Abstract
The interaction of bacteria with surfaces has important implications in a range of areas, including bioenergy, biofouling, biofilm formation, and the infection of plants and animals. Many of the interactions of bacteria with surfaces produce changes in the expression of genes that influence cell morphology and behavior, including genes essential for motility and surface attachment. Despite the attention that these phenotypes have garnered, the bacterial systems used for sensing and responding to surfaces are still not well understood. An understanding of these mechanisms will guide the development of new classes of materials that inhibit and promote cell growth, and complement studies of the physiology of bacteria in contact with surfaces. Recent studies from a range of fields in science and engineering are poised to guide future investigations in this area. This review summarizes recent studies on bacteria-surface interactions, discusses mechanisms of surface sensing and consequences of cell attachment, provides an overview of surfaces that have been used in bacterial studies, and highlights unanswered questions in this field.
Introduction to microbial surface sensing
Molecular phylogeny and geobiology suggest that microorganisms emerged ~3.5–3.8 billion years ago,1, 2 a mere billion years after the formation of the Earth. Since that time, microbes have done remarkably well—their total number on Earth is estimated to be 4–6 × 1030, and they are found in nearly every terrestrial environment sampled to date.3 A central hypothesis in microbiology is that the majority of bacteria in the biosphere live in communities that are associated with surfaces,4 and that this association has played an important role in the success of bacteria in colonizing different environments. In addition to nucleating cell growth into communities, surfaces have a range of characteristics that protect cells from predation and other environmental threats and facilitate the conservation of the genotype.
Bacterial attachment to surfaces has long been a topic of study. Several of the first reports on this subject came from PS Meadows, who examined the effect of salinity on attachment of both marine and freshwater organisms to glass slides.5 Many early studies focused on attachment to biotic surfaces such as teeth6 and epithelial cells,7, 8 and an interest in abiotic surfaces soon followed.9–14 The number of papers indexed by PubMed on bacterial attachment to surfaces was less than 10 per year throughout the 1970s and increased to 115 in 2011.15 Early reviews in this area, including an excellent discussion of the attachment of Staphylococcus aureus to surfaces by Katsikogianni and Missirlis,16 set the stage for many of the advances that have occurred in the past decade.
Advantages of bacterial attachment to surfaces
Adhering to surfaces provides bacteria with many advantages. Attachment to horizontal surfaces stimulates bacterial growth (particularly in nutrient-poor environments) as organic material suspended in liquid settles, is deposited on surfaces, and increases the local concentration of nutrients.17 Similarly, increasing the substrate surface area (e.g., by adding glass beads to a culture container) provides more area on which nutrients can adsorb, enabling cells to grow at nutrient concentrations that would normally be too low to support growth.18 Caulobacter crescentus is a fascinating example of a bacterium that takes advantage of surface attachment to optimize nutrient uptake. C. crescentus oscillates between stalked cells that adhere tightly to surfaces using a protein holdfast and motile cells that lack this organelle and instead have a polar flagellum. This phenotypic switch makes it possible for cells to adapt to both nutrient-rich (favoring motility) and nutrient-poor (favoring adhesion) environments.19
In addition to surface attachment facilitating nutrient capture, some bacteria obtain necessary metabolites and co-factors directly from the surfaces to which they adhere. Shewanella and other genera of bacteria that grow on metal surfaces can use metals such as iron and magnesium as terminal electron acceptors in respiration.20, 21 Extracellular organelles facilitate the transport of ions between cells and surfaces. For example, Geobacter sulfurreducens uses pili to conduct charge transport between cells and surfaces. Shewanella oneidensis uses an outer membrane protein complex to form an electron bridge between the periplasm and the extracellular environment.22
Bacteria attached to surfaces often exist as biofilms, which play several protective roles. The extracellular polymeric substance (EPS) secreted by cells in biofilms that are attached to surfaces provides protection from mechanical damage and shear caused by fluid flow.23, 24 Additionally, biofilms often exhibit resistance to antibiotic treatment.4 Several different mechanisms contribute to this resistance, including (1) the barrier function of the biofilm matrix; (2) the presence of dormant persister cells and highly resistant small colony variants; (3) and upregulation of several biofilm-specific antibiotic resistance genes. These and other mechanisms are reviewed in detail by Mah and O’Toole.25, 26
Furthermore, recent experiments have demonstrated that cells adhered to surfaces and not associated with biofilms have resistance profiles that are similar to biofilm cells.27, 28 This resistance phenotype does not require mutations in genomic DNA, as the process is reversible and cells become susceptible to antibiotics after detachment. John et al. suggest that surface attachment facilitates antibiotic resistance by two primary mechanisms: (1) reducing the net negative charge on bacterial cells; and (2) enhancing the stability of the membrane.27 Mutations in genes that produce similar phenotypes decrease the susceptibility of bacteria to antibiotics. These data suggest that the attachment of bacteria to surfaces alters their metabolic state and reduces antibiotic susceptibility, which is a common feature of bacteria during the stationary phase of cell growth.29 Even surface attachment per se may not be required for this phenotype, as very high densities of cells (similar to the densities obtained in a biofilm) also display antibiotic resistance.30 However, attachment to surfaces is one means by which bacterial communities can attain such high densities.
In addition to antibiotic resistance, cells in biofilms often gain protection from predators. When exposed to protozoa, the bacterium Serratia marcescens rapidly forms surface-associated microcolonies—an early stage in biofilm formation—that protect cells from grazing by these predators.31 Mechanisms of chemical sensing trigger the production of compounds that are toxic to protozoans during S. marcescens biofilm development, which provides a further layer of protection from predation.
Bacteria that are attached to surfaces—particularly those associated with biofilms—may become specialized in comparison to cells in other regions of the community.32 In Bacillus subtilis biofilms, motile cells, EPS-producing cells, and spore-forming cells are localized to different regions within the biofilm. Strains of B. subtilis that are unable to form structured biofilms do not sporulate, which suggests that localization/specialization is required for the formation of bacterial spores.33 Similarly, surface-associated microcolonies and biofilms of Pseudomonas aeruginosa contain groups of cells that display differential motility and susceptibility to antimicrobial agents.34, 35
Surface sensing is a precursor to swarming, an important adaptive behavior in which contact between cells and surfaces programs morphological changes that facilitate cooperative behavior, rapid community growth, and migration of communities.36, 37 Swarming motility has been reported in at least 15 different genera of bacteria from different natural habitats.36 Several mechanisms of adaptation have been reported during bacterial swarming, including reduced susceptibility to antibiotics38, 39 and mutualistic interactions with fungal spores.40 In B. subtilis swarming colonies, spatially distinct groups of cells express different levels of flagellin—the protein that assembles into the flagellar filament—and have different cell morphologies.41, 42 Similarly, Proteus mirabilis cells with distinct cell morphologies are found in different regions of swarming communities.43, 44
Cells in bacterial communities such as swarms or biofilms interact with each other in several different ways. Bacteria are able to communicate through the use of small molecule chemical messengers in a process referred to as ‘quorum sensing’.45 The dense packing of cells in bacterial communities facilitates an increase in the concentration of small molecules that transfer information between cells and trigger physiological changes.46 The shape of chemical gradients in close proximity to surfaces enhances the exchange of chemical information within biofilms and communities attached to surfaces.47–49 Lateral gene transfer is also enhanced in biofilms compared to planktonic cells freely suspended in fluids.50 Additionally, surface-associated growth induces phenotypes that promote “natural competence” in Vibrio cholerae.51 Myxobacteria cells that are associated with biofilms even exchange outer membrane proteins and lipids.52
