Abstract
Extracellular vesicles (EVs) carry signals within or at their limiting membranes, providing a mechanism by which cells can exchange more complex information than what was previously thought. In addition to mRNAs and microRNAs, there are DNA fragments in EVs. Solexa sequencing indicated the presence of at least 16434 genomic DNA (gDNA) fragments in the EVs from human plasma. Immunofluorescence study showed direct evidence that acridine orange-stained EV DNAs could be transferred into the cells and localize to and inside the nuclear membrane. However, whether the transferred EV DNAs are functional or not is not clear. We found that EV gDNAs could be homologously or heterologously transferred from donor cells to recipient cells, and increase gDNA-coding mRNA, protein expression, and function (e.g. AT1 receptor). An endogenous promoter of the AT1 receptor, NF-κB, could be recruited to the transferred DNAs in the nucleus, and increase the transcription of AT1 receptor in the recipient cells. Moreover, the transferred EV gDNAs have pathophysiological significance. BCR/ABL hybrid gene, involved in the pathogenesis of chronic myeloid leukemia, could be transferred from K562 EVs to HEK293 cells or neutrophils. Our present study shows that the gDNAs transferred from EVs to cells have physiological significance, not only to increase the gDNA-coding mRNA and protein levels, but also to influence function in recipient cells.
Keywords: extracellular vesicles, genomic DNA, AT1 receptor, BCR/ABL hybrid gene
Introduction
Cell-to-cell communication is required to guarantee proper coordination among different cell types within tissues. There are multiple types of intercellular communication, including soluble factors, tunneling nanotubules, and extracellular vesicles (EVs), which allow the transfer of surface molecules or cytoplasmic components from one cell to another. EVs are circular plasma membrane fragments that include exosomes and microparticles or shed vesicles (Trajkovic et al., 2008; Thery et al., 2009). The biological function of EVs is poorly understood, but may include secretion, immunomodulation, coagulation, and intercellular communication (Cocucci et al., 2009; Pap et al., 2009; Wang et al., 2012).
EVs may vary in their abundance, size, and composition, but they often contain materials, which include functional transmembrane proteins, mRNAs, and microRNAs (Valadi et al., 2007; Skog et al., 2008; Gibbings et al., 2009; Zen and Zhang, 2012). The components in EVs could be transferred from one cell to another by endocytosis or fusion with the recipient cell (Denzer et al., 2000; Clayton et al., 2004; Morelli et al., 2004; Ogawa et al., 2010). Moreover, the transferred components in EVs are functional. For example, glioblastoma EVs are enriched in angiogenic proteins, which stimulate tubule formation in endothelial cells (Skog et al., 2008). Besides functional proteins, Valadi et al. (2007) reported the presence of mRNA from ∼1300 genes, many of which are not present in the cytoplasm of the donor cell. The packaged mRNAs or microRNAs in EVs could be delivered to target cells that can modulate the biological functions of these cells by promoting or repressing target gene expression (Valadi et al., 2007; Al-Nedawi et al., 2008; Skog et al., 2008; Zhang et al., 2010).
Recent studies have shown that both mitochondrial DNA (mtDNA) and chromosomal DNA were found in EVs (Guescini et al., 2010; Balaj et al., 2011; Waldenstrom et al., 2012). Waldenstrom et al. (2012) reported that chromosomal DNA sequences in EVs from cardiomyocytes could be transferred to the cytosol or nuclei of target cells. However, whether the transferred EV DNAs are functional or not is unclear. In this study, we investigated the function and potential mechanisms of transferrable EV DNAs in the recipient cells. We first examined the existence of genomic DNA (gDNA) in EVs derived from human plasma and supernatants of vascular smooth muscle cells (VSMCs) in culture by Solexa sequencing and PCR.
Angiotensin II type 1 (AT1) receptors exist ubiquitously and mediate several functions, including vasoconstriction, vascular proliferation, and renal antinatriuresis (Klingbeil et al., 2000; Zhang et al., 2005). Abnormal expression and activation of AT1 receptors are associated with cardiovascular diseases (e.g. hypertension and atherosclerosis). To determine the transportability and functionality of the DNA fragments in EVs, we studied the transport of gDNAs (AT1 receptor DNA) in EVs from AT1 receptor transfected HEK293 cells or VSMCs to non-transfected HEK293 cells. To uncover the underlying mechanisms leading to the transcription of transferred EV DNA, we investigated the effect of endogenous NF-κB, a promoter of the AT1 receptor gene, on transferred DNAs including AT1 receptor DNA.
Previous studies have shown an increased quantity of EVs in the body fluids of patients with cancer and provided increasing evidence that EVs might play a pivotal role in tumorigenesis (Skog et al., 2008; Balaj et al., 2011). Chronic myeloid leukemia (CML) is a clonal myeloproliferative disease, characterized by the oncogenic Philadelphia chromosome, which is formed by a reciprocal translocation between chromosomes 9 and 22 (Pluk et al., 2002). This translocation causes 50 portions of the breakpoint cluster region (BCR) gene from chromosome 22 to juxtapose with the 30 tyrosine kinase domains of the v-abl Abelson murine leukemia viral oncogene (ABL) from chromosome 9 (Pluk et al., 2002). The newly encoded chimeric protein BCR/ABL dictates the pathophysiology of CML and is recognized as the main target for tyrosine kinase inhibitors, such as imatinib, to treat CML patients (Pluk et al., 2002; Shibata et al., 2010; Puissant et al., 2012). To determine the pathophysiological significance of transferred gDNAs in EVs between cells, we examined the transfer of BCR/ABL hybrid gene in EVs from K562 cells to normal human neutrophils isolated from human peripheral blood.
Results
Isolation and identification of EVs
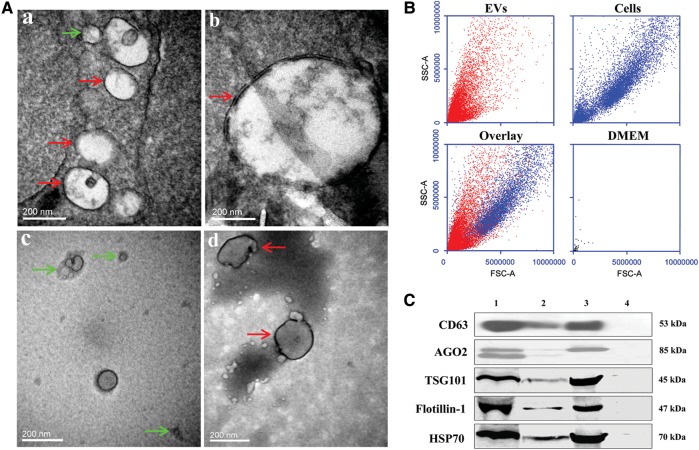
EVs from human plasma or cell culture supernatants were isolated through a series of ultracentrifugation steps as previously described (Valadi et al., 2007; Skog et al., 2008; Zhang et al., 2010). To ensure that the EVs were correctly identified, electron microscopic, immunoblotting, and flow cytometry (FCM) analyses were used. The electron micrographs of the EVs revealed rounded double-layer membranous vesicles of ∼30–1000 nm in size, similar to previously described EVs (Valadi et al., 2007; Skog et al., 2008; Zhang et al., 2010; Figure 1A). EVs more than 200 nm in size were confirmed by FCM analysis as this technology is unable to distinguish or enumerate single particles below 200 nm in size (Zwicker, 2010; Figure 1B). Immunoblotting showed the presence of CD63, argonaute 2 (AGO2), tumor susceptibility gene 101 (TSG101), flotillin-1, and heat shock protein 70 (HSP70) proteins, commonly used markers for EVs (Figure 1C).
Figure 1.
Identification and characterization of EVs from plasma of normal human subjects or VSMCs. (A) Electron micrographs of EVs. Isolated EVs were fixed, with or without negative staining with Na-phosphotungstate, and observed by TEM. Regular EVs with different sizes from plasma (a and b) of normal human subjects and supernatants of VSMC cultures (negative staining, c and d) were observed under TEM. The sizes of the EVs are consistent with the size of shed vesicles (100–1000 nm in size, red arrows) and exosomes (30–100 nm in size, green arrows). (B) FCM analysis. Samples, including VSMC EVs and VSMCs, were analyzed by BD Accuri C6 FCM. DMEM cell culture medium was used as negative control. The lower-left figure shows the overlay of the upper figures showing VSMC EVs in VSMCs (cells). (C) CD63, AGO2, TSG101, flotillin-1, and HSP70 expressions in EVs. EVs (1 × 107/ml, 200 μl), isolated from plasma of normal human subjects or VSMCs, were separated on SDS–PAGE, electroblotted onto nitrocellulose membrane, and subjected to immunoblotting with antibodies against CD63 (1:500), AGO2 (1:2000), TSG101 (1:400), flotillin-1 (1:400), or HSP70 (1:400) (lane 1: plasma EV; lane 2: EVs of VSMCs; lane 3: cellular lysate of VSMCs as positive control; lane 4: DMEM cell culture medium as negative control).
