Abstract
Degradation of proteins that, because of improper or suboptimal processing, are retained in the endoplasmic reticulum (ER) involves retrotranslocation to reach the cytosolic ubiquitin-proteasome machinery. We found that substrates of this pathway, the precursor of human asialoglycoprotein receptor H2a and free heavy chains of murine class I major histocompatibility complex (MHC), accumulate in a novel preGolgi compartment that is adjacent to but not overlapping with the centrosome, the Golgi complex, and the ER-to-Golgi intermediate compartment (ERGIC). On its way to degradation, H2a associated increasingly after synthesis with the ER translocon Sec61. Nevertheless, it remained in the secretory pathway upon proteasomal inhibition, suggesting that its retrotranslocation must be tightly coupled to the degradation process. In the presence of proteasomal inhibitors, the ER chaperones calreticulin and calnexin, but not BiP, PDI, or glycoprotein glucosyltransferase, concentrate in the subcellular region of the novel compartment. The “quality control” compartment is possibly a subcompartment of the ER. It depends on microtubules but is insensitive to brefeldin A. We discuss the possibility that it is also the site for concentration and retrotranslocation of proteins that, like the mutant cystic fibrosis transmembrane conductance regulator, are transported to the cytosol, where they form large aggregates, the “aggresomes.”
INTRODUCTION
The ubiquitin/proteasomal pathway is involved in the degradation of substrates originated in the endoplasmic reticulum (ER) (Sommer and Wolf, 1997; Bonifacino and Weissman, 1998). However, the mechanism of delivery to the cytosolic proteasomes is still unclear. One example of protein targeting from the ER to the cytosol for degradation by the ubiquitin/proteasome machinery is the major histocompatibility complex (MHC) class I heavy chain in the presence of cytomegalovirus US11 or US2 proteins. Association of class I molecules to Sec61 suggests that the translocation complex could be involved in their delivery to the cytosol (Wiertz et al., 1996b). Genetic evidence for the involvement of Sec61 in this process was found by studying the degradation of a mutant carboxypeptidase Y (cpy) (Plemper et al., 1997) and proα-factor (Pilon et al., 1997) in yeast, carrying a temperature sensitive mutant Sec61 that stabilized these proteins. The involvement of Sec61 in protein import into the ER appears different from its function in export to the cytosol (Zhou and Schekman, 1999). Some proteins are transported from the lumen of the ER to its cytosolic face (Hiller et al., 1996) or are completely dislocated to the cytosol (Wiertz et al., 1996b; Yu et al., 1997) to be degraded by the proteasomes. However, this has not been found in other cases (Chillaron and Haas, 2000; Fisher et al., 1997; Schubert et al., 1998). To better understand the transport pathway of proteins from the ER to the proteasomes, we have analyzed the fate of asialoglycoprotein receptor (ASGPR) H2a, an established model for retention and degradation from the ER (Amara et al., 1989; Lederkremer and Lodish, 1991; Wikstrom and Lodish, 1991; Shenkman et al., 1997; Ayalon-Soffer et al., 1999a; Ayalon-Soffer et al., 1999b). The membrane-bound precursor of H2a is a type II membrane protein completely retained in the ER (Shenkman et al., 1997). After retention, it can be cleaved (probably by signal peptidase [Yuk and Lodish, 1993]) to yield a soluble carboxyterminal fragment corresponding to its ectodomain. A certain amount of this fragment can travel through the Golgi and can be secreted as a soluble form of the receptor (Tolchinsky et al., 1996). The rest of the ectodomain fragment and uncleaved membrane-bound precursor molecules are degraded from the ER (Amara et al., 1989; Neumann et al., 1996). This retention and degradation is not the result, as in mutant proteins, of global misfolding (Shenkman et al., 1997; Ayalon-Soffer et al., 1999a). H2a is a proteasomal substrate (Ayalon-Soffer et al., 1999a; Ayalon-Soffer et al., 1999b), and as we show here, upon proteasomal inhibition it is not transported to the cytosol but accumulates in a novel “quality control” compartment. In view of this finding we analyzed the fate of another established substrate for proteasomal degradation from the ER, free class I MHC heavy chains (Hughes et al., 1997). Free class I MHC heavy chains, like ASGPR H2a, are retained in the ER in an endo H sensitive state and are targeted for proteasomal degradation (Machold et al., 1995; Fromm et al., 1998). Although upon proteasomal inhibition we find a small portion of the heavy chains in the cytosol, most of them are associated with the membrane fraction in a subcellular region similar to H2a.
MATERIALS AND METHODS
Materials
Rainbow 14C-labeled methylated protein standards and Promix cell-labeling mix ([35S]methionine plus [35S]cysteine) were from Amersham (Arlington Heights, IL) (>1000 Ci/mmol; 1 Ci = 37 GBq). Protein A-Sepharose was from Repligen (Needham, MA). Endo-N-acetyl glucosaminidase H (endo H) was from New England Biolabs (Beverly, MA). N-glycosidase F, tosyl-lysylchloromethylketone (TLCK), N-acetyl-leucyl-leucyl-norleucinal (ALLN), and N-acetyl-leucyl-leucyl-methional (ALLM) were obtained from Boehringer Mannheim (Indianapolis, IN). Lactacystin (Lac) and N-carbobenzoxyl-leucinyl-leucinyl-leucinal (MG-132) were from Calbiochem (La Jolla, CA). Other common reagents and inhibitors were from Sigma (St. Louis, MO).
Cell Lines and Culture
NIH 3T3 cell lines stably expressing H2a (2–18 cells) or H2b (2C cells) (Lederkremer and Lodish, 1991) were grown in Dulbecco's modified Eagles's medium (DMEM) plus 10% calf serum under 5% CO2. Mutant ts20 cell line (kind gift from Aaron Ciechanover, Technion, Haifa, Israel) and CHO parental cells were maintained in DMEM plus 10% fetal calf serum under 5% CO2 at 31°C or 37°C, respectively. These were cotransfected with the use of a calcium phosphate protocol with a pcDNA1 expression vector containing H2a and a pBabe-puro vector (Morgenstern and Land, 1990), followed by selection with puromycin. Representative clones expressing H2a (2D- wild-type CHO and 4D- mutant ts20) were expanded and grown as described above. E1Ad5-transformed embryonal mouse fibroblasts (A505) (Fromm et al., 1998) were grown in DMEM plus 10% fetal calf serum under 5% CO2.
Antibodies
Rabbit polyclonal antipeptide antibodies anti-H2 carboxyterminal or H2a-specific (anti-H2a) were the ones used in earlier studies (Tolchinsky et al., 1996). Rabbit antibodies directed against free class I MHC heavy chains (H-2Kb and H-2Db) were a kind gift from Dr. H. Ploegh (Harvard Medical School, Boston, MA) (Machold et al., 1995). Y3 are mouse monoclonal antibodies against H-2Kb complexes (Jones and Janeway, 1981).
For immunofluorescence IgGs of primary antibodies were purified from the antisera as before (Shenkman et al., 1997). Rabbit polyclonal anti-Sec61-β antibodies were a kind gift from Tom Rapoport (Harvard Medical School, Boston, MA). Mouse monoclonals anti-β-COP and anti-γ-tubulin were from Sigma, anti-BiP from Stressgen (Victoria, BC), and antirab5 a kind gift from Marino Zerial (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany). Rabbit IgGs, antip58 and antiproteasome (Frentzel et al., 1994), and rat antilamp1, were kind gifts from Jakko Saraste (University of Bergen, Bergen, Norway), Peter-M. Kloetzel (Humboldt University, Berlin, Germany), and Hugh Rosen (Merck Research Laboratories, Rahway, NJ), respectively. Rabbit polyclonals anti-BiP, -PDI, and -Calnexin were from Stressgen, and anti-UDPGlc:glycoprotein glucosyltransferase was a kind gift from Armando Parodi (University of San Martin, Buenos Aires, Argentina). Rabbit anticalreticulin was from Affinity Bioregents (Golden, CO). Secondary Cy3- or FITC-conjugated antibodies were from The Jackson Laboratory (Bar Harbor, ME).
Metabolic Labeling and Immunoprecipitation
Subconfluent (90%) cell monolayers in 60-mm dishes were labeled with [35S] cys, lysed, and immunoprecipitated as described before with anti-H2a pentapeptide antibody for H2a and anti- H2-carboxyterminal antibody for H2b (Shenkman et al., 1997). Subconfluent A5O5 cells in 100-mm dishes were labeled with 150 μCi/ml [35S] Met, washed with PBS, and chased for the indicated intervals with complete medium. Treatments of the immunoprecipitates with endo H and N-glycosidase F were performed as described before (Shenkman et al., 1997). To analyze Triton-solubility, cells were solubilized with 1% Triton-X-100 and 0.5% sodium deoxycholate in PBS (buffer A) and centrifuged at 16,000 × g for 30 min at 4°C. Supernatants and pellets were immunoprecipitated, the latter after boiling for 5 min in 1% SDS, 2 mM DTT in PBS, followed by addition of 10 volumes of buffer A plus 2 mM oxidized glutathione.