Adhering to surfaces also has several disadvantages, including the inhibition of motility, often due to a “switch” in the activation of genes involved in motility and adhesion: for example, genes coding for flagella may be turned off by the same transcriptional regulator that turns on genes for extracellular matrix production.53–55 Inhibiting cell motility prevents cells from searching for optimal environments when nutrients become depleted. Bacterial cells may overcome this disadvantage (and others) in certain environments by sensing surfaces and triggering surface-associated phenotypes that activate motility and prevent adhesion. For example, the process of swarming provides many of the same advantages described earlier, activates cell motility, and provides a mechanism for actively capturing nutrients.38 Some pathogens also use surface sensing as a trigger to upregulate virulence factors as a prelude to invasion of the host.56
Mechanisms of bacterial attachment
Many bacteria are freely suspended in bulk fluids before attaching to surfaces. Motile bacteria occupy one of three regions of fluids: (1) bulk liquid, where the cells do not experience effects from the surface; (2) near-surface bulk liquid, where the cells experience the hydrodynamic effects of the surface; and (3) near-surface constrained, where the cells experience both the hydrodynamic and physicochemical (i.e. van der Waals and electrostatic forces) effects of the surface.57 The role of physicochemical effects in bacterial adhesion to surfaces has been reviewed in detail by Bos and colleagues.58
At low and moderate fluid velocities, non-motile bacteria adhere to surfaces. At high fluid velocity, non-motile bacteria are transported with the flow of fluids and do not attach.59 Motile bacteria attach to surfaces regardless of fluid velocity. Importantly, the difference between motile and non-motile bacteria occurs only when motile cells actuate their flagella; bacteria with non-functional flagella adhere similarly to cells lacking flagella.59 The buoyant density of bacteria (i.e. E. coli) is usually 1.06 – 1.13 g mL−1,60 and leads to the slow deposition of cells onto surfaces from suspension in bulk liquid. Sedimentation rates for different marine bacteria have been measured and range from 10 to 30 µm h−1.61 Interestingly, the buoyant density of E. coli cells increases as they enter stationary phase, which facilitates their rapid deposition on surfaces.62 Populations of cells that make contact with surfaces may thus be enriched with stationary phase cells. In contrast, the buoyant density of Vibrio parahaemolyticus cells decreases as they enter stationary phase.63
Upon contacting surfaces, cell attachment occurs in two phases. Initial attachment is reversible, occurs rapidly (on the order of ~1 min),64, 65 and involves hydrodynamic and electrostatic interactions. During this time period, the adhesive force between bacteria and surfaces increases rapidly. A similar phenomenon has been observed for the attachment of polystyrene beads to a surface,66 and several observations suggest that this phenomenon is due to physicochemical effects (and not biological effects) including: (1) the loss of interfacial water; (2) structural changes in surface molecules; and (3) repositioning of the cell body to maximize attachment to the surface.67 Most bacteria have a net negative surface charge—particularly during the early stationary phase of cell growth68—and interact preferentially with positively charged surfaces. This effect disappears in high ionic strength media due to charge screening.69 Quorum sensing in E. coli causes an increase in the negative charge on cell surfaces, which may facilitate the interaction of bacteria with surfaces during the initial stages of biofilm formation.70
The second step of attachment is irreversible, occurs on a time scale of several hours, and involves van der Waals interactions between the hydrophobic region of the outer cell wall and the surface.69 In addition, several proteins play roles in the transition from reversible to irreversible cell attachment. The cytoplasmic P. aeruginosa protein SadB is required for this transition, although its mechanism is unknown.71 In E. coli, both lipopolysaccharide (LPS) and pili increase the initial rate of cell attachment and the rate of conversion to irreversible attachment.72 In Pseudomonas fluorescens, an ABC transporter and a secreted protein are required for irreversible attachment.73 Irreversible attachment is also facilitated by the production of EPS, which anchors cells to surfaces.74
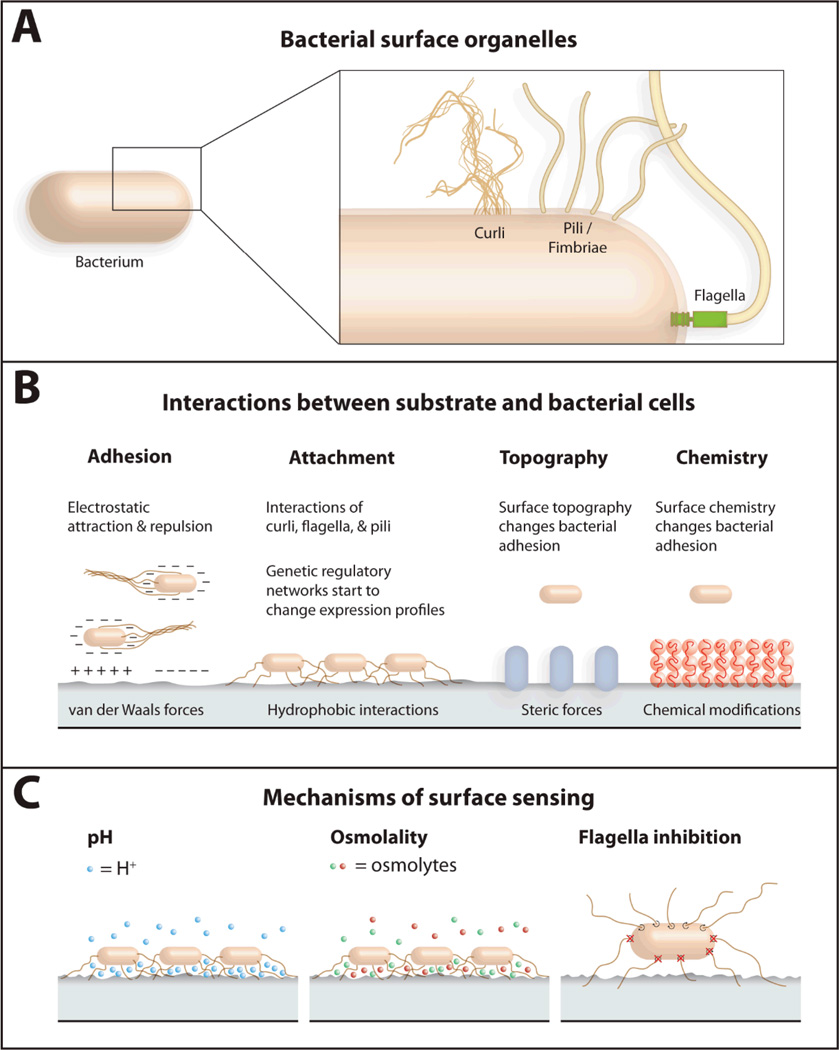
The first clue that some bacteria can interact specifically with certain surfaces came from early observations that different bacteria occupying the same niche do not necessarily interact with the same surfaces. Streptococcus mutans binds to teeth but not the tongue; Streptococcus salivarius displays an opposite preference.75 Bacteria have several different classes of extracellular organelles that mediate specific attachment to surfaces, including flagella, pili (also called fimbrae), and curli fibers (Fig. 1a).76 These organelles are frequently terminated with proteins referred to as adhesins, which bind to molecules presented on the surface of hosts. Two E. coli examples include type I pili, which bind to glycoproteins that present alpha-D-mannose, and type IV pili, which bind to phosphatidylethanolamine.77 Several species of oral bacteria adhere specifically to proline-rich proteins in saliva.78, 79 One particularly interesting example of specific attachment involves Flavobacterium johnsoniae. SprB is an adhesin that is required for F. johnsoniae motility on agar (not on glass). SprB binds to agar surfaces, is moved along the cell surface, and produces cell motion relative to the agar surface. Predicted homologs of SprB in the F. johnsoniae genome may facilitate cell attachment and motility on surfaces other than agar.80
Figure 1.
Several aspects of bacterial interaction with substrates are highlighted. (A) The bacterial surface has several organelles that facilitate interactions with substrates, including curli fibers, pili (also called fimbrae), and flagella. (B) Properties of the surface such as charge, hydrophobicity, topography, and the identity of the exposed chemical groups interact with physico-chemical properties of bacterial cells and influence attachment. (C) The proximity of bacteria to a surface causes changes in pH, osmolality, and flagella rotation that are sensed by cells. Figure adapted from Ref. 69.
Reversible cell attachment does not necessarily lead to irreversible attachment during surface colonization. In E. coli, weak cell adhesion facilitated by the pilus tip adhesin FimH enables cells to roll along surfaces. FimH mediates binding to glycoproteins that have N-linked oligosaccharides presenting terminal mannose residues.81 FimH binding to mono-mannose is dependent on shear stress, with a certain threshold level of force required to switch from weak to strong adherence.82 Anderson and co-workers observed that a wild type strain of E. coli that adhered weakly to mannosylated surfaces colonized the surface more rapidly than a mutant strain only capable of strong (irreversible) attachment.83
Cell attachment to different classes of materials
Bacterial attachment to surfaces is problematic in a wide range of areas, including medical implants,84 water purification systems,85 and industrial processes.86 The biofouling of metal ship hulls frequently begins with bacterial adhesion before progressing to larger marine organisms, and despite the use of anti-fouling coatings has been estimated to cost ~$56 bn/year (US) for a single class of Navy ships.87 Thermodynamics plays a central role in regulating the binding of bacteria to surfaces. Cells attach preferentially to hydrophilic materials (i.e., materials with a large surface energy) when the surface energy of the bacterium is larger than the surface energy of the liquid in which they are suspended. The surface energy of bacteria is typically smaller than the surface energy of liquids in which cells are suspended, and this mismatch causes cells to attach preferentially to hydrophobic materials (i.e., materials with lower surface energies).88 Bacteria are able to attach to a wide variety of different materials, including glass, aluminum, stainless steel, various organic polymers,72 and fluorinated materials such as Teflon™.89 Table 1 summarizes relevant studies in this area and the diversity of materials used to study bacterial adhesion to surfaces. Banerjee et al. have recently reviewed the materials aspects of bacterial adhesion in detail.90
Table 1.