Identification of DNA fragments in EVs
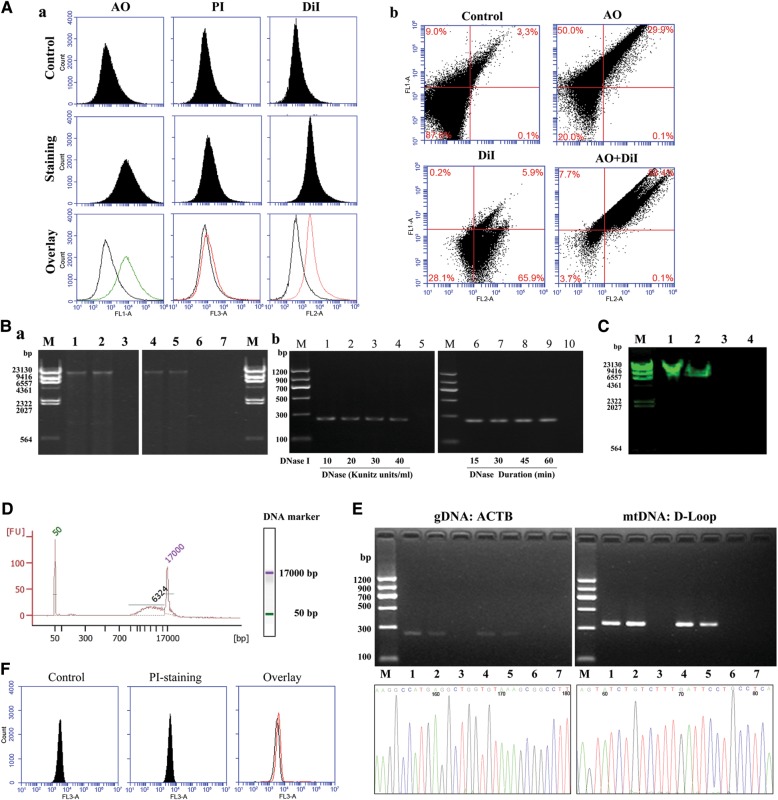
Substantial amounts of DNA were detected by fluorescence-activated cell sorter (FACS), agarose gel electrophoresis, PCR, and Agilent 2100 Bioanalyzer (Agilent Technologies) in EVs derived from human plasma and supernatants of VSMCs in culture. The single-staining of EVs with acridine orange (AO) (membrane permeable) or DiI (a lipophilic dye) showed enhanced fluorescence, while single-staining of EVs with propidium iodide (PI) (membrane impermeable) had low fluorescence, similar to the unstained control EVs (Figure 2Aa). Co-staining of EVs with AO and DiI showed an enhanced fluorescence compared with unstained (control) or single-stained EVs (Figure 2Ab), suggesting that DNA is present only inside the thoroughly washed EVs. DNase (varying concentrations and periods of incubation) was incubated with EVs to further confirm that the DNA fragments were inside the EVs. DNase treated and non-treated control EVs (Figure 2Ba and b) showed the same electrophoretic pattern, arguing against DNA being present outside the EVs.
Figure 2.
Identification of DNA fragments in EVs. (A) FCM of DNA-stained EVs. (a) Fluorescence of EVs from VSMCs that were unstained (Control) or stained with membrane permeable-AO, membrane impermeable-PI, or lipid membrane dye-DiI. Fluorescence was observed at the 530 nm channel for AO, 670 nm channel for PI, and 565 nm channel for DiI. Fluorescence of unstained EVs (Control) and EVs stained with AO, PI, or DiI are shown in the top lanes and middle lanes, respectively, and the overlay of the unstained and stained images is shown in the bottom lanes. (b) Fluorescence of EVs from VSMCs that were unstained (Control) or co-stained with membrane permeable-AO and/or lipid membrane dye-DiI. (B) Agarose gel electrophoresis of DNAs in EVs. (a) Electrophoresis on 0.7% agarose gel precasted with ethidium bromide. DNA marker (lane M), EVs without (lane 1 and lane 4) or with (lane 2 and lane 5) DNase treatment, disrupted EVs treated with DNase I (lane 3 and lane 6), and negative control (ddH2O, lane 7) are shown. Lanes 1–3 show EVs from human plasma; lanes 4–6 show EVs from VSMC supernatant. (b) EVs treated with DNase I. EVs were treated with varying concentrations (10–40 Kunitz units/ml, 30 min, lanes 1–4) and durations (15–60 min, 20 Kunitz units/ml, lanes 6–9) of incubation with DNase I and ddH2O (negative control, lanes 5 and 10). DNA marker (lane M). β-actin DNA in EVs was detected by PCR, and the PCR products were analyzed on 2% agarose gel precasted with ethidium bromide. (C) Electrophoresis on 0.7% agarose gel precasted with SYBR Green I (dsDNA stain). DNA marker (lane M), EVs DNA (lane 1), genomic DNA (dsDNA, lane 2), cDNA (ssDNA, lane 3), and negative control (ddH2O, lane 4). (D) Bioanalyzer analysis of DNAs in plasma EVs. DNAs within EVs from 30 ml human plasma mixed with DNA markers were analyzed using a bioanalyzer. The two peaks (50 bp and 17 kb) represent the DNA markers. FU, fluorescence units. (E) PCR and DNA sequencing of β-actin (ACTB) and mtDNA:D-loop in EVs. Upper two images: DNA (6.27 ng/μl) in EVs from human plasma and VSMC supernatant was amplified by PCR using β-actin or D-loop primers, and the PCR products were analyzed on 2% agarose gel precasted with ethidium bromide. DNA marker (lane M), EVs without (lane 1 and lane 4) or with (lane 2 and lane 5) DNase I digestion, followed by EDTA treatment; disrupted EVs treated with DNase I (lane 3 and lane 6), and negative control (ddH2O, lane 7). Lanes 1–3 and lanes 4–6 show EVs from human plasma and EVs from the supernatant of VSMCs in culture, respectively. Lower two figures show the sequencing results of PCR products of β-actin (ACTB) and mtDNA:D-loop fragments in EVs; the percentage identity of the sequences was more than 99.5% for each fragment. (F) The viability of VSMCs was determined by PI staining. Unstained cells served as negative control (left-hand figure). PI-stained cells are shown in the middle figure and overlay of the left-hand side and middle figures is shown in the right-hand side figure.
To show if the DNAs in EVs are double-strand (dsDNA) or single-strand (ssDNA) DNA, we used SYBR Green I, which preferentially binds to dsDNA rather than ssDNA (Dragan et al., 2012). Consistent with a previous report (Balaj et al., 2011), we found that EVs contained dsDNA (Figure 2C). Agarose gel electrophoresis showed that EVs from both human plasma and supernatants of VSMCs in culture had DNA fragments ranging in size from 1 to 20 kb, but mostly around 17 kb (Figure 2Ba). The bioanalyzer study found that the sizes of the DNA fragments within EVs ranged from 6 to 17 kb, and the DNA content in EVs from 30 ml plasma was about 300 ng (6.27 ng/μl × 50 μl; Figure 2D). Consistent with other reports (Guescini et al., 2010), after amplification of DNA fragments in EVs with specific primers, PCR showed the presence of β-actin DNA and mtDNA, D-Loop, in EVs (Figure 2E). The existence of β-actin and D-loop fragments in EVs was confirmed by sequencing (Figure 2E); the percentage identity of the sequences was more than 99.5% for each fragment. After the disrupted EVs were treated with DNase, the EV DNAs were degraded, further supporting the location of EV DNAs inside the EVs (Figure 2E). To determine whether or not the EVs are from apoptotic cells, we checked the presence of apoptosis in VSMCs by PI which is excluded by living but not by dead or dying cells; there was low fluorescence in VSMCs, excluding apoptosis as a source of DNA (Figure 2F). Solexa sequencing found at least 16434 gene fragments in the EVs from human plasma, including the AT1 receptor gene (Table 1).
Table 1.
