Abstract
Background. Studies of nontypeable Haemophilus influenzae (NTHi) have demonstrated that a number of genes associated with infectivity have long repeat regions associated with phase variation in expression of the respective gene. The purpose of this study was to determine the genes that underwent phase variation during a 6-day period of experimental human nasopharyngeal colonization.
Methods. Strain NTHi 2019StrR1 was used to colonize the nasopharynx of human subjects in a study of experimental colonization. Thirteen phase-variable genes were analyzed in NTHi 2019StrR1. Samples of NTHi 2019StrR1 were cultured from subjects during the 6-day colonization period. We used capillary electrophoresis and Roche 454 pyrosequencing to determine the number of repeats in each gene from each sample.
Results. A significant number of samples switched licA and igaB from phase off in the inoculated strain to phase on during the 4-day period of observation. lex2A also showed variability as compared to baseline, but the differences were not significant. The remaining genes showed no evidence of phase variation.
Conclusions. Our studies suggest that the phase-on genotypes of licA and igaB are important for early human nasopharynx colonization. lex2A showed a trend from phase off to phase on, suggesting a potentially important role in the colonization process.
Keywords: Haemophilus influenzae, phase variation, licA, igaB, lex2A nasopharyngeal colonization
(See the editorial commentary by Inzana on pages 713–6.)
Phase variation is described as a random process by which a clonal population of microbes can present heterogeneous phenotypes as a result of a reversible genetic event [1, 2]. The process can involve several mechanisms, including slipped-strand mispairing (SSM), site-specific recombination, and epigenetic regulation mediated by DNA methylation [2]. SSM is a mechanism found in human pathogens, including pathogenic Neisseria, Bordetella pertussis, Haemophilus influenzae, and Helicobacter pylori. The resulting frame shift causes on-off changes in gene expression [3]. The process can occur at rates as high as 2–3 × 10−4 colony forming units [3]. Nontypeable H. influenzae (NTHi), which colonizes human mucosal surfaces and can cause disease, has genes with long tandem repeats that undergo high rates of phase variation [3]. In 1989, Weiser et al identified a tetranucleotide repeat region (5′-CAAT-3′) in licA that was responsible for phase-variable expression of that gene in H. influenzae type b [4]. Subsequent work showed that a number of genes in the H. influenzae chromosome had similar characteristics and underwent phase variation. These genes are associated with iron regulation, membrane protein expression, and lipooligosaccharide (LOS) biosynthesis [5–9]. The opportunity to directly study the effect of experimental human colonization on changes in the phase-on, phase-off status of NTHi phase-varying genes was presented to us. This article describes the results of that study.
MATERIALS AND METHODS
Strains
NTHi 2019 is a clinical isolate obtained from a patient with chronic obstructive pulmonary disease (COPD) and chronic bronchitis in 1985. NTHi 2019StrR1 was created from NTHi 2019 by using a specific chromosomal point mutation at nucleotide 128, resulting in a change in amino acid 43 from lysine to arginine in the ribosomal RNA gene, rpsL, that encodes the S12 polypeptide to encode streptomycin resistance. NTHi 2019StrR1 was susceptible to amoxicillin, doxycycline, trimethoprim-sulfamethoxazole, cefuroxime-axetil, ceftriaxone, and ciprofloxacin. This organism was resistant to streptomycin (minimum inhibitory concentration, ≥256 μg/mL). This strain was used as the inoculation strain for a National Institutes of Health– and Food and Drug Administration–approved human experimental colonization study (unpublished data). Forty-eight samples of NTHi 2019StrR1 were recovered from 15 subjects who participated in the 2 studies of NTHi human experimental nasopharyngeal colonization over a 6-day colonization period (unpublished data). Pharyngeal swab samples were streaked on brain heart infusion agar containing protoporphyrin IX, nicotinamide adenine dinucleotide, and 200 μg/mL streptomycin. All of the colonies on these plates were combined, frozen at −80°C, and served as the samples used in this study. These samples were confirmed as NTHi and cultured only 1 subsequent time for DNA isolation [10].
DNA Isolation and Polymerase Chain Reaction (PCR)
DNA was isolated from each of the 48 samples, using the Masterpure DNA purification kit from EpiCentre Biotechnologies (Madison, WI). As a zero-time baseline, DNA was isolated from randomly selected NTHi 2019StrR1 freezer stock vials used to inoculate the subjects. DNA from 5 separate stocks was used in the pyrosequencing studies and in the capillary electrophoresis studies. PCR was performed using primers obtained from Integrated DNA Technologies (Iowa City, IA). Primers were labeled with 6-carboxyfluorescein for capillary electrophoresis sequencing or with MID codes to identify each subject sample for pyrosequencing shown in Supplementary Table 1. PCR was performed as previously described. The primers used are shown in Supplementary Table 2. The amplicons were purified using the Gel/PCR DNA Fragments Extraction Kit from IBI Scientific (Peosta, IA), and quantitation was performed on a Thermo Scientific NanoDrop.
Capillary Electrophoresis to Determine Nucleotide Repeat Length
Initial studies used a modified version of the capillary electrophoresis method developed by Fox et al to determine the length of the tandem repeats in PCR products for licA, lic3A, and igaB, using Peak Scanner software (Applied Biosystems International) [11]. The primers used in the capillary electrophoresis studies are shown in Supplementary Table 2.
