Abstract
The eukaryotic Y-box binding protein YB-1 is involved in various biological processes, including DNA repair, cell proliferation and the regulation of transcription and translation. YB-1 protein is abundant and expressed ubiquitously in human cells, functioning in cell proliferation and transformation. Its concentration is thought to be highly regulated at both the levels of transcription and translation. Therefore, we investigated whether or not the 5′-UTR of YB-1 mRNA affects the translation of YB-1 protein, thus influencing expression levels. Luciferase mRNA ligated to the YB-1 mRNA 5′-UTR was used as a reporter construct. Ligation of the full-length YB-1 5′-UTR (331 bases) enhanced translation as assessed by in vitro and in vivo translation assays. Deletion constructs of the YB-1 5′-UTR also resulted in a higher efficiency of translation, especially in the region mapped to +197 to +331 from the major transcription start site. RNA gel shift assays revealed that the affinity of YB-1 for various 5′-UTR probe sequences was higher for the full-length 5′-UTR than for deleted 5′-UTR sequences. An in vitro translation assay was used to demonstrate that recombinant YB-1 protein inhibited translation of the full-length 5′-UTR of YB-1 mRNA. Thus, our findings provide evidence for the autoregulation of YB-1 mRNA translation via the 5′-UTR.
INTRODUCTION
Y-box proteins function as transcriptional and translational regulators of gene expression. They are found among prokaryotes and eukaryotes and are characterized by the evolutionary conservation of a cold shock domain (CSD). Recently, it was reported that a major protein component of messenger ribonucleoprotein (mRNP) particles in somatic cells is a member of the Y-box binding transcription factor family. This protein acts either as a repressor or an activator of protein synthesis (1–4). It has been hypothesized that YB-1 might play a role in promoting cell proliferation through the transcriptional regulation of various genes, including epidermal growth factor receptor, thymidine kinase, DNA topoisomerase II and DNA polymerase (5,6). The multiple biological roles of YB-1 include the modification of chromatin, the translational masking of mRNA, participation in a redox signaling pathway, RNA chaperoning and regulation of the stress response (7).
It has also been demonstrated that eukaryotic Y-box proteins regulate gene expression at the level of translation by binding directly to RNA (8,9). The rabbit Y-box protein, p50, is found in cytoplasmic mRNP particles in somatic cells and regulates translation by interacting with mRNA (2). The murine MSY1 protein and chicken Y-box protein both regulate transcription and translation (7,10–12). Furthermore, the Y-box family proteins, Xenopus mRNP3/mRNP4 and mouse MSY2, have also been found to be mRNA-masking proteins in germinal cells (13–15). Chen et al. (16) have reported that YB-1 is involved in the mRNA stability of the cdk4 gene; this stability is achieved by the binding of YB-1 to a specific sequence of the mRNA. YB-1 also stabilizes cap-dependent mRNA, since depletion of YB-1 results in accelerated mRNA decay (17).
Previously, we identified several proteins as partners of YB-1, including YB-1 itself, iron regulatory protein 2 (IRP2) and the ribosomal proteins S3A, L18A, L5, L23A and S5. We also provided evidence that YB-1 is involved in the translational regulation of an iron-related protein (18). Y-box binding proteins thus appear to perform critical functions in both mRNA turnover and translational control. Considering the important cellular functions of YB-1, it is possible that its expression is highly regulated in eukaryotes. In fact, the YB-1 gene behaves like a primary response gene. Stimulation of mammalian cell proliferation in culture or in vivo results in increased YB-1 synthesis. The cellular level of YB-1 is usually controlled by regulating the translation of its mRNA. It is thought that an increase in the cellular YB-1 concentration could alter the translation and stability of some mRNAs. Therefore, several pathways exist to control the function of this important cellular protein.
The 5′- and 3′-untranslated regions (UTRs) of eukaryotic mRNAs are known to play a crucial role in post-transcriptional regulation that modulates nucleo-cytoplasmic mRNA transport, translation efficiency, subcellular localization and stability (19). Several regulatory signals have already been identified within the 5′- or 3′-UTR sequences (20). These signals tend to correspond to short oligonucleotide tracts, able to fold into specific secondary structures which provide binding sites for various regulatory proteins (21–23).
To examine how YB-1 mRNA translation is regulated in eukaryotic cells, we examined the possible role of its relatively long 5′-UTR. Deletion of the YB-1 mRNA 5′-UTR enhances translational activity in both in vitro and in vivo systems. The affinities of YB-1 for 5′-UTR probe sequences of various lengths were evaluated by RNA gel shift assays; the affinity of YB-1 was higher for the full-length 5′-UTR than for truncated sequences. The addition of recombinant YB-1 inhibited translation through the 5′-UTR of its mRNA; this effect was particularly marked when the full-length 5′-UTR was used. In this study, we have demonstrated for the first time that the 5′-UTR region of human YB-1 mRNA plays an important role in determining the conditions of YB-1 biosynthesis at the translational level.
MATERIALS AND METHODS
Construction of fusion protein expression plasmids
The plasmids containing full-length glutathione S-transferase (GST)–YB-1 cDNA fusions, GST–YB-1 deletion mutants and Flag–YB-1 were described previously (24–26).
Reporter gene constructs
A pT7 control plasmid, for in vitro transcription and translation experiments, was constructed by digesting luciferase cDNA of a pGL3 basic vector (Promega, Madison, WI) with EcoRI, blunting with Klenow enzyme, and ligation to pT7Blue3 (Novagen, Madison, WI). The pT7-YB5′-1 plasmid was constructed as follows. The entire length of the YB-1 5′-UTR was amplified by PCR from human YB-1 cDNA. The forward primer was 5′-AGGCAGGAACGGTTGTAGGT-3′ and the reverse primer was 5′-gtttttggcgtcttccat GGTTGCGGTGATGG-3′. The latter contains a luciferase coding sequence at the 5′-end (shown in lower case). A luciferase cDNA fragment was also amplified by PCR from a pGL3 basic vector, using the forward primer 5′-CCATCACCGCAACCatggaagacgccaaaaac-3′, complementary to the reverse primer of the YB-1 5′-UTR and the reverse primer 5′-ttacacggcgatctttcc-3′. Each PCR-amplified fragment was ligated with the complementary primer regions and amplified by PCR using the complementary primer pair. The YB-1 5′-UTR-ligated luciferase cDNA fragment was cloned into the EcoRI-digested pT7Blue3 vector in order to generate plasmid pT7-YB5′-1. To functionally characterize the 5′-UTR of the human YB-1 gene, a series of 5′-deletion plasmids (pT7-YB5′-2–pT7-YB5′-6) were amplified by PCR using the pT7-YB5′-1 plasmid as a template. The forward primers were 5′-AAGGTCCAATGAGAATGGAGGA-3′ (pT7-YB5′-2), 5′-AAGCTAGGGATTGGGGTCAG-3′ (pT7-YB5′-3), 5′-CCT AGGGCGGGTCGCTCGTA-3′ (pT7-YB5′-4), 5′-CGATCG GTAGCGGGAGCGGAG-3′ (pT7-YB5′-5) and 5′-CCG CCGCCGCCGGCC-3′ (pT7-YB5′-6). Each of the PCR- amplified fragments were cloned into the EcoRI-digested pT7Blue3 vector to generate the pT7-YB5′-2–pT7-YB5′-6 plasmids. To construct pCMV and pCMV-YB5′-1–pCMV-YB5′-6 plasmids suitable for expression in mammalian cells, the pT7 or pT7-YB5′-1–pT7-YB5′-6 plasmids were digested with EcoRI. The fragments of luciferase cDNA were ligated into the EcoRI-digested pIRES-EYFP vector (Clontech, Palo Alto, CA), using various sizes of the YB-1 5′-UTR region (pCMV-YB5′-1–pCMV-YB5′-6); a non-ligated fragment (pCMV) was used as a control.