EV-DNA signatures from normal human plasma (a small portion of the data are shown).
| Gene ID | Gene name | Length | Coverage (%) | RPKM | Description |
|---|---|---|---|---|---|
| 60 | ACTB | 1808 | 9.96 | 7.048699 | Actin, β |
| 185 | AGTR1 | 2412 | 14.93 | 13.20901 | Angiotensin II receptor, type 1 |
| 186 | AGTR2 | 2897 | 6.21 | 4.39905 | Angiotensin II receptor, type 2 |
| 1131 | CHRM3 | 2743 | 25.96 | 490.1557 | Cholinergic receptor, muscarinic 3 |
| 2250 | FGF5 | 5388 | 15.31 | 159.6554 | Fibroblast growth factor 5 |
| 9527 | GOSR1 | 5230 | 26.31 | 464.1953 | Golgi SNAP receptor complex member 1 |
| 23245 | ASTN2 | 4594 | 7.84 | 1031.952 | Astrotactin 2 |
| 23788 | MTCH2 | 2627 | 27.10 | 19.40472 | Mitochondrial carrier 2 |
| 55149 | MTPAP | 5619 | 30.08 | 27.21633 | Mitochondrial poly(A) polymerase |
| 79944 | L2HGDH | 6093 | 28.02 | 626.4307 | L-2-hydroxyglutarate dehydrogenase |
| 80298 | MTERFD3 | 1775 | 28.06 | 35.89873 | MTERF domain containing 3 |
| 122704 | MRPL52 | 1294 | 13.91 | 9.848569 | Mitochondrial ribosomal protein L52 |
| 151194 | METTL21A | 4805 | 35.80 | 754.5644 | Methyltransferase like 21A |
| 345778 | MTX3 | 7862 | 17.22 | 13.77823 | Metaxin 3 |
| 389177 | TMEM212 | 1881 | 46.36 | 1378.742 | Transmembrane protein 212 |
| … | … | … | … | … | … |
ID, identity; RPKM, reads per Kb per million reads.
Transportable and functional AT1 receptor DNA in EVs target HEK293 cells
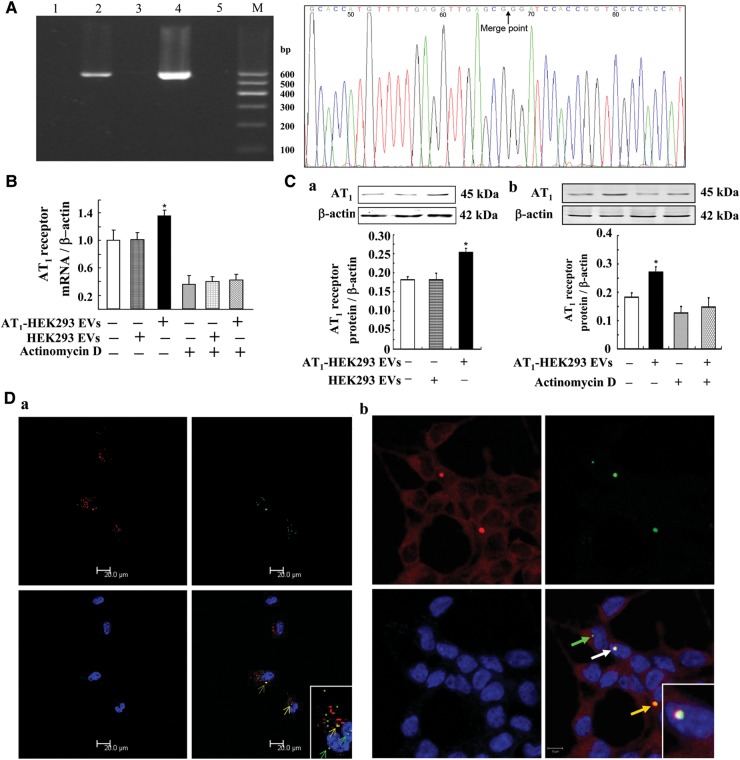
To determine the transportability and functionality of the DNA fragments in EVs, we used EVs from AT1 receptor transfected HEK293 (AT1-HEK293) cells to treat non-transfected HEK293 cells. To exclude the interference of endogenous AT1 receptor DNA, we transfected the HEK293 cells with AT1-EGFP (enhanced green fluorescent protein, EGFP) so that AT1-HEK293 cells as well as their EVs bore AT1-EGFP DNA. Therefore, the AT1-EGFP DNA in recipient cells unequivocally distinguishes the endogenous AT1 receptor from the exogenous (i.e. transferred) one. We found AT1-EGFP DNA in EVs from AT1-HEK293 cells but not in EVs from untransfected HEK293 cells (Figure 3A). The presence of AT1-EGFP DNA in AT1-HEK293 EVs was confirmed by sequencing, and the percentage sequence identity was more than 99.5% (Figure 3A). Incubation of HEK293 cells with EVs from AT1-HEK293 cells (1 × 105/ml for 24 h) increased the AT1 receptor mRNA (Figure 3B) and protein expressions (Figure 3Ca) in HEK293 cells. In the presence of actinomycin D (5.0 μg/ml), an inhibitor of de novo mRNA synthesis, the stimulatory effect of EVs from AT1-HEK293 cells on co-incubated HEK293 cells was blocked, indicating that there was de novo AT1 receptor mRNA synthesis in the HEK293 cells incubated with EVs from AT1-HEK293 cells (Figure 3B and Cb). To further elucidate the underlying mechanisms, we used laser confocal microscopy, and found that DNA (green with AO staining) in EVs (red with the DiI lipophilic dye) (co-localization = yellow), could be transferred into the cells and localize to and inside the nuclear membrane (Figure 3Da). We checked the association between endogenous NF-κB, as a promoter of AT1 receptor gene, and transferred DNAs including AT1 receptor DNA. Results showed that endogenous NF-κB could be recruited to the transferred DNAs in the nucleus (Figure 3Db), indicating that the transcription of transferred AT1 receptor gene occurs in the recipient cells.
Figure 3.
Effect of EVs from AT1-HEK293 cells on AT1 receptor expression. (A) AT1-EGFP DNA in EVs from AT1-HEK293 cells and untransfected HEK293 control cells. AT1-EGFP DNA in EVs was detected by PCR, and the PCR products were analyzed on 2% agarose gel precasted with ethidium bromide. DNA marker (lane M), untransfected HEK293 EV (lane 1), AT1-HEK293 EV (lane 2), untransfected HEK293 cells (lane 3), AT1-HEK293 cells (lane 4), and negative control (ddH2O, lane 5). The right-hand side figure shows more than 99.5% sequence identity of AT1-EGFP DNA in EVs. The black arrow indicates the merge point of the AT1 receptor gene and EGFP gene. (B) Effect of EVs from AT1-HEK293 cells on AT1 receptor mRNA expression. HEK293 cells were incubated with EVs (1 × 105/ml) from AT1-HEK293 cells or HEK293 cells for 24 h, in the absence or presence of actinomycin D (5.0 μg/ml). AT1 receptor mRNA expression was determined by quantitative RT–PCR (n=3, *P<0.05 vs. others). (C) Effect of EVs from AT1-HEK293 cells on AT1 receptor protein expression. HEK293 cells were incubated with EVs (1 × 105/ml) from AT1-HEK293 cells or HEK293 cells for 24 h in the absence (a and b) or presence of actinomycin D (5.0 μg/ml) (b). AT1 receptor protein expression was determined by immunoblotting (n=3, *P<0.05 vs. others). (D) Confocal microscopy images of HEK293 cells cultured with EVs co-stained with AO and DiI. (a) After culturing HEK293 cells with VSMC EVs co-stained with DiI (a lipophilic dye, red) and AO (a DNA dye, green) for 24 h at 37°C, the HEK293 cells were observed by confocal microscopy. The transferred EVs localized inside the cell (red) and transferred DNA inside the cell (green) and nuclei (cyan, green arrows) in HEK293 cells incubated with the VSMC EVs. Yellow fluorescence shows co-localization (yellow arrows) of membrane of EVs (DiI, red) and transferred DNA (AO, green). (b) After culturing HEK293 cells with AO-stained EVs from VSMC cells for 24 h at 37°C, HEK293 cells were observed by confocal microscopy, in which NF-κB was stained. White fluorescence shows co-localization (white arrow) of nuclei (DAPI-staining, blue), transferred DNA (AO-staining, green), and NF-κB (Cy3-staining, red).