A summary of materials that have been used to study bacteria-surface interactions.
| Material | Material properties |
Bacteria Studied† |
Type of Study | Reference |
|---|---|---|---|---|
| Metals | ||||
| Aluminum | N/A | P. aeruginosa | Biofilm formation | 168 |
| Copper | Surface energy | Marine bacteria | Cell adhesion | 169 |
| Nickel | Surface energy | Marine bacteria | Cell adhesion | 169 |
| Stainless steel | PEG coating |
L. monocytogenes Pseudomonas sp. |
Cell adhesion | 170 |
| Polymers | ||||
| Acetal polymer | Surface energy |
E. coli L. monocytogenes S. aureus S. epidermidis |
Cell adhesion in liquids of different surface energy | 88 |
| Fluoroethylene-propylene co-polymer | Surface energy |
E. coli L. monocytogenes S. aureus S. epidermidis |
Cell adhesion in liquids of different surface energy | 88 |
| Zeta potential |
A. calcoaceticus S. epidermidis S. thermophilus B |
Rate of cell attachment and bond maturation | 66 | |
| Low-density polyethylene | Surface energy |
E. coli L. monocytogenes S. aureus S. epidermidis |
Cell adhesion in liquids of different surface energy | 88 |
| N,N-dimethyl-2-morpholinone, carboxybetaine copolymer | Conversion between cationic and zwitterionic surfaces | E. coli | Cell adhesion; antibacterial properties | 171 |
| N,N-dodecyl,methyl-polyethylenemine | Surface coating | S. aureus | Biofilm formation (in vitro and in vivo on orthopedic implant surfaces) | 172 |
| N,N-hexyl methyl-polyethylenemine | Surface coating | S. aureus | Biofilm formation (in vitro and in vivo on corneal implant surfaces) | 173 |
| Poly(4-vinyl-N-alkylpyridinium bromide) | Alkyl chain length |
E. coli P. aeruginosa S. aureus S. epidermidis |
Antibacterial properties | 94 |
| Poly(carboxybetaine methacrylate) | Surface coating |
P. aeruginosa P. putida |
Protein deposition and biofilm formation | 174 |
| Poly(dimethylsiloxane) | Surface topology | S. aureus | Biofilm formation | 105 |
| Poly(allylamine) hydrochloride and poly(acrylic acid) PEMs | Surface stiffness |
E. coli S. epidermidis |
Cell adhesion | 149 |
| Polyethylene | N/A | P. fluorescens | Biofilm formation and polysaccharide production | 175 |
| Poly(ethylene oxide) | Polymer chain length |
P. aeruginosa S. epidermidis |
Cell adhesion at different temperatures | 176 |
| N/A | S. epidermidis | Cell adhesion and coating stability | 177 | |
| Polyethylene oxide/polypropylene oxide copolymer | Brush coating |
P. aeruginosa S. aureus S. epidermidis |
Cell adhesion; biofilm formation; biofilm removal | 103 |
| Poly(L-lysine)-graft-poly (2-methyl-2-oxazoline) | Polymer graft density | E. coli | Bacterial adhesion | 178 |
| Polymethyl-methacrylate | Zeta potential |
A. calcoaceticus S. epidermidis S. thermophilus B |
Rate of cell attachment and bond maturation | 66 |
| Poly(N-isopropyl-acrylamide) | Surface coating |
E. coli S. epidermidis |
Protein deposition; cell adhesion; biofilm formation | 179 |
| Surface energy | H. marina | Cell adhesion | 100 | |
| L. monocytogenes | Cell adhesion | 180 | ||
| Poly(N-isopropyl-acrylamide) / poly(sulfobetaine methacrylate) | Surface coating |
E. coli S. epidermidis |
Protein deposition; cell adhesion; biofilm formation | 179 |
| Poly(N,N-dimethyl-N-(ethylcarbonylmethyl)-N-[2-methacryloyloxy)-ethyl]ammonium salicylate) | N/A |
E. coli S. epidermidis |
Antibacterial properties | 181 |
| Poly(oligo(ethylene glycol) methyl ether methacrylate) | Thickness and chemistry of surface coating |
P. aeruginosa S. epidermidis |
Cell adhesion; biofilm formation | 97 |
| Polypropylene | Surface energy | Marine bacteria | Cell adhesion | 169 |
| N/A | P. fluorescens | Biofilm formation and polysaccharide production | 175 | |
| Polystyrene | Surface energy | Marine bacteria | Cell adhesion | 169 |
|
E. coli, L. monocytogenes S. aureus S. epidermidis |
Cell adhesion in liquids of different surface energy | 88 | ||
| Surface energy; zeta potential | E. coli | Cell/substrate interactions | 182 | |
| N/A | P. fluorescens | Biofilm formation and polysaccharide production | 175 | |
| Poly(sulfobetaine methacrylate) | Thickness and chemistry of surface coating |
P. aeruginosa S. epidermidis |
Cell adhesion; biofilm formation | 97 |
| Surface coating |
E. coli S. epidermidis |
Protein deposition; cell adhesion; biofilm formation | 179 | |
| Polyurethane | Hydrophobicity | S. epidermidis | Cell adhesion; biofilm formation | 183 |
| Polyvinylchloride | N/A | P. fluorescens | Biofilm formation and polysaccharide production | 175 |
| Polyvinylfluoride | Surface energy | Marine bacteria | Cell adhesion | 169 |
| Sulfonated polystyrene | Surface energy |
E. coli L. monocytogenes S. aureus S. epidermidis |
Cell adhesion in liquids of different surface energy | 88 |
| Teflon (polytetra-fluoroethylene) | Surface energy | Marine bacteria | Cell adhesion | 169 |
| Surface energy; zeta potential | E. coli | Cell/substrate interactions | 182 | |
| SAMs | ||||
| Methyl-terminated SAMs | Thickness and chemistry of surface coating |
P. aeruginosa S. epidermidis |
Cell adhesion; biofilm formation | 97 |
| Oligo(ethylene glycol) SAMs | Thickness and chemistry of surface coating |
P. aeruginosa S. epidermidis |
Cell adhesion; biofilm formation | 97 |
| SO3−/N+(CH3)3 SAMs | Thickness and chemistry of surface coating |
P. aeruginosa S. epidermidis |
Cell adhesion; biofilm formation | 97 |
| Various SAMs | Surface-exposed chemical groups |
S. aureus S. epidermidis |
Cell adhesion | 184 |
| Protein adsorption | S. aureus | Cell adhesion | 185 | |
| Other | ||||
| CaF2 | N/A | P. aeruginosa | Biofilm formation | 168 |
| Glass | Surface energy | Marine bacteria | Cell adhesion | 169 |
| S. epidermidis | Cell/substrate bond maturation | 64 | ||
| S. mutans | Cell adhesion on saliva-coated surfaces | 186 | ||
| Surface energy; zeta potential | E. coli D21 | Cell/substrate interactions | 182 | |
| Zeta potential |
A. calcoaceticus S. epidermidis S. thermophilus B |
Rate of cell attachment and bond maturation | 66 | |
| Mica | Surface energy; zeta potential | E. coli | Cell/substrate interactions | 182 |
| Sulfopropylbetaine | Surface coating |
E. coli S. aureus |
Antibacterial properties | 187 |
Abbreviations are as follows: A. calcoaceticus = Acinetobacter calcoaceticus; E. coli = Escherichia coli; H. marina = Halomonas marina; L. monocytogenes = Listeria monocytogenes; P. aeruginosa = Pseudomonas aeruginosa; P. fluorescens = Pseudomonas fluorescens; P. putida = Pseudomonas putida; S. aureus = Staphylococcus aureus; S. epidermidis = Staphylococcus epidermidis; S. mutans = Streptococcus mutans; S. thermophilus = Streptococcus thermophiles; SAM = self-assembled monolayer; PEM = polyelectrolyte monolayer.
Bacteria also attach to surfaces that initially resist the attachment of cells. This process occurs through the deposition of a layer of proteins—including proteins found naturally in the environment as well as those secreted by bacteria—that “condition” the surface and mask functional groups that reduce cell adhesion.91, 92 The formation of conditioning layers presents a challenge for creating surfaces that are bacteria-resistant. An excellent case in point is surfaces that present quaternary ammonium salts, which are initially bactericidal before conditioning layers are deposited.93, 94 Despite the challenges presented by biofouling, the development of surfaces that resist bacterial attachment is an active area of research. Several strategies have been described for reducing the attachment of bacteria, including: (1) controlling surface chemistry;95 and (2) controlling structural properties of surfaces (Fig. 1b).96 Zwitterionic surfaces such as betaine97–99 and silver-impregnated surfaces that slowly release silver ions are among the most effective chemical strategies for inhibiting the attachment of bacteria to surfaces. Another strategy is the use of thermoresponsive hydrogels, such as poly(N-isopropylacrylamide). Above a critical temperature, this polymer undergoes a phase transition and presents a hydrophobic surface that facilitates cell attachment and growth. When the temperature is decreased, the polymer swells and presents a hydrophilic surface, and adsorbed cells are released from the surface.100, 101
Polymer brushes, in which one end of a polymer is attached to a surface and the polymer chain is extended into solution, have been successfully used as anti-adhesive coatings.102 Surfaces coated with Pluronic F-127—a tri-block co-polymer of polyethylene oxide and polypropylene oxide—reduce the initial attachment and growth rate of Staphylococcus aureus and Staphylococcus epidermidis and enable the removal of biofilms consisting of these organisms using fluid flow.103 The same study found that Pluronic F-127-coated surfaces had no effect on Pseudomonas aeruginosa adhesion and biofilm removal. Polymer brushes can also be functionalized with antimicrobial peptides, which enhance the overall antimicrobial activity of the surface.104
Physical strategies for reducing attachment have frequently been inspired by natural materials such as shark skin and lotus leaves.96 Several examples of structured materials that are designed to reduce bacterial attachment include Sharklet™ technology,105 nanostructured surfaces with a low effective stiffness,106 and slippery liquid-infused porous surfaces.107 These and other strategies for preventing bacterial attachment have recently been reviewed in depth.84, 90, 108 A shift away from materials that prevent cell attachment to materials that enhance growth may provide insight into the properties of surfaces sensed by bacteria and the biological machinery that is involved in this process.
