Abstract
Nonsense-mediated mRNA decay (NMD) is a eukaryotic surveillance process that promotes selective degradation of imperfect messages containing premature translation termination codons (PTCs). In yeast, PTCs trigger both deadenylylation-independent mRNA decapping, thereby allowing their rapid degradation by a 5′ to 3′ exonuclease, and to a smaller extent accelerated deadenylylation. It is not clear to what extent this decay pathway is conserved in higher eukaryotes. We used a transcriptional pulse strategy relying on a tetracycline-regulated promoter to study the decay of a PTC- containing β-globin mRNA in human cells. We show that a PTC destabilizes the mRNA and decreases its half-life from >16 h to 3 h. The deadenylylation rate is increased, but not sufficiently to account for the decreased half-life on its own. Using a circularization RT–PCR (cRT–PCR) strategy, we could detect decapped degradation intermediates and measure simultaneously their poly(A) tail length. This allowed us to show that a PTC enhances the rate of mRNA decapping and that decapped products have been deadenylylated to a certain extent. Thus the major feature of the NMD pathway, enhanced decapping, is conserved from yeast to man even though the kinetic details might differ between various mRNAs and/or species.
INTRODUCTION
In eukaryotes, there is an mRNA surveillance process that identifies imperfect messages and promotes their degradation. This process is referred to as nonsense-mediated mRNA decay (NMD) as it targets essentially mRNAs containing premature translation termination codons (PTCs) (1–4). Such imperfect messages can result from either mutations in the DNA or errors in pre-mRNA processing. In addition, NMD participates in the decay of selected mRNAs with intact coding features. This happens either because ribosomes can translate short open reading frames (ORFs) found upstream or downstream of the normal initiation AUG, or because the mRNA contains an extended 3′-untranslated region (UTR), causing normal termination codons to be recognized as premature (5–7).
NMD appears to protect cells against the deleterious dominant-negative or gain-of-function effects of truncated proteins that could accumulate if PTC-containing mRNAs were stable. In humans, about one-third of genetic disorders and many forms of cancer result from mutations introducing PTCs (2,8,9). Most often, these mutations cause diseases only when the two alleles are affected, but dysfunction of the NMD pathway can also lead to disease phenotypes in heterozygote patients. This has been observed in several instances, either because the mutation is such that it allows the mutant mRNA to escape NMD, or because it affects components of the NMD pathway. Most mutations introducing PTCs in the β-globin mRNA cause selective degradation of this message (10). The corresponding β-thalassaemia heterozygote patients are healthy, and only homozygotes suffer from disease. In contrast, some nonsense mutations in the last exon of the β-globin gene cause diserythropoeisis in heterozygote patients. In these patients, the mutated mRNA escapes NMD, producing a truncated β-globin chain (11). NMD dysfunction also contributes to the severity of a dominant genetic disease known as Marfan syndrome caused by mutations in the fibrillin-1 gene. The phenotype is milder when the mutant mRNA levels are decreased than when these mRNAs escape NMD (8,12). Mutations in components of the NMD pathway could participate in a number of complex multigenic diseases by creating dominant-negative phenotypes of otherwise innocuous heterozygote mutations introducing PTCs. This has not yet been observed in humans, but in Caenorhabditis elegans, when smg genes involved in NMD are mutated (13). Finally, NMD participates in the proper function of the immune system as it eliminates non-productively rearranged immunoglobulin or T-cell receptor mRNAs, thus allowing B or T cells to express only the productively rearranged allele (14). However, despite the number of situations where NMD appears to play an important role, the underlying mechanisms are poorly understood in higher eukaryotes.
The NMD pathway is best understood in the yeast Saccharomyces cerevisiae, even though most of the data are derived from the study of a single mRNA, coding for PGK1. Imperfect messages are shunted into the normal mRNA degradation pathway occurring in the cytoplasm (3,15–17). The degradation of most normal polyadenylylated mRNAs is initiated by poly(A) tail shortening, leading to decapping that is followed by rapid 5′ to 3′ exonucleolysis [(16,18,19) and references therein]. In contrast, PTCs containing mRNAs are rapidly decapped and degraded without a requirement for prior deadenylylation (20). A minor pathway involving accelerated deadenylylation is also observed (21,22). It is not fully clear how PTCs are discriminated from normal stop codons. Sequences 3′ of the PTC, termed the ‘downstream element’ (DSE), appear to target mRNA to the NMD pathway, if not traversed by translating ribosomes (15,17,23). The hnRNP-like protein Hrp1p appears to be involved in DES recognition and NMD triggering (24). In addition, three factors, encoded by the UPF1–3 genes, are involved in the discrimination of PTCs (3,9,17,25). Upf1p is an RNA helicase that not only plays a key role in PTC recognition but also affects both translation termination and reinitiation, demonstrating that NMD and translation are intimately linked processes (3,6,17).