Site-directed mutagenesis of the YB-1 5′-UTR in pT7-YB5′-1 was performed using a PCR-based method. To obtain pT7-YBM1–pT7-YBM3, the full length of the YB-1 5′-UTR sequence was first amplified by PCR. The forward primers were 5′-GGTGGGCAGTACATCAGTACCACTGG-3′ (pT7-YBM1), 5′-GCGGGTCGCTAGAGAGGCTTATCCCGC-3′ (pT7-YBM2), 5′-CATTCTCGCTAGAACAGTCGGTAG CGGG-3′ (pT7-YBM3) and the reverse primers were 5′-CCAGTGGTACTGATGTACTGCCCACC-3′ (pT7-YBM1), 5′GCGGGATAAGCCTCTCTAGCGACCCGC-3′ (pT7-YBM2) and 5′-CCCGCTACCGACTGTTCTAGCGAGA ATG-3′ (pT7-YBM3). A second PCR was then performed with Taq polymerase using the first PCR products as templates. The PCR products were cloned into the EcoRI-digested pT7Blue3 vector in order to generate the pT7-YBM1–pT7-YBM3 plasmids. All constructs were confirmed by sequencing using a DNA sequencing system (model 377; Applied Biosystems, Foster City, CA).
Cell lines
A human epidermoid cancer cell line, KB3-1, was cultured in MEM supplemented with 10% newborn calf serum. The human lung cancer cell line H1299 was cultured in RPMI supplemented with 10% fetal bovine serum (FBS). HUVECs were isolated from individual human umbilical cord veins by collagenase digestion and were routinely cultured on type 1 collagen-coated plates in endothelial cell growth medium (Clonetics, Boston, MA) supplemented with 2% FBS. Tissue samples were obtained under an Institutional Review Board approved protocol, after the subjects had provided informed consent. The cells were maintained under standard cell culture conditions at 37°C and 5% CO2 in a humid environment.
Recombinant proteins and antibodies
Recombinant proteins were expressed in Escherichia coli DH5α. YB-1 and YB-1 deletion mutants were purified as GST fusion proteins as described previously (25). Briefly, GST fusion protein expressed in bacteria was induced by incubation with isopropyl-1-thio-β-d-galactopyranoside and cells were lysed by sonication in 1 ml of binding buffer [1 mM ditiothreitol, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 200 mM NaCl, 10% v/v glycerol, 1% Triton-X, in phosphate-buffered saline (PBS) pH 7.3]. Cellular debris was removed by centrifugation and the supernatants were subjected to affinity column chromatography using glutathione–Sepharose 4B (Amersham Biosciences, Piscataway, NJ) according to the manufacturer’s recommendations. Antibody to YB-1 was generated as described previously (27).
Primer extension by reverse transcriptase
The primer extension experiments were carried out as described previously (28). Total RNA was prepared from each cell line using an RNeasy Miniprep Kit (Qiagen, Chatsworth, CA) and a QIAshredder microspin homogenizer according to the manufacturer’s recommendations. The poly(A)+ RNA was isolated from the total RNA using a Poly(A)+ Isolation Kit from Total RNA (Nippon Gene Co. Ltd, Tokyo, Japan). The primer for the primer extension analysis, 5′-GCTCATGGTTGCGGTGATGG-3′, was synthesized to hybridize the sense strand between nucleotides –14 and +6 in the first exon of the YB-1 gene. The synthetic primer was labeled at its 5′-end with [γ-32P]ATP using T4 polynucleotide kinase and hybridized to 1 µg poly(A)+ RNA in 80% formamide, 0.4 M NaCl, 40 mM PIPES (pH 6.4) and 1 mM EDTA for 4 h at 50°C. After precipitation, the nucleic acid pellet was dissolved in reverse transcriptase buffer (Invitrogen Corp., Carlsbad, CA). The primer was extended with 200 U of SuperScript II RNase H– reverse transcriptase (Invitrogen) using 1 mM each of the four deoxynucleotides. After 1 h at 37°C, the reaction was neutralized and the DNA was collected. The reaction products were analyzed on a 7 M urea–8% polyacrylamide gel to determine the size of the extended product. The gel was exposed to an imaging plate and the blots were visualized and quantified using a phosphorimaging analyzer (model BAS 2000; Fuji Photo Film Co., Tokyo, Japan) and the Image Gauge (version 3.4) program.
RNA immunoprecipitation assay
Cells (100 mm dishes) were transfected with 5 µg of Flag–YB-1 plasmid DNA using LipofectAMINE 2000 reagent (Invitrogen). After 48 h, the cell extract was preincubated with protein A/G–agarose in TNE buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 1 mM PMSF, 10 µg/ml leupeptin, 10 µg/ml aprotinin] for 1 h at 4°C with rotation. After centrifugation, the supernatant was incubated with anti-Flag M2-agarose affinity gel (Sigma Chemical Co., St Louis, MO) for 12 h at 4°C with rotation in TNE buffer. The beads were washed four times with TNE buffer. After centrifugation, total RNA was extracted from the precipitate using a RNeasy Miniprep Kit (Qiagen). Total mRNAs were reverse transcribed and amplified by PCR using the ThermoScript RT–PCR System (Invitrogen). The following YB-1- and β-actin-specific primer pairs were used: the forward primers were 5′-ACCACAGTATTCCAACCC TCCTG-3′ (YB-1) and 5′-CTGGCACCACACCTTCTA CAATG-3′ (β-actin) and the reverse primers were 5′-ATC TTCTTCATTGCCGTCCTCTC-3′ (YB-1) and 5′-ATAGC AACGTACATGGCTGGGG-3′ (β-actin). The RT–PCR amplification products were analyzed by 2% agarose gel electrophoresis.
RNA band shift assays
The RNA electrophoretic mobility shift assay (REMSA) was carried out according to established techniques (29). Briefly, 32P-labeled YB-1 5′-UTR probe was transcribed in vitro from the plasmid pT7blue3, which contains a sequence corresponding to the 5′-UTR of YB-1. An aliquot of 2 µg of the linearized plasmid was transcribed in vitro by T7 RNA polymerase in the presence of [α-32P]UTP. The DNA template was removed by digestion with DNase I and the YB-1 5′-UTR probe was then extracted by column chromatography. To form RNA–protein complexes, 1–10 µg of cytoplasmic protein or the indicated amount of purified GST–YB-1 was incubated with 32P-labeled YB-1 5′-UTR probe at 25°C for 15 min. Next, the samples underwent electrophoresis through a 4% non-denaturing polyacrylamide gel (polyacrylamide:bisacrylamide, 80:1) in Tris–borate buffer. For the supershift experiments, 2 µg of the YB-1 antibody was incubated with cytoplasmic protein or purified GST–YB-1 at 25°C for 5 min before adding the 32P-labeled YB-1 5′-UTR probe. The gels were dried, visualized and then quantified as described above for the primer extension analysis.
In vitro transcription and translation
An aliquot of 2 µg of plasmid pT7Blue3, which encodes both luciferase cDNA ligated to the YB-1 mRNA 5′-UTR region (YB-1 5′-UTR–luciferase) as well as luciferase cDNA not ligated to the YB-1 mRNA 5′-UTR region (luciferase) (see Fig. 2A), was transcribed in vitro using an In vitro Transcription System (Promega). The DNA template was removed by digestion with DNase I and the RNA was purified by phenol/chloroform extraction. The integrity of the RNA was then examined using an Agilent 2000 Bioanalizer (Yokogawa, Osaka, Japan). Then, 50 ng of each RNA was translated using a rabbit reticulocyte lysate system (Promega). The luciferase assay was performed after incubation for 1.5 h at 30°C as described previously (18). To characterize the translation inhibition of each of the reporter constructs, the indicated amounts of GST fusion proteins were added to the rabbit reticulocyte lysate system either initially or after 10 min of incubation using 50 ng of the RNA constructs. The in vitro translation was performed for 30 min at 30°C and the translation activity of each experiment was measured. Data are shown as the means ± SD from three independent experiments.
Figure 2.
Effect of deletions of the 5′-UTR of YB-1 on the expression of a luciferase reporter in vitro and in vivo. (A) Schematic representation of the reporter constructs containing luciferase cDNA ligated to the YB-1 5′-UTR region (pT7-YB5′-1–pT7-YB5′′-6) fragments of various sizes, as well as the non-ligated control construct (pT7). The YB-1 5′-UTR region (pT7-YB5′-1–pT7-YB5′′-6) fragments were enlarged to show the limits of regions of the 5′-UTR of YB-1. The right-angled arrow denotes the start site and direction of transcription. Each of the reporter plasmids was transcribed in vitro in a reaction driven by T7 RNA polymerase and then 50 ng of the RNA constructs were translated using the rabbit reticulocyte lysate system. The luciferase assay was performed after incubation for 90 min at 30°C. (B) Schematic representation of the bicistronic reporter constructs containing luciferase cDNA ligated to the YB-1 5′-UTR region (pCMV-YB5′-1–pCMV-YB5′-6) fragments of various sizes, as well as the non-ligated control construct (pCMV). Each of the reporter plasmids was transcribed under the control of the human cytomegalovirus early promoter (CMV). The right-angled arrow indicates the start site and direction of translation. H1299 cells were transfected with these constructs and their luciferase activities were measured. To standardize the translation efficiency, relative luciferase activity was expressed as the ratio of downstream cistron expression to upstream cistron expression (luciferase/EYFP). Data are shown as the means ± SD (error bars) of three independent experiments.