Mechanisms of sensing surfaces and surface properties sensed by bacteria
Cells that compare a signal input at a time interval to the total signal at a previous time interval are capable of sensing spatial gradients (e.g., a cell that moves from one position at time t1 to another position at time t2 can evaluate the relative concentrations of a given signal at the two time points).109 The canonical example of this process in bacteria is the chemotaxis system, which uses membrane-embedded receptors to detect the concentration of extracellular small molecules and ions and influences the direction of bacterial motility.110 Recent evidence demonstrates that bacteria are capable of sensing spatial changes in concentrations in certain conditions. Cells of Staphylococcus aureus sense the binding of surface ligands to receptors on one side of its cell body and respond by localizing receptors to the surface-associated region.111 This process involves the response of a cell to a step-function rather than an extracellular gradient per se; however, it indicates that bacteria can distinguish between signals arising from spatially distinct subsets of cell surface receptors.
An example of chemical gradients impacting surface-associated bacteria occurs during biofilm formation. Cells adhering to surfaces may trap ions and small molecules in the thin layer of fluid positioned between the cell body and the surface, which forms a microenvironment that has different properties than the bulk liquid (Fig. 1c). When E. coli adheres to surfaces, the pH of fluid close to the surface decreases below the pH of the bulk liquid phase and persists for at least 72 h.112 The Cpx two-component system may play a role in connecting pH sensing and cell-surface responses.113 This system is also involved in regulating the expression and assembly of pili in E. coli.114, 115 Additionally, the decrease in the pH of fluids in close proximity to surface-associated cells enhances the proton motive force and directly affects cellular bioenergetics.116 Cells adhering to surfaces have been shown to produce 2–5-fold more ATP per cell than non-adherent cells.117
Similarly to pH, the osmolality of liquids in close proximity to surfaces also changes rapidly when cells attach to substrates. Gram-negative bacteria sense osmolality using OmpA,118 and this process leads to changes in the transcription of other genes. OmpA represses cellulose production through the Cpx pathway and increases biofilm formation in E. coli.118 This role for OmpA may be conserved among bacteria (or at least among γ-proteobacteria), as CpxA-mediated sensing of pH by OmpA has been identified as a required element for biofilm formation in Sodalis glossinidius, an endosymbiont of the tsetse fly.119 OmpR is a cytoplasmic response regulator that is activated by EnvZ and derepresses the expression of genes that code for pili.115 Increases in osmolality stabilize conformations of the cytoplasmic domain of EnvZ and increase the autophosphorylation and activation of OmpR.120
As some surfaces release soluble compounds that attract bacteria, sensing may begin before bacteria come into direct contact with the surface. Chitin is a homopolymer consisting of N-acetyl-D-glucosamine (GlcNAc) and is the second-most abundant natural polysaccharide on the Earth. The degradation of chitin produces GlcNAc monomers and oligomers, which are chemoattractants of V. cholerae.121 V. cholerae cells that sense and move towards the source of GlcNAc adhere to chitin surfaces using type IV MSHA pili.121 Cells sense the presence of GlcNAc oligomers using the protein ChiS122 and respond by upregulating the transcription of numerous genes, including genes that code for chitinolytic enzymes and components of type IV pili.121
Bacteria that attach to surfaces become physically constrained by surfaces and the close proximity of adjacent, attached cells. These physical constraints can inhibit the rotation of the flagella on motile bacterial cells, which alters cell physiology. The influence of surfaces, viscous liquids, and anti-flagella antibodies on the rotation of flagella on V. parahaemolyticus,123 S. marcescens,124 and P. mirabilis125 is sensed by cells and triggers surface-related morphological changes such as those discussed in detail below. While it is unclear exactly how the signal from the flagella is transduced, it has been hypothesized that the membrane-associated Umo proteins play a role in P. mirabilis.126–128
Other membrane-based mechanisms also play a role in surface sensing. For example, the induction of the Cpx signaling pathway upon surface adhesion of E. coli requires the outer membrane lipoprotein NlpE.129 Cell contact with a hydrophobic surface may damage the outer membrane, trigger NlpE, and activate the Cpx pathway, which responds to envelope stress. As described above, the Cpx pathway also plays a role in biofilm formation.130
Changes in phenotypes upon contact of cells with surfaces
Surface sensing triggers a variety of cellular changes. Many of the changes are morphological and facilitate attachment to surfaces. For example, cells of S. aureus preferentially localize fibronectin-binding protein when presented with surfaces that display fibronectin.111 During E. coli cell adhesion, a decrease in OmpX in the membrane increases the production of EPS and alters antibiotic susceptibility.131 During biofilm formation, quorum sensing alters the charge on E. coli cells,70 which influences binding to surfaces presenting electrostatically attractive charges. Some organisms, including Helicobacter pylori132 and S. aureus111 produce organelles for adhesion only upon cell contact with host tissues. In response to surfaces, P. aeruginosa activates the Wsp system, which consists of a complex of proteins that share homology to the chemotaxis system and dynamically localize in cells. WspA is a chemoreceptor-like protein that senses surfaces and transduces a signal through the other Wsp proteins that ultimately leads to the phosphorylation of WspR.133 Phosphorylated WspR catalyzes the synthesis of cyclic-di-GMP (c-di-GMP),134 which is implicated in biofilm formation, the suppression of swarming motility in P. aeruginosa,135, 136 and the regulation of several motility- or attachment-related systems in bacteria.137, 138 In P. aeruginosa, c-di-GMP affects the activity of the transcription factor FleQ. FleQ, the master regulator of flagella gene expression, also inhibits the expression of genes required for EPS synthesis (the pel operon). FleQ binds to c-di-GMP, and elevated levels of c-di-GMP in vivo relieve the inhibition of pel gene expression by FleQ.139 The surface-induced increase and the subsequent role of c-di-GMP in P. aeruginosa is among the best-characterized mechanisms of bacterial surface sensing, and provides an example of the influence of physical interfaces on bacterial biochemistry and physiology.
Pathogenic bacteria frequently decouple their division from growth and form filaments in response to the presence of host surfaces.140 The filamentation of uropathogenic E. coli facilitates the escape of cells from the host immune response during urinary tract infections.141 Agrobacterium tumefaciens also filaments upon contact with plant host tissues.142
Filamentation also occurs during the surface sensing-associated process of swarming. During swarming, a wide variety of changes occur in the global transcription of bacterial genes.128, 143, 144 These changes produce significant alterations in cell morphology, including an increase in the surface density of flagella and cell length.44 Swarming cells also upregulate several infection-related genes, including protease, urease, hemolysin, and proteins that facilitate host invasion.56, 128
Interestingly, the physical properties of surfaces may influence cell morphology and community structure. The marine bacterium SW5 adheres to and grows on both hydrophobic and hydrophilic surfaces; however, its growth into communities is influenced by surface properties. Cells adhere uniformly to hydrophobic surfaces, form microcolonies, and grow into tightly packed multi-layer biofilms. Fewer cells attach to hydrophilic surfaces, and changes in cell division lead to the formation chains of cells that are >100 µm long. These chains become loosely entangled to form relatively unstructured and less densely packed biofilms.145
Methods of study
A challenge in the field of microbiology has been the limited range of materials that are available for mechanistic studies of cell/surface interactions. Ironically, most of the surfaces that have been designed for studying bacteria are centered upon inhibiting cell growth and stimulating cell death rather than facilitating cell attachment and stimulating cell growth. A limited number of materials have been developed for studying various types of surface interactions and complement agar and agarose. Eladium™ is a polysaccharide produced by Rhizobium that has been used to screen yeasts for their biofilm-forming abilities.146 Gellan (GELRITE™ or Gelzan™) is a heterosaccharide derived from Pseudomonas that has been used to culture a variety of clinically important bacteria.147 Polyacrylamide (PA) gels148 and polyelectrolyte multilayers (PEMs)149 provide controllable surfaces for studies of cell-surface interactions.
In contrast to the limited materials that have been used to explore microbe-surface interactions, a wide variety of materials have been implemented to study mammalian cell attachment to surfaces. These biocompatible materials may be translated into studies of bacterial cell attachment and growth, and include: PA,150 alginates,151 fibronectin-coated silicone elastomers (useful for making patterned substrates and for controlling the stiffness of surfaces),152 poly(vinyl alcohol)/chitosan (a biocompatible copolymer),153 and a wide variety of other synthetic and natural polymers.154 Composite polymeric materials have expanded the available repertoire of surfaces and substrates. For example, gels consisting of hyaluronic acid (HA) and heparin are better substrates for the adhesion of endothelial progenitor cells than pure HA gels.155 New classes of polymers for studying bacteria interactions with surfaces could facilitate a renaissance in this area.