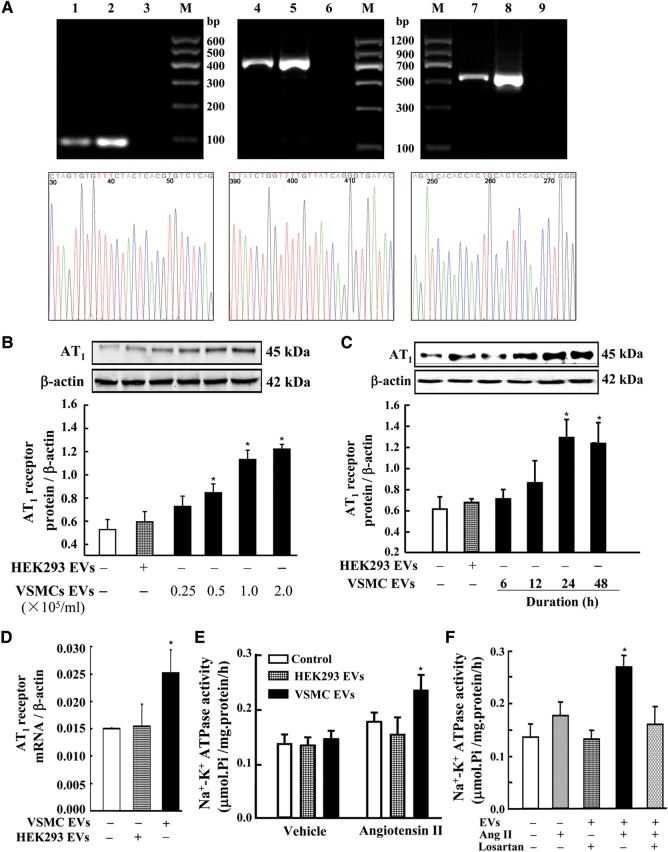
In addition to the studies on the transfer of gDNA in EVs between same originated cell lines (e.g. AT1-HEK293 and HEK293), we also studied the transport of gDNAs in EVs between different cell lines (VSMC and HEK293). We found, by PCR, the presence of AT1 receptor gene coding region, 5′ promoter region, and 3′ untranslated region in EVs from VSMCs (Figure 4A). The presence of the three regions of the AT1 receptor gene in VSMC EVs was confirmed by sequencing, and the percentage sequence identity was more than 99.5% for each fragment (Figure 4A). We also found that EVs from VSMCs increased AT1 receptor protein expression in HEK293 cells in a concentration- (0.25 × 105–2.0 × 105/ml) and time (6–48 h)-dependent manner (Figure 4B and C), and also increased AT1 receptor mRNA levels in HEK293 recipient cells (Figure 4D). The transportable AT1 receptor was functional because treatment of HEK293 cells with VSMC EVs (1 × 105/ml) for 24 h increased the stimulatory effect of angiotensin II (1 × 10−6 M for 30 min) on Na+–K+ ATPase activity (Figure 4E), an effect that was blocked by losartan (1 × 10−6 M), an AT1 receptor blocker (Figure 4F). In contrast, in HEK293 cells treated with its own EVs, the stimulatory effect of angiotensin II on Na+–K+ ATPase activity was not affected (Figure 4E).
Figure 4.
Effect of EVs from VSMCs on AT1 receptor expression and function in HEK293 cells. (A) Several regions of AT1 receptor gene in EVs from VSMCs. AT1 receptor gene coding region, 5′ promoter region, and 3′ untranslated region in EVs were detected by PCR, and the PCR products were analyzed on 2% agarose gel precasted with ethidium bromide. DNA marker (lane M), VSMC EVs (lanes 1, 4, and 7), VSMC cells (lanes 2, 5, and 8), negative control (ddH2O, lanes 3, 6, and 9). Lanes 1–3, lanes 4–6, and lanes 7–9 show AT1 receptor gene coding region, 5′ promoter region, and 3′ untranslated region, respectively. The lower three figures show the sequencing results of PCR products of the respective regions of AT1 receptor gene in EVs, and the percentage sequence identity was more than 99.5%. (B and C) Effect of EVs from VSMCs on AT1 receptor expression in HEK293 cells. HEK293 cells were incubated with EVs from VSMCs at the indicated concentrations (0.25 × 105–2.0 × 105/ml) (B) and durations of incubation (1 × 105/ml for 6–48 h) (C). The expression of AT1 receptor protein expression was determined by immunoblotting (n=4, *P<0.05 vs. control, i.e. HEK293 cells treated with EVs from control HEK293 cells). (D) Effect of EVs from VSMCs on AT1 receptor mRNA expression in HEK293 recipient cells. HEK293 cells were incubated with EVs (1 × 105/ml) from VSMCs for 24 h. The expression of AT1 receptor mRNA expression was determined by quantitative RT–PCR (n=3, *P<0.05 vs. others). (E and F) Effect of VSMC EVs on angiotensin II-stimulated Na+–K+ ATPase activity in HEK293 cells. HEK293 cells were incubated with EVs (1 × 105/ml) from HEK293 cells or VSMCs for 24 h (E and F). HEK293 cells were treated with VSMC EVs (1 × 105/ml), and then treated with angiotensin II (Ang II, 1 × 10−6 M) alone (E and F) or in the presence of the AT1 receptor blocker, losartan (1 × 10−6 M) (F), for 30 min (n=9–12, *P<0.05 vs. others).
Transportable and functional BCR/ABL hybrid gene in EVs from K562 cells to HEK293 cells or neutrophils
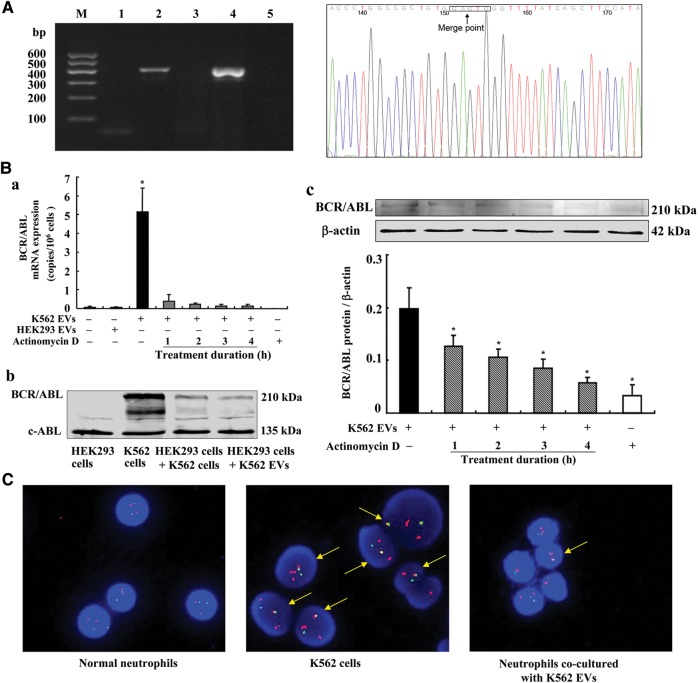
The VSMC and HEK293 studies only provided indirect evidence that the increased AT1 mRNA is due to the incorporation of DNA from EVs. To overcome this limitation, we studied the transport of the BCR/ABL hybrid gene, which is not normally expressed in HEK293 cells. PCR showed that the BCR/ABL hybrid gene is present in the EVs from K562 cells, but not from HEK293 cells, suggesting that the presence of BCR/ABL hybrid gene is unique for K562 cells (Figure 5A). Incubating HEK293 cells with EVs (1 × 105/ml) from K562 cells for 24 h resulted in the expression of BCR/ABL hybrid gene mRNA and protein in the recipient HEK293 cells (Figure 5Ba and b). The heterologous expression of BCR/ABL hybrid gene mRNA and protein in the HEK293 cells incubated with K562 EVs was prevented by the concurrent incubation with actinomycin D (5 μg/ml; Figure 5Ba and c).
Figure 5.
