Abstract
Background
Use of drug-eluting stents (DES) has reduced in-stent restenosis after percutaneous coronary intervention (PCI); however, DES are associated with late stent thrombosis. There is no accurate way to predict in-stent restenosis, although risk factors for atherosclerosis overlap those for in-stent restenosis. Therefore, we evaluated atherosclerosis candidate genes for association with in-stent restenosis.
Methods
We identified 46 consecutive cases that had undergone PCI with bare-metal stents who subsequently developed symptomatic in-stent restenosis of the target lesion (≥75% luminal narrowing) within six months. Forty-six age-, race-, vessel-diameter- and sex-matched controls without in-stent restenosis after PCI with bare-metal stent were also identified. Single-nucleotide polymorphisms (SNPs, N=82) from 39 candidate atherosclerosis genes were genotyped. Multivariable logistic regression models were used to test for association.
Results
Five SNPs were associated with in-stent restenosis. Three ALOX5AP SNPs were most strongly associated, two with increased risk (OR 3.74, p=0.01; OR 3.46, p=0.02), and the third with decreased risk of in-stent restenosis (OR 0.09, p=0.004). Two ALOX5AP haplotypes were associated with in-stent restenosis (HapB: OR 3.13, p=0.03); and a haplotype similar to HapA: OR 0.14, p=0.0009).
Conclusions
ALOX5AP, a gene within the inflammatory leukotriene pathway linked to and associated with coronary atherosclerosis, is also associated with in-stent restenosis. Genotyping these variants may help identify those at risk for in-stent restenosis who would benefit most from use of DES.
Keywords: Genetics, Stent Restenosis, Coronary Artery Disease, Inflammation
Percutaneous coronary intervention (PCI) has become a mainstay of treatment for coronary atherosclerosis. Unfortunately 30% to 50% of patients undergoing balloon angioplasty soon develop restenosis. The routine use of stents has decreased the incidence, but it remains a significant problem, with 10–50% of patients receiving stents developing restenosis1. Clinical predictors of restenosis have been described and include diabetes mellitus, multiple stents, and minimal luminal diameter2. However, even applying these criteria in an ideal population, 16% of patients with none of these characteristics may have restenosis.
Clinical trials have shown that drug-eluting stents (DES) can reduce in-stent restenosis rates to very low levels3–5, resulting in widespread use of DES for PCI. More recently, however, several reports have highlighted the increased risk of late stent thrombosis with DES6,7, and the proportional use of DES versus bare metal stents has fallen substantially (CRUSADE database, accessed at Duke Clinical Research Institute, May 2007). Furthermore, routine use of DES in all patients undergoing PCI incurs a substantial incremental cost8.
Ideally, clinicians would be able to identify a subgroup of patients in which the risk-benefit assessment favors DES implantation. Given the lack of a strong clinical predictive model for restenosis, elucidation of non-conventional markers may help with identification of this subgroup. Genetic polymorphisms have been associated with development of coronary artery disease (CAD) and atherosclerosis risk factors (i.e. diabetes), and pathophysiologic mechanisms (i.e. inflammation and thrombosis) overlap those for restenosis. Therefore, in this study, we pursued a candidate gene association design to test the hypothesis that atherosclerosis susceptibility genetic variants are associated with risk of in-stent restenosis after PCI with bare-metal stents.
Methods
Study Population
The CATHGEN biorepository consists of subjects recruited sequentially through the cardiac catheterization laboratories at Duke University Medical Center (Durham, NC, USA). We restricted years of enrollment to 2001–2003, to ensure a time during which patients routinely and uniformly received a bare-metal stent. Cases were individuals who had undergone successful PCI with bare-metal stenting during or related to their index catheterization at enrollment in CATHGEN, who subsequently presented with symptomatic restenosis of the target lesion (≥75% luminal narrowing), documented by coronary angiography within six months post-PCI. Cases for which coronary stenting was performed in an emergent setting (i.e. ST-segment elevation MI) were excluded, as well as those receiving DES. Controls were defined as individuals who had undergone successful PCI with bare-metal stenting during or related to their index catheterization at enrollment in CATHGEN, who did not present with clinical restenosis within six months of PCI, who had at least six months of follow-up available, and who had no previous history of restenosis. Controls were matched as carefully as possible to cases on age, race, sex, and vessel-diameter (±0.5 mm). Review of medical records was performed by the same cardiologist to verify phenotypes. The Duke University Institutional Review Board approved study protocols and informed consent was obtained from each subject.
