Abstract
Barrier surfaces are home to a vast population of commensal organisms that together encode millions of proteins, each of them possessing several potential foreign antigens. Regulation of immune responses to this enormous antigenic load represents a tremendous challenge for the immune system. Tissues exposed to commensals have developed elaborate systems of regulation including specialized populations of resident lymphocytes that maintain barrier function and limit potential responses to commensal antigens. However, in settings of infection and inflammation these regulatory mechanisms are compromised and specific effector responses against commensal bacteria can develop. This review discusses the circumstances controlling the fate of commensal specific T cells and how dysregulation of these responses could lead to severe pathological outcomes.
Commensal microbiota shape T cell resident homeostasis
The body's epithelial surfaces act as a scaffold to sustain diverse communities of commensal organisms that include bacteria, archaea, fungi, protozoa, and viruses 1-5. With an estimated composition of 100 trillion cells, commensals outnumber host cells by at least a factor of 10 and encode 100 fold more genes than their host's genome 6. All barrier sites, including the genital mucosa, skin, airways and gut are constitutively colonized by highly diverse and site-specific flora. The GI tract represents the most abundant commensal niche with a population of more than 1000 individual strains that contain some 3 million unique genes 7,8. While commensalism is defined in ecology as a relationship where only one party benefits while the other is neutral, many of the bacteria of the GI tract are better described as mutualists, adding tremendous enzymatic and protective capability to the host while taking advantage of the nutrient-rich environment the host provides 9,10. In particular, commensal bacteria can prevent colonization by pathogenic organisms 11 and control many aspects of host physiology, not the least of which are lymphocytes of the immune system.
T cells are found in large numbers lining barrier surfaces where they are tasked with surveillance against infection while maintaining diplomatic relations with the resident commensal microbiota 12. As a result, CD4 T cells at these surfaces can adopt multiple inter-related fates associated with the expression of characteristic cytokines and transcription factors 13,14. Under steady state conditions, the GI tract and gut-associated lymphoid tissue (GALT) is dominated by IL-17A producing Th17 cells, IFNγ producing Th1 cells and Foxp3+ regulatory T (Treg) cells 15,16. The balance between these populations is tightly controlled by the cytokine milieu, which at barrier surfaces is in part dependent upon dietary elements and the microbiota 17-20. Th17 cells are largely limited to barrier surfaces and have been an area of particular interest in the study of mucosal immunology. The protective role of IL-17A is associated with its capacity to induce neutrophil granulopoiesis by stimulating epithelial cells to secrete granulocyte colony-stimulating factor (G-CSF) and drive the recruitment of neutrophils by local stromal cells 21,22. Additionally, via their capacity to also produce both IL-22 and IL-17A, Th17 cells can bolster innate epithelial defense mechanisms and reinforce tight junctions 23-26. The function of mucosal Th1 cells under steady state conditions remains unclear, but we might speculate that these cells also contribute to the promotion of various aspects of innate mucosal responses. Treg cells also represent a prominent population of resident cells at barrier sites. Treg cells are required for the maintenance of tolerance to both self antigens and innocuous antigens derived from food, commensal bacteria and other environmental sources 27. Treg cells that line the GI tract can arise from the thymus or be induced locally in response to oral antigen, a process required for the acquisition of oral tolerance 28,29.
The differentiation of T cells at barrier sites into each of these different fates has been associated with the presence of signals derived from the commensal microbiota30-33 .Notably, the capacity of T cells to produce IL-17A and IFN-γ is severely compromised in absence of commensals in both the gut and skin 16,32,34. Germ-free mice also tend to harbor higher frequencies of Th2 cells and this too can be reversed by colonization of the mice with a single species of commensal bacteria 35. In the GI tract, microbial products such as bacterial DNA or defined group of bacteria such as Segmented filamentous bacteria (SFB) can play a dominant role in the promotion of steady-state GI resident Th1 and Th17 cells 16,32. In the skin reconstitution of germ free mice with Staphylococcus epidermidis restored dermal IL-17A34. The frequencies, origin and activation of Foxp3+ Treg cells in the skin and gastrointestinal tract are also influenced by the microbiota. In the skin for instance, the absence of commensals is associated with enhanced frequencies of regulatory T cells 34. In contrast, in the GI tract B. fragilis via expression of Polysaccharide A can expand IL-10 producing CD4+T cells and Treg cells at the expense of the differentiation of Th17 cells 33, 36-38. Bacteria of the Clostridium cluster XIV species also promote Treg cell accumulation in the colon, but not small intestine via an increased capacity to promote TGF-β 31. The gut microbiota can also set the tone of immune responses at distal sites during infection and in some cases contribute to the induction of autoimmune disorders. 31,39-42
Antigen-specific responses to commensal organisms at steady state
Independent of differentiation state and function, a critical question associated with T cells residing at barrier sites is their antigen specificity. The microbiota encodes millions of proteins each of them expressing several potential foreign antigens directly associated with inflammatory Pathogen-Associated Molecular Patterns. This enormous antigenic load represents a tremendous challenge for barrier immunity as unwanted responses against these antigens could lead to severe pathological consequences. A central strategy utilized by the mucosal immune system to maintain its homeostatic relationship with the microbiota is to limit contact between luminal microorganisms and the epithelial cell surface. This is accomplished by the establishment of a structural and immunological barrier resulting from the combined action of mucus, IgA, and antimicrobial proteins 12 (Figure 1, panel 1). Further, commensals can promote their own containment and thus mucosal tissue homeostasis by enhancing various aspects of this physical and immunological shield 43-45.