The NMD pathway in higher eukaryotes is not as clearly understood. In contrast to yeast, PTCs are discriminated from normal stop codons in mammals according to the spatial relationship between the termination codon and the most 3′ exon–exon junction. A termination codon would be recognized as normal if located downstream, or within about 50 nucleotides upstream of this junction (26–28). The exon junction complex (EJC) is deposited at the exon boundaries during splicing, recruits homologues of the Upf proteins and must be displaced by translating ribosomes to prevent mRNA degradation by the NMD pathway (29–33). The intracellular site of decay of PTC-containing mRNAs in higher eukaryotes is not clear. Nuclear-associated mRNAs would be subject to NMD in some cases, but cytoplasmic translation appears necessary to trigger NMD, and cytoplasmic decay events have been observed (28,33–35). A current hypothesis is that normal and imperfect mRNAs are discriminated by translation occurring while the mRNA is exported from the nucleus (1,2), but there is other evidence in favour of a nuclear mRNA surveillance pathway (33).
In higher eukaryotes, the subsequent degradation steps of PTC-containing mRNAs have not been fully elucidated. As in yeast, the normal mRNA degradation pathway appears to involve deadenylylation-triggered decapping (36). However, in contrast to the yeast paradigm of NMD, recent results indicate that a mammalian PTC-containing mRNA is rapidly deadenylylated before being degraded (35).
Here we show, by analysing the degradation kinetics of a PTC-containing mRNA, conservation from yeast to human of the major features of the NMD pathway. Using a transcriptional pulse strategy (37), coupled with a sensitive circularization RT–PCR (cRT–PCR) method that allows us to detect traces of decapped mRNA (36), we analysed the degradation of a PTC-containing β-globin mRNA in human cells and show that it is decapped more actively than its wild-type counterpart.
MATERIALS AND METHODS
Plasmid construction
Plasmid pTetβ-glob contains a PvuI–PvuI fragment originating from the rabbit β-globin gene (from –9 to +1655) inserted in pUHD10-3 (http://www.zmbh.uni-heidelberg.de/bujard/reporter/pUHD10-3.html) in between the XbaI and HindIII sites downstream of the tetracycline (Tet)-regulated promoter. Plasmid pTetPTC39-β-glob was created by inserting in pTetβ-glob a termination codon at position 39 (CAG→TAG) using site-directed mutagenesis by PCR. The whole subcloned fragment was verified by sequencing.
Cell culture, transfection and transcriptional pulse
The HeLa cell clone H-tTa-1 (38) was cultured and transfected by the calcium phosphate co-precipitation method using standard procedures (39). Transcriptional pulse experiments were performed similarly to those in Xu et al. (37). Briefly, 1 ml of DNA co-precipitate suspension containing 1 µg of the Tet-regulated reporter plasmid, 0.5 µg of a luciferase expression vector as an internal control of transfection efficiency and 18.5 µg of carrier plasmid was added to cells grown at 30% confluence on a 100 mm dish in 10 ml of medium containing 1 ng/ml doxycycline (Dox; Sigma). The precipitate was left on the cells for 16 h, the Dox-containing medium was renewed and cells were further grown for 24 h. To induce reporter gene transcription, cells were washed three times with phosphate-buffered saline (PBS) and incubated with Dox-free medium for 3 h. Reporter gene transcription was blocked following addition to the medium of Dox to a final concentration of 50 ng/ml.
RNA preparation and analysis
Total RNA was prepared using the single-step guanidinium method (40) with some modifications. The cell pellet from a 100 mm dish was homogenized in 750 µl of GTC solution (4 M guanidinium thiocyanate, 0.1 M sodium acetate pH 5.0, 5 mM EDTA pH 8.0, 2% sarkosyl, 0.14 M 2-mercaptoethanol) with a 19-gauge needle. The homogenate was phenol extracted, isopropanol precipitated and further processed as described (40) except that the RNA pellet was resuspended in H2O. RNA samples were then treated with DNase I as described (36). When indicated, poly(A)– mRNAs were obtained following oligo(dT)/RNase H treatment performed as described (36). Northern blot analysis was performed using an agarose–formaldehyde gel as described (39).