Candidate Gene Selection
Candidate genes were selected based on (1) review of the literature showing strong association with in-stent restenosis and/or atherosclerosis (7 genes); or (2) identification as a atherosclerosis susceptibility gene from two studies of gene expression: one performed in human aortas harvested from heart donors9, and the second in a mouse model of atherosclerosis10 (32 genes). Single-nucleotide polymorphisms (SNPs) within each gene were selected based on previous reports in the literature, or based on an average of two SNPs per gene, focusing on functional SNPs and SNPs with a minor allele frequency of >0.05 (Supplement I).
Genotyping
DNA was extracted using PureGene (Gentra Systems, Minneapolis, MN). SNPs were genotyped using either Taqman or Illumina BeadArray systems. The 7900HT Taqman genotyping system (Applied Biosystems, Foster City, CA) incorporates a standard PCR-based, dual fluor, allelic discrimination assay. QC samples, composed of 12 reference controls, were included in each quadrant of the plate. Illumina BeadStation genotyping was performed using the 500G system (Illumina, San Diego, CA). Within each individual experiment four QC samples were included. SNPs showing mismatches on QC samples were reviewed by an independent supervisor. All SNPs were successfully genotyped for 95% or more of the individuals in the study. Error rate estimates for SNPs meeting QC benchmarks were <0.2%.
Statistical Analysis
Association of genetic variants with restenosis was assessed using logistic regression, assuming dominant (allele) and additive (genotype) models. Models were adjusted for hypertension, diabetes, dyslipidemia, smoking, and body-mass-index (BMI). Because of concerns of confounding by extent of CAD, a second model was also constructed adjusting for CAD-index, a numerical summary of the extent of angiographic CAD11. The Graphical Overview of Linkage Disequilibrium (GOLD) program was used to assess linkage disequilibrium. Haplotype analysis was performed using HaploStats 1.1.0 (Mayo Clinic, Rochester, MN). Power calculations used QUANTO software (http://hydra.usc.edu/gxe). As all analyses were exploratory in nature, nominal two-sided p-values unadjusted for multiple comparisons are presented. Significance was defined as p-value ≤0.05. Statistical analyses used SAS version 9.1 (SAS Institute, Cary NC).
Results
Clinical Characteristics
Forty-six cases with clinical in-stent restenosis within six months of PCI were identified, as well as 46 matched controls. Consistent with previous reports, the only strong clinical factor that differentiated cases and controls was diabetes (Table I).
Table I.
Baseline characteristics.
| Variable | Overall (N=92) | Cases (N=46) | Controls (N=46) | p-value |
|---|---|---|---|---|
| Age (mean, SD) | 61.88 (10.64) | 62.09 (10.52) | 61.67 (10.88) | 0.8 |
| Sex (% female) | 33.7% | 34.8% | 32.6% | 0.8 |
| Dyslipidemia | 65.2% | 73.9% | 56.5% | 0.08 |
| Lipids (mean, SD) | ||||
| Total cholesterol | 188.42 (49.99) | 186.92 (57.41) | 190.23 (40.20) | 0.6 |
| Triglyerides | 190.18 (144.18) | 210.36 (172.26) | 165.97 (98.34) | 0.7 |
| HDL cholesterol | 41.69 (11.99) | 40.58 (13.70) | 43.03 (9.61) | 0.2 |
| LDL cholesterol | 104.56 (40.42) | 91.45 (30.32) | 123.29 (46.49) | 0.06 |
| Hypertension | 78.3% | 80.43% | 76.1% | 0.6 |
| Diabetes mellitus | 33.7% | 47.8% | 19.6% | 0.004 |
| CAD-index (mean, SD) | 44.27 (17.46) | 48.62 (19.24) | 40.02 (14.51) | 0.02 |
| Number of diseased vessels | ||||
| 1 | 42.9% | 33.3% | 52.2% | 0.09 |
| 2 | 33.0% | 33.3% | 32.6% | |
| 3 | 24.2% | 33.3% | 15.2% | |
| Vessel-diameter mm (mean, SD)* | 2.74 (0.48) | 2.64 (0.49) | 2.85 (0.45) | 0.04 |
| Vessel-length mm (mean, SD)* | 15.44 (5.64) | 15.07 (5.27) | 15.82 (6.04) | 0.6 |
| BMI (mean, SD) | 30.68 (5.56) | 30.90 (5.83) | 30.45 (5.34) | 0.7 |
| Smoking | 63.0% | 58.7% | 67.4% | 0.4 |
| History of MI | 50.0% | 43.5% | 56.5% | 0.2 |
| Race (% Caucasian) | 69.6% | 67.4% | 71.7% | 0.4 |
| Family history CAD | 45.7% | 41.3% | 50.0% | 0.4 |
for vessel that underwent PCI.