Effect of K562 EVs on the BCR/ABL hybrid gene in HEK293 cells or neutrophils. (A) BCR/ABL hybrid gene in the EVs from K562 cells. BCR/ABL hybrid gene in the EVs from K562 cells was detected by qualitative PCR, and the PCR products were analyzed on 2% agarose gel precasted with ethidium bromide. DNA marker (lane M), HEK293 EVs (lane 1), K562 EVs (lane 2), HEK293 cells (lane 3), K562 cells (lane 4), negative control (ddH2O, lane 5). The right-hand side figure shows the sequencing result of PCR product of BCR/ABL hybrid gene in K562 EVs, and the percentage sequence identity was more than 99.5%. The black frame indicates the merge point of the BCR gene and ABL gene. (B) Effect of K562 EVs on BCR/ABL hybrid gene mRNA (a) and protein (b and c) expressions in the HEK293 cells. HEK293 cells were incubated with EVs (1 × 105/ml) from K562 cells for 24 h with or without the co-incubation with actinomycin D (5 μg/ml) at varying times (1–4 h) (*P<0.01 vs. others, n=3–12). (b) One immunoblot from three experiments is shown. (c) Protein expression was determined by immunoblotting (*P<0.01 vs. the group treated with K562 EVs, n=3). (C) D-FISH study. Neutrophils were incubated with EVs (1 × 105/ml) from K562 cells for 24 h. The D-FISH probe is composed of two directly labeled probes; the BCR gene was labeled with a red fluorochrome, and the ABL-probe labeled with a green fluorochrome. BCR/ABL hybrid gene is therefore detected as yellow in the merged images.
To determine the pathophysiological significance of transferred gDNAs in EVs between cells, we investigated the effect of BCR/ABL hybrid gene in EVs transferred from K562 cells to normal human neutrophils isolated from human peripheral blood. Using a BCR/ABL D-FISH (dual-fluorescence in situ hybridization) method, we found that normal neutrophils did not contain the BCR/ABL hybrid gene. However, incubation of normal human neutrophils with EVs (1 × 105/ml) from K562 cells for 24 h resulted in the expression of BCR/ABL hybrid gene in 20% of the neutrophils (Figure 5C).
Discussion
EVs can carry signals within or at their limiting membrane, providing a mechanism by which cells can exchange more complex information than what was previously thought (Trajkovic et al., 2008; Thery et al., 2009). The packaged mRNAs or microRNAs in EVs can be delivered into target cells, and modulate the biological functions of these cells by gene-regulated promotion or repression of target gene expression (Skog et al., 2008; Cocucci et al., 2009; Pap et al., 2009). In the present study, we found that in addition to mRNA, microRNA, and protein, EVs contain numerous gDNA fragments. These gDNAs in EVs are transportable between the same or different types of cells, increase the gDNA-coding mRNA and protein expressions in the recipient cells, and have physiological significance to influence function in recipient cells. In this study, the AT1 receptor DNA in EVs could increase AT1 receptor expression and AT1 receptor-stimulated Na+–K+ ATPase activity in recipient HEK293 cells. The underlying mechanisms are not completely clear, or at least, could not be explained by the existence of mRNA in the EVs, because the level of AT1 mRNA in EVs is much lower than the increased AT1 receptor mRNA level in the recipient cells (data not shown). Moreover, actinomycin D, an inhibitor of DNA transcription, blocks the stimulatory effect of EVs from the donor cells on AT1 receptor mRNA expression in the recipient HEK293 cells, indicating that the transcription of DNAs from the EVs occurs in the recipient cells. It is known that for a DNA fragment to have the potential for expression it should, apart from the open reading frame, include regulatory 5′ and 3′ elements necessary for the recruitment and proper function of the cellular machinery involved in transcription (Balaj et al., 2011). Our current study found and confirmed the presence of AT1 receptor gene coding region, 5′ promoter region, and 3′ untranslated region in EVs from VSMC cells. The view that DNAs in EVs could be transferred into the recipient cells and localize to and inside the nuclear membrane is supported by the previous study (Waldenstrom et al., 2012). Our present study provides direct evidence that transferred gene can be transcribed in the recipient cells.
Owing to the presence of endogenous AT1 receptor DNA in the HEK293 cells, it is possible that the endogenous DNA was activated by the EVs, causing the increase in AT1 receptor mRNA and protein expressions in HEK293 recipient cells. To overcome this limitation, we treated the HEK293 cells with EVs bearing AT1-EGFP DNA derived from AT1-HEK293 cells, by which the endogenous AT1 receptor can be distinguished from exogenous (i.e. transferred) AT1-EGFP DNA transcribed receptor. Moreover, we chose the BCR/ABL hybrid gene, which is unique in a CML cell line K562 but not endogenously present in the HEK293 cells or neutrophils. By this study, we provided further direct evidence that EV gDNAs can be transferred to and expressed in recipient cells.
CML is characterized by a reciprocal translocation between chromosomes 9 and 22, which encodes the chimeric tyrosine kinase BCR/ABL that is responsible to affect malignant cell behavior and pathophysiology, and thus a therapeutic target of CML (Pluk et al., 2002; Shibata et al., 2010; Puissant et al., 2012). Although the detailed actions of EVs in CML are not clear, our present study found that incubation of K562 EVs with normal neutrophils makes them express BCR/ABL, indicating a possible role of EVs in tumorigenesis.
Gene therapy has an enormous potential to treat genetic diseases. Two types of vectors, viruses and liposomes, have been used in clinical and research work (Wahlgren et al., 2012). However, their side-effects, including toxicity and immunogenic concerns, hamper their use (Wahlgren et al., 2012). Therefore, finding a biocompatible method to introduce exogenous gene into target cells becomes very urgent. Our findings in this study suggest that exogenous gene can be packaged into EVs of certain cells. Further, DNA-containing EVs can be delivered into target cells and these exogenous genes can alter the cellular functions of the recipient cells. EVs are proved to be safe and effective gene delivery tools (Cocucci et al., 2009; Wahlgren et al., 2012). From this point of view, secreted DNA may represent a class of signaling molecules that may play an important role in mediating intercellular communication. Moreover, the selective secretion and targeting of DNA among different cells provide a highly regulated complex network under various physiological and pathophysiological conditions.
In conclusion, we have shown that EV DNAs, which could be delivered from one cell to another, can regulate the gDNA-coding mRNA and protein expressions in the recipient cells, and affect the physiological function in the recipient cells. EV-mediated transfer of gDNAs may represent a new method of gene delivery and novel way of signal transduction among cells. These findings would help in the discovery of novel mechanisms of disease and development of new therapeutic strategies.
Materials and methods
Blood collection
Blood was drawn from healthy donors at Daping Hospital (Chongqing, China), and stored in test tubes containing 3.8% trisodium citrate. Plasma was isolated by centrifugation at 1800× g for 15 min. All donors or their guardians provided signed informed consent forms, and the protocols for handling human blood and tissues were approved by the Ethics Committee of Daping Hospital.
Isolation of EVs
EVs were isolated from the plasma of healthy donors or cell culture supernatants by differential centrifugation according to previous reports (Valadi et al., 2007; Skog et al., 2008; Zhang et al., 2010). When cells have grown to about 90% confluence, cell culture supernatants were collected. The plasma or cell culture supernatants were centrifuged at 500× g for 20 min, and the initial pellets were discarded to remove residual cells. Then, the supernatants were centrifuged again at 1500× g for 20 min and the pellets were also discarded to remove other debris. After re-centrifugation at 110000× g for 70 min, the final pellets containing the EVs were resuspended in fetal bovine serum-free medium (for cell EV) or phosphate-buffered saline (PBS) (for plasma EV). All steps were performed at 4°C. The isolated EVs were then subjected to DNase I (20 Kunitz units/ml, Sigma-Aldrich Co.) digestion (30 min at 37°C) to remove any DNA outside of the EVs. Ethylenediaminetetraacetic acid (EDTA) (10 mmol/L EVs) was then added to the EVs, and incubated for 5 min at 65°C to inactivate any residual DNase. These EVs were used in the experiments.
Transmission electron microscopy
For negative-staining transmission electron microscopy (TEM), EVs were adsorbed to copper-coated mesh-grids for 2 min, and rinsed in filtered PBS. EVs on the grids were immediately fixed with 4% glutaraldehyde for 1 min and then negatively stained with 2% (wt/vol) Na-phosphotungstate for 1 min. Microscopic examinations were then carried out using Hitachi-7500 TEM (Hitachi, Ltd.), operated at 80 kV.
For conventional TEM, the EVs were placed in a droplet of 2.5% glutaraldehyde in PBS and fixed overnight at 4°C. The samples were rinsed with PBS (10 min × 3) and fixed in 1% osmium tetroxide for 60 min at room temperature. The samples were then embedded in 10% gelatin, fixed in glutaraldehyde at 4°C, and cut into several blocks (<1.0 mm3). The samples were dehydrated with sequential increasing concentrations of alcohol (30%, 50%, 70%, 90%, 95%, and 100% × 3) for 10 min per step. Absolute alcohol was then finally changed to propylene oxide, and the specimens were exposed sequentially to increasing concentrations of Quetol-812 epoxy resin (25%, 50%, 75%, and 100%) mixed with propylene oxide for a minimum of 3 h per step. Samples were embedded in pure, fresh Quetol-812 epoxy resin and polymerized at 35°C for 12 h, 45°C for 12 h and 60°C for 24 h. Ultrathin sections (100 nm) were cut using a Leica-UC6 ultra-microtome (Leica Co.) and post-stained with uranyl acetate for 10 min and then with lead citrate for 5 min at room temperature before observation by a Hitachi-7500 TEM, operated at 80 kV.