Northern blot analysis
To detect in vitro transcribed RNA, a luciferase cDNA was used as the probe. The luciferase cDNA was obtained by digesting the pGL3-Basic vector (Promega) with NarI and XbaI. Reaction mixtures (20 µl, as described above) containing 50 ng of in vitro transcribed RNA were preincubated or treated after 10 min incubation with 5 pmol of GST–YB-1. After incubation for 30 min in a rabbit reticulocyte lysate system, total RNA was extracted using an RNeasy Miniprep Kit (Qiagen). RNA samples (0.5 µg/lane) were separated on a 1% formaldehyde–agarose gel and transferred onto Biodyne B membranes (Pall, Port Washington, NY). The membrane was prehybridized and hybridized with [α-32P]dCTP-labeled probe. Radioactivity was analyzed by autoradiograpy.
Transfections and luciferase assays
Cells underwent transient transfection using the LipofectAMINE method. Human lung cancer H1299 cells were plated at a density of 1 × 105 cells/35 mm well the day before transfection. At ∼80–90% confluence, the cells were transfected with 1 µg of reporter plasmid DNA or control vector using LipofectAMINE 2000 reagent (Invitrogen). Three hours later, the cells were washed twice with PBS and placed in fresh medium. Twenty-four hours post-transfection, the luciferase activity was measured as described below. The luciferase activity of the transfected cells was measured using the Dual Luciferase Assay System (Promega). Briefly, cells were lysed with 250 µl of 1× Passive Lysis Buffer (Promega). After a brief centrifugation, 10 µl of each supernatant was assayed for luciferase activity. Light emission was measured for 15 s with a luminometer. To standardize translation efficiency, the relative luciferase activity was expressed as the ratio of downstream cistron expression to upstream cistron expression (luciferase/EYFP). The fluorescence of enhanced yellow fluorescent protein (EYFP) was excited at 488 nm and measured for 1 s using a fluorometer.
RESULTS
Analysis of the multiple transcription initiation sites of the YB-1 gene
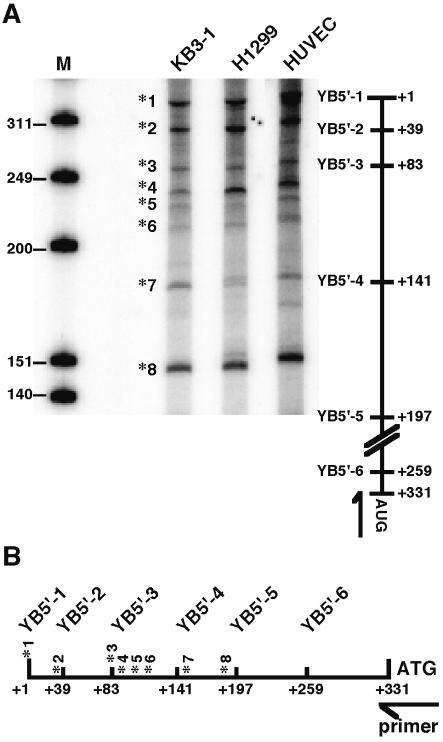
To investigate the mechanisms of translational control regulating YB-1 protein levels, the possible involvement of YB-1 mRNA was investigated. The 5′-UTR has a high G+C content (61%), suggesting that it could assume a high level of secondary structure in vivo. Computer modeling of potential secondary structures suggested that structures with free energies (ΔG values) lower than –190 kcal/mol could be formed (data not shown). We previously identified several transcription initiation sites for the YB-1 gene in KB3-1 and T24 cells (28). To compare the major transcription initiation sites in other cell lines, primer extension analysis was performed (Fig. 1A). The lung cancer cell line H1299 and endothelial cells (HUVECs), were compared with KB3-1 cells. The transcription initiation sites (*) were identical in all of the cells and eight transcripts (*1–*8) were observed in the region mapped to +1 to +197 (Fig. 1B). The ratio of the transcripts differed in each cell line. The proportion of transcripts starting at initiation site *1 was significantly higher in HUVECs, compared to the other cancer cells. These results suggest that the multiple transcription start sites of YB-1 mRNA observed in cell lines may be involved in the regulation of YB-1 protein expression.
Figure 1.
Analysis of the transcription start site of the YB-1 gene by primer extension assay. (A) Primer extension assay. Hybridization of the primer extension was performed with 5′-labeled oligonucleotide and 1 µg poly(A)+ RNA of each cell line. The markers shown on the left are end-labeled HinfI fragments of φ-X174 DNA. Black asterisks (*) indicate transcription initiation start sites. The 5′ furthest transcription initiation start site is indicated as +1 and the first AUG codon is indicated at +331. The arrow indicates the position of the primer that was used for extension. The primer hybridizes the sense strand between nucleotides +317 and +337 of the YB-1 gene. (B) Schematic distribution of the transcription initiation site in the YB-1 5′-UTR.
The 5′-UTR of YB-1 mRNA increases the expression of a luciferase reporter in vitro and in vivo
To study the possible involvement of the 5′-UTR of YB-1 mRNA in translation control, we generated two types of reporter constructs. (i) Those containing luciferase cDNA ligated to the full-length 5′-UTR of YB-1 gene (pT7-YB5′-1) and (ii) the non-ligated luciferase control construct (pT7) (Fig. 2A). The predominant 5′ transcription initiation site is designated +1 in the text and figures. Additional experiments were performed to investigate the regulatory region of the 5′-UTR of YB-1 mRNA. A total of five serial deletion mutants (pT7-YB-5′-2–pT7-YB5′-6) were constructed, each corresponding to a particular transcription initiation site. We first cloned transcripts *1, *2, *4, *7 and *8, which were each expressed at up to 10% of the total. These transcripts corresponded to YB5′-1–YB5′-5. We made an additional probe, YB5′-6, for detection of the shortest 5′-UTR fragment of YB-1. In vitro transcription reactions were performed on each of these constructs using T7 RNA polymerase and the integrity of the transcripts was confirmed by gel electrophoresis (data not shown). The differences in the transcript sizes were consistent with the length of the 5′-UTR (Fig. 2A). Equal amounts of the transcripts were translated in rabbit reticulocyte lysate and the luciferase activity was measured. The presence of the full-length 5′-UTR of YB-1 mRNA by itself increased the level of luciferase activity ∼2-fold relative to that of the control mRNA (pT7). Furthermore, each of the YB-1 5′-UTR deletion constructs showed higher translational activity than did the pT7 control. Of all the constructs, pT7-YB5′-5 showed the highest activity, which was ∼4-fold greater than that of the control construct (Fig. 2A).
We next determined whether a similar increase in activity could also be induced in cultured cells by the 5′-UTR. We constructed eukaryotic expression vectors containing luciferase cDNA ligated to various regions of the YB-1 5′-UTR (pCMV-YB5′-1–pCMV-YB5′-6), as described for the in vitro experiment (Fig. 2A). The reporter constructs were transcribed under control of the human cytomegalovirus early promoter. After transfection into a H1299 lung carcinoma cell line, the levels of luciferase activity were compared (Fig. 2B). As observed with the in vitro translation assays (Fig. 2A), the YB-1 5′-UTR increased the level of luciferase expression. The full-length YB-1 5′-UTR by itself increased luciferase activity ∼2-fold compared to the control construct (pCMV). Furthermore, pCMV-YB5′-5 also showed the highest activity, which was 4–5-fold that of the control. The mRNAs encoded by the reporter constructs were equally expressed, as determined by northern blot analysis (data not shown). These results suggest that the YB-1 5′-UTR enables more efficient translation of mRNA in vitro and in vivo and the short-length construct (YB5′-5) facilitates the most efficient translational activity.