Figure 1.
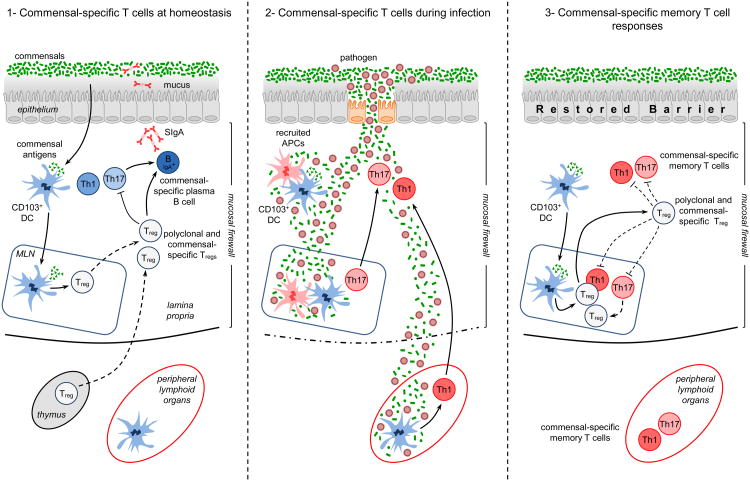
Commensal-specific T cells at homeostasis and during infection: 1) At homeostasis – commensal and food antigens are presented to T cells by CD103+ T cells that have trafficked to the mLN from the lamina propria. Presentation by these DCs to commensal-specific T cells may lead to the differentiation of commensal-specific T regulatory cells (Tregs). Commensal-specific Treg cells traffic to the lamina propria and Peyer's Patches, where they, along with polyclonal Tregs, can regulate effector T cell responses and induce class switching and IgA production from resident commensal-specific B cells, reinforcing the commensal barrier. Critically, the combination of the epithelial barrier, mucus layer, IgA and regulatory DCs and T cells comprises the ‘mucosal firewall’, which limits the passage of commensal and food-derived antigens to the Gut-Associated Lymphoid Tissue (GALT), preventing untoward activation and pathology. 2) During GI infection – Invasion of the intestinal barrier can cause inflammation and tissue damage. This pathology can disrupt the mucosal firewall and allows for systemic translocation of commensal organisms and their associated antigens. During times of translocation, the host immune system becomes unable to discriminate between commensal and pathogen-derived antigens and therefore commensal-specific T cell responses mimic responses to the invasive pathogen. 3) After clearance of the infection –the intestinal barrier reforms and the mucosal firewall is restored perhaps preventing the chronic activation of commensal-specific memory T cells. Treg cells may regulate commensal specific responses either directly or via the modulation of resident dendritic cells.
The physical segregation between the host and microbiota is not absolute and the GI epithelium is semi-permeable to allow for the passage of nutrients. Antigens that bypass the mucosal barrier are prevented from systemic traffic and confined to the GALT by the ‘mucosal firewall’ 46,47. In the GALT, active mechanisms of tolerance regulate T cell responses and as a result the GI tract represents a privileged site for the induction of tolerance (Figure 1, panel 1).. The regulatory nature of the GI tract is perhaps best demonstrated by experiments examining oral tolerance to food 48. This phenomenon has long been recognized in both rodent models and humans and is clinically important, as dysregulated T cell responses against food-derived antigens are associated with both Celiac disease and gastrointestinal allergies. Experiments have shown that under steady-state conditions, the predominant antigen-presenting cells of food-derived antigens are dendritic cells (DC) that express the integrin CD103 28,29. CD103+ DCs drive the differentiation of Foxp3+Treg cells via their capacity to produce TGF-β [ET1]and to convert retinol to retinoic acid (RA) 28,29,49-52. Further CD103+ DCs can also induce the expression of gut homing receptors on immune cells, including Tregs, favoring their migration to the GI tract 53. Because commensal antigens are likely encountered in the context of these tolerogenic responses, it has been proposed that the GI tract also promotes the induction of Treg cells specific for commensal antigens (Figure 1, panel 1). Indeed, some types of commensal bacteria seem to support the differentiation and maintenance of colonic regulatory T cells, though whether these Tregs are specific to commensal-derived antigens is not known 31,54. T cell receptor transgenic mice made specific to commensal-derived antigens can differentiate to become Treg cells after transfer to lymphopenic hosts 55,56 and Foxp3+ Treg cells specific for commensals have been identified in the GI tract of healthy mice 55. Aside from the direct maintenance of T cell tolerance, commensal-specific Treg cells may reinforce the mucosal firewall. Treg cells present in the Peyer's Patches of the small intestine have been shown to promote class-switching to IgA in an antigen-specific manner 45,57. As IgA can directly modulate expression of commensal antigen and mucosal association, this implies that Treg cells may play multiple and complementary roles in controlling the host relationship with the microbiota 58,59. Commensal-specific T cells transferred into lymphopenic hosts can also differentiate into Th1 and Th17 cells 56, but Th1, Th17 and Treg cells resident in the GI tract are by not by necessity specific to commensal-derived antigens 60,61. For example, Th17 cells develop in the absence of cognate antigen in mice expressing a single TCR 60,61. Further, Treg cells can be found in the GI tract of germ-free mice, and albeit significantly reduced, Th17 and Th1 cells are also present in the absence of live commensals 62. It is worth noting that the diet of germ free mice contains microbial products, including antigens, that can provide surrogate signals to the ones normally provided by the flora and may be responsible for the presence of these cells without live commensals 63. Alternatively, T cell differentiation and gut homing may occur at a low level independently of signals derived from the commensal flora. Although these data support the notion that the flora promotes the induction and/or maintenance of T cells in the GI tract independent of antigen specificity, they do not exclude that a significant portion of mucosal T cells recognize commensal antigens and the specificity of Th1, Th17 or Treg cells that reside at barrier site under physiological settings remains an important open question.
