Abstract
The bacteriophage λ CII protein stimulates the activity of three phage promoters, pE, pI and paQ, upon binding to a site overlapping the –35 element at each promoter. Here we used preparations of RNA polymerase carrying a DNA cleavage reagent attached to specific residues in the C-terminal domain of the RNA polymerase α subunit (αCTD) to demonstrate that one αCTD binds near position –41 at pE, whilst the other αCTD binds further upstream. The αCTD bound near position –41 is oriented such that its 261 determinant is in close proximity to σ70. The location of αCTD in CII-dependent complexes at the pE promoter is very similar to that found at many activator-independent promoters, and represents an alternative configuration for αCTD at promoters where activators bind sites overlapping the –35 region. We also used an in vivo alanine scan analysis to show that the DNA-binding determinant of αCTD is involved in stimulation of the pE promoter by CII, and this was confirmed by in vitro transcription assays. We also show that whereas the K271E substitution in αCTD results in a drastic decrease in CII-dependent activation of pE, the pI and paQ promoters are less sensitive to this substitution, suggesting that the role of αCTD at the three lysogenic promoters may be different.
INTRODUCTION
The temperate bacteriophage, λ, is one of the simplest model organisms for the study of developmental regulation. The decision between lytic and lysogenic growth is based on the activity of two phage-encoded transcriptional activators, CI and CII, both of which are required for lysogenization. Establishment of lysogeny depends on the CII-dependent pE promoter (also known as pRE) that directs the expression of cI necessary for maintenance of the lysogenic state. CII also stimulates pI, which directs expression of the λ int gene, and paQ, which directs synthesis of an antisense mRNA that regulates late gene expression [for a review, see Echols (1)].
The mechanism by which CII activates transcription is of special interest. The CII homotetramer regulates all three lysogenic promoters co-ordinately by binding to a tetrad repeat (TTGC) flanking the –35 element, and it was the first regulatory protein suggested to recognize direct repeats [for a review, see Ho et al. (2)]. The location of the CII-binding site at the λ lysogenic promoters is consistent with the idea that it is a class II activator (3). Class II activators bind to target sites that overlap the promoter –35 region and, in most cases, make contact with domain 4 of the RNA polymerase (RNAP) σ subunit, that is bound to the –35 element (4). However, CII binds on the opposite face of the DNA helix to RNAP, and thus, target promoter –35 elements are sandwiched between σ and the activator. Binding of CII to the direct repeats appears to distort the intervening –35 hexamer in some way, although it remains unclear whether this distortion facilitates recognition of the –35 region by σ70 or whether CII makes direct contact with σ (2).
At many bacterial promoters, the C-terminal domain of the α subunit (αCTD) interacts with upstream promoter DNA, the RNAP σ subunit and/or transcription activator proteins (5,6). These interactions are mediated by determinants on the surface of αCTD. For example, residue 265 and neigbouring residues contribute to the αCTD 265 determinant, which is responsible for interactions with DNA (7–10). Similarly, residue 261 and neighbouring residues contribute to the 261 determinant, that can contact σ (11–13). Previous experiments demonstrated that deletion of αCTD greatly reduces CII-dependent activation of pE (14). We have shown that the rpoA341 mutation, specifying the K271E substitution in αCTD, blocks lysogenization of Escherichia coli by phage λ (15). Analysis of reporter gene fusions revealed that this substitution abolishes activation of pE by CII (16). At many class II or class II-like promoters, both αCTDs bind immediately upstream of the bound activator and, in some cases, one or both of them make interactions with the activator that contribute to activation. However, CII-dependent promoters present an interesting situation, as CII binds to the opposite face of the DNA to RNAP. Here we have carried out a genetic and biochemical analysis of the positioning and role of αCTD at the CII-dependent pE promoter. Our results show that αCTD contacts the DNA immediately upstream of the –35 region at pE, on the opposite face of the DNA helix to CII, and that these contacts are important for CII-dependent activation.