Association Results
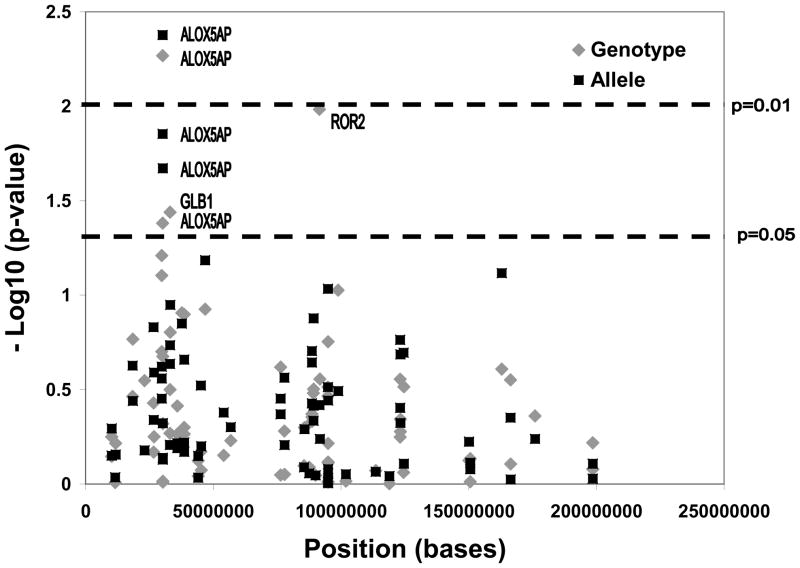
All SNPs were in Hardy-Weinberg equilibrium. A total of 82 SNPs within 39 genes were genotyped. Five SNPs within three genes were associated with in-stent restenosis in at least one of the models (Figure 1, Table II). Three of the SNPs (rs17222814 (G→A, minor allele frequency (MAF) 0.07); rs17216473 (G→A, MAF 0.19), and rs10507391 (T→A, MAF 0.49)), which were also the most significant SNPs, reside within the ALOX5AP gene. Rs17222814 was protective (allelic OR 0.089, p=0.004), whereas the other two ALOX5AP SNPs were associated with increased risk of in-stent restenosis (OR 3.743, p=0.01 and OR 3.436, p=0.02). After adjusting for CAD-index, all results remained significant suggesting that these results are specific for restenosis, and not merely reflective of association with CAD. No SNPs were in linkage disequilibrium (pairwise R2<0.3). One SNP within the ALOX5 gene (upstream in the leukotriene pathway) was included in this study, but was not significant. One SNP each in the genes ROR2 and GLB1 were also weakly associated with in-stent restenosis.
Figure 1. Association results, all SNPs.
Results for all SNPs for association with restenosis are displayed. Genomic position (X-axis) and negative log10 of the p-value (Y-axis) are noted. Gene annotations are made for SNPs meeting statistical significance.
Table II.
Odds ratios for significant SNPs.
| Gene | SNP | Type of SNP | Nucleotide | Adjusted* | Adjusted for CAD-index† | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Genotype | Allele | Genotype | Allele | ||||||||
|
| |||||||||||
| OR (95% CI) | p-val | OR (95% CI) | p-val | OR (95% CI) | p-val | OR (95% CI) | p-val | ||||
| ALOX5AP | RS17222814 | Upstream | G→A | 0.089 (0.016–0.506) | 0.004 | 0.089 (0.016–0.506) | 0.004 | 0.058 (0.009–0.383) | 0.003 | 0.058 (0.009–0.383) | 0.003 |
| ALOX5AP | RS17216473 | Upstream | G→A | 2.566 (1.054–6.251) | 0.04 | 3.743 (1.331–10.518) | 0.01 | 3.082 (1.205–7.881) | 0.02 | 5.097 (1.673–15.528) | 0.004 |
| ALOX5AP | RS10507391 | Upstream | T→A | 1.452 (0.791–2.666) | 0.21 | 3.436 (1.197–9.861) | 0.02 | 1.644 (0.864–3.129) | 0.13 | 3.605 (1.190–10.924) | 0.02 |
| ROR2 | RS10820899 | Intronic | G→A | 2.341 (1.215–4.510) | 0.01 | 1.537 (0.575–4.106) | 0.39 | 3.095 (1.467–6.532) | 0.003 | 1.838 (0.646–5.228) | 0.25 |
| GLB1 | RS9861960 | Intronic | G→A | 0.513 (0.275–0.957) | 0.04 | 0.525 (0.204–1.352) | 0.18 | 0.464 (0.242–0.890) | 0.02 | 0.406 (0.147–1.089) | 0.07 |
adjusted for smoking, body-mass-index, diabetes, hypertension and dyslipidemia, and
also adjusted for CADindex.
