PRISM-SRM assay can be extended to detect and quantify 50–100 pg/mL level of proteins in human plasma/serum when IgY14 immunoaffinity depletion is applied.32 However, using depletion columns to remove high-abundance proteins from human plasma is often associated with significant losses of proteins of interest. For example, PSA recovery following depletion of albumin and IgG was around 40% to 65%.27 When IgY12/IgY14 depletion column was used, the recovery was only ~25%8, 32 due to PSA binding to other abundant proteins such as α-2-macroglobulin and α-1-antichymotrypsin. Moreover, there may not be prior knowledge about how target proteins might be lost in the depletion process. Thus, it is highly desirable to develop SRM assays that avoid immunoaffinity depletion for plasma protein quantification. In this work, we demonstrated that the application of PRISM-SRM enabled detection and quantification of target proteins including a number of endogenous proteins at low ng/mL levels in non-depleted human serum using ~10 µL serum as the starting material. We note that all 5 PRISM-quantified endogenous proteins except POSTN were reported to be undetectable by coupling MARS Hu-14 immunoaffinity depletion with conventional LC-SRM in a recently reported large-scale quantification of human plasma proteins,46 further illustrating the superior sensitivity of PRISM-SRM.
The workflow of PRISM-SRM is mainly built upon LC separations for analyte enrichment/focusing and the robustness of this process in terms of reproducibility and accuracy has been demonstrated. PRISM-SRM is also relatively easy to implement with commercially available instruments and reagents, and offers a good multiplexing capability for simultaneous quantification of many proteins. Since the assay development process does not require affinity reagents, this approach offers the advantages of fast assay developments with low costs. For establishing PRISM-SRM assays for large-scale clinical studies, external calibration may be necessary to reduce day-to-day variations from trypsin digestion and sample processing.47
The main limitation of PRISM-SRM is its reduced analytical throughput as a result of reversed-phase cLC fractionation. Nevertheless, the two dimensional separations in PRISM-SRM workflow is only partially orthogonal, which provides an advantage for fraction concatenation to alleviate the throughput inadequacy.48 In our previous study,32 we have preliminarily illustrated that 96 fractions can be concatenated into 12 fractions based on peptide elution times to achieve a moderate throughput (~50 sample analyses/week) depending upon experimental details without loss of sensitivity. Ideally, the post-concatenation should be performed based on informed pooling based on the use of target fractions only and the retention time of target fractions. Further optimization of fraction multiplexing process is necessary to evaluate sensitivity and reproducibility for large-scale multiplexed quantification using the PRISM workflow. Alternatively, fast nanoLC-SRM analyses using short LC gradients (e.g., 5 min) can be employed to enhance the overall throughput since PRISM fractionation significantly reduces the complexity of individual target peptide fractions.
In summary, our results demonstrate that PRISM-SRM produces reliable quantification of target proteins at low ng/mL levels in non-depleted human plasma/serum. The ability to use fraction multiplexing or concatenation provides a moderate analytical throughput of PRISMSRM. The antibody-free, high sensitivity, and high reproducibility features of PRISM-SRM make it useful for quantification of low-abundance proteins (and protein modifications) in any type of biological samples including biofluids, cells, or tissues in biomarker pre-verification and systems biology studies.2, 21
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Lori Sokoll and Daniel Chan at the Johns Hopkins University for providing the clinical serum samples. Portions of this work were supported by the NIH New Innovator Award Program DP2OD006668, NCI Early Detection Research Network Interagency Agreement Y01-CN-05013-29, R33CA155252, U01CA111244, U24CA160019, and P41GM103493. The experimental work described herein was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the DOE and located at Pacific Northwest National Laboratory, which is operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL0 1830.