Sec61 Coimmunoprecipitation
Cells were lysed in 1% digitonin with 20 mM HEPES, pH 7.6 in the presence of 2 mM PMSF, 5 μg/ml aprotinin and 5 μg/ml leupeptin (protease inhibitor mix) and immunoprecipitated with anti-Sec61-β antibodies. Immunoprecipitates were washed with lysis buffer and boiled for 5 min in 1% SDS and 2 mM DTT. The supernatants were diluted with 10 volumes of buffer A containing 2 mM oxidized glutathione and reimmunoprecipitated with anti-H2 carboxyterminal antibodies.
Gel Electrophoresis, Fluorography, and Quantitation
Reducing SDS-PAGE was performed on 10% Laemmli gels, except where stated otherwise. The gels were analyzed by fluorography with the use of 20% 2,5-diphenyloxazole and were exposed to Kodak Biomax MR film (Vancouver, BC) except for the Sec61 coimmunoprecipitation experiment where Biomax MS film and a Transcreen-LE from Kodak were used. Quantitations were performed in a Fuji BAS 1000 phosphorimager.
Protein Transfer and Immunoblotting
Cells were lysed in a buffer containing 100 mM NaCl, 20 mM Tris-HCl pH 7.4, 5 mM EDTA, 15% glycerol, 0.2% Triton X-100, and protease inhibitor mix. Aliquots (corresponding to ∼2 × 105 cells) were boiled in SDS sample buffer for 5 min before loading for SDS-PAGE. Proteins were transferred to a nitrocellulose membrane. Blocking was in 5% low-fat milk and 0.1% Brij-35 in PBS for 2 h, and incubation with the primary antibody was overnight at 4°C. After wash in 0.1% Brij-35 in PBS, incubation with the appropriate secondary antibody was for 2 h at room temperature. After washing, ECL was performed, and the blot was exposed to Agfa CP-BU film.
Trypsin Digestion with Digitonin Permeabilization
After incubation at either the permissive or restrictive temperature (ts20 cells) or at the end of a pulse-chase procedure (2–18 cells), cells were trypsinized and diluted in KH buffer (100 mM potassium acetate, 20 mM HEPES, titrated to pH 7.2 with KOH). Cells were then pelleted by centrifugation at 1200 rpm for 5 min and were resuspended in KH buffer or in immunoblot lysis buffer. Different concentrations of digitonin and 0.5 mg/ml trypsin were added to the samples, which were incubated for 30 min at 4°C. Soybean trypsin inhibitor (2 mg/ml) was then added for 5 min before solubilization with 0.2% Triton-X-100 (plus protease inhibitor mix) for 30 min at 4°C. Digitonin concentrations were chosen after titration and cell examination with trypan blue.
Subcellular Fractionation
Metabolically labeled A505 cells were washed with PBS and were resuspended in homogenization buffer (0.25 M sucrose, 10 mM HEPES pH = 7.2 with addition of protease inhibitor mix). Cells were homogenized on ice with the use of a Dounce homogenizer (40 strokes). The homogenate was spun at 1000 × g for 10 min (giving the 1000 × g pellet), and the supernatant was subjected to 10,000 × g for 30 min (giving the 10,000 × g pellet). Finally the 10,000 × g supernatant was spun for an hour at 100,000 × g resulting in the 100,000 × g pellet and the 100,000 × g supernatant. Lysis mix (0.5% Triton X-100, 50 mM Tris-HCl pH = 7.4 and 150 mM NaCl in the presence of protease inhibitors) was added to the pellets. The 100,000 × g supernatants were adjusted to 0.5% Triton X-100 and 150 mM NaCl. Lysates were immunoprecipitated with antifree heavy chain antibodies (HC) and washed with 0.1% Triton X-100, 50 mM Tris-HCl pH = 7.4 and 150 mM NaCl in the presence of protease inhibitor mix.
Immunofluorescence Microscopy
The procedures used were as described previously (Shenkman et al., 1997) except that digital photography was done on an imaging system on a Leica fluorescence microscope. Confocal microscopy was performed on a Perkin Elmer-Cetus/Wallac (Norwalk, CT) confocal microscope. Staining of nuclei was with DAPI (Sigma). Incubation of cells with nocodazole was as in Shenkman et al. (1997). Cells permeabilized by breakage were prepared by treating cells suspended in 0.25 M sucrose, 1 mM EDTA, and 20 mM HEPES pH 7.2, with 7 strokes in a Dounce homogenizer and were then adhered to coverslips with 5 μg/ml polylysine before fixation for immunofluorescence. Permeabilization was analyzed with trypan blue. Some samples were treated with 0.2% Triton-X-100 for 30 min at 4°C before adhering to coverslips.
RESULTS
Degradation of ASGPR H2a, H2b, and the Soluble 35 kDa H2a Ectodomain Fragment Is Blocked by Proteasomal Inhibitors, and They Remain in the Secretory Pathway
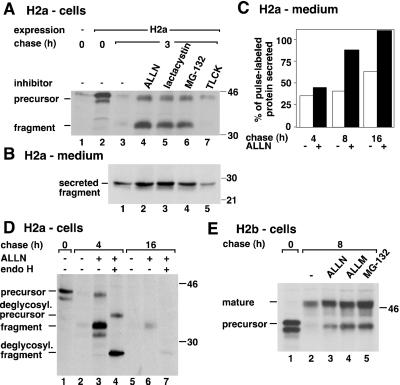
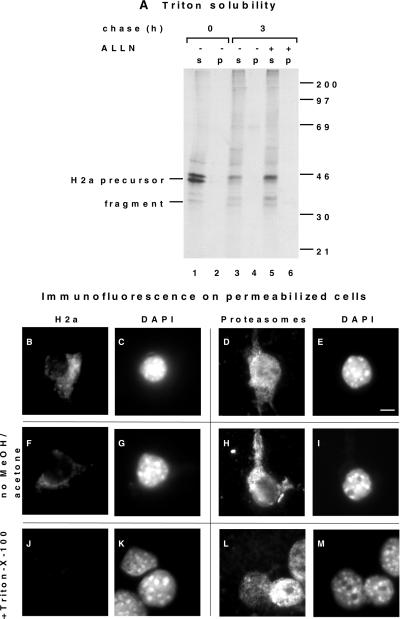
To analyze the involvement of the proteasome in the degradation of H2a, we tested several inhibitors. We then looked at the fate of H2a membrane-bound precursor and its cleavage product, a soluble 35-kDa carboxyterminal ectodomain fragment. Proteasome inhibitors strongly protected the precursor and the soluble fragment from degradation (Figure 1 A). The levels of precursor plus fragment remaining after 3 h of chase increased ∼6-fold with ALLN or lactacystin and ∼5-fold with MG-132. The serine and cysteine protease inhibitor TLCK that had been used before to inhibit the degradation of H2a (Wikstrom and Lodish, 1991) showed much less potency (1.5-fold increase) and did not protect the fragment. The cleavage to yield ectodomain fragment was not inhibited by the proteasomal inhibitors (Figure 1A, lanes 4–6). A similar block in the degradation of H2a by ALLN and lactacystin and not by PMSF (our unpublished results) suggests that the proteasome is indeed responsible and not another protease complex, TPPII, which was reported to be inhibited by the 2 latter but not by ALLN (Geier et al., 1999). A shift of H2a bands to a faster mobility can be seen upon chase (compare Figure 1A, lanes 2 and 4). This shift is due to mannose trimming of the sugar chains (Z. Frenkel and G. Lederkremer, unpublished observations). Secretion of H2a ectodomain fragment was significantly increased by the action of the proteasomal inhibitors (>2-fold with lactacystin after 3 h chase, Figure 1B), and it slowly reached completion after 16 h chase (Figure 1C). Thus, H2a does not seem to be transported to the cytosol like some other substrates (Hiller et al., 1996; Wiertz et al., 1996b; Yu et al., 1997), but it remains instead in the secretory pathway, where it can be cleaved to yield the ectodomain fragment, which is in a transport-competent state. The intracellular 35-kDa fragment is always endo H sensitive (Figure 1D, lanes 4 and 7), indicating rapid secretion after exit from the ER.
Figure 1.