cRT–PCR (36) was performed as follows: 4 µg of RNAs were circularized by incubation with 60 U of T4 RNA ligase (New England Biolabs) and 20 U of human placental RNase inhibitor (Amersham) in 400 µl of 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 20 mM dithiothreitol (DTT), 100 mM ATP, 100 µg/ml acetylated bovine serum albumin (BSA) for 16 h at 16°C. The ligation products were then phenol/CHCl3 extracted, ethanol precipitated and resuspended in 12 µl of H2O. Primer 1 (AACCAGCAGCCTGCCCA) was hybridized with the RNA for 45 min prior to reverse transcription as follows: 6 µl of the ligation products were mixed with 1 µl of primer 1 (10 ng/µl) and 1 µl of 10× Taq buffer [650 mM Tris–HCl pH 8.8, 100 mM 2-mercaptoethanol, 165 mM (NH4)2SO4]. The sample was incubated for 30 s at 95°C, cooled down to 42°C, and 1 µl of 60 mM MgCl2 was added to the reaction, which was further incubated for 45 min at 42°C. A 1 µl aliquot of a mixture of the four dNTPs (5 mM each) and 3 U of AMV reverse transcriptase (Stratagene) were subsequently added and the incubation continued at 42°C for 40 min. The reaction was stopped by a 5 min incubation at 95°C. The following reagents were added to the reverse transcription reaction: 1 µl of 10× Taq buffer, 1 µl of a mixture of the four dNTPs (5 mM each), 2 µl of DNase-free BSA (2 mg/ml; Pharmacia), 2 µl (100 ng) of primer 2 (CCCTTGAGC ATCTGACTTCTG), 2 µl (100 ng) of primer 3 (AGGTCAAAACAGCGTGGATGG), 2 µl of H2O and 1 U of Taq DNA polymerase (Cetus). After 5 min at 95°C, the reaction was cycled 30 times (30 s at 94°C, 3 min at 56°C, 3 min at 74°C). Primer 4 (GATGGCGTCTCCAGGCG ATCTTT) was labelled at the 5′ end using T4 polynucleotide kinase (PNK) as follows: 50 ng of primer 4 were incubated with 10 µCi of [γ-32P]ATP and 10 U of T4 PNK in 20 µl of PNK buffer for 30 min at 37°C. The reaction was stopped by a 5 min incubation at 95°C. The PCR products were labelled as follows: 8 µl of the PCR were mixed with 5 µl of a solution containing 1 µl of the primer 4 labelling reaction, 0.5 µl of 10× Taq buffer, 0.5 µl of a mixture of the four dNTPs (5 mM each), 1 µl of DNase-free BSA (2 mg/ml), 0.5 µl of 30 mM MgCl2, 2.5 µl of H2O and 0.5 U of Taq DNA polymerase. After a 5 min denaturation step at 95°C, the reaction was cycled five times (30 s at 94°C, 3 min at 68°C, 3 min at 74°C). Following concentration on a speed-vac, half of the reaction was loaded on a 6% sequencing gel.
RESULTS
A premature termination codon destabilizes the β-globin mRNA and increases its deadenylylation rate
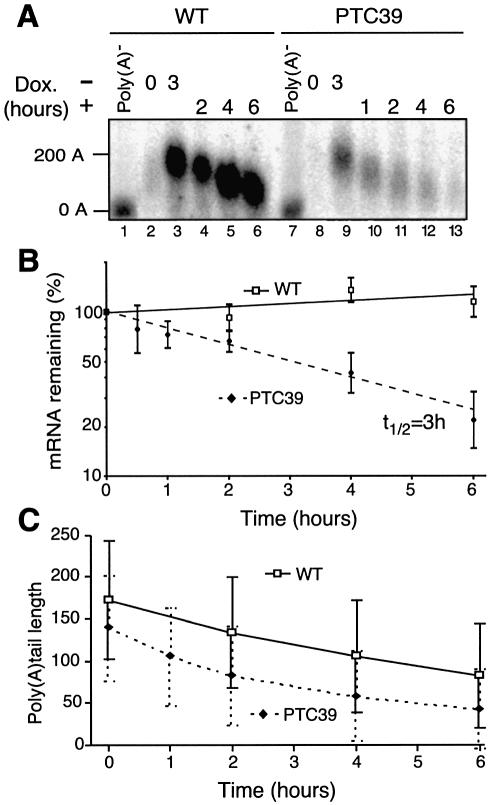
mRNA stability in human cells was analysed using a transcriptional pulse strategy with a Tet-regulated promoter. This allows us to selectively turn the transcription of the reporter gene on and off without interfering with cellular physiology (37,38,41). Using this approach, we can analyse precursor–product relationships in the mRNA decay pathway without interference from the modifications of mRNA stability that are often induced by transcriptional inhibitors (37,41). We compared the stability of a wild-type (WT) β-globin mRNA with that of a single nucleotide mutant mRNA containing a PTC at position 39 (PTC39). Such a mutation occurs naturally in the human β-globin gene, causes β-thalassaemia in homozygote individuals, and is known to reduce transcript levels (11,28). The WT and PTC39 mRNAs were synthesized from plasmids containing the promoter-less β-globin gene controlled by a Tet-regulated minimal promoter (Fig. 1). These plasmids were transiently transfected in HeLa cells expressing the tTA trans-activator (38). During transfection, cells were grown in the presence of the Tet analogue Dox, at the lowest concentration achieving optimal blocking of the interaction of the tTA activator with its target sites. A short pulse of transcription was induced by growing the cells for 3 h in the absence of Dox. The pulse was terminated by adding an excess of Dox, and RNAs were collected at various time intervals following transcriptional arrest (37). Northern blot analysis reveals that the PTC39 mRNA levels are approximately five times lower than WT upon transcriptional induction, and that the mRNA decay rate is faster (Fig. 2A). β-Globin mRNA levels were quantified, expressed as the percentage of the mRNA level obtained at the end of the transcription pulse, and plotted (Fig. 2B). For the PTC39 mRNA, results could be fitted to first-order decay kinetics, even though it is presumably an oversimplification as many eukaryotic mRNAs decay with biphasic kinetics (41). Accordingly, the PTC39 mRNA half-life is ∼3 h. This is markedly shorter than the half-life of the WT β-globin mRNA, which does not decay in the first 6 h (Fig. 2), and has been shown to have a half-life >16 h using a similar transcription pulse strategy (37).