Multivariable Modeling
We sought to understand the additive predictive capabilities of these SNPs in addition to clinical factors. First, a logistic regression model inclusive of all clinical factors (diabetes, hypertension, smoking, BMI, family history of CAD, vessel-length, vessel-diameter, MI and CAD-index) was fit. In this model, only diabetes (OR (95% CI) 3.61 (1.28–10.18), p=0.02) and CAD-index (OR 1.04 (1.004–1.070), p=0.03) were predictive of restenosis. The c-statistic for this model was 0.76. A model was subsequently fit including all clinical factors, and then all significant SNPs were assessed using a backward stepwise regression fashion. In this model, two SNPs remained significantly associated with restenosis: rs17216473 (OR 5.11 (1.31–19.92), p=0.02) and rs17222814 (OR 0.07 (0.01–0.56), p=0.01), both in the ALOX5AP gene. The c-statistic for this model was 0.85, suggesting that these two ALOX5AP SNPs contribute independent information to the capacity of a clinical model to discriminate between risk of in-stent restenosis.
ALOX5AP Haplotype Analyses
Given the strong findings for ALOX5AP, we focused further efforts on understanding this pathway in restenosis risk. Haplotypes within ALOX5AP (HapA and HapB) and within a downstream gene, LTA4H (HapK) have previously been shown to be associated with MI12,13. Therefore, we genotyped additional SNPs to complete these haplotypes (Table III). The global p-value for the haplotype of the SNPs comprising HapA was significant (p=0.02), signifying that the cluster of SNPs overall is associated with restenosis. However, the combination of alleles for HapA itself was not significant, suggesting that the haplotype background of these specific HapA alleles is not associated with restenosis. Interestingly, a novel haplotype within the SNPs comprising HapA was associated (p=0.0009). This haplotype is composed of alleles that are the same as HapA except for rs17222814, where the allele is the opposite of HapA (A vs. G). This haplotype is protective for restenosis (OR 0.14), consistent with the prior HapA results (G allele associated with increased risk of MI). Within HapB SNPs, the global p-value was not significant; however, HapB itself was associated with increased risk of restenosis (OR 3.13, p=0.03). The SNPs comprising HapK were not associated with in-stent restenosis.
Table III.
Leukotriene pathway haplotypes.
| Haplotype | Global p-value† | SNPs | Haplotype Frequency | P-value† | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall | Cases | Controls | |||||||||||||
| ALOX5AP Three-way haplotype* | 0.003 | 1 | 2 | 3 | |||||||||||
|
| |||||||||||||||
| G | A | T | 0.06 | 0.02 | 0.11 | 0.0009 | |||||||||
| G | G | A | 0.30 | 0.31 | 0.29 | 0.81 | |||||||||
| G | G | T | 0.45 | 0.43 | 0.46 | 0.85 | |||||||||
| A | G | A | 0.19 | 0.24 | 0.14 | 0.01 | |||||||||
|
| |||||||||||||||
| ALOX5AP** | 0.02 | 1 | 2 | 3 | 4 | ||||||||||
|
| |||||||||||||||
| HapA | A | T | G | A | 0.066 | 0.109 | 0.022 | 0.0009 | |||||||
| G | T | G | C | 0.316 | 0.149 | 0.109 | 0.48 | ||||||||
| G | A | G | C | 0.129 | 0.351 | 0.280 | 0.59 | ||||||||
| G | A | A | C | 0.049 | 0.099 | 0.142 | 0.47 | ||||||||
| G | T | G | A | 0.121 | 0.033 | 0.067 | 0.28 | ||||||||
| G | A | G | A | 0.319 | 0.260 | 0.380 | 0.07 | ||||||||
|
| |||||||||||||||
| ALOX5AP** | 0.24 | 1 | 2 | 3 | 4 | ||||||||||
|
| |||||||||||||||
| HapB | G | T | A | C | 0.496 | 0.560 | 0.430 | 0.10 | |||||||