Abbreviations
- XIC
extracted ion chromatogram
- LOD
limit of detection
- LOQ
limit of quantification
- L/H
light to heavy
- PRISM
high-pressure, high-resolution separations with intelligent selection and multiplexing
- PSA
prostate-specific antigen
- SRM
selected reaction monitoring
- SISCAPA
stable isotope standards and capture by anti-peptide antibodies
- TFA
trifluoroacetic acid
Footnotes
ASSOCIATED CONTENT
Supporting Information Available
Supplemental tables and figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24(8):971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 2.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138(4):795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9(6):555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 4.Kijanka G, Ipcho S, Baars S, Chen H, Hadley K, Beveridge A, Gould E, Murphy D. Rapid characterization of binding specificity and cross-reactivity of antibodies using recombinant human protein arrays. J Immunol Methods. 2009;340(2):132–137. doi: 10.1016/j.jim.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Michaud GA, Salcius M, Zhou F, Bangham R, Bonin J, Guo H, Snyder M, Predki PF, Schweitzer BI. Analyzing antibody specificity with whole proteome microarrays. Nat Biotechnol. 2003;21(12):1509–1512. doi: 10.1038/nbt910. [DOI] [PubMed] [Google Scholar]
- 6.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5(4):573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100(12):6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6(12):2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lange V, Malmstrom JA, Didion J, King NL, Johansson BP, Schafer J, Rameseder J, Wong CH, Deutsch EW, Brusniak MY, Buhlmann P, Bjorck L, Domon B, Aebersold R. Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol Cell Proteomics. 2008;7(8):1489–1500. doi: 10.1074/mcp.M800032-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander A, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27(7):633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whiteaker JR, Zhao L, Anderson L, Paulovich AG. An automated and multiplexed method for high throughput peptide immunoaffinity enrichment and multiple reaction monitoring mass spectrometry-based quantification of protein biomarkers. Mol Cell Proteomics. 2010;9(1):184–196. doi: 10.1074/mcp.M900254-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Addona TA, Shi X, Keshishian H, Mani DR, Burgess M, Gillette MA, Clauser KR, Shen D, Lewis GD, Farrell LA, Fifer MA, Sabatine MS, Gerszten RE, Carr SA. A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease. Nat Biotechnol. 2011;29(7):635–643. doi: 10.1038/nbt.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bisson N, James DA, Ivosev G, Tate SA, Bonner R, Taylor L, Pawson T. Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nat Biotechnol. 2011;29(7):653–658. doi: 10.1038/nbt.1905. [DOI] [PubMed] [Google Scholar]
- 14.Whiteaker JR, Zhao L, Abbatiello SE, Burgess M, Kuhn E, Lin C, Pope ME, Razavi M, Anderson NL, Pearson TW, Carr SA, Paulovich AG. Evaluation of large scale quantitative proteomic assay development using peptide affinity-based mass spectrometry. Mol Cell Proteomics. 2011;10(4) doi: 10.1074/mcp.M110.005645. M110 005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuzyk MA, Smith D, Yang J, Cross TJ, Jackson AM, Hardie DB, Anderson NL, Borchers CH. Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol Cell Proteomics. 2009;8(8):1860–1877. doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malmstrom J, Beck M, Schmidt A, Lange V, Deutsch EW, Aebersold R. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature. 2009;460(7256):762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whiteaker JR, Lin C, Kennedy J, Hou L, Trute M, Sokal I, Yan P, Schoenherr RM, Zhao L, Voytovich UJ, Kelly-Spratt KS, Krasnoselsky A, Gafken PR, Hogan JM, Jones LA, Wang P, Amon L, Chodosh LA, Nelson PS, McIntosh MW, Kemp CJ, Paulovich AG. A targeted proteomics-based pipeline for verification of biomarkers in plasma. Nat Biotechnol. 2011;29(7):625–634. doi: 10.1038/nbt.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prakash A, Rezai T, Krastins B, Sarracino D, Athanas M, Russo P, Ross MM, Zhang H, Tian Y, Kulasingam V, Drabovich AP, Smith C, Batruch I, Liotta L, Petricoin E, Diamandis EP, Chan DW, Lopez MF. Platform for establishing interlaboratory reproducibility of selected reaction monitoring-based mass spectrometry peptide assays. J Proteome Res. 2010;9(12):6678–6688. doi: 10.1021/pr100821m. [DOI] [PubMed] [Google Scholar]