Amplicon Sequencing
In the second half of the study, changes in the number of repeats of 12 of the 13 phase variable genes with tandem repeats were analyzed simultaneously by pyrosequencing, using the GS FLX Titanium System (Roche, Branford, CT) according to the manufacturer's recommended protocols. Pyrosequencing was performed by the University of Iowa DNA Facility and involved a 4-region slide, in which amplicon fragments of isolated DNA from each day were placed into a separate region on the chip. Sequencing libraries, prepared using the GS FLX Titanium Rapid Library Preparation Kit (Roche), were started with a modified fragment end repair step. The modified fragment end repair reaction did not include the T4 polymerase, and only half the recommended volumes of the polynucleotide kinase and Taq polymerase were used to minimize exonuclease digestion of the MIDs that had been incorporated into the 5′ ends of the genome-specific amplification primers. Following ligation of the RL adaptors, a modified AMPure XP cleanup kit was used that excluded the sizing solution and used a 1.6:1 (v/v) ratio to DNA to target fragments longer than 200 bp. Libraries were clonally amplified by the GS FLX Titanium MV emulsion PCR kit (Lib-L; Roche), using a 0.4 copy per bead ratio. Beads with amplified libraries were loaded onto GS FLX Titanium PicoTiterPlate set up with 4-region dividers to accommodate the sample pools.
Informatics Analysis of the Expansion of the Various Repeats
After a run was completed on the 454, the sfffile tool (Roche v2.3) was used to split the sff files into separate fasta files based on bar codes and assign them to patient identification numbers on the basis of plate quadrant. Then, sequences with exact matches to the 10 bp before and after the expected repeat regions, considering both forward and reverse reads, were identified. By use of the program countRepeats.py (available at: https://bitbucket.org/tbair/published/src), the exact positions of the start and the end of the repeats were identified, and the number of repeats was identified. Sequences that did not have an exact multiple of the repeat because of sequencing errors or nontypical expansion were identified. Statistics on the minimum, maximum, and average number of repeats were calculated, as well as the number of sequences that were in or out of phase. These data were then incorporated into downstream statistical analysis.
Fluorescent Antibody Studies
Fluorescent antibody analysis was performed using monoclonal antibody 12D9, which is specific for the phosphorylcholine epitope, on strains grown in supplemented brain heart infusion broth for 6 hours as previously described [4].
Statistical Analysis
For measurements on each gene, the proportion of sequences that were phase on was calculated. For proportions that were 0 or 1, an adjustment of 10−2 was used. For each gene, a baseline confidence interval for the proportion of genes that were phase on was calculated using control DNA samples from the inoculating bacteria. The proportions were transformed to a log odds (logit) scale, and a normal distribution was fit to the logit values, using maximum likelihood and a continuity adjustment to the normal approximation that accounted for the different number of reads for each measurement. A mean and a 95% confidence interval for the mean were calculated in the logit scale and back transformed to an interval for mean baseline proportion.
The in vivo proportions for each gene were also transformed to the logit scale and were analyzed using a linear mixed model. The inherent correlation between measurements on the same subjects was accounted for by using a random effect for subject. The days after challenge were then incorporated as fixed effects. The 95% confidence interval for the mean logit for each day after challenge was calculated and also transformed back to the proportion scale. If this interval did not overlap with the baseline interval, the change from baseline was considered to be statistically significant (P < .05). The difference between technical replicates was also examined within the appropriate linear mixed models.
RESULTS
On the basis of sequence analysis of the NTHi strain 2019 genome, we identified 14 genes with ≥12 tandem repeats within their open reading frames (one gene, hgpB, was not studied). Seven of these genes are involved in LOS biosynthesis, 5 are involved in iron acquisition from hemoglobin, 1 encoded DNA methyltransferase, and 1 encoded immunoglobulin A (IgA) protease. Studies were initiated on 13 of these genes (Table 1) in strains recovered from 15 subjects during the 2 challenge studies (a multidose study and a 2-dose study) to determine whether there was on-off switching in these phase-variable genes during colonization. In our initial studies, 2 phase-variable genes, licA and lic3A, were selected for analysis by capillary electrophoresis of samples from the multidose study. Figure 1A demonstrates that licA was phase on in only a mean of 2% of the inoculate strain population and that, by day 3 after nasopharyngeal inoculation, licA phase on had increased to a mean of approximately 15%, with this level maintained in subsequent samples. In contrast, Figure 1B shows that, for lic3A, a mean of approximately 95% of the population was phase on in the inoculation samples, and this percentage did not change significantly over the next 4 days in the organisms recovered from the patients.
Table 1.
Phase-Variable Genes Analyzed in this Study, Repeat Sequence, and Number of Repeats, Based on Genome Sequencing of Nontypeable Haemophilus influenzae
| Gene | Gene Annotation | Repeat Sequence | No. of Repeats (Phase On) | Accession No. |
|---|---|---|---|---|
| Choline kinasea | licA | CAAT | 25, 27 | KC607490 |
| Glycosyltransferaseb | lic2A | CAAT | 16, 17 | KC607492 |
| Sialyltransferasec | lic3A | CAAT | 15, 16 | KC607491 |
| Glycosyltransferase | lgtC | GACA | 4 | KC607500 |
| Putative glycosyltransferase 2c | pgt2 | CCAA | 12, 13 | KC607499 |
| LOS biosynthesis | lex2A | GCAA | 21 | KC607502 |
| Hemoglobin-haptoglobin binding protein Cd | hbpC | CCAA | 46, 47 | KC607495 |
| Hemoglobin-haptoglobin binding protein B2b | hbpB2 | CCAA | 48, 49 | KC607497 |
| Hemoglobin-haptoglobin binding protein B3d | hbpB3 | CCAA | 31, 33 | KC607496 |
| Hemoglobin-haptoglobin binding protein C2c | hbpC2 | CCAA | 40, 41 | KC607494 |
| Sialyltransferasec | siaA2 | CAAT | 20, 21 | KC607493 |
| Mod methylase TIIIR/M system | Mod-like | GAGAC | 6 | KC607501 |
| IgA1 protease | igaB | AAATTCA | 11 | KC607498 |
a Three possible starts in 2 reading frames.; gene is out of frame, so add or subtract 1 repeat to have an in-frame start.