Environmentally-induced shifts in the microbiota
Active regulation of immune tolerance to the commensal microbiota is a life-long process, because, in contrast to the host genome, the commensal microbiota is not fixed. Recent studies have shown that while the core metagenome of the gut microbiota is quite stable, species composition and consequently antigenic composition of the flora varies over time in response to a variety of factors such as diet, sanitation, infection and antibiotic use 64,65. Therefore, the adaptive immune system of the gastrointestinal mucosa must be flexible and constantly reset itself to take into account novel commensal antigens. Changes in the microbiota are particularly pervasive during gastrointestinal infection and inflammation, where the commensal microbiota becomes dominated by commensals with enhanced invasive properties 66. For instance, the γ-proteobacteria class of bacteria are particularly selected for growth during a diverse set of inflammatory conditions that ranges from Crohn's Disease (CD), colon cancer and Type 2 Diabetes in humans to T. gondii infections and IBD induction in mice 67-73. Since the γ-proteobacteria family is mostly composed of genera that are opportunistic or obligate pathogens, this effect could be due to the unique ability of these organisms to colonize the inflamed gastrointestinal tract. Indeed, E. coli that dominates the GI tract in T. gondii infection are skewed toward inflammatory and invasive strains that can contribute to the pathological process during inflammatory responses and infection 70,71,74,75. This phenomenon is not limited to γ-proteobacteria as many other clinically relevant species, such as E. faecalis and C. difficile can bloom during bouts of gastrointestinal dysbiosis 76,77. How the immune system deals with these opportunistic residents and in particular whether tolerance to antigen associated with these bacteria is maintained during inflammation is of central importance to our understanding of mucosal inflammatory disorders.
Environmentally-induced breach of the ‘mucosal firewall’
At homeostasis and in highly controlled experimental settings, the mucosal firewall insures that bacterial translocation is strictly limited to the intestinal tissue and associated lymphoid structures 47. In mouse models, this system was proven extraordinarily robust as only complete deficiency of key innate and adaptive mucosal immune mechanisms led to systemic commensal specific antibody responses 78. Accordingly, in mice raised under pathogen free conditions, commensal-specific T cells present outside the mucosa remain naïve despite the presence of commensal antigens in the GI tract 45,79. However, controlled environments that lack pathogens completely represent a highly artificial setting to understand tissue homeostasis and local immune responses. Indeed, we are now beginning to appreciate that under physiological conditions, systemic translocation of commensals and microbial products beyond the mucosa and GALT is a more common occurrence than initially postulated. Translocation of bacterial products to the systemic circulation has been associated clinically with a diverse set of circumstances such as alcohol abuse and cirrhosis, chronic NSAID use, malnutrition, chronic inflammation, extreme exercise regimens and in particular infections 80-86. In a number of murine models of gastrointestinal infections, such as Toxoplasma gondii and Yersinia pseudotuberculosis, immunopathology can induce the translocation of commensal bacteria 70,79,87. Chronic GI barrier dysfunction is also observed during HIV infection in humans and in SIV infection of rhesus macaques 88-90. Barrier sites including the gut, skin and airway, are primary sites for infections. It is estimated that a child will suffer 10- 15 diarrheal episodes on average before the age of 5, all of them potentially associated with transient commensal translocation 91,92, which if added together with common skin and lung infections, provides ample opportunity for exposure of the immune system to commensal antigens under inflammatory conditions.