- 19.Shi T, Su D, Liu T, Tang K, Camp DG, 2nd, Qian WJ, Smith RD. Advancing the sensitivity of selected reaction monitoring-based targeted quantitative proteomics. Proteomics. 2012;12(8):1074–1092. doi: 10.1002/pmic.201100436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiyonami R, Schoen A, Prakash A, Peterman S, Zabrouskov V, Picotti P, Aebersold R, Huhmer A, Domon B. Increased selectivity, analytical precision, and throughput in targeted proteomics. Mol Cell Proteomics. 2011;10(2) doi: 10.1074/mcp.M110.002931. M110 002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn E, Whiteaker JR, Mani DR, Jackson AM, Zhao L, Pope ME, Smith D, Rivera KD, Anderson NL, Skates SJ, Pearson TW, Paulovich AG, Carr SA. Inter-laboratory evaluation of automated, multiplexed peptide immunoaffinity enrichment coupled to multiple reaction monitoring mass spectrometry for quantifying proteins in plasma. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.M111.013854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 23.Domanski D, Percy AJ, Yang J, Chambers AG, Hill JS, Freue GV, Borchers CH. MRM-based multiplexed quantitation of 67 putative cardiovascular disease biomarkers in human plasma. Proteomics. 2012;12(8):1222–1243. doi: 10.1002/pmic.201100568. [DOI] [PubMed] [Google Scholar]
- 24.Huttenhain R, Soste M, Selevsek N, Rost H, Sethi A, Carapito C, Farrah T, Deutsch EW, Kusebauch U, Moritz RL, Nimeus-Malmstrom E, Rinner O, Aebersold R. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci Transl Med. 2012;4(142):142ra94. doi: 10.1126/scitranslmed.3003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Percy AJ, Chambers AG, Yang J, Borchers CH. Multiplexed MRM-based Quantitation of Candidate Cancer Biomarker Proteins in Undepleted and Non-enriched Human Plasma. Proteomics. 2013 doi: 10.1002/pmic.201200316. [DOI] [PubMed] [Google Scholar]
- 26.Keshishian H, Addona T, Burgess M, Mani DR, Shi X, Kuhn E, Sabatine MS, Gerszten RE, Carr SA. Quantification of cardiovascular biomarkers in patient plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2009;8(10):2339–2349. doi: 10.1074/mcp.M900140-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rafalko A, Dai SJ, Hancock WS, Karger BL, Hincapie M. Development of a Chip/Chip/SRM Platform Using Digital Chip Isoelectric Focusing and LC-Chip Mass Spectrometry for Enrichment and Quantitation of Low Abundance Protein Biomarkers in Human Plasma. Journal of Proteome Research. 2012;11(2):808–817. doi: 10.1021/pr2006704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hossain M, Kaleta DT, Robinson EW, Liu T, Zhao R, Page JS, Kelly RT, Moore RJ, Tang K, Camp DG, 2nd, Qian WJ, Smith RD. Enhanced Sensitivity for Selected Reaction Monitoring Mass Spectrometry-based Targeted Proteomics Using a Dual Stage Electrodynamic Ion Funnel Interface. Mol Cell Proteomics. 2011;10(2) doi: 10.1074/mcp.M000062-MCP201. M000062MCP000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicol GR, Han M, Kim J, Birse CE, Brand E, Nguyen A, Mesri M, FitzHugh W, Kaminker P, Moore PA, Ruben SM, He T. Use of an Immunoaffinity-Mass Spectrometry-based Approach for the Quantification of Protein Biomarkers from Serum Samples of Lung Cancer Patients. Molecular & Cellular Proteomics. 2008;7(10):1974–1982. doi: 10.1074/mcp.M700476-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Hoofnagle AN, Becker JO, Wener MH, Heinecke JW. Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin Chem. 2008;54(11):1796–1804. doi: 10.1373/clinchem.2008.109652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whiteaker JR, Zhao L, Lin C, Yan P, Wang P, Paulovich AG. Sequential multiplexed analyte quantification using peptide immunoaffinity enrichment coupled to mass spectrometry. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.M111.015347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi T, Fillmore TL, Sun X, Zhao R, Schepmoes AA, Hossain M, Xie F, Wu S, Kim JS, Jones N, Moore RJ, Pasa-Tolic L, Kagan J, Rodland KD, Liu T, Tang K, Camp DG, 2nd, Smith RD, Qian WJ. Antibody-free, targeted mass-spectrometric approach for quantification of proteins at low picogram per milliliter levels in human plasma/serum. Proc Natl Acad Sci U S A. 2012;109(38):15395–15400. doi: 10.1073/pnas.1204366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu T, Hossain M, Schepmoes AA, Fillmore TL, Sokoll LJ, Kronewitter SR, Izmirlian G, Shi T, Qian WJ, Leach RJ, Thompson IM, Chan DW, Smith RD, Kagan J, Srivastava S, Rodland KD, Camp DG., 2nd Analysis of serum total and free PSA using immunoaffinity depletion coupled to SRM: correlation with clinical immunoassay tests. J Proteomics. 