Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1) has become an important pharmacological target in the treatment of cancer due to its cellular role as a ‘DNA-strand break sensor’, which leads in part to resistance to some existing chemo- and radiological treatments. Inhibitors have now been developed which prevent PARP-1 from synthesizing poly(ADP-ribose) in response to DNA-breaks and potentiate the cytotoxicity of DNA damaging agents. However, with the recent discoveries of PARP-2, which has a similar DNA-damage dependent catalytic activity, and additional members containing the ‘PARP catalytic’ signature, the isoform selectivity and resultant pharmacological effects of existing inhibitors are brought into question. We present here the crystal structure of the catalytic fragment of murine PARP-2, at 2.8 Å resolution, and compare this to the catalytic fragment of PARP-1, with an emphasis on providing a possible framework for rational drug design in order to develop future isoform-specific inhibitors.
INTRODUCTION
Poly(ADP-ribose) polymerase-2 (PARP-2) (1–3) is one member of a growing family (18 proteins to date) related by a highly conserved catalytic fragment (CF) (J.-C.Amé et al., submitted), capable of synthesizing polymers of ADP-ribose, utilizing NAD+ as a substrate. Characterized family members currently include the proteins PARP-1, PARP-3 (2,4), Tankyrase 1 (5), Tankyrase 2 (6,7), TiPARP (8) and vPARP (9). To date, the only proteins of this family known to generate poly(ADP-ribose) (PAR) as a direct response to DNA breaks are PARP-1 and -2 (10). These proteins bind directly to single-strand DNA breaks (PARP-1) or gaps (PARP-1 and -2) and as such are enzymatically hyper-activated (∼500-fold over basal levels for PARP-1); the ADP-ribose moiety of bound NAD+ is then transferred to specific acceptor proteins, and to PARP-1 and -2 themselves via an auto-modification reaction. With successive enzymatic rounds, this initial post-translational modification can be extended to yield long, branched, polymeric chains of ADP-ribose [reviewed in de Murcia and Shall (11) and Murkle (12)].
Given that all currently identified PAR acceptors are DNA-binding proteins, and that PAR itself has considerable chemical and biophysical similarity to nucleic acids, PAR covalently attached to a DNA-binding protein could compete for the DNA-binding site on the modified protein and displace it from DNA, allowing DNA repair proteins access to the DNA break. Known polymer acceptors include histones (mainly H1 and H2B) (13), DNA topoisomerases I and II (13,14), DNA polymerase β (15), p53 (16) and XRCC1 (17). Immunoprecipitation and co-purification experiments have demonstrated that both PARP-1 and PARP-2 are directly associated with a short-patch base excision repair (BER) complex, comprising the proteins XRCC1, DNA polymerase β and DNA Ligase IIIα (10,17–20). Moreover, the importance of both PARP-1 and -2 in BER and single-strand break repair (SSBR) has been supported by a wide range of experimental data; knock-out mice deficient in either protein, show hypersensitivity to both alkylating agents and γ-irradiation (both strong inducers of SSBR) (21,22), and repair of alkylated bases in cell lines derived from these mice, shows a significant delay or defect (10,23,24). Additional evidence to support a role for the PARPs in BER and SSBR, arises from the observation that short oligomeric lengths of ADP-ribose, attached to PARP-1 or -2, are necessary for either the recruitment or enhanced binding of BER/SSBR associated protein partners, principally XRCC1 (10,17,20,25). Further more, in living cells, PAR production in the vicinity of localized DNA strand breaks has recently been shown to be essential for the rapid recruitment of XRCC1 to the damage sites [Okano et al. (26) and J.-C.Amé et al., submitted].
Because it is implicated in the resolution of DNA breaks, PARP-1 has attracted a great deal of attention as a therapeutic drug target in the treatment of cancer. A number of methodologies have been proposed; including the use of PARP-1 inhibitors to obstruct DNA repair, thus increasing the cytotoxicity of certain anticancer agents (both in chemo- and radiotherapies) (27,28). It has also been selected as a target for medical conditions involving oxidant-mediated cell death, such as ischaemia-reperfusion injuries, neurodegenerative disorders and endotoxic shock, due to its involvement in other cellular pathways that are not discussed here [reviewed in (29,30)].
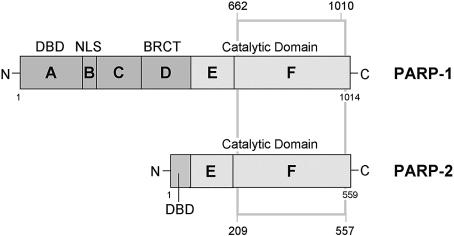
Until relatively recently, PARP-1 was believed to be responsible for all the DNA-damage dependent PAR synthesis in mammalian cells. However, a second DNA-damage dependent activity, PARP-2, was subsequently identified (1). Like PARP-1, PARP-2 interacts with components of the BER and single-strand gap-repair pathways, and can probably function as both a homo- and heterodimer with PARP-1 (10). PARP-2 lacks the N-terminal tandem zinc fingers and BRCT domain of PARP-1, which are replaced by a small highly basic N-terminal DNA-binding domain, with the E domain acting both as a dimerization and automodification domain (10), but shares the C-terminal catalytic domain, which is the unifying feature of the wider PARP family (Fig. 1). The identification of 18 PARPs in mammalian cells potentially complicates the development of inhibitors intended to achieve targeted pharmacological effects. With this in mind, we have now determined the crystal structure of the catalytic fragment of murine PARP-2 (PARP-2-CF) at 2.8 Å resolution. Comparison with the known structure of chicken PARP-1-CF provides a rational framework for the future development of type-specific PARP inhibitors.
