Abstract
In response to Catani et al., we show that corticospinal pathways adhere via sharp turns to two local grid orientations; that our studies have three times the diffusion resolution of those compared; and that the noted technical concerns, including crossing angles, do not challenge the evidence of mathematically specific geometric structure. Thus, the geometric thesis gives the best account of the available evidence.
In a Technical Comment, Catani et al. (1) question our findings of geometric structure of the pathways of the brain (2), suggesting that they may be technical artifacts. First, Catani et al.’s imaging and analysis of the example of the corticospinal tract is not completely accurate. Classic degeneration studies show that corticospinal pathways make exceedingly sharp turns between local expressions of two cardinal axes, confirming the grid thesis rather than refuting it (3). This feature is not reflected in the image presented by Catani et al., but is reflected in our images, presumably owing to improved technical characteristics ex vivo. Second, the account of resolution in diffusion magnetic resonance imaging (MRI) given by Catani et al. is incomplete. We show that the resolution for tissue microstructure in our ex vivo experiments is approximately threefold higher than the in vivo studies compared. Third, Catani et al. do not address the central observation of the study—the sheet structure of crossing pathways—or the evidence that this structure is both pervasive and a priori exceedingly improbable, hence equally unlikely to be an artifact. Thus, the thesis of geometric structure best accounts for the available evidence.
Catani et al. present the corticospinal tract as a counterexample to our model of geometric structure, showing diffusion MRI that depicts corticospinal and callosal paths as crossing in a continuum of angles from 60° down to 0° angles, identical orientations, at the vertex. Even if we allowed that the 60° crossing was consistent with developmental deformation of cardinal orientations at this location per our thesis, a continuous series of crossings down to 0° would be inconsistent with continuous deformation (2). Specifically, if the callosal paths express the transverse direction of a coordinate system and the projection paths as they appear in Catani et al. express the rostrocaudal, then they would need to remain distinct.
More detailed analysis of the corticospinal tract resolves this difficulty consistent with the geometric hypothesis. In a classic degeneration study, Krieg (3) showed that corticospinal fibers have extremely sharp turns; after an initial segment from the cortex parallel to callosal paths, they turn caudally, forming a rostrocaudal orientation field at each point nearly perpendicular to that of the callosal fibers (Fig. 1, A and B). Thus, corticospinal trajectories adhere to the local expressions of the transverse and rostrocaudal cardinal axes down to an exceedingly small scale, one below the resolution limits of these MRI studies. This observation lends our geometric thesis new and independent support and scope. We add that Krieg also finds equivalent structure in humans (3).
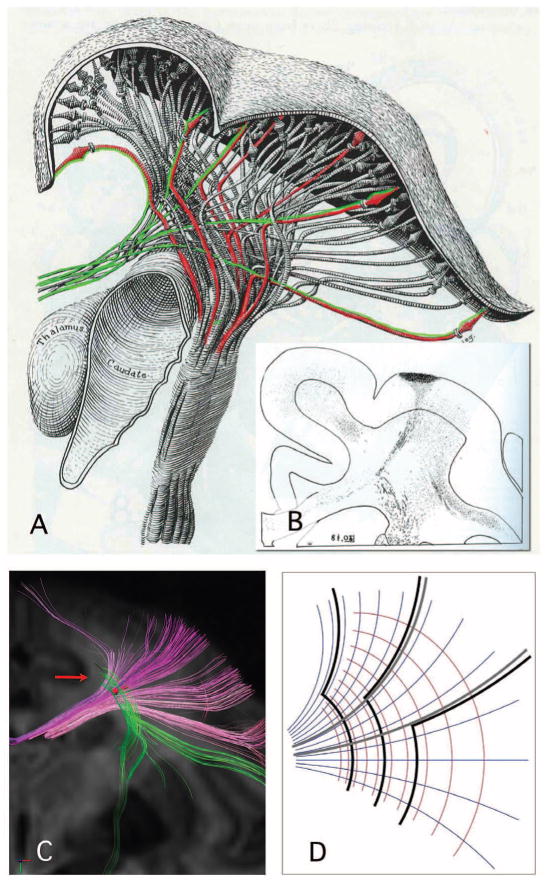
Fig. 1.