b Four possible starts in 2 reading frames; gene is in frame.
c Two possible starts in 2 reading frames; gene is in frame.
d Three possible starts in 2 reading frames; gene is out of frame.
Figure 1.
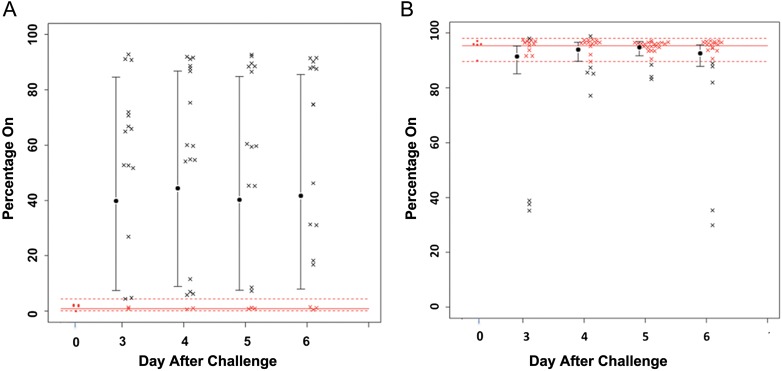
Changes in the expression status (percentage phase on) for licA (A) and lic3A (B) undergoing phase variation in samples during the experimental human nasopharyngeal colonization study. Samples from 9 subjects in the multidose study were examined in this analysis. Each data point represents measurement of an individual sample. Percentage on was determined by capillary electrophoresis of polymerase chain reaction products. As can be seen, licA shows a shift from predominantly phase off in the inoculation strain (day 0) to significantly phase on at each time point over this period, while lic3A stays phase on in the inoculation strain and during the colonization period. The red open circles indicate the percentage phase on in the inoculation strain. The dotted lines represent the 95% confidence interval of the inoculation strain phase-on value. Each black × represents a sample with a phase-on value outside the interval, and each red × represents a sample within the interval.
To study whether phase variation was occurring in the samples from both colonization studies, we used 454 pyrosequencing to simultaneously analyze all of these genes except igaB from the 48 samples recovered from the 15 subjects who participated in both studies. The samples were isolated over the last 4 days of the 6-day colonization period. To accomplish this, we used primers that were bar coded for each patient, primers that were specific for each gene, and 4-region picotiter plates, with each region defining a specific day. This analysis confirmed the results of the licA and lic3A capillary fragment analysis, and the results are shown in Table 2. Figure 2 shows the results obtained on phase variation of licA obtained by pyrosequencing. The average phase-on status of the inoculating strain was approximately 3.8% of the population; this increased to approximately 18.8% on day 3 and to 25% on day 6. Fluorescent antibody studies were performed on the sample isolated on day 6 from subject 10 (Figure 3A and 3B). Genotyping indicated that >60% of the organisms in this sample population had shifted to phase on. These studies used monoclonal antibody 12D9, which is specific for the phosphorylcholine epitope on the LOS. As can be seen, there was a marked increase in fluorescence of the bacteria in the sample recovered on day 6 after inoculation. lex2A showed more variability in the percentage of the population in which it was phase on as compared to baseline, but the change in mean values was not significant (Figure 4). Analysis of the remaining phase-variable genes showed minimal changes between the challenge strain and the sample recovered from days 3 through 6. Pyrosequencing studies of lic3A and the remaining 10 genes showed that the phase status changed in only a few instances during the period of colonization, compared with the inoculation strain (Table 2).
Table 2.