MATERIALS AND METHODS
Bacterial strains, plasmids and gene fusions
The rpoA+ and rpoA341 strains, WAM106 and WAM105, respectively, have been previously described (17). Derivatives of these strains containing single copy pE–lacZ, pI–lacZ and paQ–lacZ fusions were constructed by lysogenization with λ299 derivatives containing the respective fusions. The fusions were constructed according to Giladi et al. (18) and have been reported previously (16,19). Plasmid pJMH1 is a pSC101-based replicon carrying the lacIQ and kanamycin resistance genes (17). Plasmid pMO23 is a p15A-derived replicon bearing a chloramphenicol resistance gene and the cII and cIII genes, each under the control of ptac (16). Plasmid pTJSpE was constructed by cloning the EcoRI–DraI fragment of pHG86 (20), containing the pE–lacZ promoter fusion, into the RK2 minireplicon, pTJS42, which harbours a tetracycline resistance gene (21). For the expression of mutant rpoA alleles for the alanine scan analysis, derivatives of plasmids pHTf1α, encoding alanine substitutions at position 255–271 and 302 in αCTD (7,22), pREIIα, encoding alanine substitutions at remaining positions in αCTD (7,22–25), or pLAW2phs, encoding the K271E substitution (17), were used. The control plasmid encoding wild-type rpoA was pLAW2 (26). All the plasmids which encode α and mutant derivatives thereof specify resistance to ampicillin.
Measurement of β-galactosidase activity.
β-Galactosidase assays were performed on mid-logarithmic phase cultures according to the method of Miller (27). Results presented are averages of at least three independent experiments and are shown with standard deviations.
Protein purification and reconstitution of RNA polymerase.
Plasmid pET-CII (28) was used for over-production of N-terminally His-tagged CII protein, which was purified as described previously (28). For the reconstitution of RNAP, inclusion bodies of RNAP β, β′ and σ subunits from strains XL1-Blue (pMKSe2), BL21(DE3)(pT7β′) and BL21(DE3) (pLHN12σ), respectively, were prepared as described previously (29). His-tagged RNAP α subunits were prepared using plasmid pHTT7f1NHα. Derivatives of pHTT7f1NHα carrying mutant rpoA alleles were constructed by replacing the HindIII–BamHI fragment, which encodes wild-type αCTD, with the corresponding fragments from plasmids pHTf1α (258A, 261A, 265A, 271A) (7,22,30) or pLAW2phs (271E) (17). Over-expression of the α subunits in strain BL21(DE3) and purification of α by Ni2+-affinity chromatography and reconstitution into RNAP were performed essentially as described previously (7,29). Purification of α subunits with single cysteine residues at positions 273 and 302, conjugation with iron [S]-1-[p-bromoacetamidobenzyl] ethylenediaminetetraacetate (Fe·BABE), and reconstitution into RNAP were performed as described in Lee et al. (31).
In vitro transcription
Single round in vitro transcription reactions were performed in a total volume of 20 µl in buffer containing 50 mM KCl, 40 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 1 mM dithiothreitol (DTT), 100 µg/ml bovine serum albumin (BSA) and 150 ng of linear template DNA. Template was prepared by PCR amplification of λ DNA using primers 5′-TGGCTGATGGTGCGATAGTC-3′ and 5′-ACGTGCGTCCTCAAGCTG-3′, for 35 cycles of denaturation at 95°C for 30 s, annealing at 53.2°C for 30 s and extension at 72°C for 1 min. The resultant PCR product (1433 bp) was digested by BsuRI to obtain a fragment of 1172 bp, containing the λ pE and poop promoters. The binding reaction of CII (40 ng) with the DNA (150 ng) was carried out at 37°C for 10 min, after which RNA polymerase was added and the incubation continued for a further 10 min. After the addition of nucleotides (CTP, GTP and ATP each to a final concentration of 150 µM, UTP to 15 µM, and 0.6 µCi of [α-32P]UTP per reaction) and 50 µg heparin/ml, the samples were incubated at 37°C for 15 min and the reactions were stopped by the addition of an equal volume of 95% formamide containing 20 mM EDTA, 0.05% bromophenol blue and 0.05% xylene cyanol. The samples were separated by electrophoresis in 6% polyacrylamide gels containing 46% urea in TBE buffer (32). The gel was dried, and RNA bands were visualized and quantified, following background subtraction, using a PhosphorImager (Bio-Rad). Concentrations of RNAP, calibrated to give the same amount of transcription from the activator-independent poop promoter, were: 46 nM wild-type RNAP, 34 nM RNAP αK271E, 54 nM RNAP αK271A, 13 nM RNAP D258A, 28 nM RNAP αE261A and 35 nM RNAP αR265A. Transcriptional activities were calculated from at least three independent experiments and are presented as a percentage (with standard deviation) of transcripts obtained with wild-type RNAP.