Proteasomal inhibitors cause accumulation of ASGPR H2a membrane-bound precursor, its soluble 35-kDa fragment, and membrane-bound H2b inside the secretory pathway. (A) NIH 3T3 cells stably expressing H2a or untransfected (lane 1) were metabolically labeled with [35S] Cys for 20 min and chased with complete medium in the absence or in the presence of the indicated protease inhibitors. Incubations with the protease inhibitors were only during the chase period, with the following concentrations: 100 μM ALLN, ALLM, and TLCK, 25 μM lactacystin, and 10 μM MG-132. Cell lysates were immunoprecipitated, and immunoprecipitates were analyzed by SDS-PAGE followed by fluorography. On the left are indicated the positions for migration of H2a precursor and of its 35-kDa ectodomain fragment. On the right are molecular masses of protein standards in kilodaltons. (B) At the end of each chase period supernatants from cells in (A) were immunoprecipitated, and immunoprecipitates were treated with N-glycosidase F before SDS-PAGE and fluorography. The gel was exposed for 3 wk compared with 1 week in (A). (C) Phosphorimager quantitation of an experiment similar to that in (B). H2a ectodomain fragment secreted to the cell supernatants in the absence (white bars) or in the presence (black bars) of 100 μM ALLN were plotted as a percent of pulse-labeled H2a. (D) In a similar experiment to that in (A), the immunoprecipitates were treated in some cases with endo H before SDS-PAGE. (E) An experiment similar to that in (A) was done but with cells stably expressing H2b. On the left are indicated the positions for migration of H2b precursor and its mature Golgi-processed form.
We found a similar fate when we looked at the alternatively spliced variant H2b. The membrane-bound form of H2a is completely retained in the ER. In contrast, ∼30% of membrane-bound H2b is Golgi-processed and reaches the cell surface in singly transfected fibroblasts, while the rest is degraded in the ER (Lederkremer and Lodish, 1991). When we treated cells that express H2b with proteasomal inhibitors, the degradation of the precursor was inhibited and Golgi-processing increased (Figure 1E, lanes 2–5). In the presence of MG-132, almost all (94%) of H2b was saved from degradation after 8-h chase, and the level of Golgi-processed mature molecules increased by almost 2-fold as compared with untreated (Figure 1E, compare lanes 2 and 5). Thus, upon proteasomal inhibition, both soluble H2a ectodomain fragment and membrane-bound H2b remain in the secretory pathway.
H2a Precursor and Ectodomain Fragment Are Not Transported to the Cytosol or Exposed to the Cytosolic Side of ER Membranes upon Inhibition of Ubiquitination or of the Proteasome
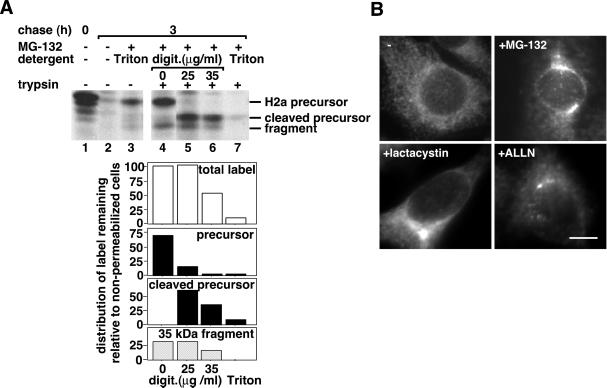
To confirm that H2a is not transported to the cytosol, we analyzed its susceptibility to trypsin digestion in semipermeabilized cells after inhibition of the degradation with MG-132. We chose to examine the transport to the cytosol after permeabilization under very mild conditions and not by subcellular fractionation, because the latter method frequently leads to leakiness of the organelles (Chillaron and Haas, 2000). At a concentration of digitonin that led to permeabilization of 100% of the cells (as judged by staining with trypan blue), trypsin caused trimming of the precursor's cytoplasmic tail, converting it to a 38-kDa fragment, the carboxyterminal transmembrane plus luminal domains remained protected from trypsin (Figure 2 A, lane 5). This suggests that H2a indeed remains membrane-bound and is not dislocated to the cytosol. The naturally occuring 35-kDa soluble ectodomain fragment was completely protected, suggesting that it is accumulated in an enclosed compartment and is inaccessible to trypsin. A larger concentration of digitonin caused partial permeabilization of internal membranes and digestion of 50% of the precursor and ectodomain fragment (Figure 2A, lane 6). In a control lysis with 0.2% Triton X-100, trypsin digested extensively H2a precursor and ectodomain fragment; only ∼5% of the initial label remained (Figure 2A, lane 7). After lysis with Triton X-100, even in the absence of trypsin, some label disappeared (Figure 2A, lane 3), probably due to exposure to a solubilized endogenous protease.
Figure 2.
H2a precursor and soluble ectodomain fragment accumulate in a membrane-enclosed compartment upon proteasomal inhibition. (A) After metabolic labeling similar to that in Figure 1 (A) and chase in the absence or presence of 10 μM MG-132, cells were incubated with or without trypsin in Triton X-100 or in low of H2aconcentrations of digitonin, as detailed in MATERIALS AND of H2a METHODS. Cleaved precursor of H2arefers to H2a precursor cleaved by trypsin. The lower panels are of H2aphosphorimager quantitations of this experiment, where the value of 100 represents the total amount of H2a after 3 h chase in the absence of digitonin (lane 4). (B) H2a immunofluorescence on fixed and permeabilized cells expressing H2a preincubated for 5 h with or without proteasome inhibitors at concentrations similar to those in Figure 1. Anti-H2 carboxyterminal were the primary antibodies and Cy3-conjugated goat antirabbit IgG the secondary. Bar = 10 μm.
The accumulation of H2a precursor and ectodomain fragment in an endo H-sensitive state (Figure 1D) suggested a preGolgi localization, probably the ER. Nevertheless, after accumulation with proteasomal inhibitors, H2a appeared surprisingly by immunofluorescence in a juxtanuclear pattern, as opposed to its ER pattern without treatment (Figure 2B).
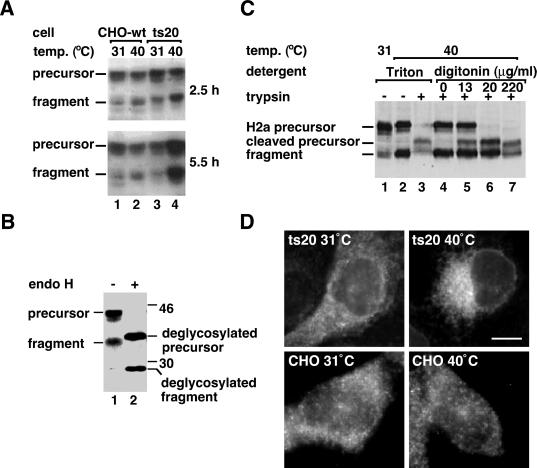
To analyze if the ubiquitination machinery is involved in the degradation of H2a, we stably transfected it into the ts20 cell line, a CHO cell line with a mutant thermosensitive ubiquitin-activating enzyme, E1 (Kulka et al., 1988). Incubation of the ts20 cells, but not of the wild-type cells at the restrictive temperature, caused a marked accumulation of H2a precursor and ectodomain fragment (Figure 3A, lane 4 compared with 2). Accumulated H2a remained endo H-sensitive, even after long incubations at the nonpermissive temperature (Figure 3B). As with proteasomal inhibition, digitonin permeabilization in the presence of trypsin led to a complete cleavage of the precursor to a cytoplasmic-tailless species, while the naturally occurring ectodomain fragment was completely protected (Figure 3C, compare lanes 4 and 6). Thus, also upon inhibition of ubiquitination, H2a precursor remains membrane-bound and the 35-kDa fragment in a membrane-enclosed compartment, and they are not transported to the cytosol. As they also remain in a high mannose, endo H-sensitive form, the compartment must be preGolgi. The subcellular localization in the ts20 cell line at the restrictive temperature also appears juxtanuclear, although less compact than after proteasomal inhibition (Figure 3D).
Figure 3.
Inhibition of ubiquitination causes H2a precursor and soluble ectodomain fragment to accumulate in a membrane-enclosed compartment. (A) A cell line carrying a thermosensitive mutant E1 (ts20 cells) and wild-type cells were both stably transfected with H2a cDNA. Representative clones of transfected cells were grown at 31°C and then incubated for the indicated times at 31°C (permissive temperature) or 40°C (restrictive), followed by lysis, SDS-PAGE, and immunoblotting with anti-H2 carboxyterminal antibodies. A secondary HRP-conjugated goat anti-rabbit IgG was used followed by ECL. (B) Ts20 cells expressing H2a were incubated for 8 h at the restrictive temperature, followed by lysis, treatment without (lane 1) or with endo H (lane 2), SDS-PAGE, and immunoblotting as in (A). (C) Ts20 cells expressing H2a were incubated for 5 h at the indicated temperatures. Before lysis cells were treated with Triton-X-100 or with low concentrations of digitonin in the presence or absence of trypsin as in Figure 2 A. SDS-PAGE and immunoblotting was as in (A). (D) H2a immunofluorescence as in Figure 2 B was visualized on ts20 cells expressing H2a incubated at 31°C or at 40°C for 5 h before fixation. Bar = 10 μm.