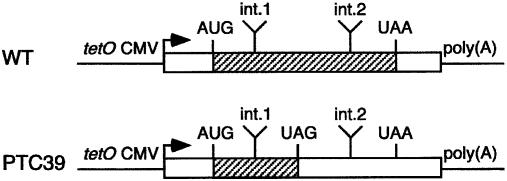
Figure 1.
Tetracycline-regulated chimeric β-globin genes used for transfection. The mature mRNA is represented by a box that includes the ORF (hatched box). The Tet-regulated minimal CMV promoter (tetO CMV) and polyadenylylation site [poly(A)] are represented by lines. The locations of introns 1 and 2 (int.1, int.2) are indicated above the box; the introns themselves are not represented. The initiation (AUG) and the normal and mutant termination codons (UAA and UAG, respectively) are represented above the ORF.
Figure 2.
A premature termination codon enhances β-globin mRNA decay without affecting its deadenylylation rate. (A) Northern blot analysis of the decay of both WT and mutant PTC-containing β-globin mRNAs following a pulse of transcription. tTA-expressing HeLa cells (38) grown in the presence of doxycycline (Dox) were transiently transfected with constructs containing the Tet-regulated CMV promoter driving expression of either the WT or PTC39 β-globin genes. One day after transfection (lanes 2 and 8), transcription was induced by growing the cells for 3 h in Dox-depleted medium (lanes 3 and 9) and then stopped by addition of Dox. mRNA was extracted 1–6 h later as indicated (lanes 4–6 and 10–13). The amount of total RNA loaded on the gel is twice as much for the PTC39-transfected cells (lanes 7–13) to compensate for the lower absolute level of β-globin mRNA produced. Poly(A)– mRNA was obtained after oligo(dT)/RNase H treatment of the induced RNA preparations (lanes 1 and 7). The estimated size of the poly(A) tail is represented on the left-hand side (0 and 200 As). This estimation was performed using as size references the poly(A)– β-globin mRNA, rRNA and the mRNAs used as internal controls of transfection efficiency and RNA loading [luciferase and poly(A)-binding protein mRNAs]. (B) Semi-log plot showing decay of WT and PTC39 β-globin mRNAs. Data from three independent transfection experiments were quantified using a phosphorimager (Molecular Dynamics) and are expressed relative to the β-globin mRNA level measured at the end of the transcription pulse. The mean ± SD is represented as well as the exponential regression curve that allows us to estimate mRNA half-lives (WT, continuous line; PTC39, broken line). (C) Comparison of the deadenylylation rates of WT and PTC39 mRNAs. The average distributions of the poly(A) tail length measured on four independent gels are represented.
In both yeast and mammals, normal mRNAs are deadenylylated before being decapped and degraded (3,36). Newly synthesized WT and PTC39 β-globin mRNAs have a mean poly(A) tail of about 175 and 145 residues, respectively [compare lanes 3 and 9 with the corresponding poly(A)– mRNA in lanes 1 and 7, Fig. 2A and recapitulation of four independent measurements in Fig. 2C]. The bulk of the tail of the WT mRNA is shortened in the first 6 h at a rather constant rate of about 15 A residues/h. In contrast, deadenylylation of the bulk of the PTC39 mRNA is twice as fast in the first 2 h following transcription arrest. Then, after the poly(A) tail has reached an average length of about 80 residues, deadenylylation slows down and continues at a rate similar to that of the WT mRNA. As a consequence of this faster initial deadenylylation, the poly(A) tail of the PTC39 mRNA, 2 h after transcription termination, has a similar length to that of the 6-h-old WT mRNA (Fig. 2A–C). Degradation of the PTC39 mRNA has already been initiated at 2 h, whereas the WT mRNA levels have not yet decreased at 6 h (Fig. 2B). Thus, increased deadenylylation of the PTC39-containing mRNA is not sufficient to account for its enhanced decay.