YB-1 binds its own mRNA in the cytoplasm
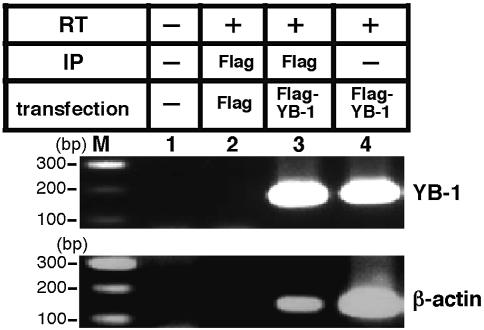
YB-1, which posseses RNA binding activity (26), has been reported to be involved in translational regulation and in the regulation of mRNA stability (16). In order to identify whether or not YB-1 protein interacts with its own mRNA in the cytoplasm, we performed RT–PCR using mRNA isolated by co-immunoprecipitation with YB-1 immunoprecipitant (Fig. 3). Flag–YB-1 or an empty Flag expression vector were transfected into an H1299 lung carcinoma cell line. After 48 h, cells were lysed and YB-1 proteins were immunoprecipitated with Flag antibody. The mRNA was purified after immunoprecipitation and amplified by RT–PCR using YB-1- and β-actin-specific primers. YB-1 mRNA and β-actin mRNA were both expressed in the H1299 cell lines (Fig. 3, lane 4). Flag–YB-1 immunoprecipitates contained YB-1 mRNA (Fig. 3, lane 3), while the control Flag immnoprecipitate did not (Fig. 3, lane 2). Furthermore, the YB-1 immunoprecipitate also contained β-actin mRNA (Fig. 3, lane 3). These data provide evidence that YB-1 interacts with its own mRNA and β-actin mRNA in the cytoplasm of cultured cells.
Figure 3.
YB-1 interacts with its own mRNA in the cytoplasm. Flag–YB-1 or Flag expression vector was transfected into H1299 cells. After 48 h, cells were lysed and the YB-1 proteins were immunoprecipitated (IP) with Flag antibody. The mRNA were purified after immunoprecipitation and amplified by RT–PCR with YB-1- (upper panel) and β-actin-specific (lower panel) primers. The RT–PCR amplification products were analyzed by 2% agarose gel electrophoresis.
YB-1 binds to the 5′-UTR of its cognate mRNA through a C-terminal domain
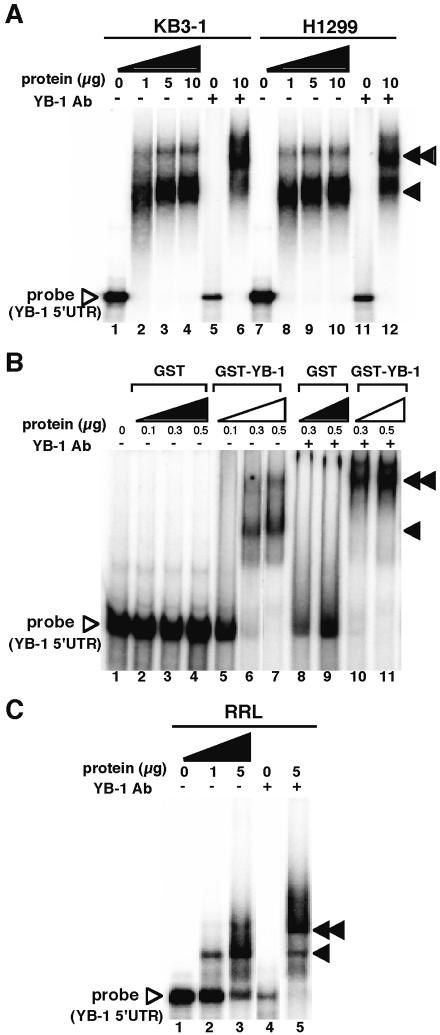
We next investigated the interaction of YB-1 protein with the 5′-UTR of its own mRNA in vitro. An RNA gel shift assay (REMSA) was performed by using KB3-1 and H1299 cell lysates and an in vitro synthesized mRNA probe corresponding to the full-length 5′-UTR of YB-1 mRNA (Fig. 4A). YB-1 5′-UTR formed an RNA–protein complex with lysates made using KB3-1 and H1299 cell lysates (Fig. 4A, lanes 2–4 and 8–10). The presence of endogenous YB-1 protein in the major complex was confirmed by the ability of a YB-1-specific antibody to supershift most of the complex (Fig. 4A, lanes 6 and 12). To determine whether YB-1 protein is able to directly interact with the 5′-UTR of YB-1 mRNA, we performed a REMSA using purified recombinant YB-1 (Fig. 4B). Recombinant YB-1 also clearly bound to the 5′-UTR of YB-1 mRNA (Fig. 4B, lanes 5–7), while control GST protein did not (Fig. 4B, lanes 2–4). The interaction with YB-1 protein was also confirmed by the use of YB-1-specific antibody (Fig. 4B, lanes 10 and 11).
Figure 4.
YB-1 binds to the 5′-UTR region of YB-1mRNA. (A) Endogenous YB-1 protein binds to the 5′-UTR region of YB-1 mRNA in the cytoplasm. The indicated amounts of KB3-1 and H1299 cell lysate were incubated with 32P-labeled YB-1 5′-UTR RNA at 25°C for 15 min. An aliquot of 2 µg YB-1-specific antibody (Ab) was added to lanes 5, 6, 11 and 12. An arrow indicates the YB-1/YB-1 5′-UTR RNA complex and a double-headed arrow indicates supershifted complexes. (B) Purified YB-1 binds to YB-1 5′-UTR RNA. The indicated amount of GST or GST–YB-1 fusion was incubated with 32P-labeled YB-1 5′-UTR RNA at 25°C for 15 min. An aliquot of 2 µg YB-1-specific antibody (Ab) was added to lanes 8–11. An arrow indicates the YB-1/YB-1 5′-UTR RNA complex and a double-headed arrow indicates supershifted complexes. (C) Rabbit YB-1 (p50) binds to the 5′-UTR region of YB-1 mRNA in the in vitro translation system. The indicated amounts of rabbit reticulocyte lysate (RRL) were incubated with 32P-labeled YB-1 5′-UTR RNA at 25°C for 15 min. An aliquot of 2 µg YB-1-specific antibody (Ab) was added to lanes 4 and 5. An arrow indicates the rabbit YB-1/YB-1 5′-UTR RNA complex and a double-headed arrow indicates supershifted complexes. YB-1/YB-1 5′-UTR RNA probe complexes were separated as described above.
We previously observed that rabbit p50, a homolog of human YB-1 protein, was present in rabbit reticulocyte lysate. To assess the nature of the rabbit p50 interaction with the 5′-UTR of YB-1 mRNA, we performed a REMSA using rabbit reticulocyte lysate (Fig. 4C). When added to rabbit reticulocyte lysate, the YB-1 5′-UTR formed an RNA–protein complex (Fig. 4C, lanes 2 and 3). The presence of rabbit p50 in the major complex was confirmed by the ability of a YB-1-specific antibody to supershift most of the complex (Fig. 4C, lane 5).
YB-1 protein consists of three major domains, each of which has the potential to bind nucleic acids (30): the alanine/proline-rich N-terminal domain, the highly conserved nucleic acid binding domain and the C-tail domain. To identify which domains of YB-1 protein are responsible for this interaction, we performed a REMSA using a series of GST fusion proteins containing either full-length YB-1 (FL) or the mutant derivatives GST–YB-1 N-ter, CSD and C-tail (Fig. 5A). A strong interaction was observed using the full-length YB-1 (Fig. 5B, lanes 3 and 4) and the C-tail of YB-1 (Fig. 5B, lanes 9 and 10). The central CSD, containing the RNP1 motif, was not able to bind to the 5′-UTR of YB-1 mRNA (Fig. 5B, lanes 7 and 8). The binding activity of both constructs resulted in the appearance of a supershifted band in the presence of a YB-1-specific antibody that recognizes the C-terminal tail of YB-1 (Fig. 5B, lanes 12 and 15). These results suggest that the C-tail domain of YB-1 protein interacts with the 5′-UTR of its own mRNA, independent of other domains.
Figure 5.