H2a Accumulates in a Juxtanuclear Compartment, Different than the Golgi and the ERGIC
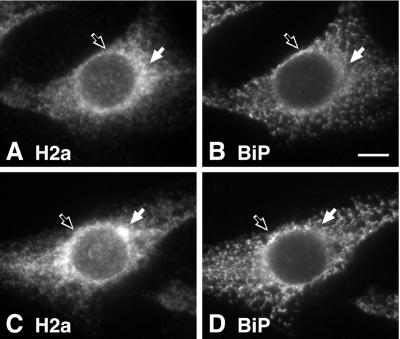
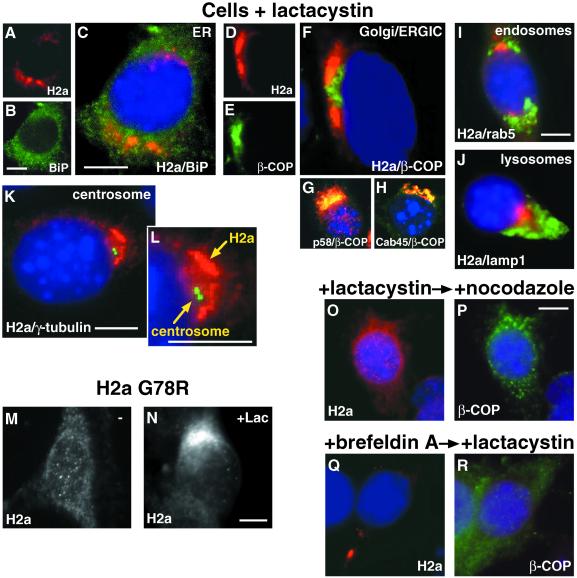
To identify the compartment where H2a accumulates, we performed double- and triple-label immunofluorescence. In the absence of any treatment, most cells showed H2a in an ER-like pattern. However, this pattern was not identical to that of an ER marker, BiP (Figure 4, A and B). H2a concentrated in some perinuclear regions where BiP was reduced (Figure 4, white arrows), and conversely, BiP concentration was higher where H2a concentration was reduced (Figure 4, black arrows). Some cells (∼5%) showed a stronger localized accumulation of H2a in a region where BiP was reduced (Figure 4, C and D, white arrows). In the presence of lactacystin, H2a appeared in most cells in a tight juxtanuclear pattern next to, but not overlapping with, β-COP, a marker of the Golgi and the ERGIC (Figure 5, A-F). We could not perform a direct comparison of H2a with separate Golgi and ERGIC markers as the available antibodies were rabbit, the same as our anti-H2a antibodies, precluding a double labeling. However, in contrast to the nonoverlap of H2a with β-COP, the latter (labeled with a mouse antibody) overlapped extensively with Golgi (Cab45; Scherer et al., 1996) and ERGIC (p58; Saraste and Svensson, 1991) markers (Figure 5, D-H). H2a staining did not colocalize with markers for endosomes or lysosomes either (Figure 5, I and J). H2a accumulated near to but not overlapping with the centrosomes as visualized by staining of γ-tubulin (Figure 5, K and L). To eliminate the possibility that by immunofluorescence we were seeing mainly the H2a soluble ectodomain fragment, we looked at a mutant H2a (G78R) that is not cleaved (Yuk and Lodish, 1993; Neumann et al., 1996). Membrane-bound G78R-H2a accumulated in the presence of lactacystin in the same way as wild-type H2a in the juxtanuclear compartment (Figure 5, M and N).
Figure 4.
H2a is partially concentrated in a juxtanuclear region in some untreated cells. Cells expressing H2a were fixed and permeabilized, and double label immunofluorescence was performed with the use of anti-H2 carboxyterminal antibodies and Cy3-conjugated goat anti-rabbit IgG (A and C) and mouse anti-BiP antibodies with FITC-conjugated goat-anti-mouse IgG (B and D). A and B show a cell representative of the general population. C and D show a cell with an H2a immunofluorescence pattern representing ∼5% of the cells. The white arrows show regions with higher H2a concentration and not BiP and black outlined arrows show regions with higher BiP concentration and not H2a. Bar = 10 μm.
Figure 5.
On proteasomal inhibition accumulated H2a is localized in a nocodazole-sensitive compartment, next to the centrosome, the Golgi and the ERGIC. Cells were incubated for 5 h with 25 μM lactacystin. Triple label immunofluorescence on fixed and permeabilized cells was done with secondary goat-antirabbit IgG conjugated to Cy3 with anti-H2a (panels A, C, D, F, I-O, Q), antip58 (ERGIC) (G) and anti-Cab45 (Golgi; Scherer et al., 1996) (H). FITC-conjugated goat-antimouse IgG was used with anti-BiP (B,C), anti-β-COP (E-H, P, R), antirab5 (I), and anti-γ-tubulin (K, L). FITC-conjugated goat-antirat IgG was used with antilamp1 (J). Nuclei were stained with DAPI. Overlap of Cy3 and FITC in panels G and H appears yellow. Immunofluorescence with anti-H2a antibodies was also done on fixed and permeabilized cells expressing the mutant H2a G78R (uncleavable; Yuk and Lodish, 1993; Neumann et al., 1996) (M) or on the same cells treated with lactacystin (Lac) (N). (O, P) Cells were treated for 3 h with 25 μM lactacystin with the addition of 20 μM nocodazole for an extra 2 h before fixation, permeabilization, and immunofluorescence (Q, R) Cells were treated for 2 h with 5 μg/ml brefeldin A, then with 25 μM lactacystin for an extra 3 h before fixation, permeabilization, and immunofluorescence. Bars = 10 μm.
The Quality Control Compartment Is Dependent on Microtubules and Is Insensitive to Brefeldin A
To see if the accumulation of H2a is reversible, we treated the cells with nocodazole. This treatment dispersed the juxtanuclear pattern of H2a, showing that the compartment is dependent on microtubules and that the accumulation of H2a can be reversed by disrupting them, even in the presence of a proteasome inhibitor (Figure 5, O and P). On the other hand, treatment with brefeldin A changed only slightly the pattern of H2a. The accumulation appeared to be restricted to a more compact area even if cells were first incubated with brefeldin A for 2 h and only then lactacystin was added (Figure 5, Q and R).
Altogether, the results suggest that H2a accumulates in a novel compartment, which we propose to call the “quality control” (QC) compartment.
Triton Solubility and Topology of H2a Compartmentalization Compared with that of Proteasomes
Although we showed that H2a is not transported to the cytosol (Figure 2), the proximity to the centrosome resembles that reported for cytosolic aggregates of mutant cystic fibrosis transmembrane conductance regulator (CFTR), named aggresomes (Johnston et al., 1998). Therefore, we analyzed if H2a forms Triton-insoluble aggregates like mutant CFTR. Figure 6A shows that only an insignificant amount (<1%) of total H2a remains insoluble in 1% Triton-X-100.
Figure 6.
H2a is not aggregated, and it is enclosed in juxtanuclear membranes, while proteasomes are on the cytosolic side. (A) Cells stably expressing H2a were metabolically labeled with [35S] Cys for 20 min and chased with complete medium for 3 h in the absence or in the presence of 100 μM ALLN. They were then lysed and immunoprecipitated as in Figure 1 (A), but immunoprecipitation was done also on material insoluble in Triton-containing buffer (“p”) and not only on the soluble (“s”) as described in MATERIALS AND METHODS. (B-E) Cells were treated for 5 h with 25 μM lactacystin. They were then permeabilized by breakage with a few strokes in a Dounce homogenizer, then adhered to coverslips with polylysine as described in MATERIALS AND METHODS. They were then fixed with methanol/acetone, and immunofluorescence was performed as in Figure 5, with the use of anti-H2a and DAPI (B, C) or antiproteasome antibody and DAPI (D, E). Cy3-conjugated secondary goat antirabbit IgG was used. Bar = 10 μm. (F-I) The same procedure was done as in (B-E), except that incubations with antibodies were done before fixation with methanol/acetone. (J-M) The same procedure was done as in (B-E), except that cells were treated with 0.2% Triton-X-100 before adhering to the coverslips.