Percentage of Phase-Variable Genes in Nontypeable Haemophilus influenzae 2019StrRR Isolates That Were Phase On During the Last 4 Days of the 6-Day Experimental Colonization Period
| Percentage Phase On (95% Confidence Interval) |
|||||
|---|---|---|---|---|---|
| Gene | Inoculum | Day 3 | Day 4 | Day 5 | Day 6 |
| hgpC | 46 (33.3–59.1) | 5.8 (1.3–22.2) | 43.4 (14.2–78.0) | 8.9 (2.4–28.0) | 38.6 (13.0–72.6) |
| hgpB2 | 30.4 (21.2–41.4) | 37.6 (21.0–57.7) | 40.1 (23.8–59.1) | 21.0 (11.4–35.4) | 36.4 (21.0–55.2) |
| hgpB3 | 33.8 (23.7–45.8) | 24.1 (11.7–43.2) | 39.2 (21.9–59.8) | 24.2 (12.8–40.8) | 26.5 (13.9–44.6) |
| hgpC2 | 35.4 (24.8–47.5) | 11.5 (3.3–33) | 31.3 (11.6–61.3) | 23.8 (8.7–50.4) | 16.7 (5.6–40.5) |
| lex2A | 14.3 (9.4–21.1) | 20.5 (14.8–27.8) | 27.4 (20.4–35.7) | 24.0 (17.9–31.3) | 16.5 (11.9–22.4) |
| lic2A | 1.3 (.8–2.1) | 0.7 (.3–1.8) | 0.9 (.4–2) | 2.1 (.9–4.6) | 3.0 (1.3–6.7) |
| lic3A | 90.7 (86.1–93.9) | 80.3 (65.2–89.9) | 88.5 (78.3–94.30) | 93.0 (86.3–96.5) | 85.0 (72.2–92.4) |
| licA | 3.8 (2.4–6) | 18.8 (7.1–40.9) | 13.4 (5.0–31.4) | 18.5 (7.3–39.6) | 25.2 (10.3–49.6) |
| Mod-like | 7.4 (4.7–11.3) | 2.6 (1.2–5.8) | 2.5 (1.2–5.8) | 1.6 (.8–3.3) | 6.3 (3–12.7) |
| lgtC | 6.8 (4.3–10.8) | 4.9 (2.2–11) | 8.7 (4–17.8) | 3.0 (1.4–6.4) | 5.5 (2.5–11.4) |
| pgt2 | 0.4 (.2–.9) | 0.1 (.0–.2) | 0.2 (.1–.4) | 0.1 (.0–.2) | 0.1 (.0–.2) |
| siaA2 | 2.5 (1.6–3.9) | 0.6 (.3–1.4) | 1.3 (.6–2.8) | 1.3 (.6–2.8) | 1.3 (.6–2.8) |
| igaB | 3.8 (2.0–7.3) | 8.3 (4.7–14.2) | 11.3 (6.8–18.3) | 10.3 (6.1–16.9) | 12.5 (7.5–20.2) |
Figure 2.
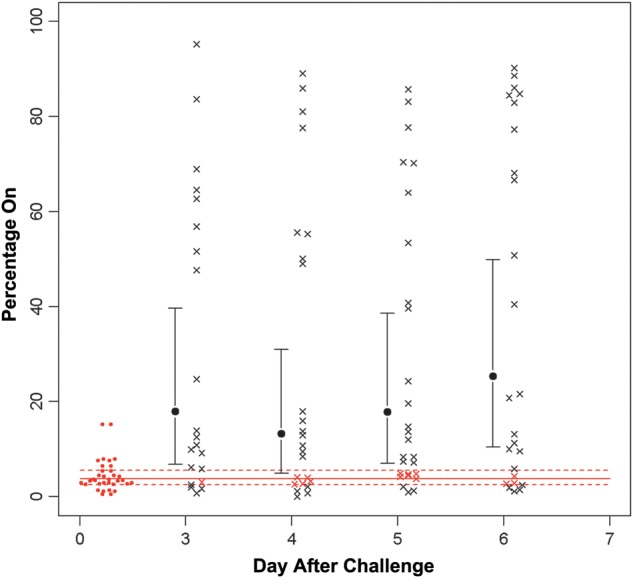
Results of 454 pyrosequencing analysis of phase variation of licA during the colonization period. Samples from all 15 subjects were analyzed by pyrosequencing. The solid red line is the mean phase on in the inoculation samples, and the dotted lines represent the 95% confidence intervals. As can be seen, there is a significant shift in the samples to phase on in the inoculation strain (day 0) over the colonization period. The red open circles indicate the percentage phase on in the inoculation strain. Each black × represents a sample outside of the 95% confidence interval of the inoculation strain, whereas each red × represents a sample within that interval. The differences seen between the inoculation strain and the sample were statistically significant except on day 3.
Figure 3.
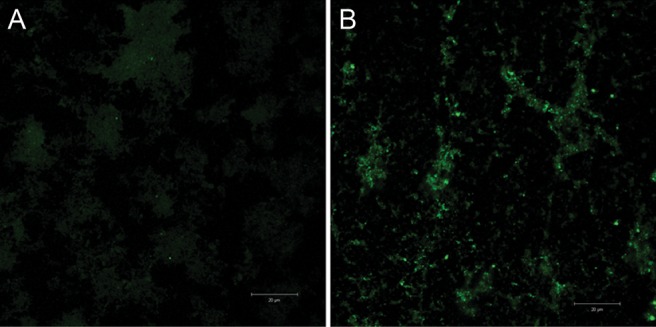
Fluorescent antibody study (original magnification, ×100) showing differences in expression of phosphorylcholine on the inoculation strain, nontypeable Haemophilus influenzae 2019StrR1, and the sample recovered from subject 10 on day 6. Monoclonal antibody 12D9, which is specific for the phosphorylcholine, was used. This study demonstrates the phenotypic change consistent with a phase-on genotype. The white lines indicate 20 microns.
Figure 4.