Figure 1.
Schematic representation of the functional domains of chicken PARP-1 and mouse PARP-2. The grey box indicates the crystallized region of PARP-2 and compares it to the previous crystal structure of chicken PARP-1 (PARP-1-CF, PDB: 2PAW). Protein domains (A–F) are labelled according to de Murcia and Shall (11). DBD = DNA binding domain; NLS = nuclear localization sequence; BRCT = BRCA1 C-terminus.
MATERIALS AND METHODS
Expression and purification
Full-length recombinant murine PARP-2 (PARP-2-FL) was expressed in insect cells (Spodoptera frugiperda, Sf9). The baculovirus encoding the full-length protein, was generated as described previously (1). Insect cells were cultured in shaker flasks, containing SF900II media (supplemented with penicillin and streptomycin), with growth conditions of 27°C and 150 r.p.m., as per the manufacturer’s instructions (Life Technologies). Virus was amplified from cells infected at a cell density of 2.0 × 106 cells/ml, a multiplicity of infection (MOI) of 0.1, and a 3-day incubation period. For expression, Sf9 cells were infected at the same cell density, but with an MOI of 2, and a 3-day incubation period. Cells were harvested by centrifugation at 1000 g for 5 min, and the resulting cell pellet stored at –80°C until required.
The cell pellet resulting from 1.25 l of expression culture was resuspended, on ice, in 45 ml of PBS A; 171 mM NaCl, 10.6 mM KH2PO4, 3.35 mM KCl, 1.76 mM Na2HPO4, pH 7.2, supplemented with protease inhibitors. Cells were lysed through a combination of the thawing process, hand-homogenization and a brief sonication step. Cell debris was then removed by high-speed centrifugation at 40 000 g for 45 min. The resulting supernatant was additionally clarified by the addition of protamine sulphate to a final concentration of 1 mg/ml. Precipitated material was again removed by high-speed centrifugation.
Heparin Sepharose 6 resin (Pharmacia) was added to the clarified supernatant and incubated, with mixing, at 4°C for 30 min (∼16 ml resin per 45 ml cell extract). The resin slurry was then distributed between a number of disposable plastic chromatography columns, and washed with successive volumes of PBS A + 100 mM NaCl. Partially purified protein was eluted with PBS A + 450 mM NaCl. Fractions containing PARP-2-FL were identified and pooled, then diluted ∼6-fold with the addition of 100 mM Tris–HCl pH 7.5, 1 mM DTT, 0.5 mM EDTA (to reduce the overall NaCl concentration to ∼100 mM). This was then applied to an ECH-Sepharose 4B (Pharmacia)/3-aminobenzamide coupled affinity column. The column was washed with a linear salt gradient from 0.1 to 1 M NaCl in 100 mM Tris–HCl pH 7.5, 1 mM DTT, 0.5 mM EDTA, and bound protein eluted with 100 mM Tris–HCl, 400 mM NaCl, 3 mM 3-methoxybenzamide, 1 mM DTT, 0.5 mM EDTA. The protein was buffer exchanged into 10 mM Tris–HCl pH 8.0, 100 mM NaCl, and concentrated to 24 mg/ml using Vivaspin 500 concentrators (10 kDa cut-off). Purified PARP-2-FL was stored at 4°C.
Crystallization and data collection
Crystallization trials were carried out at 24 mg/ml in hanging-drop experiments using Structure Screen I (MDL). Small orthorhombic crystals were observed in condition 38. This condition was optimized, again in hanging-drop experiments, to mixing 1 µl of protein (24 mg/ml in 10 mM Tris–HCl pH 8.0, 100 mM NaCl) with 1 µl of precipitant containing 9% PEG 8000, and 100 mM Tris–HCl pH 7.5. Data to 2.8 Å were collected from a single crystal at 100 K at the SRS, Grenoble, and recorded on an ACSD scanner. Images were integrated using MOSFLM (31) and reduced/scaled using programs of the CCP4 suite (32). The protein crystallized in space-group P212121 with cell dimensions of a = 83.65 Å, b= 85.82 Å and c = 139.46 Å. Statistics for the data collection are given in Table 1.
Table 1. Crystallographic statistics.
| Data collection | All data (outer shell) |
|---|---|
|
Rmerge |
0.073 (0.255) |
|
I/σ(I) |
4.2 (2.4) |
| Completeness (%) |
99.9 (99.9) |
| Multiplicity |
4.0 (4.1) |
| No. of unique reflections |
25 407 |
| Structure refinement |
|
| No. of atoms (protein) |
5461 |
| No. of atoms (All) |
5558 |
| Resolution range (Å) |
29.95–2.8 |
|
Rcryst |
0.244 |
| Rfree | 0.268 |
Analysis of crystal content
Calculations of crystal volume were not commensurate with integral numbers of the full-length protein in the asymmetric unit, so that direct analysis of crystal content was required. Several crystals (≈30) were harvested from a crystallization drop, and washed in successive volumes of Ultra-pure water. After washing, crystals were pooled, then dissolved in 10 mM Tris–HCl pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM DTT and analysed on a 12.5% SDS–PAGE gel. Crystallized material corresponded to a polypeptide migrating with an apparent molecular mass of ≈50 kDa, indicating that a proteolytic or break-down product had crystallized, rather than the full-length protein. N-terminal sequencing and mass-spectrometry confirmed the crystalline protein as a C-terminal fragment (residues 209–557). On the basis of this, the asymmetric unit was expected to contain two molecules with a solvent content of 61% (v/v).