Commensal-specific T cell responses during infection and inflammation
Infections represent highly volatile situations for the mucosal immune system, as pathogens and commensals will share the same inflamed environment. The potential danger of this situation is illustrated by studies that suggest that oral tolerance to newly introduced food antigens breaks down during acute GI infection 93,94. Recent evidence suggests that in similar manner, tolerance to commensal derived antigens may be lost during acute infections. In a mouse model of T. gondii infection, Treg cells are lost, commensals translocate and the immune system becomes unable to discriminate between commensals and pathogen-derived antigens 79 (Figure 1, panel 2). During this highly Th1 polarized infection, commensal specific T cells also develop as Th1 cells according to cues provided by the inflammatory milieu, rather than Th17 cells or Treg cells, as previous studies have associated with commensal-driven responses 31,32. The idea that innate inflammatory cues drive the fate of commensal specific T cells rather than their specificity is further supported by the observation that physical breakdown of the intestinal barrier via the administration of DSS resulted in the activation of commensal-specific T cells that differentiated towards the Th17 fate 79. Thus, the immune response to gastrointestinal pathogens is associated with parallel responses against commensal-derived antigens that develop according to the inflammatory milieu. The CBir1 commensal antigen followed in this study, is expressed by the Clostridia cluster XIV class of bacteria, known to live in the mucus layer of the intestinal mucosa and is a dominant antigen in Crohn's Disease 31,95. It will be important to determine with future studies whether these responses are directed against all commensal antigens or are limited to the most prevalent or accessible antigens.
Commensal-specific memory T cell responses
One remarkable feature of all barrier sites is their ability to efficiently repair after inflammation or breach. In the GI tract, this implies that after acute tissue damage and transiently increased exposure to commensals, physical segregation between the flora and the immune system is rapidly restored. In absence of chronic exposure to antigen, activated lymphocytes can survive long–term as memory populations capable of rapid re-activation and proliferation. Indeed, following gastrointestinal infection with T. gondii, commensal-specific CD4 T cells can persist long-term in both the GI tract and secondary lymphoid tissue and maintain the ability to become activated, express Th1 inflammatory cytokines and proliferate upon secondary encounter with their cognate antigen 79.
Much like pathogen-specific CD4 T cells and in contrast to virus-specific CD8 memory T cells, commensal-specific memory T cells declined steadily over time 79,96,97. As CD4 T cells carry out the complex task of discriminating pathogenic organisms from benign organisms in the face of a constantly changing environment, perhaps development of CD4 memory reflects this necessity for flexibility. An evolving pool of specificities within the Treg and CD4 effector compartment may allow for the maintenance of tolerance and barrier function in the context of fluctuating commensal populations and intermittent infection. Such flexible repertoire has been proposed for mucosal IgA responses that lack the memory characteristics associated with CD8 T cells and are able to respond to flux in the commensal microbiota composition. Indeed, established IgA producing clones are outcompeted by novel anti-bacterial responses allowing the mucosal immune system to respond to a constantly changing microbiota 98.
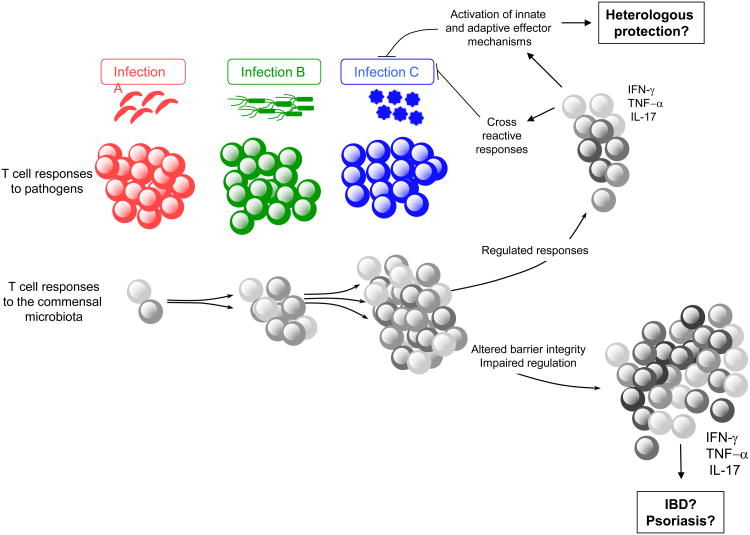
The physiological consequence of long-term CD4+T cell memory against commensals remains to be addressed. Due to the extraordinary antigenic diversity of the host microbiota at all body surfaces and the prevalence of infections, a significant fraction of memory cells are expected to be commensal specific and could develop over time in response to successive infections and/or various barrier breaches (Figure 2). In support of this hypothesis, healthy human serum contains antibodies specific to skin and intestinal microbiota 82. Thus, primary exposure to a pathogen in the skin, lung and GI tract is likely to occur in the context of a much broader recall response against commensal bacteria. One possible consequence of these responses may be the induction of heterologous memory wherein antigen-specific responses against previously encountered commensal bacteria could drive the rapid production of inflammatory cytokines, leading to increased protection against secondary infection and associated translocation of commensal bacteria (Figure 2). Colonic resident Th17 cells have been shown to contribute to early protection against enteric pathogens, though whether these cells are specific to commensal antigens is not clear 99. Further contributing to the possibility of heterologous protection against infection by commensal-specific T cells, a recent study suggests that CD4 T cell clones that are cross-reactive to commensals and viruses are common in healthy patients 100. On the other hand, aberrant accumulation of commensal specific T cells under defined settings may lead to several pathogenic consequences, such as IBD and psoriasis 101. Exploration of the antigen-specificity of the memory cell compartment of lymphocytes residing at all barrier sites would inform us of the potential impact of these commensal-specific T cell responses on tissue physiology and subsequent pathologies. It would be of particular interest to address how responses to conserved bacterial antigens across barrier surfaces impact local and systemic tissue responses over time.