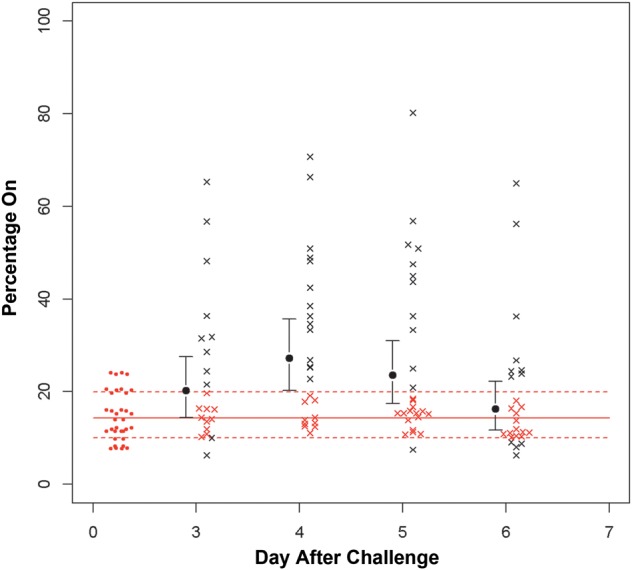
Results of 454 pyrosequencing analysis of phase variation of lex2A during the colonization period. The solid red line is the mean phase on in the inoculation samples, and the dotted lines represent the 95% confidence intervals. As can be seen, there is a shift in the samples to phase on on day 4 of the colonization period, which does not persist into day 6. The red open circles indicate the percentage phase on in the inoculation strain. Each black × represents a sample outside of the 95% confidence interval of the inoculation strain, while each red × represents a sample within that interval.
The phase variation of igaB was studied by capillary electrophoresis alone. Samples from all 15 subjects were studied. We found that, over the 6-day colonization period, igaB was initially predominantly phase off, with a shift to phase on in a substantial percentage of the population (Figure 5).
Figure 5.
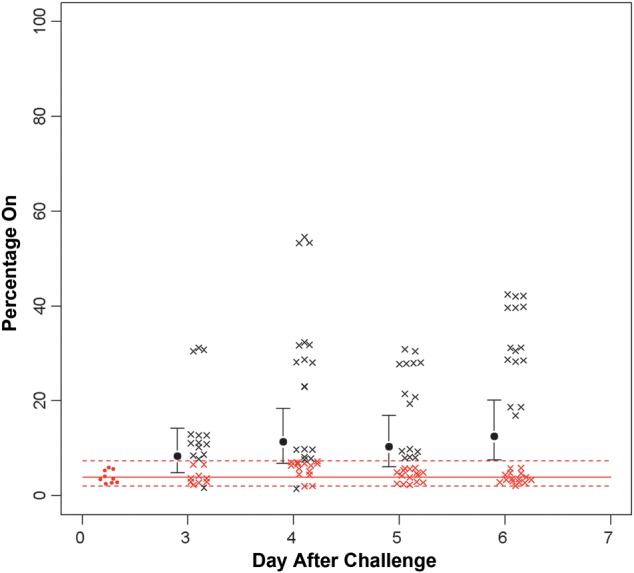
Results of capillary electrophoresis analysis of phase variation of iga during the colonization period. The solid red line is the mean phase on in the inoculation samples, and the dotted lines represent the 95% confidence intervals. The majority of samples within the inoculation strain (day 0) are phase off. As can be seen, there is a progressive shift in the samples to phase on, and by day 6 this difference is significant. The red open circles indicate the percentage phase on in the inoculation strain. Each black × represents a sample outside of the 95% confidence interval of the inoculation strain, while each red × represents a sample within that interval.
In summary, both licA and igaB showed a significant shift from phase off to phase on during a 6-day colonization period, compared with the inoculation strain. The other 12 genes examined did not show significant differences in phase variation, compared with the inoculation strain.
DISCUSSION
NTHi is a commensal bacterium of the human upper airway that can cause diseases such as otitis media, acute sinusitis, and bronchitis [12]. It possesses a number of genes with multiple tandem repeats, which undergo phase variation at high rates [3]. Phase variation is the rapid, reversible switching of gene expression that creates a heterogeneous phenotypic population in a clonal population. Site-specific recombination, SSM, and epigenetic regulation by DNA methylation are all mechanisms that mediate phase variation [2]. The mechanism for phase variation in H. influenzae is SSM. SSM is an intrahelical event and involves the mispairing of complementary bases at a site of a short tandem repeat by the local denaturation and displacement of DNA strands during replication. This leads to an insertion or deletion in the short tandem repeats [13]. The resulting frame shift in the open reading frame of a gene causes premature termination of translation if the gene is out of frame for expression (ie, phase off) or synthesis of a complete protein if in frame (ie, phase on) [3]. Alteration of repeats in promoter regions can also influence gene expression by altering transcription efficiency.
In this study, we looked at 13 phase-variable genes in 48 NTHi 2019StrR1 samples recovered over 4 days during an experimental human nasopharyngeal colonization study. These genes are involved in LOS biosynthesis, membrane protein expression, IgA protease production, iron regulation, and epigenetic regulation [5–9, 14–17]. The inoculation strain had only one of these genes predominantly phase on, the sialyltransferase lic3A. The 4 iron regulation genes showed an intermediate pattern, with approximately 30%–40% of the population being phase on at inoculation, and this persisted at that level throughout the study. The LOS biosynthesis gene, lex2A, had a mean phase-on frequency of 14% in the inoculation strain, and, by days 4 and 5 of the observation period, approximately 23%–25% population had switched to phase on. At day 4, the 95% confidence intervals of the lex2A phase-on percentage was barely overlapping the confidence intervals of that for the inoculation strain. licA and igaB were the only genes that showed a significant change in the samples recovered during the course of the infection, as compared to baseline. This change began with the first isolation of studies on day 3 for licA and continued until day 6. The average phase on percentage for licA in the inoculating strain was approximately 2%. This increased to 19% on day 3, to 13% on day 4, to 19% on day 5, and to 25% on day 6. The phase-on status of igaB samples rose progressively over the colonization period and, by day 6, was significantly different than that for the inoculation strain (Figure 5). The relationship between phase variation of licA and alternative start sites has been extensively studied by Weiser et al [4] and High et al [18].