Structure determination and refinement
The structure of the crystallized fragment of murine PARP-2 was determined by molecular replacement using AMoRe (33). The structure of the catalytic domain from chicken PARP-1 was used as a search model (PDB: 2PAW), but with the amino acid sequence of PARP-2 threaded onto the PARP-1 fold. Two solutions were obtained, and improved by rigid body refinement and subsequent simulated annealing in CNS (34). Difference maps were used to rebuild the initial model in O (35). Iterative cycles of refinement and manual intervention gave the current model which consists of 5461 protein atoms and 96 solvent atoms, with R = 0.244 and Rfree = 0.268 for ≈5% of the data omitted from refinement. NCS restraints were throughout refinement, with the exclusion of amino acid groups 207–215, 235–250, 290–296, 320–335, 343–357 and 374–380, which showed deviations in conformation between the two crystallographically independent molecules. The r.m.s. deviation for all Cα atoms between the two molecules is 0.124 Å. Coordinates have been deposited in the Protein Databank (code: 1GS0). Crystallographic statistics are given in Table 1.
RESULTS
Full-length murine PARP-2 was expressed in insect cells and purified to homogeneity (see Materials and Methods). In crystallization trials, crystals took extended periods of time to appear, in some cases up to 6 months from the date of experimental set-up. Despite full-length protein being used, slow proteolysis during the time course of the experiment produced a stable fragment of ≈50 kDa (as judged by SDS–PAGE analysis) which crystallized.
N-terminal sequencing and western blots identified the crystalline protein as a C-terminal fragment (residues 194–559) encapsulating the catalytic function of the protein, but lacking the putative 7 kDa DNA-binding domain and 16 kDa dimerization domains. The structure of this PARP-2 catalytic fragment (PARP-2-CF) was determined by molecular replacement with the structure of chicken PARP-1-CF (PDB: 2PAW), and refined at 2.8 Å (see Materials and Methods). Two molecules of PARP-2-CF are present in the asymmetric unit of the crystal. Clear electron density corresponding to residues 209–557 of the full-length protein can be seen for both molecules, apart from the loops comprised of residues 325–330 which appear to be poorly ordered in these crystals.
PARP-2-CF structure
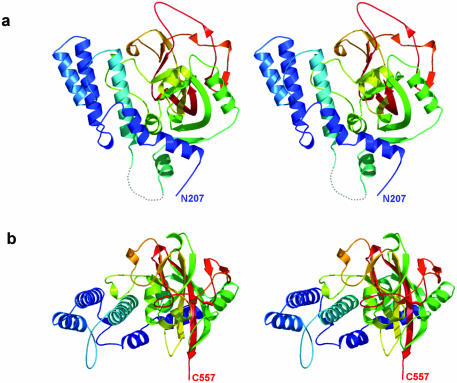
The PARP-2-CF structure consists of two main parts: an α-helical N-terminal domain comprising residues 207–324, and a mixed α/β C-terminal domain (residues 332–557) containing the catalytic glutamate residue (Glu534) (Fig. 2). As expected, the overall fold of PARP-2-CF is very similar to that of PARP-1-CF (36), with an r.m.s. deviation of 1.16 Å between 339 common Cα positions when the structures are superimposed (37) (Figs 3 and 5a).
Figure 2.
Structure of the CF of murine PARP-2. (a) Stereo-pair secondary structure cartoon of PARP-2-CF coloured blue to red from the visible N-terminus at residue 207 to the visible C-terminus at residue 557. The disordered loop comprising residues 325–331 is represented as a dotted line. (b) As (a), but rotated 90° around the horizontal axis.
Figure 3.
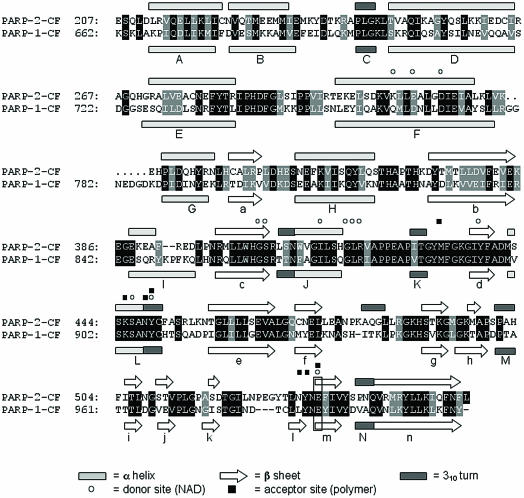
Secondary structure-based sequence alignment between PARP-2-CF and PARP-1-CF. A sequence alignment for PARP-2-CF and PARP-1-CF is shown, shaded to detail both identical residues (black) and conserved residues (grey) between the two molecules. Secondary structure elements are also shown, labelled according to Ruf et al. (36). Amino acid residues important for catalytic function are also highlighted.
Figure 5.