Enhanced decapping of the PTC39 β-globin mRNA
Decapping is the key step of mRNA degradation in both yeast and mammals (18,36). Monophosphorylated decapped 5′ ends can be ligated to 3′-OH RNA ends, and such ligation products can be detected using PCR strategies (36,42–44). Using a cRT–PCR strategy where the 5′ and 3′ ends of the same molecule are ligated, followed by reverse transcription across the junction and PCR amplification, we could detect traces of decapped mRNA and measure their poly(A) tail length simultaneously (36). The strategy is recapitulated in Figure 3A. Capped mRNA cannot be ligated, and only capless 5′ ends participate in the ligation reaction. Using the appropriate nested primers, only decapped full-length spliced mRNAs are detected (Fig. 3B). Selective reverse transcription of β-globin mRNA where at least the first intron has been spliced is performed using a primer (no. 1) overlapping the exon 1–exon 2 boundary. Selective PCR amplification of the cDNA originating from the circularized mRNA is performed using primers (nos 2 and 3) corresponding to the 3′ and 5′ ends, respectively, of the β-globin mRNA. Finally, only products issued from the full-length decapped species are visualized during the labelling step using a primer (no. 4) that overlaps the junction point, i.e. that is complementary to the 5′ most bases of the mRNA and contains in addition at its 3′ end two thymidine residues that are complementary to the end of the poly(A) tail (Fig. 3A and B). To ensure that primer 4 would indeed detect such full-length mRNA species, the start site of transcription of the transfected chimeric construct was verified by primer extension (data not shown).
Figure 3.
A premature termination codon enhances β-globin mRNA decapping. (A) Strategy used for detecting specific decapped mRNAs and measuring their poly(A) tail length. mRNA molecules are represented as dotted lines and DNA molecules as thick lines. The cap and poly(A) tail (AAAAA) are indicated by a diamond and a rectangle, respectively. The circle indicates the 5′-phosphate group of the 5′ most base of the original mRNA. (B) Arrangement of the primers used. (C) cRT–PCR analysis of the decapped β-globin mRNA degradation products. The pulse of transcription was performed as described in Figure 1 except that equal amounts of total RNA were used for analysis of each point. The size of the poly(A) tail was deduced by comparison with a size standard and subtraction of the predicted length of the poly (A)– PCR product.
Using cRT–PCR to analyse the decay products of both WT and PTC39 mRNAs, we could observe that the PTC39 mutation triggers a considerable increase in the amount of decapped products (Fig. 3C). The PTC39 decapped intermediates are more abundant than the WT ones throughout the time course analysed, but they accumulate at 2–4 h following the pulse of transcription (lanes 7 and 8) when the mRNA level has already decreased about 2-fold (Fig. 2). In contrast, the decapped WT species level rises only slightly 4–6 h following transcription arrest (lanes 3 and 4) at time points when the mRNA level has not significantly decreased (Fig. 2). The difference in the levels of decapped species is particularly striking if expressed relative to the overall β-globin mRNA level as there is about 5- to 10-fold less PTC39 mRNA than WT detected on northern blot (Fig. 2A). Thus, the PTC39 mutation enhances the decapping rate of the β-globin mRNA. The poly(A) tail length of the most abundant decapped species ranges from 10 to 50 residues. This is significantly shorter than the poly(A) tail length of the mRNA population at the corresponding time point, which, 2 h after transcription arrest, ranges from 140 to 30 residues (Fig. 2A–C). This shorter poly(A) tail length is not due to a limitation of the cRT–PCR since we have previously shown that the method allows detection of poly(A) tail length up to 250 residues (36). Thus, even though the PTC39 mutation enhances decapping, this decapping affects preferentially the subset of the mRNA population that has a short poly(A) tail.
DISCUSSION
The NMD pathway was first elucidated in yeast since this organism allows combined genetic and biochemical approaches (3,17,20). Transcriptional pulsing through the use of properly regulated promoters, subject to transient induction, was instrumental in the understanding of this mRNA degradation pathway. This pulsing approach is more reliable than the use of transcriptional inhibitors that profoundly affect the cell metabolism and artificially stabilize a number of mRNAs, presumably by inducing stabilization mechanisms that would be operative during mitosis (41,45). We used the transcriptional pulsing approach established for mammalian cells by Shyu and colleagues, which relies on the use of a Tet-regulated transcriptional activator that should not interfere with endogenous gene expression (37,41). This approach allowed us to show that a PTC reduces the β-globin mRNA half-life from >16 h to ∼3 h.
To study the mRNA decay pathway in mammalian cells, the analysis of mutants allowing accumulation of unstable decay intermediates cannot be performed as easily as in yeast (3,16). To identify such intermediates, it is necessary to use more sensitive detection methods. We have developed a PCR-based approach that permits detection of traces of full-length decapped mRNA and simultaneous measurement of their poly(A) tail length [Fig. 3A; (36)]. When combined with transcriptional pulse analyses, it allowed us to demonstrate that a PTC enhances the rate of mRNA decapping in human cells, similarly to that observed in yeast. In agreement with our observations, it has just been observed that reduction of the levels of the Dcp2 decapping enzyme increases the stability of PTC-containing mRNAs in mammalian cells (46). In yeast, the major degradation pathway for both normal and PTC-containing mRNAs involves decapping as the rate-limiting step, followed by rapid 5′→3′ exonucleolysis by the XRN1p RNase (19,20). In mammals, both normal and imperfect mRNAs apparently follow a similar pathway, as the decapped species appear when the bulk of the mRNA disappears [this study and Couttet et al. (36)]. Further degradation must occur rapidly as decay intermediates are not readily detectable. Since the XRN1p exonuclease is conserved in mammals (47), it is likely that the rapid degradation of the decapped species also occurs by 5′→3′ exonucleolysis.