Adhesion between bacteria and surfaces can be measured by a variety of methods, many of which have been reviewed by Bos et al.58 Microbial adhesion to hydrocarbons (MATH) involves vortexing mixtures of bacteria and different hydrocarbons and quantifying bacteria adhesion to the hydrocarbon droplets that form.58 The number of bacteria that adhere to different types of surfaces can be assayed using various labeling methods, including radioactivity, fluorescence, and crystal violet staining.156 Other methods include atomic force microscopy (AFM), total internal reflection microscopy (TIRM), quartz crystal microbalance (QCM), and the use of liquid flow cells.82, 83, 157–160 Coating AFM tips with confluent layers of bacteria enables the measurement of the attractive or repulsive forces between bacteria and various materials.161 An interesting extension of this technique involves attaching microbeads to an AFM cantilever and coating them with bacteria, growing the bacteria into a biofilm, and measuring the adhesive and viscoelastic properties of the biofilm at different stages.162
New opportunities for chemists, materials scientists, and engineers
Our understanding of the interaction of bacteria with surfaces is remarkably incomplete. This topic seems ideally suited for collaborations between microbiologists and materials scientists, chemists, and engineers as it is poised to benefit from multidisciplinary approaches that are formulated to penetrate into a range of areas, including: (1) identifying the properties of surfaces that are sensed by bacteria; (2) elucidating the molecular mechanisms bacteria use to sense surfaces and their biochemical responses; and (3) determining how to modulate surface properties to provoke a desired cellular response, including changes in morphology, alterations in bioenergetics, or cell death.
One area in particular that may benefit from contributions from the physical sciences is the impact of the conditioning layer of proteins that frequently precedes bacterial attachment to a surface.90 Conditioning layers often have the effect of rendering the underlying chemistry of the surface irrelevant, which translates into antimicrobial surfaces that have short lifetimes over which they are effective. Self-polishing and self-peeling anti-fouling coatings consist of exposed layers that gradually delaminate over time and have been a moderately successful approach for overcoming bacterial attachment in industrial applications.163 Laboratory studies of bacteria-surface interactions require the prevention of forming conditioning layers. Materials scientists and engineers are uniquely positioned to tackle this challenge.
One difficulty in the study of cell attachment to surfaces is the measurement of cellular responses, in particular changes in gene expression. These assays typically require removing cells from the surface before analyzing mRNA levels. However, the advent of fluorescent reporters enables the measurement of levels of gene expression in real time in cells attached to surfaces. An elegant demonstration of this technique is the measurement of yellow fluorescent protein expression controlled by promoters of cell-state specific genes such as the gene encoding the flagellin protein.33 The availability of libraries of fluorescent fusions to a large number of both promoters164 and proteins165 in E. coli may enable high-throughput screens to identify genes not previously associated with surface adhesion (e.g., by comparing broth-grown cells and biofilms). Recent advances in surface enhanced Raman scattering (SERS)—specifically the peptide-guided localization of nanoparticle probes to the bacterial membrane—may be applied to studying the chemistry of individual cells in bacterial communities and complement genetic profiling approaches described above.166, 167
Bacterial surface sensing is a phenomenon that is still not well understood at the level of physical chemistry, biochemistry, genetics, and cell biology. The use of chemically and physically defined substrates and new analytical and biochemical techniques—including those described in this review and those yet to be developed—may have a transformative impact on our understanding of bacteria-surface interactions and guide applications in a range of areas, including agriculture, biomedicine, defense, dentistry, food safety, and industrial processing.
Acknowledgements
We thank Carrie Harwood, Zemer Gitai, and Alex Persat for their discussion and helpful comments. Work on surface sensing in the Weibel lab is supported by the National Science Foundation (DMR-1121288, MCB-1120832) and the National Institutes of Health (1DP2OD008735-01).
References
- 1.Kumada Y, Benson DR, Hillemann D, Hosted TJ, Rochefort DA, Thompson CJ, Wohlleben W, Tateno Y. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3009–3013. doi: 10.1073/pnas.90.7.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schidlowski M. Nature. 1988;333:313–318. [Google Scholar]
- 3.Whitman WB, Coleman DC, Wiebe WJ. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Annu. Rev. Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 5.Meadows P. Nature. 1965;207:1108–1101. [Google Scholar]
- 6.Van Houte J, Green D. Infect. Immun. 1974;9:624–630. doi: 10.1128/iai.9.4.624-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellen R, Gibbons R. Infect. Immun. 1972;5:826–830. doi: 10.1128/iai.5.5.826-830.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams R, Gibbons R. Infect. Immun. 1975;11:711–718. doi: 10.1128/iai.11.4.711-718.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher M. Can. J. Microbiol. 1977;23:1–6. [Google Scholar]
- 10.Fletcher M. J. Gen. Microbiol. 1976;94:400–404. doi: 10.1099/00221287-94-2-400. [DOI] [PubMed] [Google Scholar]
- 11.Fletcher M, Loeb G. Appl. Environ. Microbiol. 1979;37:67–72. doi: 10.1128/aem.37.1.67-72.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orstavik D, Orstavik J. J. Oral Rehabil. 1976;3:139–144. doi: 10.1111/j.1365-2842.1976.tb00937.x. [DOI] [PubMed] [Google Scholar]
- 13.Clark W, Gibbons R. Infect. Immun. 1977;18:514–523. doi: 10.1128/iai.18.2.514-523.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark W, Bammann L, Gibbons R. Infect. Immun. 1978;19:846–853. doi: 10.1128/iai.19.3.846-853.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corlan AD. Medline trend: Automated yearly statistics of PubMed results for any query. [Accessed February 2, 2013];2012 http://dan.corlan.net/medline-trend.html. [Google Scholar]
- 16.Katsikogianni M, Missirlis YF. Eur. Cell. Mater. 2004;8:37–57. doi: 10.22203/ecm.v008a05. [DOI] [PubMed] [Google Scholar]
- 17.ZoBell CE. J. Bacteriol. 1943;46:39–56. doi: 10.1128/jb.46.1.39-56.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heukelekian H, Heller A. J. Bacteriol. 1940;40:547–558. doi: 10.1128/jb.40.4.547-558.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poindexter JS. Microbiol. Rev. 1981;45:123–179. doi: 10.1128/mr.45.1.123-179.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grantham MC, Dove PM. Geochim. Cosmochim. Acta. 1996;60:2473–2480. [Google Scholar]
- 21.Nealson KH, Finkel SE. MRS Bull. 2011;36:380–384. [Google Scholar]
- 22.Nevin KP, Kim B-C, Glaven RH, Johnson JP, Woodard TL, Methé BA, DiDonato RJ, Covalla SF, Franks AE, Liu A, Lovley DR. PLoS ONE. 2009;4:e5628. doi: 10.1371/journal.pone.0005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donlan RM, Costerton JW. Clin. Microbiol. Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simões M, Pereira MO, Vieira MJ. Water Res. 2005;39:5142–5152. doi: 10.1016/j.watres.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 25.Mah TF, O'Toole GA. Trends Microbiol. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 26.Mah T. Future Microbiol. 2012;7:1061–1072. doi: 10.2217/fmb.12.76. [DOI] [PubMed] [Google Scholar]