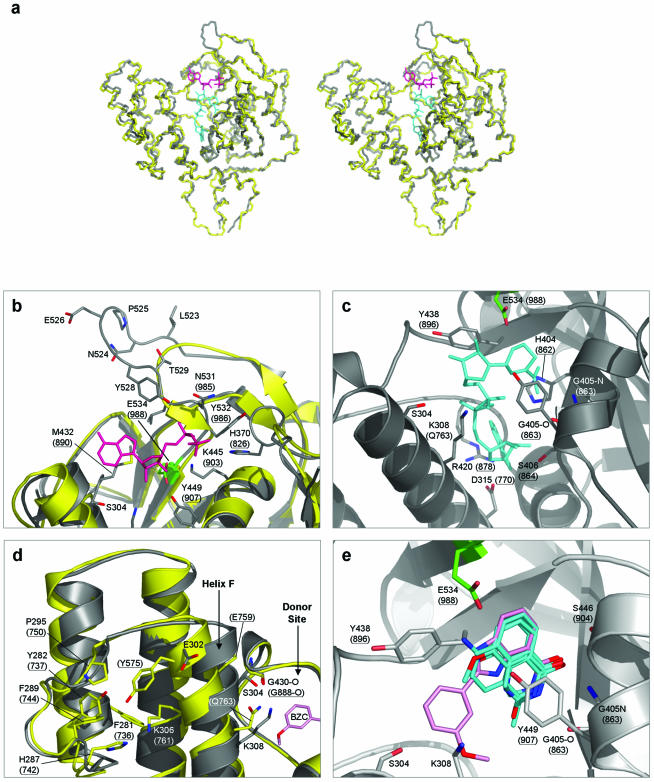
Amino acid residues shown both in parentheses and underlined are in PARP-1. All other residues are in PARP-2. In addition, cartoon representations of PARP-1 are coloured yellow, and those of PARP-2 coloured grey. The catalytic residue E534 (E988 in PARP-1) is highlighted in green, in all cases. (a) Superimposed Cα traces of PARP-1-CF and PARP-2-CF. Stereo-pair diagram showing superimposed Cα backbone traces for PARP-1-CF (PDB: 2PAW) and PARP-2-CF. Also shown are stick representations of the ADP-ribose moiety of carba-NAD (shown in magenta) and NAD+ (cyan). The positions of carba-NAD and NAD+ were modelled by comparisons with the ligand-bound crystal structures of PARP-1 (PDB: 1A26), and diphtheria toxin (PDB: 1TOX), respectively. (b) Polymer acceptor site. The ADP-ribose moiety of NAD+ (coloured magenta) is shown bound to the donor site of PARP-1 (PDB: 1EFY), along with a superimposition of the equivalent secondary structure elements in PARP-2. All residues involved in coordinating the substrate are highly conserved between the two PARP molecules. The extended loop unique to PARP-2 is clearly visible, consisting of residues Leu523–Thr529. In particular, residue Tyr528 is presented towards the acceptor site, possibly providing additional interactions with the pyrophosphate backbone of the bound substrate. (c) Polymer donor site. A model for NAD+ (shown in cyan) bound to the donor site of PARP-2 is shown. All residues involved in coordination of the substrate are again strongly conserved between PARP-1 and PARP-2, apart from residue Gln763, which is replaced by Lys308 in PARP-2. (d) Helix F in PARP-2 is displaced towards the donor site. In comparison to PARP-1, the top of helix F in PARP-2 is displaced towards the donor site of the molecule. This is due to the replacement of the side-chain of Tyr575 in PARP-1, normally buried in a hydrophobic pocket, with the solvent-exposed residue Glu302 in PARP-2. (e) PARP inhibitors bound to the donor site. PARP inhibitors (for which the structures of complexes with PARP-1 are available) are shown bound to the donor site of PARP-2. Superimposition of topologically equivalent residues, between PARP-1 and PARP-2, facilitated the modelling of these inhibitors. The inhibitor 2-(3′-methoxyphenyl) benzimidazole-4-carboxamide (BZC) is highlighted (shown in pink), demonstrating the proximity of helix F and hence residues Ser304 and Lys308 to the donor site, providing possible targets for future PARP-2 selective/discriminative inhibitors. The inhibitors shown are: 3,4-dihydro-5-methyl-isoquinolinone (DHQ, PDB: 1PAX), 4-amino-1,8-naphthalimide (4AN, PDB: 2PAX), 3-methoxybenzamide (3 MB, PDB: 3PAX) and BZC (PDB: 1EFY).
The majority of secondary structure elements are completely conserved between the two CFs; all α-helix, β-sheet and 310 helix motifs found in PARP-1-CF are also present in PARP-2-CF, with the notable exception of β-strand l which is absent from the PARP-2-CF structure due to a structural rearrangement as a result of a three-residue insertion after β-strand k (Fig. 3). An additional 310 helix (residues 481–483) is also found in PARP-2-CF, forming part of the loop connecting β-strands f and g.
Catalytic mechanism of PARP
PARP-1 is a target of substantial pharmaceutical interest (38), and a number of crystal structures of PARP-1-CF in complex with inhibitors have been reported. Although structures of PARP-2 in complex with inhibitors are not yet available, the overall similarity of the PARP-1 and PARP-2 CFs allows a detailed comparison between the ligand binding sites, which will have excellent predictive power for the future design of isoform-specific PARP inhibitors.