A PTC in the yeast PGK1 mRNA also enhances deadenylylation of the mRNA (21,22). This deadenylylation is observed when the efficiency of decapping is decreased, in particular when the PTC is at a more 3′ position. The presence of a PTC at position 36 or 39 of the β-globin mRNA increases the deadenylylation rate [this study and Chen and Shyu (35)] and decapping does not occur before deadenylylation has proceeded to a certain extent. In yeast PTC-containing PGK1 mRNA, regardless of the position of the nonsense codon, the dependence of decapping on deadenylylation is removed (22). In contrast, the PTC39 β-globin mRNA decapped species have a shorter poly(A) tail than the bulk of the population. In particular, 2 h after transcription has ceased, the decapped species have a poly(A) tail ranging from 10 to 50 A residues, whereas that of the bulk of the population ranges from 30 to 140 A residues (compare lane 11 in Fig. 2A with lane 7 in Fig. 3C). Thus, decapping affects preferentially the subset of the population that has a shorter poly(A) tail length, suggesting that, in contrast to the yeast PGK1 mRNA, the presence of a PTC does not remove the dependence of decapping on deadenylylation in the β-globin mRNA. Nevertheless, the rate of decapping is enhanced by the presence of a PTC at position 39 in the β-globin mRNA. This is particularly clear when the WT mRNA is compared with the PTC39 mRNA at 6 and 2 h, respectively, after transcription has ceased. Significantly fewer decapped products are detected with the WT population (compare lanes 4 and 7 in Fig. 3C), whereas the bulk of this population has a similar poly(A) tail at this time (compare lanes 6 and 11 in Fig. 2A). Thus, the presence of a PTC in the β-globin mRNA enhances both the rate of deadenylylation and decapping, as observed for the yeast PGK1 mRNA. The differences in the exact behaviour of the two mRNAs might rather reflect differences in the rates of the various degradation steps rather than differences in the mechanisms. Indeed, both in yeast and in mammals, the rates of the steps involved in the decay of mRNAs that are not subject to NMD vary from mRNA to mRNA, and each step appears to be regulated independently by stability determinants located in the body of the mRNA (48–50). In these species, mRNAs are deadenylylated first before optimal decapping occurs (16,19,36). Deadenylylation rates not only vary for each mRNA but also change during the life of each mRNA such that it is faster early on and slows down when the tail reaches its steady-state length (49,51). In both species, decapping occurs after deadenylylation upon reaching a certain threshold tail length that differs from mRNA to mRNA (19,36,49). As a consequence of the differences of the relative rates of deadenylylation and decapping and of the threshold poly(A) tail length that triggers decapping, the mean steady-state poly(A) tail length of both capped and decapped species can vary widely from mRNA to mRNA (36). Differences in the rates of the decay pathways are also observed for similar PTC-containing mRNAs in mammalian cells. A PTC at position 36 of the β-globin mRNA, analysed by the same transcriptional pulse strategy used herein, showed varying deadenylylation rates depending on mRNA determinants affecting translation efficiency as well as on the promoter used for transcriptional pulsing (35). The deadenylylation rate and decay kinetics measured here for the PTC39 mRNA differ slightly from those obtained with the same Tet-regulated promoter in this previous study. In particular, decay of the PTC-containing mRNA is more gradual herein. Presumably, the rate of the various decay steps is influenced by the exact structure of the mRNA and/or factors differing between the cell lines used. Thus, as mRNA- and context-specific differences in the rates of the various decay steps can be observed in both yeast and mammalian cells, it is likely that the differences in the apparent consequences of the presence of a PTC in the yeast PGK1 mRNA and the mammalian β-globin mRNA are only superficial and that most of the basic cytoplasmic NMD pathways is conserved throughout evolution.
A nuclear mechanism for NMD in mammals has been suggested, because (i) PTC-containing mRNAs are often less abundant than their WT counterparts in the nuclear fraction; (ii) pre-mRNA splicing is affecting PTC recognition; and (iii) splicing can sometimes be affected by a PTC [reviewed in Maquat (1), Nagy and Maquat (27) and Vasudevan and Peltz (33)]. However, cytoplasmic translation appears necessary to target an mRNA to NMD (28), and one current hypothesis is that the validating translation event that decides whether an mRNA is targeted to NMD occurs on mRNAs just exiting the nucleus (1,2). The slow decay kinetics observed here (2–4 h after transcriptional arrest) and the timing of poly(A) tail shortening, which precedes decapping of the PTC-containing mRNA, suggest that there is a delay between mRNA export and decapping and are more consistent with an NMD pathway occurring in the cytoplasm. The predominantly cytoplasmic localization of human decapping enzymes supports this interpretation (52). However, one aspect of our observations suggests that there could also be a degradation mechanism operating very early after transcription, earlier than the events described here. Indeed, the PTC39 β-globin mRNA visible upon a 3 h transcription pulse has a half-life of 3 h. Thus, the abundance of the PTC39 at the earliest time point measured, just when transcription is arrested, should not be less than half that of the WT mRNA; however, it is already approximately five times lower at that time point. Thus, at least half of the PTC-containing transcribed population has disappeared before we have a chance to analyse it with the procedures described here. This population must have a half-life much lower than 3 h and may have been degraded before or just when entering the cytoplasm. Given that truncated proteins can potentially be so deleterious to the cell, it should not be so surprising that multiple mechanisms acting at different levels have evolved to ensure efficient elimination of PTC-containing mRNAs.