| G | A | G | C | 0.105 | 0.187 | 0.214 | 0.86 | ||||||||
| A | A | A | T | 0.079 | 0.109 | 0.100 | 0.89 | ||||||||
| G | A | A | C | 0.201 | NA | 0.014 | 0.64 | ||||||||
| G | T | A | T | 0.006 | 0.076 | 0.079 | 0.42 | ||||||||
| A | A | G | C | 0.008 | 0.000 | 0.016 | 0.23 | ||||||||
| A | A | A | C | 0.105 | 0.068 | 0.146 | 0.03 | ||||||||
|
| |||||||||||||||
| LTA4H** | 0.44 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||||
|
| |||||||||||||||
| HapK | C | G | T | G | A | T | T | C | G | G | 0.027 | 0.070 | NA | 0.15 | |
| T | G | T | G | G | C | G | T | A | G | 0.025 | 0.039 | NA | 0.22 | ||
| C | G | T | A | A | T | T | T | A | A | 0.014 | 0.017 | 0.011 | 0.25 | ||
| C | G | T | G | A | C | G | C | G | G | 0.008 | NA | NA | 0.28 | ||
| C | G | C | G | A | T | T | T | A | G | 0.006 | 0.012 | NA | 0.34 | ||
| C | G | C | G | A | T | G | C | G | A | 0.006 | 0.011 | NA | 0.38 | ||
| C | G | T | G | A | C | T | T | A | G | 0.012 | 0.019 | NA | 0.44 | ||
| C | G | T | G | A | C | T | T | G | G | 0.020 | 0.023 | 0.024 | 0.58 | ||
| T | G | T | G | G | C | G | C | G | G | 0.058 | 0.076 | 0.064 | 0.59 | ||
| T | A | T | G | A | C | G | T | A | A | 0.104 | 0.108 | 0.100 | 0.74 | ||
| T | G | T | G | G | C | G | T | A | A | 0.050 | 0.054 | 0.046 | 0.76 | ||
| C | G | T | A | A | T | T | T | A | G | 0.032 | 0.035 | 0.000 | 0.82 | ||
| C | G | T | G | A | T | T | T | G | G | 0.324 | 0.324 | 0.312 | 0.93 | ||
| C | G | T | G | A | C | T | C | G | G | 0.012 | NA | 0.022 | 0.91 | ||
| C | G | C | G | A | T | G | T | A | G | 0.006 | NA | 0.013 | 0.71 | ||
| C | G | T | G | A | T | T | T | A | G | 0.062 | 0.054 | 0.080 | 0.60 | ||
| C | G | T | A | A | T | T | C | G | G | 0.006 | NA | 0.011 | 0.57 | ||
| C | G | T | A | A | T | T | C | A | G | 0.028 | 0.000 | 0.028 | 0.55 | ||
| C | G | C | G | A | C | T | T | G | G | 0.006 | NA | 0.011 | 0.44 | ||
| C | A | T | G | A | C | G | T | A | A | 0.158 | 0.142 | 0.188 | 0.36 | ||
| T | G | T | G | A | T | T | T | A | G | 0.006 | NA | 0.011 | 0.31 | ||
| C | G | C | G | A | T | G | T | A | A | 0.025 | NA | 0.046 | 0.01 | ||
Three-way haplotype composed of the individually associated ALOX5AP SNPs (rs17216473, rs17222814, rs10507391).
Adjusted for hypertension, dyslipidemia, diabetes, smoking, BMI and CADindex.
ALOX5AP (HapA): rs17222814 (1), rs10507391 (2), rs476874 (3), rs9551963 (4)
ALOX5AP (HapB): rs17216473 (1), rs10507391 (2), rs9315050 (3), rs17222842 (4)
LTA4H (HapK): 12p0557 (1), rs2660880 (2), rs6538697 (3), rs1978331 (4), rs17677715 (5), rs2247570 (6), rs2660898 (7), rs2540482 (8), rs2660845 (9), rs2540475 (10)
NA: numbers too small to converge on estimate
Additionally, three-way haplotypes composed of the individually associated ALOX5AP SNPs (rs17216473, rs17222814, and rs10507391) were significant (global p=0.003). The haplotype composed of alleles associated with increased risk of in-stent restenosis at each SNP (A, G, A, respectively) was significant (p=0.01). Even more significant was the background of allele G, A and T (corresponding to the opposite allele at each SNP, hence a “protective” haplotype (overall frequency 0.07; cases 0.02, controls 0.11, p=0.0009)). These results suggest that this haplotype is more strongly associated than any individual SNP, with consistency of the associated alleles. Allele frequencies are presented (Table IV).
Table IV.
Minor Allele Frequencies for significant SNPs/haplotypes and for SNPs composing the haplotypes.