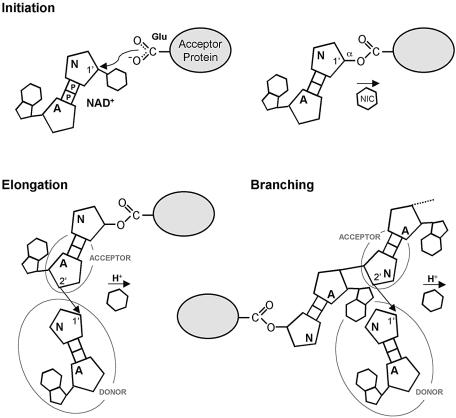
PARP-1 and PARP-2 are able to catalyze three related but distinct reactions in which the ADP-ribose moiety of NAD+ is transferred to one of three different acceptor groups, which in ordered combination generate PAR-modified proteins. ‘Initiation’ involves transfer to the carboxylate group of a glutamic acid side chain on the modified protein, while subsequent ‘elongation’ and ‘branching’ reactions transfer ADP-ribose to the 2′-hydroxyl of the adenine or nicotinamide ribose of an already attached ADP-ribose chain. The sequential nature of the reaction therefore requires adjacent binding sites for the ‘acceptor’ terminal ADP-ribose of the existing poly- or oligo-ADP-ribose chain, and for the ‘donor’ NAD+ ligand that will provide the ADP-ribose to be added (Fig. 4).
Figure 4.
Mechanism of poly-ADP-ribosylation by PARP-1 and PARP-2. PARP-1 and PARP-2 catalyse the transfer of the ADP-ribose moiety of NAD+ to a nucleophilic acceptor. In an initiation reaction, the acceptor is the carboxyl side chain of a glutamate residue on the acceptor protein, but is the 2′ hydroxyl of the adenine or nicotinamide ribose of the poly-ADP-ribose in elongation and branching reactions, respectively. A = adenine-ribose; P = phosphate; N = nicotinamide-ribose, and NIC = nicotinamide.
PARP-2 acceptor site
To analyse interactions in the PARP-2 acceptor site, we modelled ADP into the crystal structure of PARP-2-CF by superimposing the crystal structure of the PARP-1-CF complex with carba-NAD (PDB: 1A26), as it has been demonstrated previously that the ADP moiety of this ligand delineates the binding site of a poly(ADP-ribose) acceptor molecule (39). All amino acid residues involved in the coordination of the ADP-moiety are conserved in PARP-2-CF, both in their identity and in their spatial arrangement (Fig. 5b). The pyrophosphate backbone can be bound through the formation of hydrogen bonds to the Nε2 atom of His370 (826: numbers in brackets indicate the topologically equivalent residue in PARP-1), the ζ-amino group of Lys445 (903) and the backbone amides of residues 531 and 532 (985 and 986). In addition, the hydroxyl groups of the ribose sugar can hydrogen-bond to Tyr449 (907) and to the catalytic residue Glu534 (988), with the adenine moiety packed against the side-chain of Met432 (890).
The local environment of the acceptor site in PARP-2-CF is modified compared to PARP-1 however, due to a three-amino acid insertion in the loop connecting β-strands k and l (Figs 3 and 5a). Comparison of this loop in the two PARP structures identifies PARP-2 residues Leu523 and Leu530 as topologically equivalent to residues Asn980 and Leu984 of PARP-1, but with the additional PARP-2 residues 524-Asn-Pro-Glu-Gly-Tyr-Thr-530 forming a six-residue excursion from the backbone path observed in PARP-1-CF (Fig. 5b). Within the longer PARP-2 loop, the side chain phenyl of Tyr528 (which has no equivalent in PARP-1) points directly into the acceptor site, with the hydroxyl group in a position to hydrogen-bond to the Nδ2 of Asn531 and/or to interact with the pyrophosphate backbone of bound PAR. Although the loop is well-ordered in the crystals, it has different conformations in the two crystallographically independent molecules in the crystal structure, with the peptide bond preceding Pro525 in a trans conformation in one and cis in the other. As well as providing the binding site for the terminal ADP-ribose of an existing chain in elongation and branching reactions, this site must also furnish interactions to position the glutamate and adjacent polypeptide chain of a protein acceptor in the initiation reaction. The nature of these interactions, which may afford some degree of protein substrate specificity for ADP-ribosylation, are unknown in both PARP-1 and -2.
PARP-2 donor site
In the absence of an experimental structure for NAD+ bound to PARP-1 or PARP-2, we have modelled the binding of NAD+ in the donor site of the PARP-2-CF molecule, on the basis of the structure of the NAD+-bound diphtheria toxin (a mono-ADP-ribosyltransferase) (PDB: 1TOX) (40) as previously described for PARP-1 (41). As with the acceptor site, most of the residues involved in coordinating NAD+ within the donor site are conserved in PARP-1 and PARP-2 (Fig. 5c) (41). The 2′ hydroxyl residue of the donor ribose group makes a hydrogen bond with the side-chain carboxyl of the catalytic residue Glu534 (988). This interaction positions the C1′N for nucleophilic attack by the 2′ hydroxyl of the acceptor ribose group. Additional conserved hydrogen bonding interactions serve to position the donor NAD+ molecule: the backbone oxygen of Gly418 (876) to N6A, Asp315-Oδ2 (770) to N7A, the backbone nitrogen of Arg420 (878) to N1A, Ser406-Oγ (864) to O2′A, the backbone oxygen of Gly405 (863) to N7N, the backbone nitrogen of Gly405 to O7N, Ser446-Oγ (904) to O7N, and Tyr449-OH (907) to O4′N. The only difference between PARP-1 and PARP-2 in donor site interactions, is the replacement of Gln763 in PARP-1, with a lysine (308) in PARP-2. Gln763 hydrogen bonds to the pyrophosphate moiety of the donor site NAD+ via its side-chain amide group, but this interaction could be fully recapitulated by the Nε of Lys308.