Acknowledgments
ACKNOWLEDGEMENTS
We thank A. Holmes, and D. Vetrie for critical reading of the manuscript. This work was supported in part by the CNRS and grants from the Association de Recherche sur le Cancer and the Ligue Nationale contre le Cancer. P.C. was supported by fellowships from the Ministère de la Recherche et de l’Enseignement Supérieur and from the Fondation pour la Recherche Médicale.
REFERENCES
- 1.Maquat L.E. (1995) When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA, 1, 453–465. [PMC free article] [PubMed] [Google Scholar]
- 2.Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- 3.Hilleren P. and Parker,R. (1999) mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA, 5, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilusz C.J., Wang,W. and Peltz,S.W. (2001) Curbing the non-sense: the activation and regulation of mRNA surveillance. Genes Dev., 15, 2781–2785. [DOI] [PubMed] [Google Scholar]
- 5.Oliveira C.C. and McCarthy,J.E. (1995) The relationship between eukaryotic translation and mRNA stability. A short upstream open reading frame strongly inhibits translational initiation and greatly accelerates mRNA degradation in the yeast Saccharomyces cerevisiae. J. Biol. Chem., 270, 8936–8943. [DOI] [PubMed] [Google Scholar]
- 6.Muhlrad D. and Parker,R. (1999) Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA, 5, 1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welch E.M. and Jacobson,A. (1999) An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J., 18, 6134–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frischmeyer P.A. and Dietz,H.C. (1999) Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet., 8, 1893–1900. [DOI] [PubMed] [Google Scholar]
- 9.Culbertson M.R. (1999) RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet., 15, 74–80. [DOI] [PubMed] [Google Scholar]
- 10.Maquat L.E., Kinniburgh,A.J., Rachmilewitz,E.A. and Ross,J. (1981) Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell, 27, 543–553. [DOI] [PubMed] [Google Scholar]
- 11.Hall G.W. and Thein,S. (1994) Nonsense codon mutations in the terminal exon of the beta-globin gene are not associated with a reduction in beta-mRNA accumulation: a mechanism for the phenotype of dominant beta-thalassemia. Blood, 83, 2031–2037. [PubMed] [Google Scholar]
- 12.Dietz H.C., McIntosh,I., Sakai,L.Y., Corson,G.M., Chalberg,S.C., Pyeritz,R.E. and Francomano,C.A. (1993) Four novel FBN1 mutations: significance for mutant transcript level and EGF-like domain calcium binding in the pathogenesis of Marfan syndrome. Genomics, 17, 468–475. [DOI] [PubMed] [Google Scholar]
- 13.Pulak R. and Anderson,P. (1993) mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev., 7, 1885–1897. [DOI] [PubMed] [Google Scholar]
- 14.Li S. and Wilkinson,M.F. (1998) Nonsense surveillance in lymphocytes? Immunity, 8, 135–141. [DOI] [PubMed] [Google Scholar]
- 15.Peltz S.W. and Jacobson,A. (1993) mRNA turnover in Saccharomyces cerevisiae. In Belasco,J.G. and Brawerman,G. (eds), Control of Messenger RNA Stability. Academic Press Inc., San Diego, CA, pp. 291–328. [Google Scholar]
- 16.Beelman C.A. and Parker,R. (1995) Degradation of mRNA in eukaryotes. Cell, 81, 179–183. [DOI] [PubMed] [Google Scholar]
- 17.Czaplinski K., Ruiz-Echevarria,M.J., González,C.I. and Peltz,S.W. (1999) Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. Bioessays, 21, 685–696. [DOI] [PubMed] [Google Scholar]
- 18.Beelman C.A., Stevens,A., Caponigro,G., LaGrandeur,T.E., Hatfield,L., Fortner,D.M. and Parker,R. (1996) An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature, 382, 642–646. [DOI] [PubMed] [Google Scholar]
- 19.Muhlrad D., Decker,C.J. and Parker,R. (1994) Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev., 8, 855–866. [DOI] [PubMed] [Google Scholar]
- 20.Muhlrad D. and Parker,R. (1994) Premature translational termination triggers mRNA decapping. Nature, 370, 578–581. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell P. and Tollervey,D. (2003) An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′→5′ degradation. Mol. Cell, 11, 1405–1413. [DOI] [PubMed] [Google Scholar]
- 22.Cao D. and Parker,R. (2003) Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell, 113, 533–545. [DOI] [PubMed] [Google Scholar]
- 23.Peltz S.W., Brown,A.H. and Jacobson,A. (1993) mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev., 7, 1737–1754. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez C.I., Ruiz-Echevarria,M.J., Vasudevan,S., Henry,M.F. and Peltz,S.W. (2000) The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell, 5, 489–499. [DOI] [PubMed] [Google Scholar]