| Gene | SNP | Allele | MAF (overall) | MAF (cases) | MAF (controls) |
|---|---|---|---|---|---|
| ALOX5AP | RS17222814** | G→A | 0.07 | 0.02 | 0.11 |
| ALOX5AP | RS17216473†† | G→A | 0.19 | 0.23 | 0.14 |
| ALOX5AP | RS10507391† | T→A | 0.49 | 0.54 | 0.43 |
| Three-way haplotype* | -- | -- | 0.07 | 0.02 | 0.11 |
| HapB (ALOX5AP) | -- | -- | 0.11 | 0.15 | 0.07 |
| ALOX5AP novel haplotype | -- | -- | 0.07 | 0.02 | 0.11 |
| ROR2 | RS10820899 | G→A | 0.47 | 0.56 | 0.39 |
| GLB1 | RS9861960 | G→A | 0.44 | 0.37 | 0.51 |
| SNPs composing HapA (ALOX5AP) | RS4769874 | G→A | 0.05 | 0.07 | 0.03 |
| RS9551963 | A→C | 0.48 | 0.43 | 0.53 | |
| SNPs composing HapB (ALOX5AP) | RS9315050 | A→G | 0.11 | 0.11 | 0.11 |
| RS17222842 | C→T | 0.08 | 0.09 | 0.08 | |
| SNPs composing HapK (LTA4H) | 12P0557 | C→T | 0.21 | 0.19 | 0.23 |
| RS2660880 | G→A | 0.06 | 0.06 | 0.05 | |
| RS6538697 | T→C | 0.11 | 0.13 | 0.08 | |
| RS1978331 | G→A | 0.50 | 0.51 | 0.49 | |
| RS17677715 | A→G | 0.15 | 0.13 | 0.16 | |
| RS2247570 | T→C | 0.34 | 0.31 | 0.36 | |
| RS2660898 | T→G | 0.31 | 0.31 | 0.30 | |
| RS2540482 | T→C | 0.25 | 0.26 | 0.24 | |
| RS2660845 | A→G | 0.30 | 0.30 | 0.30 | |
| RS2540475 | G→A | 0.22 | 0.23 | 0.20 |
Haplotype composed of the three individually associated ALOX5AP SNPs (rs17216473 (G), rs17222814 (A), rs10507391 (T)).
SNP also included in HapA
SNP also included in both HapA and HapB
SNP also included in HapB
Discussion
In this study, we have taken a candidate gene approach to assess whether atherosclerosis susceptibility genes are also associated with risk of in-stent restenosis after PCI. We report a novel finding of association of individual variants and haplotypes within the ALOX5AP gene with in-stent restenosis, a gene previously linked to and associated with MI in multiple populations. These results persist even after adjustment for clinical factors, and two of the identified ALOX5AP SNPs add independent prognostic information and discriminative power to a model predicting restenosis inclusive of clinical factors. These results could have potentially significant clinical implications. Given the increased risk of stent thrombosis associated with DES, and lack of good clinical predictors of in-stent restenosis, identification of genetic variants for in-stent restenosis could help guide more judicious use of DES by identifying the subgroup in which the balance of risk and benefit would favor use of DES.
Previous studies have identified genes associated with in-stent restenosis14–18. In this study, we report a novel finding for strong association of ALOX5AP SNPs and haplotypes with in-stent restenosis. The individual SNPs are not known functional variants, but HapA heightens the response of 5-lipoxygenase (the protein product of the gene) to factors that stimulate inflammatory cells12. ALOX5AP was chosen for inclusion in the study based on differential expression of ALOX5 in our study of aortic atherosclerosis9 and previous linkage and association of ALOX5AP12 with MI. Although the leukotriene pathway has not been previously implicated in in-stent restenosis pathophysiology, inflammation is known to play an important role. Targeting inflammatory pathways may have utility in preventing in-stent restenosis19, and pharmacologic agents targeting the leukotriene pathway are available. Tranilast, an antiasthma drug that inhibits leukotriene C4 release from mast cells and macrophages20, and interferes with proliferation and migration of vascular SMCs21, has been shown to reduce neointimal thickening after vascular injury in animal models22. Human studies have shown varying results for tranilast on cardiovascular events23,24.
We also found that one SNP each in the ROR2 and GLB1 genes was associated with in-stent restenosis, though neither gene has any known role in cardiovascular disease. The protein encoded by ROR2 is a type I transmembrane protein and may be involved in the early formation of chondrocytes25. GLB1 encodes beta-galactosidase-1, a lysosomal hydrolase, and a variant within this gene causes GM1-gangliosidosis.
Our study has some limitations. None of the individual SNPs (lowest p=0.003) would survive a conservative Bonferroni adjustment for multiple comparisons at the gene level (p=0.001), but the HapA-related haplotype and the three-way haplotype would (p=0.0009). We do not believe this is a spurious result, as genes were carefully chosen from a comprehensive study of atherosclerosis, and the identification of three independent SNPs within the same gene lends further credence. Further, there was consistency of the associated allele both with previous studies, and across haplotypes that were associated. Power calculations demonstrate that our study has 80% power to detect an effect size of ≥2.3 for common SNPs (MAF 0.50), and effect sizes ≥4.0 for rarer SNPs (MAF 0.05.), therefore, we may not have had sufficient power for rarer SNPs or with more modest effect sizes. Our study lacks a true validation, however, prospective validation of these results are difficult given the routine use of DES. Furthermore, our results corroborate previous studies showing that these ALOX5AP SNPs and haplotypes are associated with MI, and thus our study is partially validated by other studies showing this pathway to be important in related atherosclerotic disease. Our study is remarkable for a very well-phenotyped cohort, and we took great care to adjust for potential confounders in our analyses. Finally, our results show that variants in the ALOX5AP gene can contribute significantly to predictive models of in-stent restenosis, adding independent information in addition to clinical factors.