Fe·BABE-mediated hydroxyl radical footprinting
A 275 bp DNA fragment containing the λ pE promoter was amplified from bacteriophage λ DNA by PCR using primers 5′-GCGAAGCTTCCACACCTATGGTGTATGC-3′ and 5′-GCCGAATTCCATGTCGTCGTCAACGACCC-3′, cleaved with EcoRI and HindIII restriction enzymes and cloned into the vector pSR (33). A 355 bp AatII–HindIII fragment was purified from the resultant plasmid (pSRpE) and labelled at the HindIII end with either [γ-32P]ATP and T4 polynucleotide kinase (for the template strand) or [α-32P]ATP and E.coli DNA polymerase Klenow fragment (for the non-template strand). The Fe·BABE-mediated DNA cleavage reactions were carried out in a reaction volume of 25 µl (5 mM MgCl2, 100 mM potassium glutamate, 40 mM HEPES pH 8.0, 50 µg/ml BSA, 10 µg/µl herring sperm DNA). Promoter DNA fragments were incubated with CII protein (3 µM final concentration) at 37°C for 10 min. After 10 min, RNAP holoenzyme was added (200 nM final concentration) and incubated at 37°C for 15 min. Complexes were then challenged with heparin (50 µg/ml final concentration) for 1 min at 37°C, then DNA cleavage was initiated by the addition of 3 mM sodium ascorbate and 3 mM hydrogen peroxide. The reactions were incubated for at least 2 min before being stopped by the addition of thiourea and EDTA to final concentrations of 7 and 45 mM, respectively. DNA was then extracted with phenol/chloroform, precipitated with ethanol and analysed by electrophoresis on a 6% polyacrylamide gel. The gels were calibrated with Maxam–Gilbert G + A ladders and analysed using a PhosphorImager and Quantity One software (Bio-Rad).
RESULTS
Location of αCTD–DNA interactions at the pE promoter
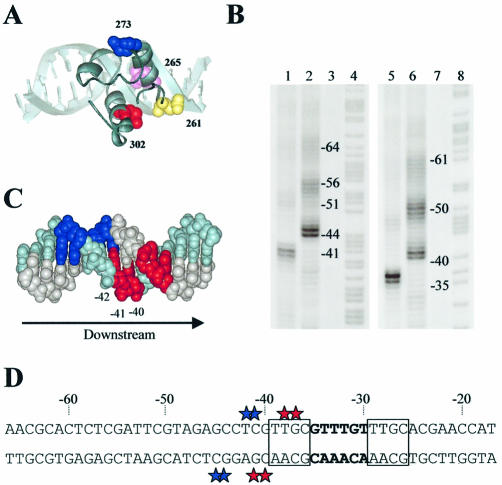
To determine the location of αCTD at the pE promoter, we exploited the DNA cleavage reagent Fe·BABE that can be attached to specific locations in RNAP α subunits (34,35). Thus, RNAP was reconstituted with purified α subunits that had been covalently modified with Fe·BABE, CII-dependent open complexes were formed at pE, and the DNA cleaving ability of Fe·BABE was triggered by the addition of ascorbate and hydrogen peroxide. In these experiments, the Fe·BABE reagent was tethered either to position 273 or to position 302, located on opposite faces of αCTD (Fig. 1A) (31). Figure 1B shows the patterns of DNA cleavage by Fe·BABE-tagged RNAP, revealed by gel electrophoresis and phosphorimager analysis. These cleavages occur in small clusters, separated by 10–11 bp, suggesting that αCTD binds to successive minor grooves on one face of the promoter DNA [see Murakami et al. (34,35)]. The strongest DNA cleavage on both strands is found near position –41, and we interpret this as due to the binding of one of the two αCTDs. For both strands, cleavage due to Fe·BABE conjugated to residue 302 occurs 4–5 bp downstream of the sites of cleavage due to Fe·BABE conjugated to residue 273. The locations of the different cleavage sites on the two strands are illustrated in Figure 1C and D. The simplest interpretation of these data, based on the model shown in Figure 1A, is that this αCTD binds to the minor groove near position –41, and is oriented such that the 261 determinant points downstream. The 261 determinant would thus be well placed to interact with region 4 (domain σ4) of the RNAP σ subunit, in agreement with results from previous authors (11–13). The results in Figure 1B show that αCTD can also bind to promoter DNA near positions –51 and –61. The signals near these positions are weaker than those near position –41, but it is clear that the cuts, at least around the –51 position, are also staggered. Therefore, we suggest that the position of the second αCTD is not fixed, and that the orientation of this αCTD, bound at –51, is the same as that of αCTD bound at –41.