- 27.John A-K, Schmaler M, Khanna N, Landmann R. Antimicrob. Agents Chemother. 2011;55:3510–3516. doi: 10.1128/AAC.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu Y, Daley AJ, Istivan TS, Rouch DA, Deighton MA. J. Antimicrob. Chemother. 2010;65:1405–1411. doi: 10.1093/jac/dkq119. [DOI] [PubMed] [Google Scholar]
- 29.Eun Y-J, Foss MH, Kiekebusch D, Pauw DA, Westler WM, Thanbichler M, Weibel DB. J. Am. Chem. Soc. 2012;134:11322–11325. doi: 10.1021/ja302542j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Connell JL, Wessel AK, Parsek MR, Ellington AD, Whiteley M, Shear JB. mBio. 2010;1:e00202–e00210. doi: 10.1128/mBio.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Queck S-Y, Weitere M, Moreno AM, Rice SA, Kjelleberg S. Environ. Microbiol. 2006;8:1017–1025. doi: 10.1111/j.1462-2920.2006.00993.x. [DOI] [PubMed] [Google Scholar]
- 32.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vlamakis H, Aguilar C, Losick R, Kolter R. Genes Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haagensen JAJ, Klausen M, Ernst RK, Miller SI, Folkesson A, Tolker-Nielsen T, Molin S. J. Bacteriol. 2007;189:28–37. doi: 10.1128/JB.00720-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purevdorj-Gage B, Costerton WJ, Stoodley P. J. Gen. Microbiol. 2005;151:1569–1576. doi: 10.1099/mic.0.27536-0. [DOI] [PubMed] [Google Scholar]
- 36.Copeland MF, Weibel DB. Soft Matter. 2009;5:1174–1187. doi: 10.1039/B812146J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patrick JE, Kearns DB. Mol. Microbiol. 2012;83:14–23. doi: 10.1111/j.1365-2958.2011.07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butler MT, Wang Q, Harshey RM. Proc. Nat. Acad. Sci. U.S.A. 2010;107:3776–3781. doi: 10.1073/pnas.0910934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Zhang C, Gong F, Li H, Xie X, Xia C, Chen J, Song Y, Shen A, Song J. Curr. Microbiol. 2011;63:377–386. doi: 10.1007/s00284-011-9979-0. [DOI] [PubMed] [Google Scholar]
- 40.Ingham CJ, Kalisman O, Finkelshtein A, Ben-Jacob E. Proc. Nat. Acad. Sci. U.S.A. 2011;108:19731–19736. doi: 10.1073/pnas.1102097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chai L, Vlamakis H, Kolter R. MRS Bull. 2011;36:374–379. [Google Scholar]
- 42.Hamze K, Autret S, Hinc K, Laalami S, Julkowska D, Briandet R, Renault M, Absalon C, Holland IB, Putzer H, Séror SJ. Microbiology. 2011;157:2456–2469. doi: 10.1099/mic.0.047159-0. [DOI] [PubMed] [Google Scholar]
- 43.Matsuyama T, Takagi Y, Nakagawa Y, Itoh H, Wakita J, Matsushita M. J. Bacteriol. 2000;182:385–393. doi: 10.1128/jb.182.2.385-393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuson HH, Copeland MF, Carey S, Sacotte R, Weibel DB. J. Bacteriol. 2012;195:368–377. doi: 10.1128/JB.01537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng WL, Bassler BL. Annu. Rev. Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frederix M, Downie JA. In: Advances in Microbial Physiology. Robert KP, editor. vol. Volume 58, ch. 2. Academic Press; 2011. pp. 23–80. [DOI] [PubMed] [Google Scholar]
- 47.Flickinger ST, Copeland MF, Downes EM, Braasch AT, Tuson HH, Eun Y-J, Weibel DB. J. Am. Chem. Soc. 2011;133:5966–5975. doi: 10.1021/ja111131f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dilanji GE, Langebrake JB, De Leenheer P, Hagen SJ. J. Am. Chem. Soc. 2012;134:5618–5626. doi: 10.1021/ja211593q. [DOI] [PubMed] [Google Scholar]
- 49.Shrout JD, Tolker-Nielsen T, Givskov M, Parsek MR. MRS Bull. 2011;36:367–373. doi: 10.1557/mrs.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madsen JS, Burmølle M, Hansen LH, Sørensen SJ. FEMS Immunol. Med. Microbiol. 2012 doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 51.Meibom KL, Blokesch M, Dolganov NA, Wu C-Y, Schoolnik GK. Science. 2005;310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 52.Pathak DT, Wei X, Bucuvalas A, Haft DH, Gerloff DL, Wall D. PLoS Genet. 2012;8:e1002626. doi: 10.1371/journal.pgen.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro MVAS, Yildiz FH, Sondermann H. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caiazza NC, Merritt JH, Brothers KM, O'Toole GA. J. Bacteriol. 2007;189:3603–3612. doi: 10.1128/JB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. Science. 2008;320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 56.Allison C, Coleman N, Jones PL, Hughes C. Infect. Immun. 1992;60:4740–4746. doi: 10.1128/iai.60.11.4740-4746.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vigeant MA-S, Ford RM, Wagner M, Tamm LK. Appl. Environ. Microbiol. 2002;68:2794–2801. doi: 10.1128/AEM.68.6.2794-2801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bos R, Mei HC, Busscher HJ. FEMS Microbiol. Rev. 1999;23:179–230. doi: 10.1111/j.1574-6976.1999.tb00396.x. [DOI] [PubMed] [Google Scholar]
- 59.McClaine JW, Ford RM. Appl. Environ. Microbiol. 2002;68:1280–1289. doi: 10.1128/AEM.68.3.1280-1289.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kubitschek HE. Crit. Rev. Microbiol. 1987;14:73–97. doi: 10.3109/10408418709104436. [DOI] [PubMed] [Google Scholar]
- 61.Inoue K, Nishimura M, Nayak BB, Kogure K. Appl. Environ. Microbiol. 2007;73:1049–1053. doi: 10.1128/AEM.01158-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Makinoshima H, Nishimura A, Ishihama A. Mol. Microbiol. 2002;43:269–279. doi: 10.1046/j.1365-2958.2002.02746.x. [DOI] [PubMed] [Google Scholar]
- 63.Nishino T, Nayak BB, Kogure K. Appl. Environ. Microbiol. 2003;69:3569–3572. doi: 10.1128/AEM.69.6.3569-3572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boks NP, Busscher HJ, van der Mei HC, Norde W. Langmuir. 2008;24:12990–12994. doi: 10.1021/la801824c. [DOI] [PubMed] [Google Scholar]
- 65.Boks NP, Kaper HJ, Norde W, Busscher HJ, van der Mei HC. Colloids Surf. B. Biointerfaces. 2008;67:276–278. doi: 10.1016/j.colsurfb.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 66.Meinders JM, Van der Mei HC, Busscher HJ. J. Colloid Interface Sci. 1995;176:329–341. [Google Scholar]
- 67.Busscher HJ, Norde W, Sharma PK, van der Mei HC. Curr. Opin. Colloid In. 2010;15:510–517. [Google Scholar]
- 68.Hayashi H, Seiki H, Tsuneda S, Hirata A, Sasaki H. J. Colloid Interface Sci. 2003;264:565–568. doi: 10.1016/S0021-9797(03)00418-1. [DOI] [PubMed] [Google Scholar]
- 69.Renner LD, Weibel DB. MRS Bull. 2011;36:347–355. doi: 10.1557/mrs.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eboigbodin KE, Newton JRA, Routh AF, Biggs CA. Appl. Microbiol. Biotechnol. 2006;73:669–675. doi: 10.1007/s00253-006-0505-4. [DOI] [PubMed] [Google Scholar]
- 71.Caiazza NC, O'Toole GA. J. Bacteriol. 2004;186:4476–4485. doi: 10.1128/JB.186.14.4476-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chao Y, Zhang T. Langmuir. 2011;27:11545–11553. doi: 10.1021/la202534p. [DOI] [PubMed] [Google Scholar]
- 73.Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. Mol. Microbiol. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 74.Marshall KC, Stout R, Mitchell R. J. Gen. Microbiol. 1971;68:337. doi: 10.1139/m71-225. [DOI] [PubMed] [Google Scholar]
- 75.Beachey EH. J. Infect. Dis. 1981;143:325–345. doi: 10.1093/infdis/143.3.325. [DOI] [PubMed] [Google Scholar]
- 76.Van Houdt R, Michiels CW. Res. Microbiol. 2005;156:626–633. doi: 10.1016/j.resmic.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 77.Spears KJ, Roe AJ, Gally DL. FEMS Microbiol. Lett. 2006;255:187–202. doi: 10.1111/j.1574-6968.2006.00119.x. [DOI] [PubMed] [Google Scholar]
- 78.Newman F, Beeley JA, MacFarlane TW. Electrophoresis. 1996;17:266–270. doi: 10.1002/elps.1150170146. [DOI] [PubMed] [Google Scholar]