LOS is a major antigenic component of the NTHi surface [15]. H. influenzae expresses host carbohydrate mimics within the oligosaccharide region of the LOS. These include phosphorylcholine (ChoP), N-acetyl-lactosamine, pK antigen, and sialylactosamine [12]. H. influenzae acquires choline from its environment, as choline is a major constituent of eukaryotic membrane lipids [19, 20]. Choline is a structure present on other human respiratory pathogens, including Neisseria meningitidis, Streptococcus pneumoniae, and Pseudomonas aeruginosa. The phosphorylcholine epitope undergoes phase variation in H. influenzae, on pili of N. meningitidis and N. gonorrhoeae, and on a protein in P. aeruginosa [20, 21]. licA in H. influenzae has a tetranucleotide repeat sequence of 5′-(CAAT)-3′ within its open reading frame, which is part of a 4-gene operon involved in deposition of phosphorylcholine on the LOS [8, 20, 21].
H. influenzae licA phase variation studies in animal models support our findings of a significant selection for licA on during the course of the human nasopharyngeal colonization [8]. This would support our finding of a significant selection for licA on during the course of the human nasopharyngeal colonization. Weiser et al showed that, in the infant rat model, there is a gradual selection for licA on (ChoP+) over 10 days in the nasopharynx and that constitutively licA-off (ChoP–) mutants were cleared faster from the nasopharynx, compared with wild-type controls [17]. Phase variation of ChoP has also been investigated in the chinchilla model. It was found that, during nasopharyngeal colonization, there was a switch to ChoP+ in as early as 3 days and that chinchillas inoculated with the ChoP+ phenotype had a greater level of NTHi 2019 per milliliter than chinchillas inoculated with a mostly ChoP– population of 2019. This suggested better colonization ability by the ChoP+ variants in chinchillas. Furthermore, there was a higher concentration of bacteria from the middle ear of chinchillas with the ChoP+ phenotype. ChoP– variants were also able to colonize the middle ear, but the infection was less severe than the infection displayed by ChoP+ variants [15].
The expression of ChoP has been linked to susceptibility to serum killing mediated by C-reactive protein (CRP). With H. influenzae, CRP-mediated killing is done through activation of complement and bacteriolysis. This is different than with pneumococcus, where CRP-complement interaction mediates opsonization by phagocytes. The position of the phosphorylcholine on LOS affects the susceptibility of H. influenzae to CRP-mediated killing [22, 23]. CRP is present in the human respiratory tract. It was shown that CRP is present in both inflamed (concentration, 0.17–42 μg/mL) and uninflamed (concentration, <0.05 to 0.88 μg/mL) human respiratory secretions. The human epithelial cells of the respiratory tract were discovered to be capable of expressing CRP, but complement is required for killing of NTHi [24]. This may suggest that the levels of functionally active complement are not high enough in the airway to make NTHi favor the ChoP– phenotype. The ability to turn off licA could be more of a factor in systemic disease. licA in H. influenzae type B was examined in the infant rat model, with comparisons in bacteria from the nasopharynx, bloodstream, and cerebrospinal fluid obtained 48 hours after infection. There was a different selection for licA on in the nasopharynx, with almost equal amounts on and off, compared with the bloodstream and cerebrospinal fluid, in which the gene was mostly off [8]. A licA mutant that is constitutively on has been constructed, and it showed a reduced ability to cause invasive disease and increased susceptibility to the bactericidal activity of serum with CRP [25]. From this, there can be a selection for licA, depending on where the bacteria are growing, that is potentially due to CRP.
The ability of NTHi to adhere to and invade the epithelial lining in the upper airway is an important step in colonization and persistence. Subsets of LOS glycoforms containing ChoP play an important role in adherence and entry into human bronchial epithelial cells, providing further evidence of why a selection for licA on might be expected. Infected monolayers were analyzed by confocal microscopy and showed that ChoP+ NTHi cells colocalized with the platelet-activating factor (PAF) receptor [26]. Further evidence demonstrated that NTHi binding to the PAF receptor initiates receptor coupling to a PTX-sensitive heterotrimeric G protein complex followed by cell entry [12].
The role of LOS phosphorylcholine in colonization and pathogenesis has also been investigated with Histophilus somni in cattle. H. somni is among the primary pathogens responsible for respiratory disease and other systemic infections in cattle. H. somni LOS also undergoes antigenic phase variation. Elswaifi et al found that ChoP expression was associated with colonization in the respiratory tract of compromised calves, whereas systemic dissemination was associated with the loss of ChoP expression [27]. ChoP was not required for adherence to bovine turbinate cells but could play a role in pathogenesis of H. somni by binding to the PAF receptor [27, 28].
The igaB gene encodes a type I IgA1 protease that cleaves human IgA1 in the hinge region of the α-heavy chain and is present in essentially all of the H. influenzae genomes [29, 30]. Some strains of H. influenzae isolated from patients with COPD have acquired another IgA protease gene. The gene, designated igaB, may have been acquired from N. meningitidis, based on flanking transposase sequences and sequence homology [29]. In strain 2019, the igaB gene is phase variable, and igaB was initially predominantly phase off. There was a progressive change to phase on over the course of the experiment; this difference was statistically significant at day 6. IgA1 is the most abundant immunoglobulin associated with human upper respiratory tract mucosal surfaces, so iga (igaA or igaB) would be important for colonization and persistence [30]. Differences in IgA1 protease levels between clinical and carriage strains have been observed, and levels were significantly higher in the clinical strains [30]. Human milk lactoferrin can inactivate the IgA1 protease by extracting the preprotein from the cell membranes [31]. The igaB variable region works well to differentiate H. influenzae and Haemophilus haemolyticus [32]. This could be important clinically because β-hemolysis by H. haemolyticus is a poor indicator of species separation [33].