Identification of the YB-1 5′-UTR binding domain in YB-1. (A) Schematic illustration of the GST–YB-1 deletion mutants used in this study. CSD indicates the cold shock domain. (B) REMSA assay. The indicated amount of each GST–YB-1 deletion mutant and GST was incubated with 32P-labeled YB-1 5′-UTR RNA at 25°C for 15 min. An aliquot of 2 µg YB-1-specific antibody (Ab) was added to lanes 11–15. An arrow indicates the YB-1/YB-1 5′-UTR RNA complex and a double-headed arrow indicates supershifted complexes. YB-1 mutant/YB-1 5′-UTR RNA probe complexes were separated using 4% native polyacrylamide gels.
Identification of the YB-1 binding region in the 5′-UTR of YB-1 mRNA
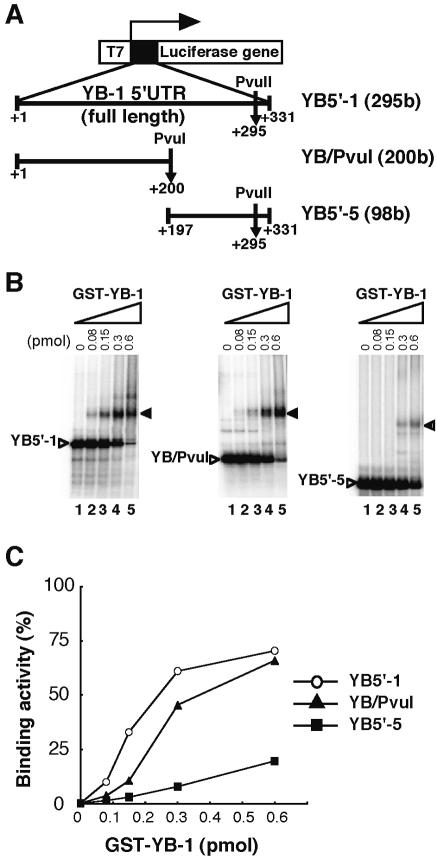
To identify which region of the YB-1 mRNA 5′-UTR interacts with YB-1 protein, we performed a REMSA using probes corresponding to YB5′-1 (295 bases), YB/PvuI (200 bases) and YB5′-5 (98 bases), as defined in Figure 1B (Fig. 6A). All three constructs formed RNA–protein complexes in a dose-dependent manner. However, the affinity of YB-1 for the three probes differed. When GST–YB-1 was added to the full-length YB-1 5′-UTR (YB5′-1) and YB/PvuI, a retarded complex was clearly visible (Fig. 6B). When 0.6 pmol of GST–YB-1 was added, up to 60% of the YB5′-1 and YB/PvuI probes were bound to GST–YB-1 (Fig. 6C). However, little retarded band was observed using the deleted YB5′-5 probe (Fig. 6B) when the same amount of YB-1 was used. When 0.6 pmol of YB-1 was added to YB5′-5, <20% YB5′-5 was bound. These results indicate that YB-1 protein binds to the full-length and the first half of the YB-1 5′-UTR more efficiently than a construct containing the second half of the YB-1 5′-UTR, YB5′-5.
Figure 6.
Identification of the YB-1 binding region in YB-1 5′-UTR mRNA. (A) Schematic illustration of YB-1 5′-UTR deletion constructs used as the probe in a REMSA. To produce RNA of defined length, restriction enzyme (PvuI or PvuII) was used to linearize the DNA templates. (B) REMSA. The indicated amount of GST–YB-1 fusion was incubated with each 32P-labeled YB-1 5′-UTR deletion mRNA at 25°C for 15 min. An arrow indicates the YB-1/YB-1 5′-UTR RNA complex. YB-1/YB-1 5′-UTR RNA probe complexes were separated using 4% native polyacrylamide gels. (C) Kinetic analysis of GST–YB-1 binding to YB5′-1, YB/PvuI and YB5′-5 probes. The GST–YB-1 binding activity to YB-1 5′-UTR fragments identical to that shown in (B) were quantified by monitoring amounts of the binding complexes.
Recombinant YB-1 protein decreases translation through its own 5′-UTR mRNA element in vitro
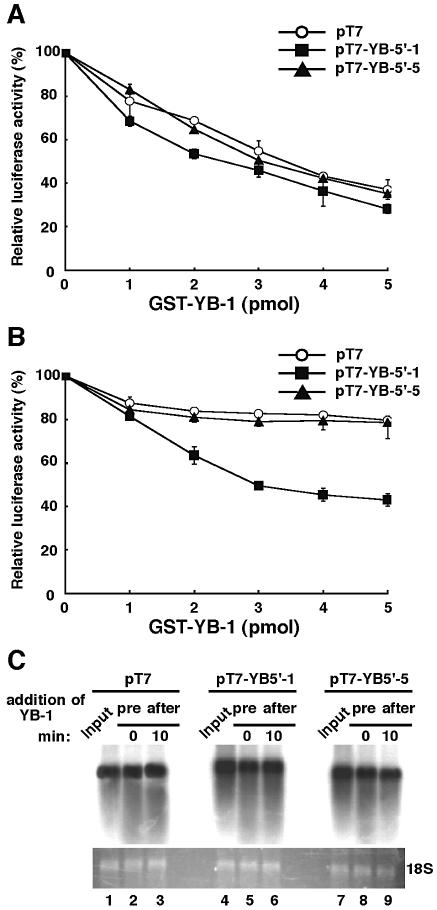
As shown in the REMSA, we observed higher affinity binding to the full-length 5′-UTR of YB-1 mRNA with recombinant YB-1. It has been shown that inhibitory concentrations of YB-1 suppressed the interaction with initiation complexes such as eIF4E, eIF4A and eIF4B, and these concentrations inhibited translation at the stage of initiation (17,31). YB-1 has also been reported to affect the translation of both cap+ poly(A)+ and cap– poly(A)– mRNAs (1,2). Skabkina et al. also demonstrated that poly(A) binding protein positively affects YB-1 mRNA translation through specific interaction with YB-1 mRNA (32). Thus, we investigated whether or not recombinant YB-1 inhibits translation in a cell-free translation system by using the full-length and deleted YB-1 5′-UTR constructs. In this system the transcript of YB-1 5′-UTR were neither capped nor polyadenylated. We therefore excluded the possibility that binding between YB-1 protein and translation initiation and elongation factors such as cap binding protein or poly(A) binding protein was involved. To test the functional activity of YB-1 protein, recombinant YB-1 protein was added to the in vitro translation system. In vitro transcripts from pT7, pT7-YB5′-1 and pT7-YB5′-5 were translated in a rabbit reticulocyte lysate system in the absence or presence of recombinant YB-1 protein and the luciferase activity was measured. As shown in Figure 7A, the addition of YB-1 at the start of translation inhibited the translation of all three constructs in a dose-dependent manner to a maximum of ∼60%.
Figure 7.
Characterization of translation inhibition by YB-1 protein through its own 5′-UTR mRNA element in vitro. The indicated amounts of GST–YB-1 fusion were added to a rabbit reticulocyte lysate system either initially (A) or after 10 min incubation (B) using 50 ng of the RNA constructs described in Figure 2A. The in vitro translation was performed for 30 min at 30°C and the translational products were directly used for the luciferase assay. Data are shown as means ± SD (error bars) of three independent experiments. (C) The effect of YB-1 on mRNA stability was examined. An aliquot of 50 ng of in vitro transcribed RNA constructs was preincubated (pre) or treated after 10 min incubation (after) with 5 pmol of GST–YB-1. After incubation for 30 min in a rabbit reticulocyte lysate system, the RNA was detected by northern blot hybridization. Lanes 1, 4 and 7 (Input) show the in vitro transcribed RNA constructs isolated from the exact at 0 min incubation.
Next, we measured the kinetics of luciferase translation in a rabbit retilculocyte lysate. After the first 10 min, almost 10% of translation had occurred and translation efficiency was saturated after 60 min (data not shown). To investigate the inhibitory effects of YB-1 protein on translation, we added various amounts of YB-1 protein 10 min after translation was started (Fig. 7B). The addition of YB-1 10 min after translation inhibited the luciferase activity in a dose-dependent manner when the construct containing the full-length 5′-UTR (pT7-YB5′-1) was used; this reduction amounted to ∼50% with 5 pmol of YB-1, implying that only 10% of the inhibition results from translation that could have occurred in the first 10 min of the 30 min reaction. On the other hand, little YB-1-dependent repression of translation was seen using pT7-YB5′-5 and pT7-luciferase. These results suggested that pretreated YB-1 inhibited translation of all constructs and the addition of YB-1 after translation also specifically inhibited translation of the construct with the full-length 5′-UTR.