Figure 2.
Potential consequences of commensal-specific memory T cells: During infection at barrier sites (gut, skin, airway, genital mucosa) immune responses against the invading agent can be associated with specific T cell responses against a large number of coincident commensal antigens. These commensal-specific effector T cell responses can persist as memory cells that upon subsequent infection will be recalled as secondary commensal-specific effectors, alongside the priming of a novel immune response to the invasive pathogen. Therefore, each infection at barrier surfaces represents an additional opportunity for the reactivation of commensal-specific T cells. Given the extraordinary number of commensal antigens, these responses may represent a significant proportion of memory T cells. If properly controlled, commensal-specific effector/memory cells could contribute to protection against infections by promoting innate and adaptive effector mechanisms that assist in the clearance of the pathogen. Further, commensal memory responses could be protective due to cross reactivity with pathogen-derived antigens. In contrast, in situations where commensal-specific T cells become dysregulated due to impaired regulatory pathways and/or barrier function, these T cells could drive chronic pathology such as IBD or psoriasis.
Commensal-specific T cells and gastrointestinal disorders
Inflammatory Bowel Diseases (IBD) refers to a group of chronic inflammatory disorders affecting the gastrointestinal tract 102. There are two main clinical forms of IBD: Crohn's disease (CD) that can affect any part of the gastrointestinal tract and ulcerative colitis (UC), in which pathology is restricted to the colonic mucosa 102. The etiology of these disorders is complex and believed to be the consequence of genetic factors, the host immune system and environmental factors such as the microbiota 103. Individual genome-wide association studies have revealed that a large number of risk factors are associated with active immune responses and altered barrier function. 104-108. In light of recent findings, commensal-specific T cells may represent an important component of the disease. In mouse models of colitis, it is well known that the commensal microbiota is necessary for the induction of disease via activation of both innate and adaptive immune mechanisms. Commensal-specific CD4 T cells have been identified in murine models of colitis and in some models are required to drive disease 109-111. For example, immortalized commensal-specific CD4 T cell clones derived from colitis-prone mice are capable of transferring disease to wild-type mice 112. Activated T cells can also induce colitis in NOD2 deficient mice in response to antigens carried by commensal bacteria 113. Studies comparing TCR transgenic cells in the Rag/SCID model of colitis indicate that although commensal specificity is unnecessary for the proliferation and accumulation of CD4 T cells at mucosal sites, cognate antigen responses are required for the induction of colitis 114. Additionally, germ-free IL-10 knockout mice monocolonized with either E.coli or E. faecalis were shown to develop CD4 T cell responses against the colonizing bacteria 109. Further, feeding IL-10 knockout mice a diet high in milk fat is associated with increased Th1 T cell responses to a particular bacteria, B. wadsworthia, though whether these are strictly antigen-specific remains unclear 115., A single genus of bacteria, Helicobacter, is sufficient for the induction of colitis in both the IL-10 and Rag/SCID models of colitis and T cells specific to Helicobacter are present and sufficient for disease in animal models of IBD 116-118. Finally, in mice where both IL-10 and TGFβ signaling is deficient in T cells spontaneous colitis occurs that is directly dependent upon the presence of bacteria of the genus Bacteroides 119. One key point taken from these studies is that multiple bacterial types comprising several distantly related bacterial phyla can promote colitis, possibly via the induction of specific CD4 T cell responses, supporting the idea that IBD may develop as a consequence of a broad loss of tolerance to the commensal microbiota 101. In support of this hypothesis, higher titers of commensal-specific antibodies are found in the serum of Crohn's patients compared to healthy donors and the measurement of antibody responses to a panel of seemingly unrelated commensal-derived antigens is commonly used as a diagnostic for IBD 82,120. However, commensal specific responses are observed in healthy individuals suggesting that on its own, immunity to commensals is not sufficient for the induction of diseases 100,121. Thus, under normal settings, effector and memory responses against commensals that have been induced by infections or injuries are likely held in check by the combined effect of the mucosal firewall and active mechanisms of tolerance. IBD, on the other hand, could be the result of the environmental activation of commensal-specific T cells in the context of a genetic predisposition for intestinal pathology and in particular defects in repair and immune regulation. This multiple hit mechanism of disease induction is supported by data that shows mucosal viral infection, the commensal microbiota and diminished Paneth cell function due to reduced expression of ATG16L1, converge to increase disease severity in experimental colitis 122. However, despite clear connections between commensal-specific T cell responses and IBD, more remains to be done to understand how immunity to commensals could be causative in disease and how commensal-specific effector T cells are regulated under homeostatic conditions (Figure 1, Panel 3).