PARP inhibitor interactions
A large number of PARP-1 enzymatic inhibitors have been developed, the majority of which mimic to some degree the nicotinamide moiety of NAD+ and bind to the donor site of the protein. The residues in the donor site of PARP-1 that provide hydrogen-bonding interactions with these inhibitors, are completely conserved in PARP-2. Hydrogen bonds can be formed between Gly405-N (863), Gly405-O (863) and Ser446-Oγ (904) and an inhibitor’s amide or lactame group with additional, non-polar interactions with Tyr438 (896) and/or Tyr449 (907). Additional inhibitor specificity has been correlated with the formation of additional hydrogen bonds either to the catalytic glutamate, Glu534 (988), or to the hydroxyl group of Tyr449 (907). Superimposition of known PARP-1/inhibitor complexes [PDB: 1EFY, 1PAX, 2PAX, 3PAX and 4PAX; (41,42)] onto the PARP-2-CF structure, shows that the binding environment in both PARPs is virtually identical, suggesting that these existing PARP-1 inhibitors would show little discrimination between PARP-1 and PARP-2 (Fig. 5e). However, the position of 2-(3′-methoxyphenyl) benzimidazole-4-carboxamide (BZC; PDB: 1EFY) within the donor site suggests that selectivity for (or against) PARP-2 inhibition could be introduced by inhibitor groups designed to interact with the positively charged terminal amino group of Lys308, which replaces the polar side chain of Gln763 found at this position in PARP-1. A second possible source of selectivity results from the displacement in PARP-2, relative to PARP1, of the top of helix F towards the catalytic site. In all the currently available PARP-1 structures (both in the presence or absence of bound inhibitors/substrates), helix F is presented in the same conformation—with no movement of the helix either towards or away from the catalytic site. However, this helix is displaced in PARP-2 due to the replacement of the buried side-chain of Tyr757 in PARP-1, by the exposed side-chain of Glu302 in PARP-2, which presents the side-chain hydroxyl of Ser304 towards the donor site (Fig. 5d). Additional iso-form specificity could also be introduced by production of inhibitors that could simultaneously occupy both the donor and acceptor sites of the protein, providing hydrogen-bonding access to Tyr528; the most apparent difference between the two CFs.
DISCUSSION
Of the 18 PARP activities identified in mammalian cells so far, only PARP-1 and PARP-2 are activated by the presence of interruptions in the DNA, and thereby are implicated in DNA damage repair. Consistent with this, both PARP-1 and -2 interact with the enzymes involved in small-patch gap repair (DNA polymerase β and DNA ligase III) and the associated XRCC1 scaffold protein (10). In addition, PARP-1 and -2 can both homo- and heterodimerize in vitro and reciprocally trans-ADP-ribosylate each other, suggesting that they may function as a unit in vivo (43). PARP-1 and PARP-2 may have redundant functions which are nonetheless essential for viability, as mice embryos homozygously defective in both genes (PARP-1 –/–, PARP-2 –/–) die with the onset of gastrulation (43).
The overall three-dimensional structure of the CF of PARP-2 is very similar to that of PARP-1, but with some differences in the vicinity of the acceptor site, which could reflect differences in the substrate proteins ADP-ribosylated by the two enzymes, and may ultimately provide the means for engineering differential specificity into PARP-inhibitors targeted at DNA repair pathways. Development of isoform-specific inhibitors would also provide a method for dissecting and elucidating the precise cellular roles for each PARP protein. Some putative PARP-2 selective inhibitors have already been identified in yeast-growth inhibition studies (44), but co-crystal structures would be necessary to provide the necessary details of the drug–protein interactions.
The most significant structural difference between the two enzymes occurs N-terminal of the common ‘E’ domain and CF, where the large zinc finger DNA-binding domain (DBD) is replaced by a highly basic 64-residue DBD in PARP-2. Although able to bind DNA in isolation from the rest of the protein, the PARP-2-DBD does not have a folded structure in solution, indicated by Circular Dichroism and 1D-NMR spectroscopy experiments which both showed spectra characteristic of an unstructured polypeptide (our unpublished observations). The structural nature of the interaction of PARP-1 and PARP-2 DBDs with DNA has yet to be defined.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to the ESRF Grenoble for access to synchrotron beam-time, and to the Institute of Cancer Research Structural Biology Initiative and Cancer Research UK (L.H.P.), and Centre National de la Recherche Scientific, Association pour la Recherche Contre le Cancer, Electricité de France, Ligue Nationale contre le Cancer and Commissariat à l’Energie Atomique (G.d.M.) for financial support.