YB-1 protein was also involved in stabilization of mRNA in rabbit reticulocyte system. We confirmed the integrity of the in vitro transcribed mRNA by northern blot analysis before and after treatment with YB-1 (Fig. 7C). No change in transcribed RNA stability was observed with up to 30 min incubation in rabbit reticulocyte system (Fig. 7C, lanes 1–3, 4–6 and 7–9).
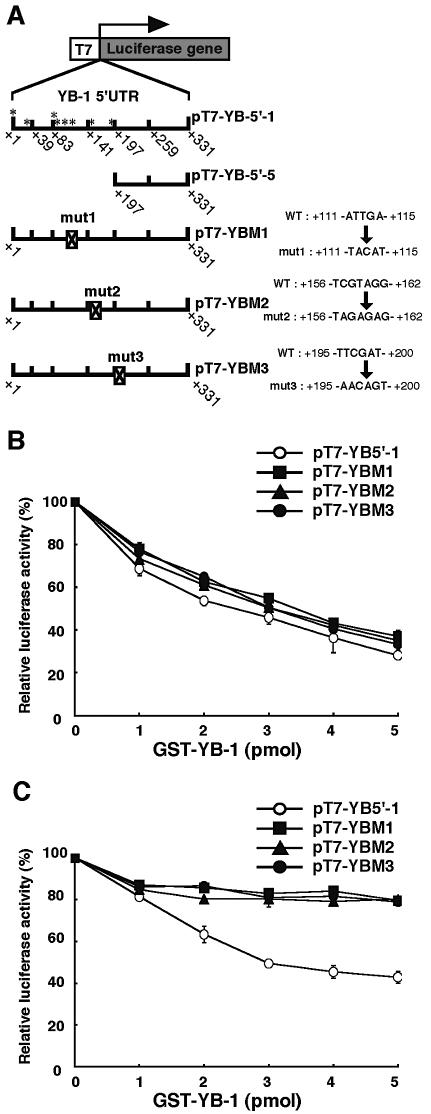
Recombinant YB-1 protein did not decrease translation through a mutant 5′-UTR element
Next, we focused on the possible secondary structure of the full-length 5′-UTR of YB-1 mRNA. In general, the stem–loop structure found in 5′-UTRs of mRNAs affect their translational efficiency as, for example, in the ferritin 5′-UTR (18). In the YB-1 5′-UTR, we observed three regions which are predicted to form possible stem–loop structures. To investigate the translational regulatory region of the 5′-UTR of YB-1 mRNA, we constructed three mutants which disrupt the stem–loop structure. These contained internal mutations (mut1–mut3) which we have designated using the nucleotide at the 5′ end corresponding to the transcription initiation site (*) (Fig. 8A). To characterize the translational inhibition observed with the addition of YB-1, we performed cell-free translational assays using mutants of the above constructs of the YB-1 5′-UTR fused to luciferase mRNA (Fig. 8B and C). The addition of YB-1 at the start of translation resulted in up to a 60% inhibition of luciferase activity in a dose-dependent manner, in the case of both the mutant and full-length 5′-UTR (Fig. 8B). However, no repression of the luciferase activity of these mutant constructs (pT7-YBM1–pT7-YBM3) was observed when YB-1 was added 10 min after translation was initiated (Fig. 8C). These results suggest that YB-1 protein regulates the translation of all constructs and also that it specifically inhibits translation when the 5′-UTR of YB-1 mRNA is present. Our findings are consistent with a model in which YB-1 protein specifically binds to the full-length 5′-UTR of YB-1 mRNA and inhibits its own translation in vitro.
Figure 8.
Effect of mutation of the YB-1 5′-UTR mRNA element on translation. (A) Schematic representation of the YB-1 5′-UTR–luciferase fusion reporter constructs used in this study. pT7-YBM1∼3 containing an internal mutation (mut1-3), designated by the nucleotide at the 5′ end that corresponded to the transcription initiation sites (*) (see Fig. 1A). (B and C) The characterization of translation inhibition by each reporter construct was compared to that of the other constructs. The indicated amounts of GST–YB-1 fusion were added to a rabbit reticulocyte lysate system either initially (B) or after 10 min incubation (C) using 50 ng of the RNA constructs described in (A). The in vitro translation was performed for 30 min at 30°C and the translation activity of each experiment was measured. Data are shown as the means ± SD (error bars) of three independent experiments.
DISCUSSION
In this report, our data demonstrate that YB-1 protein not only binds to an RNA containing the human YB-1 5′-UTR in vitro and in vivo, but that it also exhibits functional activity by specifically inhibiting the translation of a YB-1 5′-UTR–luciferase reporter mRNA in rabbit reticulocyte lysate assays. The multifunctional protein YB-1 was first identified as a transcription factor which binds to the Y-box of the MHC class II promoter sequence (33). Recent studies have demonstrated that YB-1 regulates gene expression not only at the level of transcription, but also at the level of mRNA translation. Considering these characteristics, it is possible that YB-1 might regulate its own expression at the translational level.
The present paper provides evidence that the translational control of YB-1 expression is mediated via the 5′-UTR of YB-1 mRNA. Global control of translational efficiency can be achieved by regulating the phosphorylation state of an array of initiation factors (such as eIF2 and eIF4E). However, control of translational initiation on an individual mRNA is determined primarily by its nucleotide sequence and the secondary structure of the regulatory protein. The 5′-UTR plays a particularly important role in the regulation of translation initiation via an interaction with RNA binding proteins or by secondary structure formation, which hinders the activity of the translational machinery. Analysis of the 5′-UTR sequence of YB-1 (331 bases in length) revealed several important features that are known to influence translation.
Many in vitro and in vivo studies have shown that mRNAs with a high likelihood of forming a stable secondary structure in the 5′-UTR tend to be inefficiently translated (34). However, in both in vitro and in vivo experiments we found that the 5′-UTR sequence of YB-1 mRNA, with its high G+C content, acts as a potent translational enhancer. The full-length 5′-UTR of YB-1 mRNA increased the level of translation activity and cloning a 134 base region of the YB-1 5′-UTR upstream of the coding initiation sequence was also associated with a significant increase in translation activity (Fig. 2A and B). This region might therefore be crucial for the translation activity of the 5′-UTR of YB-1 mRNA. These data suggest that elements within the 5′-UTR of YB-1 mRNA can act as enhancers of mRNA translation.
According to the scanning mechanism postulated for translation initiation, the small (40S) ribosomal subunit enters at the 5′ end of the mRNA and migrates linearly, stopping when the first AUG codon is reached (35). Nekrasov et al. (31) demonstrated that YB-1 protein inhibited the initiation step of translation, but did not inhibit the elongation step. Rabbit YB-1 displays both RNA melting and annealing activities in a dose-dependent manner; a relatively low amount of YB-1 promotes the formation of RNA duplexes, whereas an excess of YB-1 causes unwinding of double-stranded forms (36). It is also possible that YB-1 additionally facilitates movement of the small ribosomal subunit to the initiation codon complexed with initiator tRNAMet. This may occur up to the initiation codon by YB-1 melting the secondary structure in the mRNA 5′-UTR (31).
In this study, we observed that YB-1 inhibited the translation of all of the transcripts of YB-1 5′-UTR that lacked sequence specificity, suggesting that YB-1 reduced the translation efficiency of 5′-UTR elements, either at the initiation step or by binding directly to the whole RNA. These results provided evidence that YB-1 might block the first step in mRNA recruitment into translation initiation.
The addition of YB-1 10 min after initiation inhibited translational activity only when the full-length 5′-UTR of YB-1 mRNA was present (Figs 7B and 8C). No repression of the luciferase activity of the deleted or mutant constructs (pT7-YBM1–pT7-YBM3) (with disrupted stem–loop structures) was observed. We also observed that YB-1 binds to the first half-region of YB-1 5′ UTR RNA more efficiently than does the latter half construct comprising the second half (Fig. 7). The affinity of YB-1 for the mutated probes was the same as for the wild-type YB5′-1 probes (data not shown). These results suggest that ribosome scanning or ongoing translation might be required for repression by YB-1 when the full-length YB-1 5′-UTR was used. And when mutated and deleted probe were used, YB-1 did not affect ribosomal scanning due to alterations in the secondary structure of the YB-1 5′ UTR. The precise secondary structures of the YB-1 5′-UTR remains to be determined.