Corticospinal and callosal pathways of the rhesus monkey, modified from W. J. S. Krieg, drawing (A) [(3), figure 37] and degeneration study (B) [(3), figure 96], with permission. In (A), corticospinal paths (red) leave the prefrontal cortex parallel to transverse callosal paths (green), then turn sharply by approximately 90° (“drop”) to the caudal direction. Even medial corticospinal paths from the vertex show this turn as small as sharp kinks. The degeneration study shown in (B) shows an example of primary data upon which (A) was based. Here, the path turns are so sharp that they show no visible curvature at the submillimeter resolution of this optical tracing. In DSI of rhesus monkey ex vivo (C), rostrocaudal segments of corticospinal paths (vertical) are nearly perpendicular locally to curved callosal paths (horizontal). These rostrocaudal segments terminate among transverse paths (arrow), consistent with turns too sharp to resolve in this study. An illustrative model of the geometric thesis [the mapping of the complex variable z] (D). The callosal paths (gray) adhere to a transverse coordinate (blue-gray), whereas corticospinal paths (black) adhere to both transverse and rostrocaludal (red) orientations via sharp 90° turns.
Thus, diffusion spectrum imaging (DSI) in the rhesus shows terminations of corticospinal paths where they turn sharply (Fig. 1C). This should be expected with current MRI tractography predicated on orientation continuity (i.e., path curvature < ~1 rad/voxel) (4, 5). Conversely, the smooth appearance of corticospinal paths shown by Catani et al. is consistent with an artifact produced by low spatial and/or diffusion resolution, both of which will tend to smooth out sharp turns. This case indicates that grid structure has implications for the MRI tractography of connectivity (2). Closely related, these sharp turns seriously challenge hypotheses that brain pathways adhere to minimal or geodesic paths in any metric (4) or are determined by mechanical tension in any simple way (6).
In the present case, the role of resolution in diffusion MRI is to enable the differentiation of populations of fibers within a voxel based on their orientation (4, 5, 7–9). The principal determinants of resolution of microstructure in diffusion MRI are the physical parameters of the encoding, whereas sampling pattern is secondary. Thus, for example, diffusion tensor MRI, however sampled, has little or no ability to resolve crossing (7–10).
We have proposed a model of resolution in diffusion MRI that includes the combined effects of the spatial frequency and mixing time, q and Δ, of the diffusion encoding (11). To map unknown microstructure, the resolution Reff of diffusion MRI may be considered the sum of the encoding resolution r = 2πq−1 and a spatial blur due to diffusion itself, whose radius is the Einstein length (2DΔ)1/2 for diffusivity D. As these are uncorrelated,
| (1) |
Assuming brain diffusivity D ≥ 0.2 Dwater, then the in vivo studies of Catani et al. would have Reff ≈ 20 μm and our ex vivo studies with Reff ≈ 6.5 μm, or more than threefold improved resolution. As shown in the supporting materials, improved diffusion resolution yields improved detection of crossing fibers (12).
Regarding the choice of DSI over Q-ball imaging or high angular resolution diffusion imaging, these latter methods effectively sample only a subset of the DSI; setting aside noise effects, they are equivalent to DSI with a band-pass filter (8–10). Hence, we believe that DSI should present the lower risk of bias (5) and consider this more conservative choice appropriate when microstructure is uncertain or at issue.
We have claimed that the pathways of the brain adhere to a three-dimensional (3D) coordinate system derived by smooth deformation from the three cardinal axes of development. The clearest evidence for this structure in the mature brain is the sheet structure. Although the body axes in the early embryo were orthogonal, they do not in general remain so in the mature brain owing to plastic deformations of development and growth that lead to curvatures in the lobes, sulci, and gyri in the mature brain structure. This follows from the theorem that the only conformal, or angle-preserving, mappings in three dimensions are rigid motions or isotropic scaling. Such deviations from orthogonality were qualitatively observed in all species [figure 4 in (2)] and measured in rhesus, where they were found to increase from cerebral center to mantle to surface [figure S2D in (2)], consistent with known characteristics of developmental deformation (13, 14). We fully agree that non–right-angle crossings exist, but this is consistent with and indeed required by our thesis that brain pathways adhere to local expressions of curved cardinal coordinates.