- 79.Amano A, Shizukuishi S, Horie H, Kimura S, Morisaki I, Hamada S. Infect. Immun. 1998;66:2072–2077. doi: 10.1128/iai.66.5.2072-2077.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nelson SS, Bollampalli S, McBride MJ. J. Bacteriol. 2008;190:2851–2857. doi: 10.1128/JB.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bouckaert J, Mackenzie J, de Paz JL, Chipwaza B, Choudhury D, Zavialov A, Mannerstedt K, Anderson J, Pierard D, Wyns L, Seeberger PH, Oscarson S, De Greve H, Knight SD. Mol. Microbiol. 2006;61:1556–1568. doi: 10.1111/j.1365-2958.2006.05352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nilsson LM, Thomas WE, Trintchina E, Vogel V, Sokurenko EV. J. Biol. Chem. 2006;281:16656–16663. doi: 10.1074/jbc.M511496200. [DOI] [PubMed] [Google Scholar]
- 83.Anderson BN, Ding AM, Nilsson LM, Kusuma K, Tchesnokova V, Vogel V, Sokurenko EV, Thomas WE. J. Bacteriol. 2007;189:1794–1802. doi: 10.1128/JB.00899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bruellhoff K, Fiedler J, Möller M, Groll J, Brenner RE. Int. J. Artif. Organs. 2010;33:646–653. doi: 10.1177/039139881003300910. [DOI] [PubMed] [Google Scholar]
- 85.Kang G-d, Cao Y-m. Water Res. 2012;46:584–600. doi: 10.1016/j.watres.2011.11.041. [DOI] [PubMed] [Google Scholar]
- 86.Marcato-Romain CE, Pechaud Y, Paul E, Girbal-Neuhauser E, Dossat-Létisse V. Biofouling. 2012;28:305–314. doi: 10.1080/08927014.2012.673122. [DOI] [PubMed] [Google Scholar]
- 87.Schultz MP, Bendick JA, Holm ER, Hertel WM. Biofouling. 2010;27:87–98. doi: 10.1080/08927014.2010.542809. [DOI] [PubMed] [Google Scholar]
- 88.Absolom DR, Lamberti FV, Policova Z, Zingg W, van Oss CJ, Neumann AW. Appl. Environ. Microbiol. 1983;46:90–97. doi: 10.1128/aem.46.1.90-97.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goulter-Thorsen RM, Taran E, Gentle IR, Gobius KS, Dykes GA. Appl. Environ. Microbiol. 2011;77:7339–7344. doi: 10.1128/AEM.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Banerjee I, Pangule RC, Kane RS. Adv. Mater. 2011;23:690–718. doi: 10.1002/adma.201001215. [DOI] [PubMed] [Google Scholar]
- 91.Lejeune P. Trends Microbiol. 2003;11:179–184. doi: 10.1016/s0966-842x(03)00047-7. [DOI] [PubMed] [Google Scholar]
- 92.Wong SY, Han L, Timachova K, Veselinovic J, Hyder MN, Ortiz C, Klibanov AM, Hammond PT. Biomacromolecules. 2012;13:719–726. doi: 10.1021/bm201637e. [DOI] [PubMed] [Google Scholar]
- 93.Melo LD, Palombo RR, Petri DFS, Bruns M, Pereira EMA, Carmona-Ribeiro AM. ACS Appl. Mater. Interfaces. 2011;3:1933–1939. doi: 10.1021/am200150t. [DOI] [PubMed] [Google Scholar]
- 94.Tiller JC, Liao C-J, Lewis K, Klibanov AM. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5981–5985. doi: 10.1073/pnas.111143098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hook AL, Chang C-Y, Yang J, Luckett J, Cockayne A, Atkinson S, Mei Y, Bayston R, Irvine DJ, Langer R, Anderson DG, Williams P, Davies MC, Alexander MR. Nat. Biotechnol. 2012 doi: 10.1038/nbt.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bazaka K, Crawford RJ, Ivanova EP. Biotechnol. J. 2011;6:1103–1114. doi: 10.1002/biot.201100027. [DOI] [PubMed] [Google Scholar]
- 97.Cheng G, Zhang Z, Chen S, Bryers JD, Jiang S. Biomaterials. 2007;28:4192–4199. doi: 10.1016/j.biomaterials.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Z, Chao T, Chen S, Jiang S. Langmuir. 2006;22:10072–10077. doi: 10.1021/la062175d. [DOI] [PubMed] [Google Scholar]
- 99.Zhang Z, Chen S, Chang Y, Jiang S. J. Phys. Chem. B. 2006;110:10799–10804. doi: 10.1021/jp057266i. [DOI] [PubMed] [Google Scholar]
- 100.Ista LK, Mendez S, Pérez-Luna VH, López GP. Langmuir. 2001;17:2552–2555. [Google Scholar]
- 101.Okano T, Kikuchi A, Sakurai Y, Takei Y, Ogata N. J. Control. Release. 1995;36:125–133. [Google Scholar]
- 102.Senaratne W, Andruzzi L, Ober CK. Biomacromolecules. 2005;6:2427–2448. doi: 10.1021/bm050180a. [DOI] [PubMed] [Google Scholar]
- 103.Nejadnik MR, van der Mei HC, Norde W, Busscher HJ. Biomaterials. 2008;29:4117–4121. doi: 10.1016/j.biomaterials.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 104.Gao G, Yu K, Kindrachuk J, Brooks DE, Hancock REW, Kizhakkedathu JN. Biomacromolecules. 2011;12:3715–3727. doi: 10.1021/bm2009697. [DOI] [PubMed] [Google Scholar]
- 105.Chung KK, Schumacher JF, Sampson EM, Burne RA, Antonelli PJ, Brennan AB. Biointerphases. 2007;2:89–94. doi: 10.1116/1.2751405. [DOI] [PubMed] [Google Scholar]
- 106.Epstein AK, Hochbaum AI, Kim P, Aizenberg J. Nanotechnology. 2011;22:494007. doi: 10.1088/0957-4484/22/49/494007. [DOI] [PubMed] [Google Scholar]
- 107.Epstein AK, Wong T-S, Belisle RA, Boggs EM, Aizenberg J. Proc. Nat. Acad. Sci. U.S.A. 2012;109:13182–13187. doi: 10.1073/pnas.1201973109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khoo X, Grinstaff MW. MRS Bull. 2011;36:357–366. [Google Scholar]
- 109.Segall JE, Block SM, Berg HC. Proc. Natl. Acad. Sci. U.S.A. 1986;83:8987–8991. doi: 10.1073/pnas.83.23.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sourjik V, Armitage JP. EMBO J. 2010;29:2724–2733. doi: 10.1038/emboj.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lower SK, Yongsunthon R, Casillas-Ituarte NN, Taylor ES, DiBartola AC, Lower BH, Beveridge TJ, Buck AW, Fowler VG. Biophys. J. 2010;99:2803–2811. doi: 10.1016/j.bpj.2010.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ponsonnet L, Boureanu M, Jaffrezic N, Othmane A, Dorel C, Lejeune P. Mater. Sci. Eng. C. 2008;28:896–900. [Google Scholar]
- 113.Nakayama S, Watanabe H. J. Bacteriol. 1995;177:5062–5069. doi: 10.1128/jb.177.17.5062-5069.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hung DL, Raivio TL, Jones CH, Silhavy TJ, Hultgren SJ. EMBO J. 2001;20:1508–1518. doi: 10.1093/emboj/20.7.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schwan WR, Lee JL, Lenard FA, Matthews BT, Beck MT. Infect. Immun. 2002;70:1391–1402. doi: 10.1128/IAI.70.3.1391-1402.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brown DG, Hong Y. J. Adhes. Sci. Technol. 2011;25:2199–2218. [Google Scholar]
- 117.Hong Y, Brown DG. Appl. Environ. Microbiol. 2009;75:2346–2353. doi: 10.1128/AEM.02671-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ma Q, Wood TK. Environ. Microbiol. 2009;11:2735–2746. doi: 10.1111/j.1462-2920.2009.02000.x. [DOI] [PubMed] [Google Scholar]
- 119.Maltz MA, Weiss BL, O'Neill M, Wu Y, Aksoy S. Appl. Environ. Microbiol. 2012 doi: 10.1128/AEM.01858-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang LC, Morgan LK, Godakumbura P, Kenney LJ, Anand GS. EMBO J. 2012;31:2648–2659. doi: 10.1038/emboj.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Meibom KL, Li XB, Nielsen AT, Wu C-Y, Roseman S, Schoolnik GK. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li X, Roseman S. Proc. Natl. Acad. Sci. U.S.A. 2004;101:627–631. doi: 10.1073/pnas.0307645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McCarter L, Hilmen M, Silverman M. Cell. 1988;54:345–351. doi: 10.1016/0092-8674(88)90197-3. [DOI] [PubMed] [Google Scholar]
- 124.Alberti L, Harshey RM. J. Bacteriol. 1990;172:4322–4328. doi: 10.1128/jb.172.8.4322-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Belas R, Suvanasuthi R. J. Bacteriol. 2005;187:6789–6803. doi: 10.1128/JB.187.19.6789-6803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cusick K, Lee Y-Y, Youchak B, Belas R. J. Bacteriol. 2012;194:437–447. doi: 10.1128/JB.05998-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dufour A, Furness RB, Hughes C. Mol. Microbiol. 1998;29:741–751. doi: 10.1046/j.1365-2958.1998.00967.x. [DOI] [PubMed] [Google Scholar]
- 128.Pearson MM, Rasko DA, Smith SN, Mobley HLT. Infect. Immun. 2010;78:2834–2845. doi: 10.1128/IAI.01222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Otto K, Silhavy TJ. Proc. Nat. Acad. Sci. U.S.A. 2002;99:2287–2292. doi: 10.1073/pnas.042521699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dorel C, Lejeune P, Rodrigue A. Res. Microbiol. 2006;157:306–314. doi: 10.1016/j.resmic.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 131.Otto K, Hermansson M. J. Bacteriol. 2004;186:226–234. doi: 10.1128/JB.186.1.226-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Couturier MR, Stein M. Can. J. Microbiol. 2008;54:537–548. doi: 10.1139/w08-042. [DOI] [PubMed] [Google Scholar]