PDB no. 1GS0
REFERENCES
- 1.Ame J.C., Rolli,V., Schreiber,V., Niedergang,C., Apiou,F., Decker,P., Muller,S., Hoger,T., Menissier-de Murcia,J. and de Murcia,G. (1999) PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem., 274, 17860–17868. [DOI] [PubMed] [Google Scholar]
- 2.Johansson M. (1999) A human poly(ADP-ribose) polymerase gene family (ADPRTL): cDNA cloning of two novel poly(ADP-ribose) polymerase homologues. Genomics, 57, 442–445. [DOI] [PubMed] [Google Scholar]
- 3.Berghammer H., Ebner,M., Marksteiner,R. and Auer,B. (1999) pADPRT-2: a novel mammalian polymerizing(ADP-ribosyl)transferase gene related to truncated pADPRT homologues in plants and Caenorhabditis elegans. FEBS Lett., 449, 259–263. [DOI] [PubMed] [Google Scholar]
- 4.Augustin A., Spenlehauer,C., Dumond,H., Menissier-de Murcia,J., Piel,M., Schmit,A.C., Apiou,F., Vonesch,J.L., Kock,M., Bornens,M. et al. (2003) PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J. Cell Sci., 116, 1551–1562. [DOI] [PubMed] [Google Scholar]
- 5.Smith S., Giriat,I., Schmitt,A. and de Lange,T. (1998) Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science, 282, 1484–1487. [DOI] [PubMed] [Google Scholar]
- 6.Kaminker P.G., Kim,S.H., Taylor,R.D., Zebarjadian,Y., Funk,W.D., Morin,G.B., Yaswen,P. and Campisi,J. (2001) TANK2, a new TRF1-associated poly(ADP-ribose) polymerase, causes rapid induction of cell death upon overexpression. J. Biol. Chem., 276, 35891–35899. [DOI] [PubMed] [Google Scholar]
- 7.Lyons R.J., Deane,R., Lynch,D.K., Ye,Z.S., Sanderson,G.M., Eyre,H.J., Sutherland,G.R. and Daly,R.J. (2001) Identification of a novel human tankyrase through its interaction with the adaptor protein Grb14. J. Biol. Chem., 276, 17172–17180. [DOI] [PubMed] [Google Scholar]
- 8.Ma Q., Baldwin,K.T., Renzelli,A.J., McDaniel,A. and Dong,L. (2001) TCDD-inducible poly(ADP-ribose) polymerase: a novel response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem. Biophys. Res. Commun., 289, 499–506. [DOI] [PubMed] [Google Scholar]
- 9.Kickhoefer V.A., Siva,A.C., Kedersha,N.L., Inman,E.M., Ruland,C., Streuli,M. and Rome,L.H. (1999) The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J. Cell Biol., 146, 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schreiber V., Ame,J.C., Dolle,P., Schultz,I., Rinaldi,B., Fraulob,V., Menissier-de Murcia,J. and de Murcia,G. (2002) Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem., 277, 23028–23036. [DOI] [PubMed] [Google Scholar]
- 11.de Murcia G. and Shall,S. (2000) From DNA Damage and Stress Signalling to Cell Death. Oxford University Press, Oxford, UK. [Google Scholar]
- 12.Burkle A. (2001) Physiology and pathophysiology of poly(ADP-ribosyl)ation. Bioessays, 23, 795–806. [DOI] [PubMed] [Google Scholar]
- 13.Krupitza G. and Cerutti,P. (1989) Poly(ADP-ribosylation) of histones in intact human keratinocytes. Biochemistry, 28, 4054–4060. [DOI] [PubMed] [Google Scholar]
- 14.Darby M.K., Schmitt,B., Jongstra-Bilen,J. and Vosberg,H.P. (1985) Inhibition of calf thymus type II DNA topoisomerase by poly(ADP-ribosylation). EMBO J., 4, 2129–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshihara K., Itaya,A., Tanaka,Y., Ohashi,Y., Ito,K., Teraoka,H., Tsukada,K., Matsukage,A. and Kamiya,T. (1985) Inhibition of DNA polymerase alpha, DNA polymerase beta, terminal deoxynucleotidyl transferase and DNA ligase II by poly(ADP-ribosyl)ation reaction in vitro. Biochem. Biophys. Res. Commun., 128, 61–67. [DOI] [PubMed] [Google Scholar]
- 16.Kumari S.R., Mendoza-Alvarez,H. and Alvarez-Gonzalez,R. (1998) Functional interactions of p53 with poly(ADP-ribose) polymerase (PARP) during apoptosis following DNA damage: covalent poly(ADP-ribosyl)ation of p53 by exogenous PARP and noncovalent binding of p53 to the M(r) 85,000 proteolytic fragment. Cancer Res., 58, 5075–5078. [PubMed] [Google Scholar]
- 17.Masson M., Niedergang,C., Schreiber,V., Muller,S., Menissier-de Murcia,J. and de Murcia,G. (1998) XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol., 18, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caldecott K.W., Aoufouchi,S., Johnson,P. and Shall,S. (1996) XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res., 24, 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubota Y., Nash,R.A., Klungland,A., Schar,P., Barnes,D.E. and Lindahl,T. (1996) Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J., 15, 6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 20.Dantzer F., de La Rubia,G., Menissier-de Murcia,J., Hostomsky,Z., de Murcia,G. and Schreiber,V. (2000) Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1. Biochemistry, 39, 7559–7569. [DOI] [PubMed] [Google Scholar]