Central to our thesis is the finding of sheet structure in cerebral fibers. We have shown that the pathways of the brain are equivalent to coordinate functions because they form in crossing parallel 2D sheets that fill 3D space like pages of a book. As we emphasize, this is mathematizcally specific and highly atypical, entailing long-range correlations between paths that are as nonrandom as a lock and key (having prior probability ≈ 0). This property does not depend on fiber orthogonally or the absence thereof—the concern of Catani et al.—but rather on a 3D relationship among crossing planes at different locations (the Frobenius integrability condition). As we have shown, this can be represented as an angle between subsheets of fibers, which must be as close to zero as noise allows [figure S2C in (2)], or by the topology of the embedding of the reconstructed paths in 3D, which must be interwoven rather than mutually helical [figure S2A in (2)]. Whereas this sheet structure may be obscured by low resolution or other technical limitations, no mechanisms are known whereby these limitations will create it as an artifact. Because we have observed this structure to be pervasive in cerebral white matter, homologous across species, consistent with embryogenesis, and consistent across diffusion contrast mechanisms, with and without Fourier-Radon reconstruction, we conclude that this organization is real and characteristic of the brain pathways (2). The thesis that brain pathways adhere to a simple geometric system best accounts for the available evidence—not like London, but Manhattan; not unfathomable, but unlimited.
Supplementary Material
Acknowledgments
We thank K. B. Blagoev, B. R. Rosen, H. E. Stanley, J. W. Belliveau, D. K. Jones, and L. L. Wald for helpful discussions, and S. A. Wedeen and L. C. Abbate for reviewing this paper. This work is directly supported by NSF PHY-0855161 and by NIH R01-MH652456, P41 RR-023953, P41 RR-14075, and U01 MH093765, the Human Connectome Project. Massachusetts General Hospital holds patent no. U.S. 6,614,226 on the DSI method of magnetic resonance imaging.
Footnotes
References and Notes
- 1.Catani M, Bodi I, Dell’Acqua F. Science. 2012;337:1605. doi: 10.1126/science.1223425. www.sciencemag.org/cgi/content/full/337/6102/1605-d. [DOI] [PubMed] [Google Scholar]
- 2.Wedeen VJ, et al. Science. 2012;335:1628. doi: 10.1126/science.1215280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krieg WJS. Connections of the Frontal Cortex of the Monkey. Charles C. Thomas; Springfield Il: 1954. figures 37 and 96. [Google Scholar]
- 4.Wedeen VJ, et al. Proc 1st Intl Conf Funct Map Human Brain. 1995;P1:69. [Google Scholar]
- 5.Wedeen VJ, et al. Neuroimage. 2008;41:1267. doi: 10.1016/j.neuroimage.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 6.Van Essen DC. Nature. 1997;385:313. doi: 10.1038/385313a0. [DOI] [PubMed] [Google Scholar]
- 7.Wiegell MR, Larsson HB, Wedeen VJ. Radiology. 2000;217:897. doi: 10.1148/radiology.217.3.r00nv43897. [DOI] [PubMed] [Google Scholar]
- 8.Tuch DS, et al. Magn Reson Med. 2002;48:577. doi: 10.1002/mrm.10268. [DOI] [PubMed] [Google Scholar]
- 9.Tuch DS, Reese TG, Wiegell MR, Wedeen VJ. Neuron. 2003;40:885. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 10.Wedeen VJ, Hagmann P, Tseng WYI, Reese TG, Weisskoff RM. Magn Reson Med. 2005;54:1377. doi: 10.1002/mrm.20642. [DOI] [PubMed] [Google Scholar]
- 11.Wedeen VJ, Dai G. Intl Soc Mag Reson Med. 2011;19:1965. [Google Scholar]
- 12.Wedeen VJ, et al. Intl Soc Mag Reson Med. 2012;20:1876. [Google Scholar]
- 13.Burnside MB, Jacobsen AG. Dev Biol. 1968;18:537. doi: 10.1016/0012-1606(68)90025-0. [DOI] [PubMed] [Google Scholar]
- 14.Nieuwenhuys R. Brain Struct Funct. 2009;214:79. doi: 10.1007/s00429-009-0223-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.