By combining the results presented here with previous data, we are able to describe the effect of YB-1 on translation. First, YB-1 blocks the initial step in mRNA recruitment to translation initiation, YB-1 then turns off the interaction of the mRNA with eIF4F (31). Second, YB-1 promotes movement of the small ribosomal subunit within the complex by melting the mRNA 5′-UTR secondary structure, suggesting that YB-1 affects ribosome scanning through the 5′-UTR.
The 5′-UTR plays an important role in regulation of the expression of a number of genes. For instance, the UTRs of mRNAs encoding heat shock proteins (HSPs), known to protect cells against a wide variety of stresses, including heat shock, viral infection and exposure to oxidative free radicals and toxic metal ions (37), have been reported to contain elements important for the post-transcriptional regulation of these key components of the stress response (38,39).
YB-1 has been reported to be involved in the regulation of stress-inducible target genes (40,41), suggesting that YB-1 itself is a stress-activated protein. YB-1 mRNA accumulates in cells when they are treated with UV irradiation or anticancer agents. We previously reported that c-myc and p73 activate YB-1 transcription and may regulate important biological processes via their effects on YB-1 gene expression (42). Additional experiments are needed to confirm the role of YB-1 in regulating translation in vivo under various stress conditions. It would be of interest to determine how the 5′-UTR of YB-1 mRNA affects the translation of YB-1 in response to extracellular stimuli or environmental stress.
The translational control of gene expression has been identified as an important regulatory mechanism for many gene products involved in the regulation of proliferation [e.g. c-mos (43), FGF-2 (44,45), PDGF-B (46), p27 (47) and cdk4 (48)]. The tumor suppressor p53 has been implicated in the translational regulation of both p53 and cdk4 mRNAs (49). Similarly, several distinct components of the translation apparatus have been shown to be deregulated or overexpressed in human tumors. So, it is of note that the level of the YB-1 mRNA from the 5′ furthest transcription initiation start site was significantly higher in HUVEC cells, compared to the other cancer cell lines examined (Fig. 1). The deregulation of the translational control of YB-1 might therefore also play a role in tumor progression.
In conclusion, YB-1 protein binds to an RNA containing the 5′-UTR of human YB-1 mRNA in vitro and in vivo and exhibits functional activity by inhibiting the translation of a YB-1 5′-UTR–luciferase reporter mRNA through the aid of its own 5′-UTR mRNA. We propose a model in which YB-1 protein autoregulates its own translation by binding to the 5′-UTR of its own mRNA. Our results support a trans-acting repressor hypothesis, in which a repressor protein specifically binds to its own 5′-UTR element, resulting in translational repression and the maintenance of constant protein levels.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Prof. R.G. Deeley for helpful discussions and for critically reading this manuscript. This work was supported by the Second-Term Comprehensive Ten-Year Strategy for Cancer Control from the Ministry of Health and Welfare of Japan and by the Cancer Research Fund from the Ministry of Education, Culture, Sports, Science and Technology.
REFERENCES
- 1.Pisarev A.V., Skabkin,M.A., Thomas,A.A., Merrick,W.C., Ovchinnikov,L.P. and Shatsky,I.N. (2002) Positive and negative effects of the major mammalian messenger ribonucleoprotein p50 on binding of 40 S ribosomal subunits to the initiation codon of beta-globin mRNA. J. Biol. Chem., 277, 15445–15451. [DOI] [PubMed] [Google Scholar]
- 2.Evdokimova V.M., Kovrigina,E.A., Nashchekin,D.V., Davydova,E.K., Hershey,J.W. and Ovchinnikov,L.P. (1998) The major core protein of messenger ribonucleoprotein particles (p50) promotes initiation of protein biosynthesis in vitro. J. Biol. Chem., 273, 3574–3581. [DOI] [PubMed] [Google Scholar]
- 3.Davydova E.K., Evdokimova,V.M., Ovchinnikov,L.P. and Hershey,J.W. (1997) Overexpression in COS cells of p50, the major core protein associated with mRNA, results in translation inhibition. Nucleic Acids Res., 25, 2911–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohno K., Izumi,H., Uchiumi,T. and Kuwano,M. (2003) The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays, 25, 691–698. [DOI] [PubMed] [Google Scholar]
- 5.Wolffe A.P. (1994) Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays, 16, 245–251. [DOI] [PubMed] [Google Scholar]
- 6.Ladomery M. and Sommerville,J. (1995) A role for Y-box proteins in cell proliferation. Bioessays, 17, 9–11. [DOI] [PubMed] [Google Scholar]
- 7.Swamynathan S.K., Nambiar,A. and Guntaka,R.V. (1998) Role of single-stranded DNA regions and Y-box proteins in transcriptional regulation of viral and cellular genes. FASEB J., 12, 515–522. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto K. and Wolffe,A.P. (1998) Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol., 8, 318–323. [DOI] [PubMed] [Google Scholar]
- 9.Sommerville J. (1999) Activities of cold-shock domain proteins in translation control. Bioessays, 21, 319–325. [DOI] [PubMed] [Google Scholar]
- 10.Swamynathan S.K., Nambiar,A. and Guntaka,R.V. (2000) Chicken Y-box proteins chk-YB-1b and chk-YB-2 repress translation by sequence-specific interaction with single-stranded RNA. Biochem. J., 348, 297–305. [PMC free article] [PubMed] [Google Scholar]
- 11.Kelm R.J. Jr, Cogan,J.G., Elder,P.K., Strauch,A.R. and Getz,M.J. (1999) Molecular interactions between single-stranded DNA-binding proteins associated with an essential MCAT element in the mouse smooth muscle alpha-actin promoter. J. Biol. Chem., 274, 14238–14245. [DOI] [PubMed] [Google Scholar]
- 12.Kelm R.J. Jr, Elder,P.K. and Getz,M.J. (1999) The single-stranded DNA-binding proteins, Puralpha, Purbeta and MSY1 specifically interact with an exon 3-derived mouse vascular smooth muscle alpha-actin messenger RNA sequence. J. Biol. Chem., 274, 38268–38275. [DOI] [PubMed] [Google Scholar]
- 13.Gu W., Tekur,S., Reinbold,R., Eppig,J.J., Choi,Y.C., Zheng,J.Z., Murray,M.T. and Hecht,N.B. (1998) Mammalian male and female germ cells express a germ cell-specific Y-Box protein, MSY2. Biol. Reprod., 59, 1266–1274. [DOI] [PubMed] [Google Scholar]
- 14.Herbert T.P. and Hecht,N.B. (1999) The mouse Y-box protein, MSY2, is associated with a kinase on non-polysomal mouse testicular mRNAs. Nucleic Acids Res., 27, 1747–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsumoto K., Meric,F. and Wolffe,A.P. (1996) Translational repression dependent on the interaction of the Xenopus Y-box protein FRGY2 with mRNA. Role of the cold shock domain, tail domain and selective RNA sequence recognition. J. Biol. Chem., 271, 22706–22712. [DOI] [PubMed] [Google Scholar]
- 16.Chen C.Y., Gherzi,R., Andersen,J.S., Gaietta,G., Jurchott,K., Royer,H.D., Mann,M. and Karin,M. (2000) Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev., 14, 1236–1248. [PMC free article] [PubMed] [Google Scholar]