In conclusion, we report herein a novel finding of association of ALOX5AP variants with risk of in-stent restenosis after PCI. These results could have significant clinical implications. In conjunction with clinical factors, genotyping of these ALOX5AP SNPs could help guide clinical decision-making and directed use of DES for PCI. Further studies and validation are warranted.
Supplementary Material
Acknowledgments
This work was supported by the American Heart Association (FTF, Shah) and the NIH (P01 HL73042, Goldschmidt-Clermont, Kraus). We are grateful for the time and effort devoted by study participants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Lowe HC, Oesterle SN, Khachigian LM. Coronary in-stent restenosis: current status and future strategies. J Am Coll Cardiol. 2002;39:183–193. doi: 10.1016/s0735-1097(01)01742-9. [DOI] [PubMed] [Google Scholar]
- 2.Kastrati A, Schomig A, Elezi S, et al. Predictive factors of restenosis after coronary stent placement. J Am Coll Cardiol. 1997;30:1428–1436. doi: 10.1016/s0735-1097(97)00334-3. [DOI] [PubMed] [Google Scholar]
- 3.Grube E, Silber S, Hauptmann KE, et al. TAXUS I: six- and twelve-month results from a randomized, double-blind trial on a slow-release paclitaxel-eluting stent for de novo coronary lesions. Circulation. 2003;107:38–42. doi: 10.1161/01.cir.0000047700.58683.a1. [DOI] [PubMed] [Google Scholar]
- 4.Holmes DR, Jr, Leon MB, Moses JW, et al. Analysis of 1-year clinical outcomes in the SIRIUS trial: a randomized trial of a sirolimus-eluting stent versus a standard stent in patients at high risk for coronary restenosis. Circulation. 2004;109:634–640. doi: 10.1161/01.CIR.0000112572.57794.22. [DOI] [PubMed] [Google Scholar]
- 5.Morice MC, Serruys PW, Sousa JE, et al. A randomized comparison of a sirolimus-eluting stent with a standard stent for coronary revascularization. N Engl J Med. 2002;346:1773–1780. doi: 10.1056/NEJMoa012843. [DOI] [PubMed] [Google Scholar]
- 6.Eisenstein EL, Anstrom KJ, Kong DF, et al. Clopidogrel use and long-term clinical outcomes after drug-eluting stent implantation. JAMA. 2007;297:159–168. doi: 10.1001/jama.297.2.joc60179. [DOI] [PubMed] [Google Scholar]
- 7.Stone GW, Moses JW, Ellis SG, et al. Safety and efficacy of sirolimus- and paclitaxel-eluting coronary stents. N Engl J Med. 2007;356:998–1008. doi: 10.1056/NEJMoa067193. [DOI] [PubMed] [Google Scholar]
- 8.Kong DF, Eisenstein EL. Decision models for assessing the cost effectiveness of drug-eluting stents. Expert Opin Pharmacother. 2005;6:965–974. doi: 10.1517/14656566.6.6.965. [DOI] [PubMed] [Google Scholar]
- 9.Seo D, Wang T, Dressman H, et al. Gene Expression Phenotypes of Atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24:1922–1927. doi: 10.1161/01.ATV.0000141358.65242.1f. [DOI] [PubMed] [Google Scholar]
- 10.Karra R, Vemullapalli S, Dong CM, et al. Molecular evidence for arterial repair in atherosclerosis. Proc Natl Acad Sci (USA) 2005;102:16789–16794. doi: 10.1073/pnas.0507718102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith LR, Harrell FE, Jr, Rankin JS, et al. Determinants of early versus late cardiac death in patients undergoing coronary artery bypass graft surgery. Circulation. 1991;84:III245–III253. [PubMed] [Google Scholar]
- 12.Helgadottir A, Manolescu A, Thorleifsson G, et al. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat Genet. 2004;36:233–239. doi: 10.1038/ng1311. [DOI] [PubMed] [Google Scholar]
- 13.Helgadottir A, Manolescu A, Helgason A, et al. A variant of the gene encoding leukotriene A4 hydrolase confers ethnicity-specific risk of myocardial infarction 1. Nat Genet. 2006;38:68–74. doi: 10.1038/ng1692. [DOI] [PubMed] [Google Scholar]
- 14.Agema WR, Jukema JW, Zwinderman AH, et al. A meta-analysis of the angiotensin-converting enzyme gene polymorphism and restenosis after percutaneous transluminal coronary revascularization: evidence for publication bias. Am Heart J. 2002;144:760–768. doi: 10.1067/mhj.2002.125509. [DOI] [PubMed] [Google Scholar]