Proteasomes are located in the cytosol and the nucleus and were recently also found on centrosomes (Wigley et al., 1999) and on the cytosolic side of the ER membrane (Enenkel et al., 1998; Rivett, 1998; Hori et al., 1999). These proteasomes could possibly provide the energy required for the retrotranslocation of ER substrates, through the ATPases that exist at their 19S subunits. To analyze the topology of proteasomal localization, compared with that of H2a, we studied the immunofluorescence pattern in cells permeabilized by mild breakage in a Dounce homogenizer. After lactacystin treatment H2a as well as proteasomes appeared in membranes close to the nucleus (Figure 6, B and D). In addition, proteasomes appeared inside the nucleus as expected (Figure 6D). Without permeabilizing these internal membranes with methanol/acetone, staining of H2a with an anticarboxyterminal peptide (luminal) antibody was very faint. (Figure 6F). This suggests that the C terminus is not accessible from the cytosolic side and must be in the luminal side of the membranes. The faint residual staining may be the result of incomplete resealing of the internal membranes after breakage in the homogenizer. In contrast, proteasomes were readily accessible, as they must be on the cytosolic side of the membranes (Figure 6H). After treatment of the broken cells with Triton-X-100, staining of H2a disappeared, as well as extranuclear staining for proteasomes, suggesting that the localization of both depends on a membrane structure (Figure 6, J and L). Nuclear staining of proteasomes was not affected by Triton (Figure 6L).
H2a Associates Increasingly with Sec61-β after Synthesis
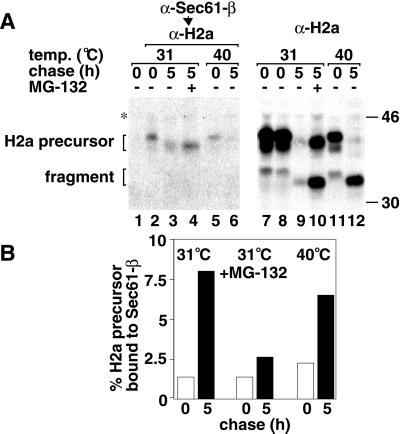
Degradation of H2a by the proteasome must involve movement to the cytosol. As we could not find direct evidence of dislocation of H2a to the cytosol, we analyzed whether it associates strongly with Sec61-β subunit, as was seen with other proteins (Wiertz et al., 1996b; Bebok et al., 1998). Immunoprecipitation with anti-Sec61-β antibodies, elution, and recapture with anti-H2a resulted in a specific interaction of H2a with Sec61-β in ts20 cells at the permissive or restrictive (ubiquitination inhibited) temperatures (Figure 7A, lanes 2 and 5). Replacement of anti-Sec61-β with a control preimmune antibody showed no coprecipitation of H2a (Figure 7A, lane 1). The fraction of H2a bound to Sec61 (Figure 7A, left panel) vs. the total amount of H2a present in each sample (Figure 7A, right panel), increased with chase time, even as the total amount of H2a decreased by degradation (Figure 7B). The increase in the fraction of H2a bound to Sec61 after 5-h chase was 5-fold compared with that bound after the pulse (Figure 7B). Association to Sec61 increased with time even when proteasomal activity or ubiquitination were inhibited, by ∼2 and 3-fold respectively (Figure 7B), suggesting that these processes are not necessary for the tight interaction to occur. In Figure 7, more clearly than in the previous figures, a shift of H2a bands to a faster mobility can be seen upon chase, which, as mentioned before, is due to mannose trimming of the sugar chains. No 35-kDa soluble H2a fragment was found in association with Sec61 (Figure 7A), even though there is a large accumulation of total fragment in the presence of MG-132 or after chase at the restrictive temperature (Figure 7A, lanes 10 and 12).
Figure 7.
H2a interacts with Sec61-β increasingly with chase time. (A) Ts20 cells expressing H2a were metabolically labeled, as in Figure 1 (A), at the indicated temperatures, in the absence or presence of 10 μM MG-132, solubilized with 1% digitonin in 20 mM HEPES, pH 7.6, and each sample was divided into 2 identical halves. These were immunoprecipitated with anti-Sec61-β antibodies (left panel) or with anti-H2a antibodies (right panel). Lane 1 is a control with preimmune antibodies instead of anti-Sec61-β. Immunoprecipitates in the right panel were run on SDS-PAGE, whereas those in the left panel were reimmunoprecipitated with anti-H2a after boiling in SDS containing buffer and addition of excess buffer A as described in Materials and Methods. The change in migration of H2a bands after chase is due to mannose trimming (Frenkel and Lederkremer, unpublished results). (B) Quotients of phosphorimager quantitation of H2a precursor bands in the left panel in (A) (Sec61-associated H2a) over those on the right panel (total H2a) were plotted for each treatment after pulse-labeling (white bars) or after 5 h chase (black bars).
On Proteasomal Inhibition, Free Class I MHC Heavy Chains Are Compartmentalized in the Region of the Quality Control Compartment
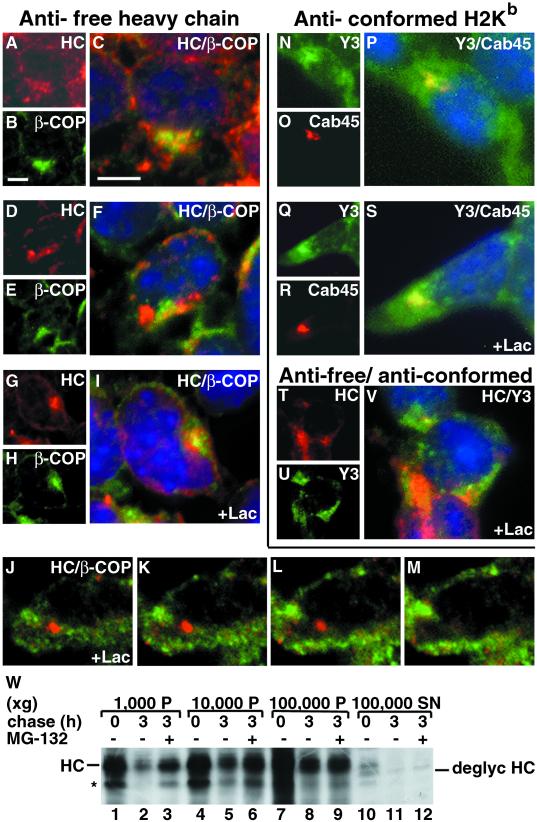
To investigate if another protein substrate that associates with Sec61 on the way to proteasomal degradation shows a localization similar to H2a, we studied the fate of free class I MHC heavy chains. Free class I heavy chains remain in all circumstances endo H- sensitive until degradation (Fromm et al., 1998). With the use of an antibody specific for mouse free class I heavy chains (HC), we observed that most untreated cells show a distributed ER-like pattern with partial overlap with β-COP staining (Figure 8, A-C). However, ∼10% of the cells show a local concentration next to, but not overlapping with, β-COP (Figure 8, D-F), similar to what we saw with H2a (Figure 5). After treatment with lactacystin for 5 h, most cells showed a strong accumulation in this region (Figure 8, G-I). As in some cells, the heavy chains seem to partially overlap with β-COP (Figure 8, I) we studied optical sections with a confocal microscope. Panels J to M show representative sequential optical sections in which the heavy chain accumulation is seen at a higher plane in the cell (Figure 8, J), whereas β-COP staining is stronger in a lower plane (Figure 8, M), where heavy chains are absent. Therefore, the apparent partial overlap of heavy chains and β-COP in images from the conventional fluorescence microscope are due to the juxtaposition of the 2 distinct compartments. In contrast, antibodies directed against assembled conformed class I complexes showed staining in the plasma membrane, the ER, and, most prominently, in the Golgi region, entirely overlapping with a Golgi marker, Cab45 (Figure 8, N-P). This pattern did not change in the presence of lactacystin (Figure 8, Q-S). Double labeling with the antifree heavy chain plus the anti-class I conformed complex antibodies showed a different pattern for the peak fluorescence for each of these proteins, indicating a differential compartmentalization (Figure 8, T-V).
Figure 8.