Concluding remarks
Adaptive immunity, as defined by the presence of lymphocytes with re-arranged antigen receptors of near infinite specificity, is a characteristic of organisms that carry complex populations of microbial symbionts upon their mucosal surfaces. One might speculate that the co-evolution between the adaptive immune system and commensal microbiota was primarily driven by the difficulty of maintaining and controlling such a complex relationship. However, barrier surfaces are not static and are often perturbed by environmental or infectious challenges, causing changes to the commensal microbiota and increasing tissue permeability. In Westernized countries, increased used of antibiotics, reduced worm infections and drastic changes in nutrition have imposed massive changes in our relationship with these organisms. Our understanding of commensal-immune interactions under these highly fluctuating circumstances is still in its infancy and much remains to be understood about commensal-specific responses and their consequences for human health.
Highlights.
The GI tract prevents systemic T cell responses against commensal bacteria
Infection can induce changes in the composition of commensals and translocation
Infection induces differentiation of effector/memory commensal-specific T cells
Commensal-specific T cells are present and may contribute to IBD
Acknowledgments
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. We would like to thank the members of the Belkaid laboratory for helpful discussions and in particular Drs. A. Poholek, E. Wohlfert and S. Naik for critical reading.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Foulongne V, et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One. 2012;7(6):e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iliev ID, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336(6086):1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466(7304):334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckburg PB, et al. Archaea and their potential role in human disease. Infect Immun. 2003;71(2):591–596. doi: 10.1128/IAI.71.2.591-596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grice EA, Segre JA. The human microbiome: our second genome. Annu Rev Genomics Hum Genet. 2012;13:151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley RE, et al. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 7.Clemente JC, et al. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Backhed F, et al. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 10.Costello EK, et al. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336(6086):1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stecher B, Hardt WD. Mechanisms controlling pathogen colonization of the gut. Current opinion in microbiology. 2011;14(1):82–91. doi: 10.1016/j.mib.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Hooper LV, et al. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou L, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453(7192):236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy KM, Stockinger B. Effector T cell plasticity: flexibility in the face of changing circumstances. Nature immunology. 2010;11(8):674–680. doi: 10.1038/ni.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maynard CL, Weaver CT. Intestinal effector T cells in health and disease. Immunity. 2009;31(3):389–400. doi: 10.1016/j.immuni.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall JA, et al. Commensal DNA Limits Regulatory T Cell Conversion and Is a Natural Adjuvant of Intestinal Immune Responses. Immunity. 2008 doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konkel JE, Chen W. Balancing acts: the role of TGF-beta in the mucosal immune system. Trends in molecular medicine. 2011;17(11):668–676. doi: 10.1016/j.molmed.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441(7090):235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 19.Hu W, et al. Priming microenvironments dictate cytokine requirements for T helper 17 cell lineage commitment. Immunity. 2011;35(6):1010–1022. doi: 10.1016/j.immuni.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall JA, et al. Essential role for retinoic acid in the promotion of CD4(+) T cell effector responses via retinoic acid receptor alpha. Immunity. 2011;34(3):435–447. doi: 10.1016/j.immuni.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fossiez F, et al. T cell interleukin-17 induces stromal cells to produce proinflammatory and hematopoietic cytokines. The Journal of experimental medicine. 1996;183(6):2593–2603. doi: 10.1084/jem.183.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korn T, et al. IL-17 and Th17 Cells. Annual review of immunology. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 23.Sonnenberg GF, et al. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nature immunology. 2011;12(5):383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 24.Ishigame H, et al. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity. 2009;30(1):108–119. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nature medicine. 2008;14(3):282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 26.Sonnenberg GF, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336(6086):1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bilate AM, Lafaille JJ. Induced CD4+Foxp3+ regulatory T cells in immune tolerance. Annual review of immunology. 2012;30:733–758. doi: 10.1146/annurev-immunol-020711-075043. [DOI] [PubMed] [Google Scholar]
- 28.Coombes JL, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-{beta}- and retinoic acid-dependent mechanism. The Journal of experimental medicine. 2007 doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun CM, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. The Journal of experimental medicine. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 31.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mazmanian SK, et al. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 34.Naik S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337(6098):1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazmanian SK, et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 36.O'Mahony C, et al. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathog. 2008;4(8):e1000112. doi: 10.1371/journal.ppat.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochoa-Reparaz J, et al. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol. 2010;185(7):4101–4108. doi: 10.4049/jimmunol.1001443. [DOI] [PubMed] [Google Scholar]