- 21.Mennisier-de Murcia J., Niedergang,C., Trucco,C., Ricoul,M., Dutrillaux,B., Mark,M., Oliver,F.J., Masson,M., Dierich,A., LeMeur,M. et al. (1997) Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl Acad. Sci. USA, 94, 7303–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z.Q., Stingl,L., Morrison,C., Jantsch,M., Los,M., Schulze-Osthoff,K. and Wagner,E.F. (1997) PARP is important for genomic stability but dispensable in apoptosis. Genes Dev., 11, 2347–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trucco C., Oliver,F.J., de Murcia,G. and Menissier-de Murcia,J. (1998) DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res., 26, 2644–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beneke R., Geisen,C., Zevnik,B., Bauch,T., Muller,W.U., Kupper,J.H. and Moroy,T. (2000) DNA excision repair and DNA damage-induced apoptosis are linked to Poly(ADP-ribosyl)ation but have different requirements for p53. Mol. Cell. Biol., 20, 6695–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pleschke J.M., Kleczkowska,H.E., Strohm,M. and Althaus,F.R. (2000) Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem., 275, 40974–40980. [DOI] [PubMed] [Google Scholar]
- 26.Okano S., Lan,L., Caldecott,K.W., Mori,T. and Yasui,A. (2003) Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol., 23, 3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffin R.J., Curtin,N.J., Newell,D.R., Golding,B.T., Durkacz,B.W. and Calvert,A.H. (1995) The role of inhibitors of poly(ADP-ribose) polymerase as resistance-modifying agents in cancer therapy. Biochimie, 77, 408–422. [DOI] [PubMed] [Google Scholar]
- 28.Veuger S.J., Curtin,N.J., Richardson,C.J., Smith,G.C. and Durkacz,B.W. (2003) Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res., 63, 6008–6015. [PubMed] [Google Scholar]
- 29.Szabo C. (2000) Activation of poly(ADP-ribose) polymerase in the pathogenesis of ischaemia-reperfusion injury. In de Murcia,G. and Shall,S. (eds), From DNA Damage and Stress Signalling to Cell Death. Oxford University Press, Oxford, UK, pp. 151–176. [Google Scholar]
- 30.Pieper A.A., Verma,A., Zhang,J. and Snyder,S.H. (1999) Poly (ADP-ribose) polymerase, nitric oxide and cell death. Trends Pharmacol. Sci., 20, 171–181. [DOI] [PubMed] [Google Scholar]
- 31.Leslie A.G.W. (1995) MOSFLM Users Guide. MRC Laboratory of Molecular Biology, Cambridge, UK. [Google Scholar]
- 32.Collaborative Computational Project, No. 4. (1994) The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- 33.Navaza J. (1994) Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- 34.Brunger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P., Grosse-Kunstleve,R.W., Jiang,J.S., Kuszewski,J., Nilges,M. and Pannu,N.S. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 35.Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 36.Ruf A., Menissier-de Murcia,J., de Murcia,G. and Schulz,G.E. (1996) Structure of the catalytic fragment of poly(ADP-ribose) polymerase from chicken. Proc. Natl Acad. Sci. USA, 93, 7481–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orengo C.A. and Taylor,W.R. (1996) SSAP: sequential structure alignment program for protein structure comparison. Methods Enzymol., 266, 617–635. [DOI] [PubMed] [Google Scholar]
- 38.Virag L. and Szabo,C. (2002) The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol. Rev., 54, 375–429. [DOI] [PubMed] [Google Scholar]
- 39.Ruf A., Rolli,V., de Murcia,G. and Schulz,G.E. (1998) The mechanism of the elongation and branching reaction of poly(ADP-ribose) polymerase as derived from crystal structures and mutagenesis. J. Mol. Biol., 278, 57–65. [DOI] [PubMed] [Google Scholar]
- 40.Bell C.E. and Eisenberg,D. (1996) Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide. Biochemistry, 35, 1137–1149. [DOI] [PubMed] [Google Scholar]
- 41.Ruf A., de Murcia,G. and Schulz,G.E. (1998) Inhibitor and NAD+ binding to poly(ADP-ribose) polymerase as derived from crystal structures and homology modeling. Biochemistry, 37, 3893–3900. [DOI] [PubMed] [Google Scholar]
- 42.White A.W., Almassy,R., Calvert,A.H., Curtin,N.J., Griffin,R.J., Hostomsky,Z., Maegley,K., Newell,D.R., Srinivasan,S. and Golding,B.T. (2000) Resistance-modifying agents. 9. Synthesis and biological properties of benzimidazole inhibitors of the DNA repair enzyme poly(ADP-ribose) polymerase. J. Med. Chem., 43, 4084–4097. [DOI] [PubMed] [Google Scholar]
- 43.Menissier-de Murcia J., Ricoul,M., Tartier,L., Niedergang,C., Huber,A., Dantzer,F., Schreiber,V., Ame,J.C., Dierich,A., LeMeur,M. et al. (2003) Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J., 22, 2255–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perkins E., Sun,D., Nguyen,A., Tulac,S., Francesco,M., Tavana,H., Nguyen,H., Tugendreich,S., Barthmaier,P., Couto,J. et al. (2001) Novel inhibitors of poly(ADP-ribose) polymerase/PARP1 and PARP2 identified using a cell-based screen in yeast. Cancer Res., 61, 4175–4183. [PubMed] [Google Scholar]