- 15.Gomma AH, Elrayess MA, Knight CJ, et al. The endothelial nitric oxide synthase (Glu298Asp and −786T>C) gene polymorphisms are associated with coronary in-stent restenosis. Eur Heart J. 2002;23:1955–1962. doi: 10.1053/euhj.2002.3400. [DOI] [PubMed] [Google Scholar]
- 16.Humphries S, Bauters C, Meirhaeghe A, et al. The 5A6A polymorphism in the promoter of the stromelysin-1 (MMP3) gene as a risk factor for restenosis. Eur Heart J. 2002;23:721–725. doi: 10.1053/euhj.2001.2895. [DOI] [PubMed] [Google Scholar]
- 17.Koch W, Bottiger C, Mehilli J, et al. Association of a CD18 gene polymorphism with a reduced risk of restenosis after coronary stenting. Am J Cardiol. 2001;88:1120–1124. doi: 10.1016/s0002-9149(01)02045-8. [DOI] [PubMed] [Google Scholar]
- 18.Rauchhaus M, Gross M, Schulz S, et al. The E-selectin SER128ARG gene polymorphism and restenosis after successful coronary angioplasty. Int J Cardiol. 2002;83:249–257. doi: 10.1016/s0167-5273(02)00073-6. [DOI] [PubMed] [Google Scholar]
- 19.Ohtani K, Egashira K, Nakano K, et al. Stent-based local delivery of nuclear factor-kappaB decoy attenuates in-stent restenosis in hypercholesterolemic rabbits. Circulation. 2006;114:2773–2779. doi: 10.1161/CIRCULATIONAHA.105.582254. [DOI] [PubMed] [Google Scholar]
- 20.Komatsu H, Kojima M, Tsutsumi N, et al. Mechanism of inhibitory action of tranilast on the release of slow reacting substance of anaphylaxis (SRS-A) in vitro: effect of tranilast on the release of arachidonic acid and its metabolites. Jpn J Pharmacol. 1988;46:53–60. doi: 10.1254/jjp.46.53. [DOI] [PubMed] [Google Scholar]
- 21.Capper EA, Roshak AK, Bolognese BJ, et al. Modulation of human monocyte activities by tranilast, SB 252218, a compound demonstrating efficacy in restenosis. J Pharmacol Exp Ther. 2000;295:1061–1069. [PubMed] [Google Scholar]
- 22.Fukuyama J, Ichikawa K, Miyazawa K, et al. Tranilast suppresses intimal hyperplasia in the balloon injury model and cuff treatment model in rabbits. Jpn J Pharmacol. 1996;70:321–327. doi: 10.1254/jjp.70.321. [DOI] [PubMed] [Google Scholar]
- 23.Hsu YS, Tamai H, Lleda K. Efficacy of tranilast on restenosis after coronary stenting. Circulation. 1996;94:I-620. [Google Scholar]
- 24.Holmes DR, Jr, Savage M, Lablanche JM, et al. Results of Prevention of REStenosis with Tranilast and its Outcomes (PRESTO) trial. Circulation. 2002;106:1243–1250. doi: 10.1161/01.cir.0000028335.31300.da. [DOI] [PubMed] [Google Scholar]
- 25.DeChiara TM, Kimble RB, Poueymirou WT, et al. Ror2, encoding a receptor-like tyrosine kinase, is required for cartilage and growth plate development. Nat Genet. 2000;24:271–274. doi: 10.1038/73488. [DOI] [PubMed] [Google Scholar]
- 26.Kastrati A, Koch W, Berger PB, et al. Protective role against restenosis from an interleukin-1 receptor antagonist gene polymorphism in patients treated with coronary stenting. J Am Coll Cardiol. 2000;36:2168–2173. doi: 10.1016/s0735-1097(00)01014-7. [DOI] [PubMed] [Google Scholar]
- 27.Feldman LJ, Mazighi M, Scheuble A, et al. Differential expression of matrix metalloproteinases after stent implantation and balloon angioplasty in the hypercholesterolemic rabbit. Circulation. 2001;103:3117–3122. doi: 10.1161/01.cir.103.25.3117. [DOI] [PubMed] [Google Scholar]
- 28.Kosokabe T, Okumura K, Sone T, et al. Relation of a common methylenetetrahydrofolate reductase mutation and plasma homocysteine with intimal hyperplasia after coronary stenting. Circulation. 2001;103:2048–2054. doi: 10.1161/01.cir.103.16.2048. [DOI] [PubMed] [Google Scholar]
- 29.Zheng F, Chevalier JA, Zhang LQ, et al. An HphI polymorphism in the E-selectin gene is associated with premature coronary artery disease. Clin Genet. 2001;59:58–64. doi: 10.1034/j.1399-0004.2001.590110.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.