FCM analysis
The cell EVs and cells without pretreatment with latex beads were directly analyzed for the absolute counts using BD Accuri C6 FCM (Becton, Dickinson and Company; Bruno et al., 2009). DMEM cell culture medium was used as negative control.
To evaluate whether DNAs are present inside or outside the EVs, FACS was used for analysis of DNA-stained EVs (Waldenstrom et al., 2012). DNA dyes AO is membrane permeable, while PI is membrane impermeable and excluded from living cells. In addition, DiI (1,1′-dioctadecyl-3,3,3′,3′-tetramethyl-indocarbocyanine perchlorate), a lipid membrane dye, was used for staining membranes of EVs (Trajkovic et al., 2008). EVs (2 × 106) in 200 μl PBS were incubated with 10 μl AO (KeyGen Biotech) for 5 min, 10 μl PI (KeyGen Biotech) for 30 min, or 2 μM DiI (Beyotime Biotech) for 10 min dark at room temperature according to the manufacturer's recommendations. The AO- or PI-incubated EVs were ultracentrifuged, and washed twice with PBS. The same amounts of EVs and cells (about 100000 EVs or VSMCs) were analyzed by BD Accuri C6 FCM at the appropriate fluorescence emitting wavelengths (530 nm for AO, 565 nm for DiI, 670 nm for PI). For AO positive control, normal VSMCs (2 × 105) in 200 μl PBS incubated with AO were used, while for PI positive control, the same amount of VSMCs pretreated by freezing and thawing were used.
DNA extraction from EVs
The isolated EVs were subjected to DNase digestion to remove the DNA exterior to the EV, as described above. Total DNA was extracted from EVs with the TIANamp Genomic DNA Kit, following the manufacturer's recommendations (Tiangen Biotech Co. Ltd.). The quality and quantity of extracted DNA were measured by spectrophotometry (A260 nm) and electrophoresis on agarose gel precasted with ethidium bromide or SYBR Green I stain (dsDNA stain; Cai et al., 2011).
The disruption of EVs was performed using TIANamp Genomic DNA Kit, following the manufacturer's recommendations, as aforementioned. EVs (2 × 106) in 200 μl PBS were lysed with proteinase K (2 mg/ml) and 200 μl lysis buffer and incubated at 70°C for 10 min. Then the DNAs were precipitated with 200 μl ethanol and adsorbed with DNA-adsorbing column. Finally, EV DNAs were eluted with sterilized deionized water, followed by DNase I digestion.
Solexa sequencing
For Solexa sequencing, we used a DNA sample equally pooled from 30 independent samples extracted from plasma-derived EVs of healthy subjects. The characteristics of the human samples are listed in Table 2. The sequencing procedure for DNA samples was mostly conducted as previously described for RNA samples (Chen et al., 2008; Zhang et al., 2010), except of reverse transcription of RNA to cDNA. DNA samples were randomly fragmented from long to small DNA (∼200 bps). Then, the 5′ and 3′ ends of these small DNAs were repaired using end-repair enzyme, followed by adenylation of 3′-ends, agarose gel purification, and ligation of a pair of Solexa adaptors to their 5′ and 3′ ends. The small DNA molecules were amplified using the adaptor primers for 17 cycles, and these fragments around 200 bp (small DNA + adaptors) were isolated from agarose gels. The purified DNAs were used directly for cluster generation and sequencing analysis using the Illumina Genome Analyzer (Illumina), according to the manufacturer's recommendations. The image files generated by the sequencer were then processed to produce digital-quality data. In these raw data, we removed adaptor sequences, poor quality reads, in which the number of base pairs with lower quality value (<5) was more than 50%, and contaminated reads, in which the percent of N was more than 5%. Finally, clean reads were processed for computational analysis, comparing them with the National Center for Biotechnology Information (NCBI) Reference Sequence project (RefSeq) database.
Table 2.
Characteristics of the human samples.
| Patient number | Gender | Age (years) | Height (cm) | Body weight (kg) | Blood pressure (mmHg) |
Triglyceride (mM) | Total Cholesterin (mM) | HDL (mM) | LDL (mM) | Blood Glucose (mM) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Systolic pressure | Diastolic pressure | ||||||||||
| 1 | Female | 69 | 158 | 57 | 130 | 68 | 0.92 | 4.07 | 1.23 | 1.86 | 7.09 |
| 2 | Male | 52 | 170 | 74 | 142 | 94 | 1.49 | 3.63 | 1.13 | 1.73 | 11.66 |
| 3 | Male | 61 | 172 | 70 | 135 | 80 | 0.94 | 4.25 | 1.51 | 2.05 | 5.01 |
| 4 | Male | 69 | 160 | 52 | 103 | 70 | 0.49 | 3.13 | 1.47 | 1.46 | 6.54 |
| 5 | Female | 52 | 152 | 60 | 130 | 78 | 0.98 | 5.23 | 1.36 | 2.53 | 4.79 |
| 6 | Female | 62 | 155 | 62 | 120 | 80 | 1.14 | 3.3 | 1.36 | 1.43 | 5.68 |
| 7 | Female | 79 | 150 | 59 | 138 | 58 | 0.49 | 3.78 | 1.45 | 1.88 | 5.31 |
| 8 | Male | 64 | 165 | 70 | 118 | 80 | 1.01 | 4.6 | 1.56 | 2.54 | 5.19 |
| 9 | Male | 53 | 169 | 66 | 138 | 86 | 2.62 | 3.66 | 1.23 | 1.83 | 9.18 |
| 10 | Female | 60 | 158 | 69 | 172 | 92 | 1.12 | 4.19 | 1.87 | 2 | 4.92 |
| 11 | Female | 76 | 156 | 62 | 150 | 92 | 1.95 | 3.73 | 1.43 | 1.87 | 7.45 |
| 12 | Female | 53 | 158 | 60 | 158 | 100 | 0.89 | 5.08 | 1.78 | 2.5 | 7.03 |
| 13 | Female | 53 | 161 | 56.5 | 120 | 68 | 0.87 | 5.21 | 1.85 | 2.48 | 4.56 |
| 14 | Female | 73 | 147 | 57 | 120 | 68 | 1.25 | 6.88 | 1.87 | 3.49 | 4.77 |
| 15 | Female | 54 | 160 | 60 | 170 | 90 | 0.85 | 5.67 | 1.68 | 2.79 | 9.79 |
| 16 | Female | 62 | 153 | 45 | 126 | 76 | 1.15 | 3.4 | 1.32 | 1.4 | 5.28 |
| 17 | Female | 59 | 162 | 74 | 130 | 82 | 0.68 | 2.43 | 1.03 | 1.24 | 4.95 |
| 18 | Male | 56 | 168 | 62 | 110 | 75 | 1.51 | 5.06 | 1.37 | 2.73 | 5.12 |
| 19 | Female | 56 | 168 | 70 | 130 | 80 | 0.36 | 3.88 | 1.8 | 2.01 | 4.75 |
| 20 | Male | 62 | 175 | 79 | 126 | 78 | 0.34 | 3.52 | 1.6 | 1.69 | 6.07 |
| 21 | Female | 60 | 150 | 48 | 120 | 85 | 1.19 | 4.9 | 1.29 | 3.06 | 9.59 |
| 22 | Male | 58 | 168 | 65 | 120 | 70 | 1.72 | 5.26 | 2.37 | 2.62 | 5.53 |
| 23 | Female | 54 | 157 | 60 | 110 | 65 | 6.79 | 5.65 | 1.42 | 3.87 | 7.59 |
| 24 | Male | 62 | 171 | 60 | 130 | 85 | 0.61 | 2.49 | 0.98 | 1.25 | 4.92 |
| 25 | Female | 63 | 165 | 68 | 120 | 75 | 1.67 | 4.6 | 1.13 | 2.76 | 5.49 |
| 26 | Male | 63 | 172 | 64 | 140 | 70 | 1.73 | 5.6 | 1.6 | 2.73 | 4.67 |
| 27 | Male | 50 | 166 | 64 | 123 | 60 | 5.21 | 4.76 | 1.6 | 2.34 | 5.28 |
| 28 | Male | 59 | 175 | 75 | 120 | 80 | 2.12 | 5.18 | 1.42 | 2.55 | 5.35 |
| 29 | Male | 62 | 173 | 63 | 120 | 60 | 1.23 | 3.42 | 1.15 | 1.69 | 4.91 |
| 30 | Male | 73 | 176 | 70 | 120 | 68 | 1.05 | 4.64 | 1.5 | 2.89 | 5.63 |
| Average | 61.0 | 163.0 | 63.4 | 129.6 | 77.1 | 1.48 | 4.37 | 1.48 | 2.24 | 6.14 | |
Laser confocal microscopy
To observe the transferred EV DNA to the recipient cells, VSMC-derived EVs (2 × 106) in 200 μl PBS were stained with AO and then washed with PBS, to eliminate contamination of unincorporated AO, as described previously. HEK293 cells were incubated with EVs, co-stained with AO and DiI, from VSMCs (1 × 105/ml) for 24 h on a cell culture microscope slide (Nest Biotechnology Co. Ltd.; Waldenstrom et al., 2012). DAPI (4,6-diamidino-2-phenylindole) was used to stain the nucleus. Localization of EVs, co-stained with AO and DiI, was determined by laser confocal microscopy.