- 38.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332(6032):974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molloy MJ, et al. Intestinal microbiota: shaping local and systemic immune responses. Seminars in immunology. 2012;24(1):58–66. doi: 10.1016/j.smim.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32(6):815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37(1):158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108(13):5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brandl K, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455(7214):804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334(6053):255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cong Y, et al. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci U S A. 2009;106(46):19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macpherson AJ, et al. The mucosal firewalls against commensal intestinal microbes. Semin Immunopathol. 2009;31(2):145–149. doi: 10.1007/s00281-009-0174-3. [DOI] [PubMed] [Google Scholar]
- 47.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303(5664):1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 48.Weiner HL, et al. Oral tolerance. Immunological reviews. 2011;241(1):241–259. doi: 10.1111/j.1600-065X.2011.01017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mucida D, et al. Retinoic acid can directly promote TGF-beta-mediated Foxp3(+) Treg cell conversion of naive T cells. Immunity. 2009;30(4):471–472. doi: 10.1016/j.immuni.2009.03.008. author reply 472-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mucida D, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317(5835):256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 51.Benson MJ, et al. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. The Journal of experimental medicine. 2007;204(8):1765–1774. doi: 10.1084/jem.20070719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Denning TL, et al. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J Immunol. 2011;187(2):733–747. doi: 10.4049/jimmunol.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mora JR, et al. Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science. 2006;314(5802):1157–1160. doi: 10.1126/science.1132742. [DOI] [PubMed] [Google Scholar]
- 54.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107(27):12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478(7368):250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng T, et al. Interleukin-12 converts Foxp3+ regulatory T cells to interferon-gamma-producing Foxp3+ T cells that inhibit colitis. Gastroenterology. 2011;140(7):2031–2043. doi: 10.1053/j.gastro.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsuji M, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer's patches. Science. 2009;323(5920):1488–1492. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki K, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101(7):1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterson DA, et al. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell host & microbe. 2007;2(5):328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 60.Lochner M, et al. Restricted microbiota and absence of cognate TCR antigen leads to an unbalanced generation of Th17 cells. J Immunol. 2011;186(3):1531–1537. doi: 10.4049/jimmunol.1001723. [DOI] [PubMed] [Google Scholar]
- 61.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34(5):794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 62.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell host & microbe. 2008;4(4):337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hill DA, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2009 doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lozupone CA, et al. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chow J, et al. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Current opinion in immunology. 2011;23(4):473–480. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rolhion N, Darfeuille-Michaud A. Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13(10):1277–1283. doi: 10.1002/ibd.20176. [DOI] [PubMed] [Google Scholar]
- 68.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 69.Arthur JC, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heimesaat MM, et al. Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J Immunol. 2006;177(12):8785–8795. doi: 10.4049/jimmunol.177.12.8785. [DOI] [PubMed] [Google Scholar]
- 71.Craven M, et al. Inflammation drives dysbiosis and bacterial invasion in murine models of ileal Crohn's disease. PLoS One. 2012;7(7):e41594. doi: 10.1371/journal.pone.0041594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell host & microbe. 2010;8(3):292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lupp C, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell host & microbe. 2007;2(2):119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 74.Egan CE, et al. Synergy between intraepithelial lymphocytes and lamina propria T cells drives intestinal inflammation during infection. Mucosal Immunol. 2011;4(6):658–670. doi: 10.1038/mi.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benson A, et al. Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell host & microbe. 2009;6(2):187–196. doi: 10.1016/j.chom.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ubeda C, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. The Journal of clinical investigation. 2010;120(12):4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Britton RA, Young VB. Interaction between the intestinal microbiota and host in Clostridium difficile colonization resistance. Trends in microbiology. 2012;20(7):313–319. doi: 10.1016/j.tim.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Slack E, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325(5940):617–620. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hand TW, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337(6101):1553–1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wong F. Recent advances in our understanding of hepatorenal syndrome. Nature reviews Gastroenterology & hepatology. 2012;9(7):382–391. doi: 10.1038/nrgastro.2012.96. [DOI] [PubMed] [Google Scholar]
- 81.Nieman DC, et al. Ibuprofen use, endotoxemia, inflammation, and plasma cytokines during ultramarathon competition. Brain, behavior, and immunity. 2006;20(6):578–584. doi: 10.1016/j.bbi.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 82.Zimmermann K, et al. Systemic antibody responses to gut microbes in health and disease. Gut microbes. 2012;3(1):42–47. doi: 10.4161/gmic.19344. [DOI] [PubMed] [Google Scholar]
- 83.Purohit V, et al. Alcohol, intestinal bacterial growth, intestinal permeability to endotoxin, and medical consequences: summary of a symposium. Alcohol. 2008;42(5):349–361. doi: 10.1016/j.alcohol.2008.03.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arakawa T, et al. Small intestinal injury caused by NSAIDs/aspirin: finding new from old. Current medicinal chemistry. 2012;19(1):77–81. doi: 10.2174/092986712803414105. [DOI] [PubMed] [Google Scholar]
- 85.Hughes SM, et al. Dendritic cell anergy results from endotoxemia in severe malnutrition. J Immunol. 2009;183(4):2818–2826. doi: 10.4049/jimmunol.0803518. [DOI] [PubMed] [Google Scholar]