- 17.Evdokimova V., Ruzanov,P., Imataka,H., Raught,B., Svitkin,Y., Ovchinnikov,L.P. and Sonenberg,N. (2001) The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J., 20, 5491–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashizuka M., Fukuda,T., Nakamura,T., Shirasuna,K., Iwai,K., Izumi,H., Kohno,K., Kuwano,M. and Uchiumi,T. (2002) Novel translational control through an iron-responsive element by interaction of multifunctional protein YB-1 and IRP2. Mol. Cell. Biol., 22, 6375–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkie G.S., Dickson,K.S. and Gray,N.S. (2003) Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci., 4, 182–188. [DOI] [PubMed] [Google Scholar]
- 20.Pesole G., Liuni,S., Grillo,G., Licciulli,F., Mignone,F., Gissi,C. and Saccone,G. (2002) UTRdb and UTRsite: specialized databases of sequences and functional elements of 5′ and 3′ untranslated region of eukaryotic mRNAs. Update 2002. Nucleic Acids Res., 1, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy J.E. and Kollmus,H. (1995) Cytoplasmic mRNA-protein interactions in eukaryotic gene expression. Trends Biochem. Sci., 20, 191–197. [DOI] [PubMed] [Google Scholar]
- 22.Macdonald P. (2001) Diversity in translational regulation. Curr. Opin. Cell Biol., 13, 326–331. [DOI] [PubMed] [Google Scholar]
- 23.Sonenberg N. (1994) mRNA translation: influence of the 5′ and 3′ untranslated regions. Curr. Opin. Genet. Dev., 4, 310–315. [DOI] [PubMed] [Google Scholar]
- 24.Ise T., Nagatani,G., Imamura,T., Kato,K., Takano,H., Nomoto,M., Izumi,H., Ohmori,H., Okamoto,T., Ohga,T., Uchiumi,T., Kuwano,M. and Kohno,K. (1999) Transcription factor Y-box binding protein 1 binds preferentially to cisplatin-modified DNA and interacts with proliferating cell nuclear antigen. Cancer Res., 59, 342–346. [PubMed] [Google Scholar]
- 25.Okamoto T., Izumi,H., Imamura,T., Takano,H., Ise,T., Uchiumi,T., Kuwano,M. and Kohno,K. (2000) Direct interaction of p53 with the Y_-box binding protein, YB-1: a mechanism for regulation of human gene expression. Oncogene, 19, 6194–6202. [DOI] [PubMed] [Google Scholar]
- 26.Izumi H., Imamura,T., Nagatani,G., Ise,T., Murakami,T., Uramoto,H., Torigoe,T., Ishiguchi,H., Yoshida,Y., Nomoto,M., Okamoto,T., Uchiumi,T., Kuwano,M., Funa,K. and Kohno,K. (2001) Y box-binding protein-1 binds preferentially to single-stranded nucleic acids and exhibits 3′→5′ exonuclease activity. Nucleic Acids Res., 29, 1200–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohga T., Koike,K., Ono,M., Makino,Y., Itagaki,Y., Tanimoto,M., Kuwano,M. and Kohno,K. (1996) Role of the human Y box-binding protein YB-1 in cellular sensitivity to the DNA-damaging agents cisplatin, mitomycin C and ultraviolet light. Cancer Res., 56, 4224–4228. [PubMed] [Google Scholar]
- 28.Makino Y., Ohga,T., Toh,S., Koike,K., Okumura,K., Wada,M., Kuwano,M. and Kohno,K. (1996) Structural and functional analysis of the human Y-box binding protein (YB-1) gene promoter. Nucleic Acids Res., 24, 1873–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullner E.W., Neupert,B. and Kuhn,L.C. (1989) A specific mRNA binding factor regulates the iron-dependent stability of cytoplasmic transferrin receptor mRNA. Cell, 58, 373–382. [DOI] [PubMed] [Google Scholar]
- 30.Ladomery M. and Sommerville,J. (1994) Binding of Y-box proteins to RNA: involvement of different protein domains. Nucleic Acids Res., 22, 5582–5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nekrasov M.P., Ivshina,M.P., Chernov,K.G., Kovrigina,E.A., Evdokimova,V.M., Thomas,A.A., Hershey,J.W. and Ovchinnikov,L.P. (2003) The mRNA-binding protein YB-1 (p50) prevents association of the initiation factor eIF4G with mRNA and inhibits protein synthesis at the initiation stage. J. Biol. Chem., 278, 13936–13943. [DOI] [PubMed] [Google Scholar]
- 32.Skabkina O.V., Skabkin,M.A., Popova,N.V., Lyabin,D.N., Penalva,L.O. and Ovchinnikov,L.P. (2003) Poly(A)-binding protein positively affects YB-1 mRNA translation through specific interaction with YB-1 mRNA. J. Biol. Chem., 278, 18191–18198. [DOI] [PubMed] [Google Scholar]
- 33.Didier D.K., Schiffenbauer,J., Woulfe,S.L., Zacheis,M. and Schwartz,B.D. (1988) Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc. Natl Acad. Sci. USA, 85, 7322–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozak M. (1991) Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem., 266, 19867–19870. [PubMed] [Google Scholar]
- 35.Kozak M. (2002) Pushing the limits of the scanning mechanism for initiation of translation. Gene, 299, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skabkin M.A., Evdokimova,V., Thomas,A.A. and Ovchinnikov,L.P. (2001) The major messenger ribonucleoprotein particle protein p50 (YB-1) promotes nucleic acid strand annealing. J. Biol. Chem., 276, 44841–44847. [DOI] [PubMed] [Google Scholar]
- 37.Kiang J.G. and Tsokos,G.C. (1998) Heat shock protein 70 kDa: molecular biology, biochemistry and physiology. Pharmacol. Ther., 80, 183–201. [DOI] [PubMed] [Google Scholar]
- 38.Klemenz R., Hultmark,D. and Gehring,W.J. (1985) Selective translation of heat shock mRNA in Drosophila melanogaster depends on sequence information in the leader. EMBO J., 4, 2053–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGarry T.J. and Lindquist,S. (1985) Cytoplasmic mRNA–protein interactions in eukaryotic gene expression. Cell, 42, 903–911.4053186 [Google Scholar]
- 40.Bargou R.C., Jurchott,K., Wagener,C., Bergmann,S., Metzner,S., Bommert,K., Mapara,M.Y., Winzer,K.J., Dietel,M., Dorken,B. and Royer,H.D. (1997) Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nature Med., 3, 447–450. [DOI] [PubMed] [Google Scholar]
- 41.Oda Y., Sakamoto,A., Shinohara,N., Ohga,T., Uchiumi,T., Kohno,K., Tsuneyoshi,M., Kuwano,M. and Iwamoto,Y. (1998) Nuclear expression of YB-1 protein correlates with P-glycoprotein expression in human osteosarcoma. Clin. Cancer Res., 4, 2273–2277. [PubMed] [Google Scholar]
- 42.Uramoto H., Izumi,H., Ise,T., Tada,M., Uchiumi,T., Kuwano,M., Yasumoto,K., Funa,K. and Kohno,K. (2002) p73 Interacts with c-Myc to regulate Y-box-binding protein-1 expression. J. Biol. Chem., 277, 31694–31702. [DOI] [PubMed] [Google Scholar]
- 43.Steel L.F., Telly,D.L., Leonard,J., Rice,B.A., Monks,B. and Sawicki,J.A. (1996) Elements in the murine c-mos messenger RNA 5′-untranslated region repress translation of downstream coding sequences. Cell Growth Differ., 7, 1415–1424. [PubMed] [Google Scholar]
- 44.Kevil C., Carter,P., Hu,B. and DeBenedetti,A. (1995) Translational enhancement of FGF-2 by eIF-4 factors and alternate utilization of CUG and AUG codons for translation initiation. Oncogene, 11, 2339–2348. [PubMed] [Google Scholar]
- 45.Vagner S., Gensac,M.C., Maret,A., Bayard,F., Amalric,F., Prats,H. and Prats,A.C. (1995) Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol. Cell. Biol., 15, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernstein J., Shefler,I. and Elroy-Stein,O. (1995) The translational repression mediated by the platelet-derived growth factor 2/c-sis mRNA leader is relieved during megakaryocytic differentiation. J. Biol. Chem., 270, 10559–10565. [DOI] [PubMed] [Google Scholar]
- 47.Hengst L. and Reed,S.I. (1996) Translational control of p27Kip1 accumulation during the cell cycle. Science, 271, 1861–1864. [DOI] [PubMed] [Google Scholar]
- 48.Sonenberg N. (1993) Translation factors as effectors of cell growth and tumorigenesis. Curr. Opin. Cell Biol., 5, 955–960. [DOI] [PubMed] [Google Scholar]
- 49.Miller S.J., Suthiphongchai,T., Zambetti,G.P. and Ewen,M.E. (2000) p53 binds selectively to the 5′ untranslated region of cdk4, an RNA element necessary and sufficient for transforming growth factor beta- and p53-mediated translational inhibition of cdk4. Mol. Cell. Biol., 20, 8420–8431. [DOI] [PMC free article] [PubMed] [Google Scholar]