2012;75(15):4747–4757. doi: 10.1016/j.jprot.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T, Qian WJ, Mottaz HM, Gritsenko MA, Norbeck AD, Moore RJ, Purvine SO, Camp DG, 2nd, Smith RD. Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol Cell Proteomics. 2006;5(11):2167–2174. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riviere LR, Tempst P. Enzymatic digestion of proteins in solution. Curr Protoc Protein Sci. 2001;Chapter 11(Unit 11):1. doi: 10.1002/0471140864.ps1101s00. [DOI] [PubMed] [Google Scholar]
- 36.Fusaro VA, Mani DR, Mesirov JP, Carr SA. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat Biotechnol. 2009;27(2):190–198. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eyers CE, Lawless C, Wedge DC, Lau KW, Gaskell SJ, Hubbard SJ. CONSeQuence: Prediction of Reference Peptides for Absolute Quantitative Proteomics Using Consensus Machine Learning Approaches. Molecular & Cellular Proteomics. 2011;10(11) doi: 10.1074/mcp.M110.003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abbatiello SE, Mani DR, Keshishian H, Carr SA. Automated detection of inaccurate and imprecise transitions in peptide quantification by multiple reaction monitoring mass spectrometry. Clin. Chem. 2010;56(2):291–305. doi: 10.1373/clinchem.2009.138420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian WJ, Kaleta DT, Petritis BO, Jiang H, Liu T, Zhang X, Mottaz HM, Varnum SM, Camp DG, 2nd, Huang L, Fang X, Zhang WW, Smith RD. Enhanced detection of low abundance human plasma proteins using a tandem IgY12-SuperMix immunoaffinity separation strategy. Mol Cell Proteomics. 2008;7(10):1963–1973. doi: 10.1074/mcp.M800008-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diamandis EP, Scorilas A, Fracchioli S, Van Gramberen M, De Bruijn H, Henrik A, Soosaipillai A, Grass L, Yousef GM, Stenman UH, Massobrio M, Van Der Zee AG, Vergote I, Katsaros D. Human kallikrein 6 (hK6): a new potential serum biomarker for diagnosis and prognosis of ovarian carcinoma. J Clin Oncol. 2003;21(6):1035–1043. doi: 10.1200/JCO.2003.02.022. [DOI] [PubMed] [Google Scholar]
- 42.Havekes B, van Manen JG, Krediet RT, Boeschoten EW, Vandenbroucke JP, Dekker FW, Grp NS. Serum troponin T concentration as a predictor of mortality in hemodialysis and peritoneal dialysis patients. Am J Kidney Dis. 2006;47(5):823–829. doi: 10.1053/j.ajkd.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 43.Asgeirsson KS, Agrawal A, Allen C, Hitch A, Ellis IO, Chapman C, Cheung KL, Robertson JFR. Serum epidermal growth factor receptor and HER2 expression in primary and metastatic breast cancer patients. Breast Cancer Res. 2007;9(6) doi: 10.1186/bcr1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu CY, Wu MS, Chiang EP, Chen YJ, Chen CJ, Chi NH, Shih YT, Chen GH, Lin JT. Plasma matrix metalloproteinase-9 level is better than serum matrix metalloproteinase-9 level to predict gastric cancer evolution. Clin Cancer Res. 2007;13(7):2054–2060. doi: 10.1158/1078-0432.CCR-06-2299. [DOI] [PubMed] [Google Scholar]
- 45.Fujimoto K, Kawaguchi T, Nakashima O, Ono J, Ohta S, Kawaguchi A, Tonan T, Ohshima K, Yano H, Hayabuchi N, Izuhara K, Sata M. Periostin, a matrix protein, has potential as a novel serodiagnostic marker for cholangiocarcinoma. Oncol Rep. 2011;25(5):1211–1216. doi: 10.3892/or.2011.1194. [DOI] [PubMed] [Google Scholar]
- 46.Huttenhain R, Soste M, Selevsek N, Rost H, Sethi A, Carapito C, Farrah T, Deutsch EW, Kusebauch U, Moritz RL, Nimeus-Malmstrom E, Rinner O, Aebersold R. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci Transl Med. 2012;4(142):142ra94. doi: 10.1126/scitranslmed.3003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agger SA, Marney LC, Hoofnagle AN. Simultaneous quantification of apolipoprotein A–I and apolipoprotein B by liquid-chromatography-multiple- reaction-monitoring mass spectrometry. Clin Chem. 2010;56(12):1804–1813. doi: 10.1373/clinchem.2010.152264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Yang F, Gritsenko MA, Clauss T, Liu T, Shen Y, Monroe ME, Lopez-Ferrer D, Reno T, Moore RJ, Klemke RL, Camp DG, 2nd, Smith RD. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics. 2011;11(10):2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.