Misfolded unassembled MHC class I heavy chains accumulate in a compartment, next to the Golgi and the ERGIC. (A) Triple label immunofluorescence on fixed and permeabilized A5O5 cells was done with secondary goat-antirabbit IgG conjugated to Cy3 with rabbit antimouse MHC class I free heavy chain antibodies (HC) (panels A, C, D, F, G, I-M, T, V) and anti-Cab45 (O, P, R, S). FITC-conjugated goat-antimouse IgG was used with anti-β-COP (B, C, E, F, H, I-M) and mouse antibodies against conformed assembled MHC class I H2Kb complexes (Y3) (N, P, Q, S, U, V). Nuclei were stained with DAPI in C, F, I, V. Cells in G-M and Q-V were incubated for 5 h with 25 μM lactacystin before fixation and immunofluorescence. Bars = 10 μm. (W) A5O5 cells were metabolically labeled with [35S] Met for 30 min and chased with complete medium in the absence or presence of 30 μM MG-132. Cells were then homogenized and fractionated, as detailed in MATERIALS AND METHODS. Lysates of each fraction pellet (P) or supernatant (SN) were immunoprecipitated with antifree heavy chain antibodies. Immunoprecipitates were analyzed by SDS-PAGE followed by fluorography. The bands corresponding to heavy chains (HC), deglycosylated heavy chains (deglyc HC) and a possible degradation product (*) are indicated.
To analyze if indeed most of the free HCs are compartmentalized, we performed a subcellular fractionation experiment. After pulse-labeling or chases in the absence or presence of MG-132, most HCs appeared in the membrane-bound fractions corresponding to the 10,000 × g and 100,000 × g pellets (93% of the total for the pulse, 91.7% for the chase without MG-132, and 93.1% with MG-132) (Figure 8W, lanes 4–9). Only a small portion appeared in the cytosolic 100,000 × g supernatants (7% of the total for the pulse, 8.3% for the chase without MG-132, and 6.9% with MG-132) (Figure 8W, lanes 10–12). This small amount of protein transported to the cytosol appeared to be partially deglycosylated as was reported for MHC class I heavy chains in cells expressing CMV proteins (Wiertz et al., 1996a) or in β(2)-microglobulin-deficient and TAP-deficient cell lines (Hughes et al., 1997).
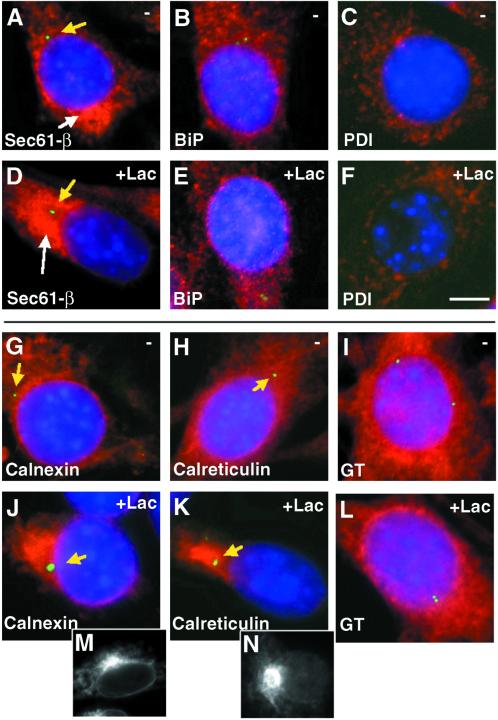
Sec61-β, Calnexin, and Calreticulin Accumulate Near the Centrosome upon Proteasomal Inhibition
The pattern of Sec61 immunofluorescence showed an ER distribution, but with a stronger accumulation on 1 side of the nucleus, not near the centrosome (Figure 9A). In the presence of lactacystin there was also a region where Sec61 accumulated, but in this case it was near the centrosome, possibly in the QC compartment (Figure 9D). This redistribution of Sec61 might reflect its active role in the retrotranslocation.
Figure 9.
Calnexin, calreticulin, and also partially Sec61-β accumulate next to the centrosome in the presence of lactacystin. Triple-label immunofluorescence (as in Figure 5) was performed on 3T3 cells expressing H2a preincubated without (A-C and G-I) or with 25 μM lactacystin (D-F and J-L), with the use of DAPI, FITC-conjugated goat antimouse IgG with anti-γ-tubulin, and Cy3-conjugated goat antirabbit IgG with anti-Sec61-β (A, D), anti-BiP (B, E), anti-PDI (C, F), anticalnexin (G, J), anticalreticulin (H, K), and anti-UDPGlc:glycoprotein glucosyltransferase (GT) (I, L). Bar = 10 μm. The yellow arrows point out the location of the centrosomes. Single label immunofluorescence is shown in the bottom panels for calnexin (M) and calreticulin (N) in cells incubated with lactacystin.
We investigated the distribution of the ER chaperones BiP, PDI, calnexin, and calreticulin after treatment of cells with proteasome inhibitors. While all these proteins showed a scattered ER pattern under normal conditions, treatment with lactacystin caused only calnexin and calreticulin to concentrate dramatically to a region near the centrosome, probably the QC compartment (Figure 9, J, K, M, and N). The relocalization of Sec61 and the 2 chaperones must be a response to the accumulation of proteins on the way to degradation. On the other hand, UDPGlc:glycoprotein glucosyltransferase, (the enzyme in charge of reglucosylation of misfolded glycoproteins [Labriola et al., 1995]) did not redistribute upon incubation of the cells with lactacystin (Figure 9, I and L).
DISCUSSION
Some substrates of the ER/proteasomal pathway had been found in the cytosol before degradation occurs, e.g., TCR-α (Huppa and Ploegh, 1997; Yu et al., 1997). The fact that we cannot detect H2a in the cytosol or on the cytosolic side of the membranes is probably the result of very rapid degradation after retrotranslocation. Deglycosylated cytosolic intermediates had been found during the degradation of other proteins from the ER (Yu et al., 1997). We did not observe deglycosylation of H2a (Figure 1), which is consistent with the fact that we cannot find molecules transported to the cytosol. Movement of a protein from the ER to the cytosol for degradation was seen for a few proteins (Hiller et al., 1996; Wiertz et al., 1996b; Yu et al., 1997; de Virgilio et al., 1998) and not for others (Chillaron and Haas, 2000; Fisher et al., 1997; McGee et al., 1996; Schubert et al., 1998). In the case of free class I MHC heavy chains, we can see a small amount in the cytosol when blocking degradation, although most are confined to a membrane-bound compartment (Figure 8). To unify the differing results with different proteins we could envisage a mechanism of reverse translocation that is coupled to the ubiquitin/proteasome degradation, as has recently been hypothesized (Hirsch and Ploegh, 2000). This coupling is sometimes so tight that it does not allow detection of the protein substrate in the cytosol before degradation occurs. Consistent with this coupling model, it was seen that inhibition of ubiquitination blocked the retrotranslocation of mutant cpy to the cytosol in yeast (Biederer et al., 1997) and of mutant ribophorin I and TCR-α in mammalian cells (de Virgilio et al., 1998; Yu and Kopito, 1999), whereas proteasomal inhibition prevented the dislocation of a membrane chimeric protein to the cytosol in yeast (Mayer et al., 1998). Our results show that in mammalian cells, after inhibition of ubiquitination or of proteasomal activity, neither the membrane-bound precursor nor the soluble ectodomain fragment of H2a are transported to the cytosol. Thus, the retrotranslocation depends on the process of degradation. One possibility is that the proteasomes responsible for this degradation are located at the cytosolic surface of the membrane of the quality control compartment. This is consistent with the fact that, like this compartment, proteasomes are also found near the centrosome (Wigley et al., 1999). Proteasomes are also on the cytosolic side of the ER membrane (Enenkel et al., 1998; Rivett, 1998; Hori et al., 1999), and there is evidence that degradation of other proteins (e.g., CFTR) starts before dislocation to the cytosol, presumably by proteasomes recruited to the membrane (Xiong et al., 1999).
The QC compartment is characterized by a juxtanuclear concentration of the substrates for ER/proteasomal degradation upon inhibition of the proteasomes or of ubiquitination (Figures 2–5, 8). The existence of cells that show the same type of juxtanuclear protein accumulation without any treatment (Figures 4C and 8F) suggests that this process occurs naturally. Cells that show this accumulation without treatment may be expressing higher levels of the proteins or alternatively may be degrading them more slowly. The concentration of Sec61, calreticulin, and calnexin in the region of the QC compartment in the presence of proteasome inhibitor (Figure 9) suggests that it could be a subcompartment of the ER. However, in contrast to the bulk of the ER, it is dependent on microtubules (sensitive to nocodazole, Figure 5O). It might therefore be a separate compartment/organelle, and future work should address this interesting possibility. The insensitivity to brefeldin A (Figure 5Q) suggests that protein accumulation in the QC compartment is an ARF-independent process. Raposo et al. had observed that free human class I MHC HCs localize (without proteasomal inhibition) to a region of the cell that was interpreted as an expanded ERGIC (Raposo et al., 1995). The QC compartment is, in fact, in an ERGIC-adjacent region, but it does not overlap with β-COP. On the other hand, β-COP shows extensive overlap with the ERGIC marker p58 (rodent homologue of ERGIC-53) (Figure 5K). No overlap of the QC compartment with β-COP was seen even after incubation of cells at 15°C (our unpublished results), which is known to cause protein accumulation in the ERGIC. The segregation between the biosynthetic ERGIC and Golgi compartments and the QC compartment can be seen most interestingly with the same protein in 2 different states, when visualizing the localization of conformed class I complexes that are concentrated in the ERGIC and Golgi on the way to the plasma membrane in comparison to the localization of free class I HC, possibly on the way to the proteasomes (Figure 8, P-R).