- 86.Berkley JA, et al. Bacteremia among children admitted to a rural hospital in Kenya. The New England journal of medicine. 2005;352(1):39–47. doi: 10.1056/NEJMoa040275. [DOI] [PubMed] [Google Scholar]
- 87.Meinzer U, et al. Yersinia pseudotuberculosis effector YopJ subverts the Nod2/RICK/TAK1 pathway and activates caspase-1 to induce intestinal barrier dysfunction. Cell host & microbe. 2012;11(4):337–351. doi: 10.1016/j.chom.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 88.Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nature medicine. 2006;12(12):1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 89.Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annual review of immunology. 2012;30:149–173. doi: 10.1146/annurev-immunol-020711-075001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Estes JD, et al. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog. 2010;6(8):e1001052. doi: 10.1371/journal.ppat.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vernacchio L, et al. Diarrhea in American infants and young children in the community setting: incidence, clinical presentation and microbiology. Pediatr Infect Dis J. 2006;25(1):2–7. doi: 10.1097/01.inf.0000195623.57945.87. [DOI] [PubMed] [Google Scholar]
- 92.Kosek M, et al. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull World Health Organ. 2003;81(3):197–204. [PMC free article] [PubMed] [Google Scholar]
- 93.Oldenhove G, et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 2009;31(5):772–786. doi: 10.1016/j.immuni.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Severance EG, et al. Anti-gluten immune response following Toxoplasma gondii infection in mice. PLoS One. 2012;7(11):e50991. doi: 10.1371/journal.pone.0050991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lodes MJ, et al. Bacterial flagellin is a dominant antigen in Crohn disease. The Journal of clinical investigation. 2004;113(9):1296–1306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pepper M, et al. Different routes of bacterial infection induce long-lived TH1 memory cells and short-lived TH17 cells. Nature immunology. 2009;11(1):83–89. doi: 10.1038/ni.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Homann D, et al. Differential regulation of antiviral T-cell immunity results in stable CD8+ but declining CD4+ T-cell memory. Nature medicine. 2001;7(8):913–919. doi: 10.1038/90950. [DOI] [PubMed] [Google Scholar]
- 98.Hapfelmeier S, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328(5986):1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Geddes K, et al. Identification of an innate T helper type 17 response to intestinal bacterial pathogens. Nature medicine. 2011;17(7):837–844. doi: 10.1038/nm.2391. [DOI] [PubMed] [Google Scholar]
- 100.Su LF, et al. Virus-Specific CD4(+) Memory-Phenotype T Cells Are Abundant in Unexposed Adults. Immunity. 2013 doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sartor RB. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nature clinical practice Gastroenterology & hepatology. 2006;3(7):390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 102.Kaser A, et al. Inflammatory bowel disease. Annual review of immunology. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474(7351):298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 104.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nature genetics. 2008;40(8):955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nature genetics. 2010;42(12):1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McGovern DP, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nature genetics. 2010;42(4):332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nature genetics. 2011;43(3):246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim SC, et al. Dual-association of gnotobiotic IL-10-/- mice with 2 nonpathogenic commensal bacteria induces aggressive pancolitis. Inflamm Bowel Dis. 2007;13(12):1457–1466. doi: 10.1002/ibd.20246. [DOI] [PubMed] [Google Scholar]
- 110.Kullberg MC, et al. Bacteria-triggered CD4(+) T regulatory cells suppress Helicobacter hepaticus-induced colitis. The Journal of experimental medicine. 2002;196(4):505–515. doi: 10.1084/jem.20020556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yoshida M, et al. Differential localization of colitogenic Th1 and Th2 cells monospecific to a microflora-associated antigen in mice. Gastroenterology. 2002;123(6):1949–1961. doi: 10.1053/gast.2002.37049. [DOI] [PubMed] [Google Scholar]
- 112.Cong Y, et al. CD4+ T cells reactive to enteric bacterial antigens in spontaneously colitic C3H/HeJBir mice: increased T helper cell type 1 response and ability to transfer disease. The Journal of experimental medicine. 1998;187(6):855–864. doi: 10.1084/jem.187.6.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Watanabe T, et al. Nucleotide binding oligomerization domain 2 deficiency leads to dysregulated TLR2 signaling and induction of antigen-specific colitis. Immunity. 2006;25(3):473–485. doi: 10.1016/j.immuni.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 114.Feng T, et al. Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. The Journal of experimental medicine. 2010;207(6):1321–1332. doi: 10.1084/jem.20092253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487(7405):104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kullberg MC, et al. Induction of colitis by a CD4+ T cell clone specific for a bacterial epitope. Proc Natl Acad Sci U S A. 2003;100(26):15830–15835. doi: 10.1073/pnas.2534546100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kullberg MC, et al. Helicobacter hepaticus-induced colitis in interleukin-10-deficient mice: cytokine requirements for the induction and maintenance of intestinal inflammation. Infect Immun. 2001;69(7):4232–4241. doi: 10.1128/IAI.69.7.4232-4241.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Powrie F, Uhlig H. Animal models of intestinal inflammation: clues to the pathogenesis of inflammatory bowel disease. Novartis Found Symp. 2004;263:164–174. discussion 174-168,211-168. [PubMed] [Google Scholar]
- 119.Bloom SM, et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell host & microbe. 2011;9(5):390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Iskandar HN, Ciorba MA. Biomarkers in inflammatory bowel disease: current practices and recent advances. Translational research : the journal of laboratory and clinical medicine. 2012;159(4):313–325. doi: 10.1016/j.trsl.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Haas A, et al. Systemic antibody responses to gut commensal bacteria during chronic HIV-1 infection. Gut. 2011;60(11):1506–1519. doi: 10.1136/gut.2010.224774. [DOI] [PubMed] [Google Scholar]
- 122.Cadwell K, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141(7):1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]