To observe the co-localization of NF-κB with transferred EV DNA, HEK293 cells were incubated with AO-stained DNA EVs, as described above, in which nuclei were also stained with DAPI. NF-κB was stained using Cellular NF-κB Translocation Kit (Beyotime Biotech; Xu et al., 2008). Nuclei (blue), transferred EV DNA (green), and NF-κB (red) were viewed through a Zeiss LSM 510 META laser confocal microscope (Carl Zeiss) at individual excitation wavelengths (350 nm for DAPI, 488 nm for AO, 540 nm for NF-κB, 549 nm for DiI). The co-localization of NF-κB, EV DNA, and nucleus appear white.
Real-time PCR of DNA
DNAs within EVs were amplified by real-time PCR, using SYBR® premix Ex Taq™ II according to the manufacturer's recommendations (TaKaRa Biotechnology Co. Ltd.). Twenty microliters of final reaction mixture contained 10 μl of Ex Taq, 0.8 μl of sense primer, 0.8 μl of antisense primer, 6.4 μl of sterile deionized water, and 2.0 μl of DNA extract. Thermocycling was conducted using a CFX96™ Real-Time PCR Detection System (Bio-Rad Laboratories Inc.) initiated by a 30 sec incubation at 95°C, followed by 40 cycles at 94°C for 5 sec and 60°C for 45 sec with a single fluorescent reading taken at the end of each cycle. Each reaction was conducted in triplicate. All the runs were completed with a melt curve analysis to confirm the specificity of amplification and lack of primer dimers. PCR products were analyzed by agarose gel electrophoresis. All primers used are listed in Table 3.
Table 3.
Sequence of PCR amplification primers.
| Primer name | Sequence (5′–3′) | Product length (bp) | Amplified gene |
|---|---|---|---|
| ACTB-gD-FP | 5′-TCTTCTGCCGTTTTCCGTAGG-3′ | 255 | Human ACTB gDNA |
| ACTB-gD-RP | 5′-TTGGGATGGGGAGTCTGTTCA-3′ | ||
| D-loop-gD-FP | 5′-TTAGGCTGGTGTTAGGGTTCTT-3′ | 339 | Human D-loop (mtDNA) |
| D-loop-gD-RP | 5′-GCATTTGGTATTTTCGTCTGG-3′ | ||
| BCR/ABL-gD-FP | 5′-TCCACTCAGCCACTGGATTTAAGCA-3′ | 418 | K562 BCR/ABL gDNA (Shibata et al., 2010) |
| BCR/ABL-gD-RP | 5′-GGTGAATTGGAAAGAAGCAGCAGGT-3′ | ||
| AT1-584FP | 5′-CCACTCAAACCTTTCAACAAAA-3′ | 603 | AT1-EGFP gDNA |
| EGFP-205RP | 5′-TGCCGTTCTTCTGCTTGTC-3′ | ||
| AT1-gD-1641FP | 5′-AAATTCGGAGCTGCCTCCTC-3′ | 788 | 5′ promotor region of human AT1 gene |
| AT1-gD-2428RP | 5′-CCTGCGGAGATTCGTGTTCT-3′ | ||
| AT1-gD-48500FP | 5′-ACTGAGATGAGCATTCAAGTATGT-3′ | 550 | 3′ untranslated region of human AT1 gene |
| AT1-gD-49049RP | 5′-ATGTGGCAGCTGGATCCTTA-3′ | ||
| AT1-cD-FP | 5′-ATTGCTTCAGCCAGCGTCAGTT-3′ | 93 | Human AT1 mRNA/gDNA |
| AT1-cD-RP | 5′-TGGGTGAACAATAGCCAGGTATCG-3′ | ||
| ACTB-cD-FP | 5′-CCACGAAACTACCTTCAACTCC-3′ | 132 | Human ACTB mRNA |
| ACTB-cD-RP | 5′-GTGATCTCCTTCTGCATCCTGT-3′ |
RNA isolation and quantitative RT–PCR of mRNA
Total RNA of EVs or cells was extracted using TRIzol Reagent (Life Technologies Co.). Residual DNA was removed by DNase I digestion following RNA isolation, as described below. The following reagents were mixed in an RNase-free microcentrifuge tube: 1 μg RNA, 1 μl 10 × DNase buffer (500 mM Tris–HCl, pH 8.0, 50 mM MgCl2, 10 mM DTT), 1 μl of 1 Kunitz units/μl RNase-free DNase I (Sigma) in a volume up to 10 μl with RNase-free water, and then incubated for 30 min at 37°C to remove contaminating gDNA. One microliter of 2.5 mM EDTA was added into the tubes containing the EVs, and incubated for 5 min at 65°C to inactivate the DNase. The mixture of 11 µl purified RNA and 1 μl of 25 pmol/μl randomized primer (Toyobo Co.) was denatured by boiling at 65°C for 5 min, followed by immediate cooling on ice. Twelve microliters of denatured RNA, 4 μl of 5× RT buffer (Toyobo), 2 μl dNTP mixture (10 mM each of dNTPs, Toyobo), 1 μl of 10 U/µl RNase inhibitor (Toyobo), and 1 μl ReverTre Ace® (a reverse transcriptase) (Toyobo) were mixed to a total volume of 20 μl transcriptase reagents. The mixture was incubated at 30°C for 10 min, 42°C for 30 min, 85°C for 5 min, and 4°C for 5 min to allow for the synthesis of first-strand complimentary DNA (cDNA). Subsequently, real-time quantification of cDNA was performed using SYBR® premix Ex Taq™ II (TaKaRa) by the CFX96™ Real-Time PCR Detection System as described above. Additionally, absolute quantitative RT–PCR of BCR/ABL mRNA was performed with BCR/ABL mRNA detection kit (Shanghai Shenyou Co. Ltd.).
Cell isolation and culture
VSMCs, isolated from human mesenteric arteries, were characterized by smooth muscle cell morphology (multilayer sheets, ‘hills and valleys’) and expression of smooth muscle α-actin by immunostaining with a specific α-actin monoclonal antibody (Li et al., 2008). K562 and HEK293 cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). HEK293 cells were transfected with human AT1 receptor using EGFP expression vector pEGFP-N1 to generate monoclonal HEK293 cells that include AT1-EGFP DNA and stably express AT1 receptors (AT1-HEK293 cells). These cells were cultured at 37°C in 95% air/5% CO2 atmosphere in Dulbecco's Modified Eagle's Medium (DMEM, Gibco, Life Technologies), supplemented with 10% fetal bovine serum (Gibco, Life Technologies). AT1-HEK293 cells were selected by adding 200 μg/ml of G418 into the culture medium.
Immunoblotting
The antibodies used in our experiments included: monoclonal mouse anti-human CD63 antibody (Abcam PLC.), monoclonal rat anti-human AGO2 antibody (Sigma-Aldrich), polyclonal rabbit anti-human AT1 receptor antibody (Santa Cruz Biotechnology, Inc.), polyclonal rabbit anti-human or rat c-ABL (BCR-ABL) antibody (Cell Signaling Technology, Inc.), polyclonal mouse anti-human β-actin antibody (Santa Cruz Biotechnology), and polyclonal rabbit anti-human actin antibody (Sigma-Aldrich). Immunoblotting was performed as previously reported except that the transblots were probed with the antibodies against CD63 (1:500) (for experiments visualizing CD63, the samples were run under non-reducing conditions;Ashiru et al., 2010), AGO2 (1:2000) (Zhang et al., 2010), AT1 receptor (1:600) (Zeng et al., 2003, 2006), TSG101 (1:400), flotillin-1 (1:400), HSP70 (1:400), c-ABL (BCR/ABL) antibody (1:1000) (Pluk et al., 2002), β-actin (1:600), or anti-actin antibody (1:400). Proteins were visualized using the enhanced chemiluminescence system. Normalization was performed by blotting the same samples with an antibody against β-actin.