The accumulation of calnexin and calreticulin upon proteasomal inhibition in a subcellular region that is probably the QC compartment (Figure 9) is consistent with the proposed role of these chaperones in protein quality control from the ER (Hammond and Helenius, 1995). We also showed extended association of H2a to calnexin (Shenkman et al., 1997). Interestingly, calnexin concentration seems to be reduced and the glucosyltransferase enhanced in ER exit sites (Cannon and Helenius, 1999), the opposite of what we found in the QC compartment. It was reported that Sec61 was found in the ERGIC, in addition to the ER (Greenfield and High, 1999). The change in the pattern of Sec61 localization that we see in the presence of lactacystin (Figure 9) could reflect a partial redistribution to the QC compartment.
Association of H2a to Sec61 increased progressively with time after synthesis (Figure 7), suggesting a reassociation event as occurs with class I MHC in the presence of cytomegalovirus proteins (Wiertz et al., 1996b). The fact that the fraction of H2a associated with Sec61 increases with time while total H2a decreases because of degradation suggests that interaction with Sec61 could be the rate-limiting step during retrotranslocation. We cannot rule out that, instead of a reassociation event, a fraction of H2a remains associated to Sec61 from its initial translocation. However, to explain the results with this alternative would require that the associated fraction be very stable and not be degraded over time, an unlikely scenario. In the case of the degradation of a mutant carboxypeptidase Y in yeast, it was shown that indeed there is reassociation to Sec61 and not a persistence after import (Plemper et al., 1999). The soluble H2a ectodomain fragment was not found in association with Sec61 (Figure 7A). We could speculate that, if the soluble fragment is degraded by a mechanism similar to that followed by the membrane-bound precursor, interaction with Sec61 might take place through a membrane-bound mediator like calnexin. The ternary complex H2a ectodomain fragment/mediator/sec61 might not survive after cell lysis.
The proteasomal degradation of H2a is not determined by misfolding or aggregation as in the case of many mutant proteins. We did not find H2a accumulated upon proteasomal inhibition to be aggregated (Figure 6), and we do not see a global misfolding of H2a (Shenkman et al., 1997; Ayalon-Soffer et al., 1999a). The recognition of membrane-bound H2a precursor for ER retention is determined by a pentapeptide on the luminal side of its membrane span (Shenkman et al., 1997). Degradation follows after a certain lag period if retained precursor molecules fail to be cleaved. This step is modulated by sugar trimming events (Ayalon-Soffer et al., 1999b). As for cleaved ectodomain molecules that fail to be secreted, it is less clear how the recognition for proteasomal degradation occurs. However, inhibition of the proteasomal machinery allows cleavage of H2a precursor molecules and rescue of cleaved ectodomain fragment molecules that are then secreted, essentially reaching completion (Figure 1). This suggests a competition between cleavage and secretion on the one hand and signaling for proteasomal disposal on the other.
The similar localization of H2a, MHC class I (Figure 8), and CFTR (Johnston et al., 1998) upon proteasomal inhibition could be explained as follows. The proteins could first be transported to the QC compartment. Subsequently, there could be retrotranslocation for proteasomal degradation to occur. Three alternatives would exist when proteasomal degradation is insufficient or inhibited, depending on how tight the coupling between the retrotranslocation and the degradation is. The protein may be transported to the cytosolic side of the membranes to a small degree, as for MHC class I, or to a large degree, where it may form large aggregates as in the case of CFTR aggresomes (Johnston et al., 1998), or it may remain in the QC compartment as occurs for H2a. Notably, the action of the proteasome inhibitors leads to an important increase in secretion of H2a ectodomain fragment and of Golgi-processing of membrane-bound H2b, implying that the QC compartment must be connected to the secretory pathway. Therefore, the QC compartment could be a sort of purgatory, where the fate of a protein is decided—between its rescue and its demise.
ACKNOWLEDGMENTS
We thank Aaron Ciechanover for the ts20 cell line, Hugh Rosen, Marino Zerial, Peter-M. Kloetzel, Jakko Saraste, Tom Rapoport, and Armando Parodi for antibodies, and Orna Elroy-Stein for critically reading the manuscript. This work was supported by the Israel Science Foundation, founded by The Israeli Academy of Sciences, and by grants from the Israel Cancer Association and the Israeli Ministry of Health to G. L. and from the US-Israel and German-Israel Science Foundations to R. E.
Abbreviations used:
- ALLM
N-acetyl-leucyl-leucyl-methional
- ALLN
N-acetyl-leucyl-leucyl-norleucinal
- ASGPR
asialoglycoprotein receptor
- CFTR
cystic fibrosis transmembrane conductance regulator
- endo H
endo-N- acetylglucosaminidase H
- ER
endoplasmic reticulum
- ERGIC
ER-to-Golgi intermediate compartment
- HC
heavy chains
- MHC
major histocompatibility complex
- MG-132
N-carbobenzoxyl-leucinyl-leucinyl-leucinal
- QC
quality control
- TLCK
tosyl-lysylchloromethylketone
REFERENCES
- Amara JF, Lederkremer G, Lodish HF. Intracellular degradation of unassembled asialoglycoprotein receptor subunits: a pre-Golgi, nonlysosomal endoproteolytic cleavage. J Cell Biol. 1989;109:3315–3324. doi: 10.1083/jcb.109.6.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalon-Soffer M, Kamhi-Nesher S, Lederkremer GZ. Folding and self-assembly do not prevent ER retention and proteasomal degradation of asialoglycoprotein receptor H2a. FEBS Lett. 1999a;460:112–116. doi: 10.1016/s0014-5793(99)01321-6. [DOI] [PubMed] [Google Scholar]
- Ayalon-Soffer M, Shenkman M, Lederkremer GZ. Differential role of mannose and glucose trimming in the ER degradation of asialoglycoprotein receptor subunits. J Cell Sci. 1999b;112:3309–3318. doi: 10.1242/jcs.112.19.3309. [DOI] [PubMed] [Google Scholar]
- Bebok Z, Mazzochi C, King SA, Hong JS, Sorscher EJ. The mechanism underlying cystic fibrosis transmembrane conductance regulator transport from the endoplasmic reticulum to the proteasome includes Sec61β and a cytosolic, deglycosylated intermediary. J Biol Chem. 1998;273:29873–29878. doi: 10.1074/jbc.273.45.29873. [DOI] [PubMed] [Google Scholar]
- Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Weissman AM. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon KS, Helenius A. Trimming and readdition of glucose to N-linked oligosaccharides determines calnexin association of a substrate glycoprotein in living cells. J Biol Chem. 1999;274:7537–7544. doi: 10.1074/jbc.274.11.7537. [DOI] [PubMed] [Google Scholar]
- Chillaron J, Haas IG. Dissociation from BiP and retrotranslocation of unassembled immunoglobulin light chains are tightly coupled to proteasome activity. Mol Biol Cell. 2000;11:217–226. doi: 10.1091/mbc.11.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Virgilio M, Weninger H, Ivessa NE. Ubiquitination is required for the retro-translocation of a short-lived luminal endoplasmic reticulum glycoprotein to the cytosol for degradation by the proteasome. J Biol Chem. 1998;273:9734–9743. doi: 10.1074/jbc.273.16.9734. [DOI] [PubMed] [Google Scholar]
- Enenkel C, Lehmann A, Kloetzel PM. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 1998;17:6144–6154. doi: 10.1093/emboj/17.21.6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher EA, Zhou MY, Mitchell DM, Wu XJ, Omura S, Wang HX, Goldberg AL, Ginsberg HN. The degradation of apolipoprotein B100 is mediated by the ubiquitin-proteasome pathway and involves heat shock protein 70. J Biol Chem. 1997;272:20427–20434. doi: 10.1074/jbc.272.33.20427. [DOI] [PubMed] [Google Scholar]