- 25.Leeds P., Peltz,S.W., Jacobson,A. and Culbertson,M.R. (1991) The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev., 5, 2303–2314. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., Sun,X., Qian,Y. and Maquat,L.E. (1998) Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA, 4, 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy E. and Maquat,L.E. (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci., 23, 198–199. [DOI] [PubMed] [Google Scholar]
- 28.Thermann R., Neu-Yilik,G., Deters,A., Frede,U., Wehr,K., Hagemeier,C., Hentze,M.W. and Kulozik,A.E. (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J., 17, 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LeHir H., Izaurralde,E., Maquat,L.E. and Moore,M.J. (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J., 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim V.N., Yong,J., Kataoka,N., Abel,L., Diem,M.D. and Dreyfuss,G. (2001) The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J., 20, 2062–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lykke-Andersen J., Shu,M.D. and Steitz,J.A. (2001) Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science, 293, 1836–1839. [DOI] [PubMed] [Google Scholar]
- 32.Gehring N.H., Neu-Yilik,G., Schell,T., Hentze,M.W. and Kulozik,A.E. (2003) Y14 and hUpf3b form an NMD-activating complex. Mol. Cell, 11, 939–949. [DOI] [PubMed] [Google Scholar]
- 33.Vasudevan S. and Peltz,S.W. (2003) Nuclear mRNA surveillance. Curr. Opin. Cell Biol., 15, 332–337. [DOI] [PubMed] [Google Scholar]
- 34.Belgrader P., Cheng,J. and Maquat,L.E. (1993) Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc. Natl Acad. Sci. USA, 90, 482–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C.Y. and Shyu,A.B. (2003) Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol. Cell. Biol., 23, 4805–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Couttet P., Fromont-Racine,M., Steel,D., Pictet,R. and Grange,T. (1997) mRNA deadenylylation precedes decapping in mammalian cells. Proc. Natl Acad. Sci. USA, 94, 5628–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu N., Loflin,P., Chen,C.Y. and Shyu,A.B. (1998) A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res., 26, 558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gossen M. and Bujard,H. (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA, 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 40.Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 41.Loflin P.T., Chen,C.Y., Xu,N. and Shyu,A.B. (1999) Transcriptional pulsing approaches for analysis of mRNA turnover in mammalian cells. Methods, 17, 11–20. [DOI] [PubMed] [Google Scholar]
- 42.Bertrand E., Fromont-Racine,M., Pictet,R. and Grange,T. (1993) Visualization of the interaction of a regulatory protein with RNA in vivo. Proc. Natl Acad. Sci. USA, 90, 3496–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fromont-Racine M., Bertrand,E., Pictet,R. and Grange,T. (1993) A highly sensitive method for mapping the 5′ termini of mRNAs. Nucleic Acids Res., 21, 1683–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertrand E., Fromont-Racine,M., Pictet,R. and Grange,T. (1997) Detection of ribozyme cleavage products using reverse ligation mediated PCR (RL-PCR). In Turner,P.C. (ed.), Ribozyme Protocols. Humana Press Inc., Totowa, NJ, Vol. 74, pp. 311–323. [DOI] [PubMed] [Google Scholar]
- 45.Ross J. (1997) A hypothesis to explain why translation inhibitors stabilize mRNAs in mammalian cells: mRNA stability and mitosis. Bioessays, 19, 527–529. [DOI] [PubMed] [Google Scholar]
- 46.Lejeune F., Li,X. and Maquat,L.E. (2003) Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating and exonucleolytic activities. Mol. Cell, 12, 675–687. [DOI] [PubMed] [Google Scholar]
- 47.Bashkirov V.I., Scherthan,H., Solinger,J.A., Buerstedde,J.M. and Heyer,W.D. (1997) A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol., 136, 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shyu A.B., Belasco,J.G. and Greenberg,M.E. (1991) Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev., 5, 221–231. [DOI] [PubMed] [Google Scholar]
- 49.Decker C.J. and Parker,R. (1993) A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev., 7, 1632–1643. [DOI] [PubMed] [Google Scholar]
- 50.Chen C.Y., Chen,T.M. and Shyu,A.B. (1994) Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabilizing function. Mol. Cell. Biol., 14, 416–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker E.J. (1993) Control of poly(A) length. In Belasco,J.G. and Brawerman,G. (eds), Control of Messenger RNA Stability. Academic Press Inc., San Diego, CA, pp. 367–415. [Google Scholar]
- 52.VanDijk E., Cougot,N., Meyer,S., Babajko,S., Wahle,E. and Seraphin,B. (2002) Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J., 21, 6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]