- 133.O'Connor JR, Kuwada NJ, Huangyutitham V, Wiggins PA, Harwood CS. Mol. Microbiol. 2012 doi: 10.1111/mmi.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Güvener ZT, Harwood CS. Mol. Microbiol. 2007;66:1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hickman JW, Tifrea DF, Harwood CS. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O'Toole GA. J. Bacteriol. 2007;189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hengge R. Nat. Rev. Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 138.Simm R, Morr M, Kader A, Nimtz M, Römling U. Mol. Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 139.Hickman JW, Harwood CS. Mol. Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Justice SS, Hunstad DA, Cegelski L, Hultgren SJ. Nat. Rev. Microbiol. 2008;6:162–168. doi: 10.1038/nrmicro1820. [DOI] [PubMed] [Google Scholar]
- 141.Horvath DJ, Li B, Casper T, Partida-Sanchez S, Hunstad DA, Hultgren SJ, Justice SS. Microb. Infect. 2011;13:426–437. doi: 10.1016/j.micinf.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Finer KR, Larkin KM, Martin BJ, Finer JJ. Plant Cell Rep. 2001;20:250–255. [Google Scholar]
- 143.Salvetti S, Faegri K, Ghelardi E, Kolstø A-B, Senesi S. Appl. Environ. Microbiol. 2011;77:5149–5156. doi: 10.1128/AEM.00245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang Q, Frye JG, McClelland M, Harshey RM. Mol. Microbiol. 2004;52:169–187. doi: 10.1111/j.1365-2958.2003.03977.x. [DOI] [PubMed] [Google Scholar]
- 145.Dalton HM, Poulsen LK, Halasz P, Angles ML, Goodman AE, Marshall KC. J. Bacteriol. 1994;176:6900–6906. doi: 10.1128/jb.176.22.6900-6906.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gognies S, Belarbi A. Appl. Microbiol. Biotechnol. 2010;88:1095–1102. doi: 10.1007/s00253-010-2800-3. [DOI] [PubMed] [Google Scholar]
- 147.Shungu D, Valiant M, Tutlane V, Weinberg E, Weissberger B, Koupal L, Gadebusch H, Stapley E. Appl. Environ. Microbiol. 1983;46:840–845. doi: 10.1128/aem.46.4.840-845.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tuson HH, Renner LD, Weibel DB. Chem. Comm. 2012;48:1595–1597. doi: 10.1039/c1cc14705f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lichter JA, Thompson MT, Delgadillo M, Nishikawa T, Rubner MF, Van Vliet KJ. Biomacromolecules. 2008;9:1571–1578. doi: 10.1021/bm701430y. [DOI] [PubMed] [Google Scholar]
- 150.Pelham RJ, Jr, Wang Y-L. Proc. Nat. Acad. Sci. U.S.A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Boontheekul T, Hill EE, Kong H-J, Mooney DJ. Tissue Eng. 2007;13:1431–1442. doi: 10.1089/ten.2006.0356. [DOI] [PubMed] [Google Scholar]
- 152.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 153.Minoura N, Koyano T, Koshizaki N, Umehara H, Nagura M, Kobayashi K-i. Mater. Sci. Eng. C. 1998;6:275–280. [Google Scholar]
- 154.Drury JL, Mooney DJ. Biomaterials. 2003;24:4337–4351. doi: 10.1016/s0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- 155.Camci-Unal G, Nichol JW, Bae H, Tekin H, Bischoff J, Khademhosseini A. J. Tissue Eng. Regen. Med. 2012 doi: 10.1002/term.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vesterlund S, Paltta J, Karp M, Ouwehand AC. J. Microbiol. Methods. 2005;60:225–233. doi: 10.1016/j.mimet.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 157.Camesano TA, Liu Y, Datta M. Adv. Water Resour. 2007;30:1470–1491. [Google Scholar]
- 158.Otto K. Res. Microbiol. 2008;159:415–422. doi: 10.1016/j.resmic.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 159.Mosier AP, Cady NC. Sci. Prog. 2011;94:431–450. doi: 10.3184/003685011X13201828216868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shen Y, Siryaporn A, Lecuyer S, Gitai Z, Stone HA. Biophys. J. 2012;103:146–151. doi: 10.1016/j.bpj.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Razatos A, Ong Y-L, Sharma MM, Georgiou G. Proc. Nat. Acad. Sci. U.S.A. 1998;95:11059–11064. doi: 10.1073/pnas.95.19.11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lau PCY, Dutcher JR, Beveridge TJ, Lam JS. Biophys. J. 2009;96:2935–2948. doi: 10.1016/j.bpj.2008.12.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xie L, Hong F, He C, Ma C, Liu J, Zhang G, Wu C. Polymer. 2011 [Google Scholar]
- 164.Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U. Nat. Methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- 165.Taniguchi Y, Choi PJ, Li G-W, Chen H, Babu M, Hearn J, Emili A, Xie XS. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Athamneh AIM, Senger RS. Appl. Environ. Microbiol. 2012 doi: 10.1128/AEM.02000-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Efrima S, Zeiri L. J. Raman Spectrosc. 2009;40:277–288. [Google Scholar]
- 168.Cheung H-Y, Chan GK-L, Cheung S-H, Sun S-Q, Fong W-F. J. Appl. Microbiol. 2007;102:701–710. doi: 10.1111/j.1365-2672.2006.03137.x. [DOI] [PubMed] [Google Scholar]
- 169.Dexter SC, Sullivan JD, Williams J, Watson SW. Appl. Microbiol. 1975;30:298–308. doi: 10.1128/am.30.2.298-308.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wei J, Ravn DB, Gram L, Kingshott P. Colloids Surf. B. Biointerfaces. 2003;32:275–291. [Google Scholar]
- 171.Cao Z, Mi L, Mendiola J, Ella-Menye J-R, Zhang L, Xue H, Jiang S. Angew. Chem. 2012;51:2602–2605. doi: 10.1002/anie.201106466. [DOI] [PubMed] [Google Scholar]
- 172.Schaer TP, Stewart S, Hsu BB, Klibanov AM. Biomaterials. 2012;33:1245–1254. doi: 10.1016/j.biomaterials.2011.10.038. [DOI] [PubMed] [Google Scholar]
- 173.Behlau I, Mukherjee K, Todani A, Tisdale AS, Cade F, Wang L, Leonard EM, Zakka FR, Gilmore MS, Jakobiec FA, Dohlman CH, Klibanov AM. Biomaterials. 2011;32:8783–8796. doi: 10.1016/j.biomaterials.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cheng G, Li G, Xue H, Chen S, Bryers JD, Jiang S. Biomaterials. 2009;30:5234–5240. doi: 10.1016/j.biomaterials.2009.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Oliveira R, Melo L, Oliveira A, Salgueiro R. Colloids Surf. B. Biointerfaces. 1994;2:41–46. [Google Scholar]
- 176.Roosjen A, van der Mei HC, Busscher HJ, Norde W. Langmuir. 2004;20:10949–10955. doi: 10.1021/la048469l. [DOI] [PubMed] [Google Scholar]
- 177.Roosjen A, de Vries J, van der Mei HC, Norde W, Busscher HJ. J. Biomed. Mater. Res., Part B. 2005;73:347–354. doi: 10.1002/jbm.b.30227. [DOI] [PubMed] [Google Scholar]
- 178.Pidhatika B, Möller J, Benetti EM, Konradi R, Rakhmatullina E, Mühlebach A, Zimmermann R, Werner C, Vogel V, Textor M. Biomaterials. 2010;31:9462–9472. doi: 10.1016/j.biomaterials.2010.08.033. [DOI] [PubMed] [Google Scholar]
- 179.Chang Y, Yandi W, Chen W-Y, Shih Y-J, Yang C-C, Chang Y, Ling Q-D, Higuchi A. Biomacromolecules. 2010;11:1101–1110. doi: 10.1021/bm100093g. [DOI] [PubMed] [Google Scholar]
- 180.Cunliffe D, Smart CA, Tsibouklis J, Young S, Alexander C, Vulfson EN. Biotechnol. Lett. 2000;22:141–145. [Google Scholar]
- 181.Cheng G, Xue H, Li G, Jiang S. Langmuir. 2010;26:10425–10428. doi: 10.1021/la101542m. [DOI] [PubMed] [Google Scholar]
- 182.Ong Y-L, Razatos A, Georgiou G, Sharma MM. Langmuir. 1999;15:2719–2725. [Google Scholar]
- 183.Patel JD, Ebert M, Ward R, Anderson JM. J. Biomed. Mater. Res. A. 2007;80:742–751. doi: 10.1002/jbm.a.31103. [DOI] [PubMed] [Google Scholar]
- 184.Ostuni E, Chapman RG, Liang MN, Meluleni G, Pier G, Ingber DE, Whitesides GM. Langmuir. 2001;17:6336–6343. [Google Scholar]
- 185.Tegoulia VA, Cooper SL. Colloids Surf. B. Biointerfaces. 2002;24:217–228. [Google Scholar]
- 186.Vassilakos N, Kalfas S, Arnebrant T, Rundegren J. Colloids Surf. B. Biointerfaces. 1993;1:341–347. [Google Scholar]
- 187.Chen S, Chen S, Jiang S, Mo Y, Luo J, Tang J, Ge Z. Colloids Surf. B. Biointerfaces. 2011;85:323–329. doi: 10.1016/j.colsurfb.2011.03.004. [DOI] [PubMed] [Google Scholar]