- Frentzel S, Pesold-Hurt B, Seelig A, Kloetzel PM. 20 S proteasomes are assembled via distinct precursor complexes. Processing of LMP2 and LMP7 proproteins takes place in 13–16 S preproteasome complexes. J Mol Biol. 1994;236:975–981. doi: 10.1016/0022-2836(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Fromm SV, Mey-Tal SW, Coligan JE, Schechter C, Ehrlich R. MHC class I heavy chain mRNA must exceed a threshold level for the reconstitution of cell surface expression of class I MHC complexes in cells transformed by the highly oncogenic adenovirus 12. J Biol Chem. 1998;273:15209–15216. doi: 10.1074/jbc.273.24.15209. [DOI] [PubMed] [Google Scholar]
- Geier E, Pfeifer G, Wilm M, Lucchiari-Hartz M, Baumeister W, Eichmann K, Niedermann G. A giant protease with potential to substitute for some functions of the proteasome. Science. 1999;283:978–981. doi: 10.1126/science.283.5404.978. [DOI] [PubMed] [Google Scholar]
- Greenfield JJ, High S. The Sec61 complex is located in both the ER and the ER-Golgi intermediate compartment. J Cell Sci. 1999;112:1477–1486. doi: 10.1242/jcs.112.10.1477. [DOI] [PubMed] [Google Scholar]
- Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Hirsch C, Ploegh HL. Intracellular targeting of the proteasome. Trends Cell Biol. 2000;10:268–272. doi: 10.1016/s0962-8924(00)01768-2. [DOI] [PubMed] [Google Scholar]
- Hori H, Nembai T, Miyata Y, Hayashi T, Ueno K, Koide T. Isolation and characterization of two 20S proteasomes from the endoplasmic reticulum of rat liver microsomes. J Biochem (Tokyo) 1999;126:722–730. doi: 10.1093/oxfordjournals.jbchem.a022509. [DOI] [PubMed] [Google Scholar]
- Hughes EA, Hammond C, Cresswell P. Misfolded major histocompatibility complex class I heavy chains are translocated into the cytoplasm and degraded by the proteasome. Proc Natl Acad Sci USA. 1997;94:1896–1901. doi: 10.1073/pnas.94.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa JB, Ploegh HL. The alpha chain of the T cell antigen receptor is degraded in the cytosol. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: A cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Janeway CA., Jr Cooperative interaction of B lymphocytes with antigen-specific helper T lymphocytes is MHC restricted. Nature. 1981;292:547–549. doi: 10.1038/292547a0. [DOI] [PubMed] [Google Scholar]
- Kulka RG, Raboy B, Schuster R, Parag HA, Diamond G, Ciechanover A, Marcus M. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme, E1. J Biol Chem. 1988;263:15726–15731. [PubMed] [Google Scholar]
- Labriola C, Cazzulo JJ, Parodi AJ. Retention of glucose units added by the UDP-GLC:glycoprotein glucosyltransferase delays exit of glycoproteins from the endoplasmic reticulum. J Cell Biol. 1995;130:771–779. doi: 10.1083/jcb.130.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederkremer GZ, Lodish HF. An alternatively spliced miniexon alters the subcellular fate of the human asialoglycoprotein receptor H2 subunit. Endoplasmic reticulum retention and degradation or cell surface expression. J Biol Chem. 1991;266:1237–1244. [PubMed] [Google Scholar]
- Machold RP, Andree S, Van Kaer L, Ljunggren HG, Ploegh HL. Peptide influences the folding and intracellular transport of free major histocompatibility complex class I heavy chains. J Exp Med. 1995;181:1111–1122. doi: 10.1084/jem.181.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer TU, Braun T, Jentsch S. Role of the proteasome in membrane extraction of a short-lived ER- transmembrane protein. EMBO J. 1998;17:3251–3257. doi: 10.1093/emboj/17.12.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee TP, Cheng HH, Kumagai H, Omura S, Simoni RD. Degradation of 3-hydroxy-3-methylglutaryl-CoA reductase in endoplasmic reticulum membranes is accelerated as a result of increased susceptibility to proteolysis. J Biol Chem. 1996;271:25630–25638. doi: 10.1074/jbc.271.41.25630. [DOI] [PubMed] [Google Scholar]
- Morgenstern JP, Land H. Advanced mammalian gene transfer: high titer retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucl Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann D, Yuk MH, Lodish HF, Lederkremer Z. Blocking intracellular degradation of the erythropoietin and asialoglycoprotein receptors by calpain inhibitors does not result in the same increase in the levels of their membrane and secreted forms. Biochem J. 1996;313:391–399. doi: 10.1042/bj3130391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Schekman R, Romisch K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 1997;16:4540–4548. doi: 10.1093/emboj/16.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Plemper RK, Deak PM, Otto RT, Wolf DH. Re-entering the translocon from the lumenal side of the endoplasmic reticulum. Studies on mutated carboxypeptidase yscY species. FEBS Lett. 1999;443:241–245. doi: 10.1016/s0014-5793(98)01724-4. [DOI] [PubMed] [Google Scholar]
- Raposo G, Vansanten HM, Leijendekker R, Geuze HJ, Ploegh HL. Misfolded major histocompatibility complex class I molecules accumulate in an expanded ER-Golgi intermediate compartment. J Cell Biol. 1995;131:1403–1419. doi: 10.1083/jcb.131.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivett AJ. Intracellular distribution of proteasomes. Curr Opin Immunol. 1998;10:110–114. doi: 10.1016/s0952-7915(98)80040-x. [DOI] [PubMed] [Google Scholar]
- Saraste J, Svensson K. Distribution of the intermediate elements operating in ER to Golgi transport. J Cell Sci. 1991;100:415–30. doi: 10.1242/jcs.100.3.415. [DOI] [PubMed] [Google Scholar]
- Scherer PE, Lederkremer GZ, Williams S, Fogliano M, Baldini G, Lodish HF. Cab45, a novel Ca2+-binding protein localized to the Golgi lumen. J Cell Biol. 1996;133:257–268. doi: 10.1083/jcb.133.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Bacik I, Cox JH, Bour S, Bennink JR, Orlowski M, Strebel K, Yewdell JW. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J Virol. 1998;72:2280–2288. doi: 10.1128/jvi.72.3.2280-2288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenkman M, Ayalon M, Lederkremer GZ. Endoplasmic reticulum quality control of asialoglycoprotein receptor H2a involves a determinant for retention and not retrieval. Proc Natl Acad Sci USA. 1997;94:11363–11368. doi: 10.1073/pnas.94.21.11363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T, Wolf DH. Endoplasmic reticulum degradation: reverse protein flow of no return. FASEB J. 1997;11:1227–1233. doi: 10.1096/fasebj.11.14.9409541. [DOI] [PubMed] [Google Scholar]
- Tolchinsky S, Yuk MH, Ayalon M, Lodish HF, Lederkremer GZ. Membrane-bound versus secreted forms of human asialoglycoprotein receptor subunits: role of a juxtamembrane pentapeptide. J Biol Chem. 1996;271:14496–14503. doi: 10.1074/jbc.271.24.14496. [DOI] [PubMed] [Google Scholar]
- Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996a;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- Wiertz EJHJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, Rapoport TA, Ploegh HL. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996b;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- Wigley WC, Fabunmi RP, Lee MG, Marino CR, Muallem S, DeMartino GN, Thomas PJ. Dynamic association of proteasomal machinery with the centrosome. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikstrom L, Lodish HF. Nonlysosomal, pre-Golgi degradation of unassembled asialoglycoprotein receptor subunits: A TLCK- and TPCK-sensitive cleavage within the ER. J Cell Biol. 1991;113:997–1007. doi: 10.1083/jcb.113.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Chong E, Skach WR. Evidence That Endoplasmic Reticulum (ER)-associated Degradation of Cystic Fibrosis Transmembrane Conductance Regulator Is Linked to Retrograde Translocation from the ER Membrane. J Biol Chem. 1999;274:2616–2624. doi: 10.1074/jbc.274.5.2616. [DOI] [PubMed] [Google Scholar]
- Yu H, Kaung G, Kobayashi S, Kopito RR. Cytosolic degradation of T-cell receptor alpha chains by the proteasome. J Biol Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- Yu H, Kopito RR. The role of multiubiquitination in dislocation and degradation of the alpha subunit of the T cell antigen receptor. J Biol Chem. 1999;274:36852–36858. doi: 10.1074/jbc.274.52.36852. [DOI] [PubMed] [Google Scholar]
- Yuk MH, Lodish HF. Two Pathways for the Degradation of the H2 Subunit of the Asialoglycoprotein Receptor in the Endoplasmic Reticulum. J Cell Biol. 1993;123:1735–1749. doi: 10.1083/jcb.123.6.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Schekman R. The engagement of Sec61p in the ER dislocation process. Mol Cell. 1999;4:925–934. doi: 10.1016/s1097-2765(00)80222-1. [DOI] [PubMed] [Google Scholar]