Our study showed that there was greater variability in lex2A as compared to the inoculated strain than with other genes investigated. The difference was not significant, but a definite population of strains had shifted from phase off to phase on. There is evidence that lex2A is somehow necessary for the phenotypic expression of lex2B. Mutant constructs of lex2A and lex2B have been made. A lex2B mutant produces a truncated LOS, because lex2B is required to add the second β-glucose in the oligosaccharide extension on the first heptose. lex2A mutants gave the same LOS phenotype, observed by sodium dodecyl sulfate polyacrylamide gel electrophoresis and colony immunoblot analyses, as the lex2B mutants. This showed that lex2A must be in frame for transcription of lex2B [34].
In summary, we examined 13 phase-variable genes from samples obtained from an experimental human colonization study for a shift in their phase-on and phase-off status over the 6-day course of colonization. licA and igaB, which encode phosphorylcholine kinase and IgA protease, respectively, showed a significant difference from baseline over the course of the study. This change was from almost completely phase off to a significant portion of the population being phase on. This correlated with other studies showing there is a selection for licA on (ChoP+) and igaB on in the airway for H. influenzae to colonize and persist during infections [15, 20].
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgment. We thank Ms Margaret Ketterer for excellent technical assistance.
Financial support. This work was supported by the National Institutes of Health (grants AI024616, AI30040, and NCATS UL1 TR00442) and a Helen C. Levitt Endowed Annual Visiting Professorship (to M. P. J.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Weiser JN. The generation of diversity by Haemophilus influenzae. Trends in Microbiology. 2000;8:433–5. doi: 10.1016/s0966-842x(00)01839-4. [DOI] [PubMed] [Google Scholar]
- 2.van der Woude MW. Phase variation: how to create and coordinate population diversity. Curr Opin Microbiol. 2011;14:205–11. doi: 10.1016/j.mib.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 3.De Bolle X, Bayliss CD, Field D, et al. The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Molecular Microbiology. 2000;35:211–22. doi: 10.1046/j.1365-2958.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 4.Weiser JN, Love JM, Moxon ER. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell. 1989;59:657–65. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- 5.Weiser JN, Maskell DJ, Butler PD, Lindberg AA, Moxon ER. Characterization of repetitive sequences controlling phase variation of Haemophilus influenzae lipopolysaccharide. Journal of Bacteriology. 1990;172:3304–9. doi: 10.1128/jb.172.6.3304-3309.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peak IR, Jennings MP, Hood DW, Moxon ER. Tetranucleotide repeats identify novel virulence determinant homologues in Neisseria meningitidis. Microbial Pathogenesis. 1999;26:13–23. doi: 10.1006/mpat.1998.0243. [DOI] [PubMed] [Google Scholar]
- 7.Peak IR, Jennings MP, Hood DW, Bisercic M, Moxon ER. Tetrameric repeat units associated with virulence factor phase variation in Haemophilus also occur in Neisseria spp. and Moraxella catarrhalis. FEMS Microbiology Letters. 1996;137:109–14. doi: 10.1111/j.1574-6968.1996.tb08091.x. [DOI] [PubMed] [Google Scholar]
- 8.Hosking SL, Craig JE, High NJ. Phase variation of lic1A, lic2A and lic3A in colonization of the nasopharynx, bloodstream and cerebrospinal fluid by Haemophilus influenzae type b. Microbiology. 1999;145(Pt 11):3005–11. doi: 10.1099/00221287-145-11-3005. [DOI] [PubMed] [Google Scholar]
- 9.Dixon K, Bayliss CD, Makepeace K, Moxon ER, Hood DW. Identification of the functional initiation codons of a phase-variable gene of Haemophilus influenzae, lic2A, with the potential for differential expression. J Bacteriol. 2007;189:511–21. doi: 10.1128/JB.00815-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey WR. aSEG. Diagnostic Microbiology. 8thed. St. Louis: Mosby; 1990. [Google Scholar]
- 11.Fox KL, Dowideit SJ, Erwin AL, Srikhanta YN, Smith AL, Jennings MP. Haemophilus influenzae phasevarions have evolved from type III DNA restriction systems into epigenetic regulators of gene expression. Nucleic Acids Research. 2007;35:5242–52. doi: 10.1093/nar/gkm571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swords WE, Ketterer MR, Shao J, Campbell CA, Weiser JN, Apicella MA. Binding of the non-typeable Haemophilus influenzae lipooligosaccharide to the PAF receptor initiates host cell signalling. Cell Microbiol. 2001;3:525–36. doi: 10.1046/j.1462-5822.2001.00132.x. [DOI] [PubMed] [Google Scholar]