Dual-color fluorescent in situ hybridization
BCR/ABL D-FISH was performed using a kit according to the manufacturer's recommendation (Beijing GP Medical Technologies, Ltd.). Two-color directly labeled translocation probes were used for the detection of BCR, ABL, and BCR/ABL hybrid gene. Neutrophils isolated from peripheral blood of healthy humans with human-neutrophil separating medium (TBD Co. Ltd.) were exposed to K562 EVs (1 × 105/ml culture medium, 24 h). To evaluate the levels of BCR/ABL hybrid gene, Philadelphia (Ph) chromosome-positive neutrophils were counted among 200 cells using a fluorescence microscope (Pelz et al., 2002).
Statistical analysis
The data are expressed as mean ± SD. Comparison within groups was made by analysis of variance (ANOVA) for repeated measures, and comparison among groups was made by factorial ANOVA and Duncan's test (t test when only two groups were compared). A value of P< 0.05 was considered significant.
Funding
These studies were supported in part by grants from the National Natural Science Foundation of China (30925018, 31130029), the National Basic Research Program of China (973 Program, 2008|CB517308, 2012CB517801 and 2013CB531104), Natural Science Foundation Project of CQ CSTC (CSTC, 2009BA5044), and National Institutes of Health, USA (R37HL023081 and P01HL074940).
Conflict of interest: none declared.
References
- Al-Nedawi K., Meehan B., Micallef J., et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- Ashiru O., Boutet P., Fernandez-Messina L., et al. Natural killer cell cytotoxicity is suppressed by exposure to the human NKG2D ligand MICA*008 that is shed by tumor cells in exosomes. Cancer Res. 2010;70:481–489. doi: 10.1158/0008-5472.CAN-09-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaj L., Lessard R., Dai L., et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno S., Grange C., Deregibus M.C., et al. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J. Am. Soc. Nephrol. 2009;20:1053–1067. doi: 10.1681/ASN.2008070798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Yao C., Xia J., et al. Rapid parallelized and quantitative analysis of five pathogenic bacteria by ITS hybridization using QCM biosensor. Sens. Actuators B Chem. 2011;155:500–504. [Google Scholar]
- Chen X., Ba Y., Ma L., et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Clayton A., Turkes A., Dewitt S., et al. Adhesion and signaling by B cell-derived exosomes: the role of integrins. FASEB J. 2004;18:977–979. doi: 10.1096/fj.03-1094fje. [DOI] [PubMed] [Google Scholar]
- Cocucci E., Racchetti G., Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Denzer K., Kleijmeer M.J., Heijnen H.F., et al. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 2000;113(Pt 19):3365–3374. doi: 10.1242/jcs.113.19.3365. [DOI] [PubMed] [Google Scholar]
- Dragan A.I., Pavlovic R., McGivney J.B., et al. SYBR Green I: fluorescence properties and interaction with DNA. J. Fluoresc. 2012;22:1189–1199. doi: 10.1007/s10895-012-1059-8. [DOI] [PubMed] [Google Scholar]
- Gibbings D.J., Ciaudo C., Erhardt M., et al. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- Guescini M., Guidolin D., Vallorani L., et al. C2C12 myoblasts release micro-vesicles containing mtDNA and proteins involved in signal transduction. Exp. Cell Res. 2010;316:1977–1984. doi: 10.1016/j.yexcr.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Klingbeil A.U., Jacobi J., Langenfeld M.R., et al. Enhanced antinatriuresis in response to angiotensin II in essential hypertension. Am. J. Hypertens. 2000;13:986–993. doi: 10.1016/s0895-7061(00)01191-2. [DOI] [PubMed] [Google Scholar]
- Li Z., Yu C., Han Y., et al. Inhibitory effect of D1-like and D3 dopamine receptors on norepinephrine-induced proliferation in vascular smooth muscle cells. Am. J. Physiol. Heart Circ. Physiol. 2008;294:H2761–H2768. doi: 10.1152/ajpheart.01344.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli A.E., Larregina A.T., Shufesky W.J., et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104:3257–3266. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- Ogawa R., Tanaka C., Sato M., et al. Adipocyte-derived microvesicles contain RNA that is transported into macrophages and might be secreted into blood circulation. Biochem. Biophys. Res. Commun. 2010;398:723–729. doi: 10.1016/j.bbrc.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Pap E., Pallinger E., Pasztoi M., et al. Highlights of a new type of intercellular communication: microvesicle-based information transfer. Inflamm. Res. 2009;58:1–8. doi: 10.1007/s00011-008-8210-7. [DOI] [PubMed] [Google Scholar]
- Pelz A.F., Kroning H., Franke A., et al. High reliability and sensitivity of the BCR/ABL1 D-FISH test for the detection of BCR/ABL rearrangements. Ann. Hematol. 2002;81:147–153. doi: 10.1007/s00277-001-0424-5. [DOI] [PubMed] [Google Scholar]
- Pluk H., Dorey K., Superti-Furga G. Autoinhibition of c-Abl. Cell. 2002;108:247–259. doi: 10.1016/s0092-8674(02)00623-2. [DOI] [PubMed] [Google Scholar]
- Puissant A., Dufies M., Fenouille N., et al. Imatinib triggers mesenchymal-like conversion of CML cells associated with increased aggressiveness. J. Mol. Cell Biol. 2012;4:207–220. doi: 10.1093/jmcb/mjs010. [DOI] [PubMed] [Google Scholar]
- Shibata Y., Malhotra A., Dutta A. Detection of DNA fusion junctions for BCR-ABL translocations by Anchored ChromPET. Genome Med. 2010;2:70. doi: 10.1186/gm191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J., Wurdinger T., van Rijn S., et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C., Ostrowski M., Segura E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- Trajkovic K., Hsu C., Chiantia S., et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- Valadi H., Ekstrom K., Bossios A., et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Wahlgren J., Karlson T.D., Brisslert M., et al. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40:e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldenstrom A., Genneback N., Hellman U., et al. Cardiomyocyte microvesicles contain DNA/RNA and convey biological messages to target cells. PLoS One. 2012;7:e34653. doi: 10.1371/journal.pone.0034653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang Z.H., Kong J., et al. Oxidized low-density lipoprotein-dependent platelet-derived microvesicles trigger procoagulant effects and amplify oxidative stress. Mol. Med. 2012;18:159–166. doi: 10.2119/molmed.2011.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Lin S., Wu W., et al. Ghrelin prevents doxorubicin-induced cardiotoxicity through TNF-alpha/NF-kappaB pathways and mitochondrial protective mechanisms. Toxicology. 2008;247:133–138. doi: 10.1016/j.tox.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Zen K., Zhang C.Y. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med. Res. Rev. 2012;32:326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- Zeng C., Asico L.D., Wang X., et al. Angiotensin II regulation of AT1 and D3 dopamine receptors in renal proximal tubule cells of SHR. Hypertension. 2003;41:724–729. doi: 10.1161/01.HYP.0000047880.78462.0E. [DOI] [PubMed] [Google Scholar]
- Zeng C., Liu Y., Wang Z., et al. Activation of D3 dopamine receptor decreases angiotensin II type 1 receptor expression in rat renal proximal tubule cells. Circ. Res. 2006;99:494–500. doi: 10.1161/01.RES.0000240500.96746.ec. [DOI] [PubMed] [Google Scholar]
- Zhang C., Knudson J.D., Setty S., et al. Coronary arteriolar vasoconstriction to angiotensin II is augmented in prediabetic metabolic syndrome via activation of AT1 receptors. Am. J. Physiol. Heart Circ. Physiol. 2005;288:H2154–H2162. doi: 10.1152/ajpheart.00987.2004. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu D., Chen X., et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Zwicker J.I. Impedance-based flow cytometry for the measurement of microparticles. Semin. Thromb. Hemost. 2010;36:819–823. doi: 10.1055/s-0030-1267035. [DOI] [PubMed] [Google Scholar]