- 13.Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–21. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 14.Weiser JN, Williams A, Moxon ER. Phase-variable lipopolysaccharide structures enhance the invasive capacity of Haemophilus influenzae. Infect.Immun. 1990;58:3455–7. doi: 10.1128/iai.58.10.3455-3457.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong HH, Blue LE, James MA, Chen YP, DeMaria TF. Evaluation of phase variation of nontypeable Haemophilus influenzae lipooligosaccharide during nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect Immun. 2000;68:4593–7. doi: 10.1128/iai.68.8.4593-4597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srikhanta YN, Maguire TL, Stacey KJ, Grimmond SM, Jennings MP. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5547–51. doi: 10.1073/pnas.0501169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srikhanta YN, Fox KL, Jennings MP. The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nature Reviews. Microbiology. 2010;8:196–206. doi: 10.1038/nrmicro2283. [DOI] [PubMed] [Google Scholar]
- 18.High NJ, Jennings MP, Moxon ER. Tandem repeats of the tetramer 5′-CAAT-3′ present in lic2A are required for phase variation but not lipopolysaccharide biosynthesis in Haemophilus influenzae. Molecular Microbiology. 1996;20:165–74. doi: 10.1111/j.1365-2958.1996.tb02498.x. [DOI] [PubMed] [Google Scholar]
- 19.Weiser JN, Goldberg JB, Pan N, Wilson L, Virji M. The phosphorylcholine epitope undergoes phase variation on a 43-kilodalton protein in Pseudomonas aeruginosa and on pili of Neisseria meningitidis and Neisseria gonorrhoeae. Infect Immun. 1998;66:4263–7. doi: 10.1128/iai.66.9.4263-4267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiser JN, Pan N, McGowan KL, Musher D, Martin A, Richards J. Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J Exp Med. 1998;187:631–40. doi: 10.1084/jem.187.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiser JN, Pan N. Adaptation of Haemophilus influenzae to acquired and innate humoral immunity based on phase variation of lipopolysaccharide. Mol Microbiol. 1998;30:767–75. doi: 10.1046/j.1365-2958.1998.01108.x. [DOI] [PubMed] [Google Scholar]
- 22.Lysenko E, Richards JC, Cox AD, et al. The position of phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae affects binding and sensitivity to C-reactive protein-mediated killing. Mol Microbiol. 2000;35:234–45. doi: 10.1046/j.1365-2958.2000.01707.x. [DOI] [PubMed] [Google Scholar]
- 23.Lysenko ES, Gould J, Bals R, Wilson JM, Weiser JN. Bacterial phosphorylcholine decreases susceptibility to the antimicrobial peptide LL-37/hCAP18 expressed in the upper respiratory tract. Infect Immun. 2000;68:1664–71. doi: 10.1128/iai.68.3.1664-1671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gould JM, Weiser JN. Expression of C-reactive protein in the human respiratory tract. Infect Immun. 2001;69:1747–54. doi: 10.1128/IAI.69.3.1747-1754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humphries HE, High NJ. The role of licA phase variation in the pathogenesis of invasive disease by Haemophilus influenzae type b. FEMS Immunology and Medical Microbiology. 2002;34:221–30. doi: 10.1111/j.1574-695X.2002.tb00628.x. [DOI] [PubMed] [Google Scholar]
- 26.Swords WE, Buscher BA, Ver Steeg Ii K, et al. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol Microbiol. 2000;37:13–27. doi: 10.1046/j.1365-2958.2000.01952.x. [DOI] [PubMed] [Google Scholar]
- 27.Elswaifi SF, Scarratt WK, Inzana TJ. The role of lipooligosaccharide phosphorylcholine in colonization and pathogenesis of Histophilus somni in cattle. Vet Res. 2012;43:49. doi: 10.1186/1297-9716-43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elswaifi SF, St Michael F, Sreenivas A, Cox A, Carman GM, Inzana TJ. Molecular characterization of phosphorylcholine expression on the lipooligosaccharide of Histophilus somni. Microb Pathog. 2009;47:223–30. doi: 10.1016/j.micpath.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy TF, Lesse AJ, Kirkham C, Zhong H, Sethi S, Munson RS., Jr A clonal group of nontypeable Haemophilus influenzae with two IgA proteases is adapted to infection in chronic obstructive pulmonary disease. PLoS One. 2011;6:e25923. doi: 10.1371/journal.pone.0025923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vitovski S, Dunkin KT, Howard AJ, Sayers JR. Nontypeable Haemophilus influenzae in carriage and disease: a difference in IgA1 protease activity levels. JAMA : the Journal of the American Medical Association. 2002;287:1699–705. doi: 10.1001/jama.287.13.1699. [DOI] [PubMed] [Google Scholar]
- 31.Qiu J, Hendrixson DR, Baker EN, Murphy TF, St Geme JW, 3rd, Plaut AG. Human milk lactoferrin inactivates two putative colonization factors expressed by Haemophilus influenzae. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12641–6. doi: 10.1073/pnas.95.21.12641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandstedt SA, Zhang L, Patel M, et al. Comparison of laboratory-based and phylogenetic methods to distinguish between Haemophilus influenzae and H. haemolyticus. Journal of Microbiological Methods. 2008;75:369–71. doi: 10.1016/j.mimet.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCrea KW, Xie J, LaCross N, et al. Relationships of nontypeable Haemophilus influenzae strains to hemolytic and nonhemolytic Haemophilus haemolyticus strains. Journal of Clinical Microbiology. 2008;46:406–16. doi: 10.1128/JCM.01832-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffin R, Cox AD, Makepeace K, Richards JC, Moxon ER, Hood DW. The role of lex2 in lipopolysaccharide biosynthesis in Haemophilus influenzae strains RM7004 and RM153. Microbiology. 2003;149:3165–75. doi: 10.1099/mic.0.26387-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