Figure 1.
Location and orientation of αCTD at the pE promoter. (A) Model of αCTD bound to DNA to show the relative location of amino acid residues pertinent to this study. The model of αCTD is adapted from Benoff et al. (10). R265, located within the DNA-binding determinant, is shown in pink; residue 261, which participates in interactions with σ, is shown in yellow; and residues 273 and 302, which were derivatized with Fe·BABE, are shown in blue and red, respectively. The orientation shows the 261 determinant facing downstream, as occurs at pE. (B) Cleavage of pE promoter DNA by Fe·BABE-labelled RNAP. Phosphorimager scan of a polyacrylamide sequencing gel showing DNA cleavage at the pE promoter resulting from attack by RNAP reconstituted with Fe·BABE-derivatized α in the presence of CII. Lanes 1–4 show results for the template strand, and lanes 5–8 show results for the non-template strand. Lanes 1 and 5, RNAP containing Fe·BABE-derivatized E302C α; lanes 2 and 6, RNAP containing Fe·BABE-derivatized E273C α; lanes 3 and 7, no proteins; lanes 4 and 8, Maxam–Gilbert A + G ladder. (C) Model of DNA showing the minor groove locations of DNA cleavage at pe due to RNAP reconstituted with α subunits derivatized with Fe·BABE at position 273 (blue) or 302 (red). The template strand is shown in pale blue and the complementary strand in grey. The arrow indicates the direction of transcription. (D) DNA sequence of the pE promoter upstream region showing sites of Fe·BABE-induced cleavage indicated by coloured stars (colour coded as in A). The –35 region is in bold and the tetrad repeats recognized by CII are boxed.
Determinants in αCTD important for CII-dependent activation of pE in vivo
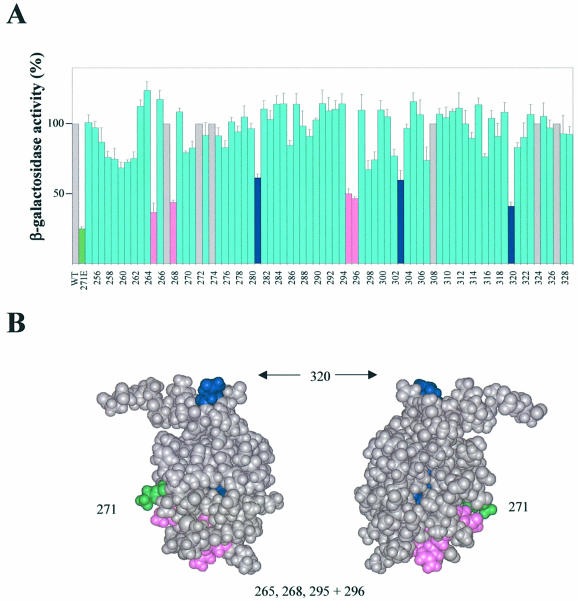
To determine whether the αCTD–DNA interactions at pE are important for CII-dependent activation, and to identify other amino acid side chains in αCTD that are important for CII-mediated activation, we used an alanine scanning approach. To do this, we exploited a set of plasmids encoding the RNAP α subunit in which residues 255–329 were each changed individually to alanine. These plasmids were introduced into a host strain carrying the pE–lacZ fusion plasmid (pTJSpE) and plasmids specifying inducible CII function (pMO23 and pJMH1). In addition, the host strain carried the rpoA341 mutant allele that encodes α subunits with the K271E substitution. Since this substitution greatly reduces expression from pE, CII-induced lacZ expression is low in the absence of plasmid-encoded wild-type α. This provides a simple system to measure the effects of different alanine substitutions in plasmid-encoded αCTD on CII-dependent activation of pE. In this experiment, the cII gene was expressed from an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter (on pMO23) and, due to the toxicity of overproduced CII (36), induction was allowed to proceed only for 75 min. In our conditions, sufficient incorporation of plasmid-encoded α into RNAP was achieved to assay the effects of the alanine substitutions, i.e. plasmid-directed synthesis of wild-type α allowed for a 4- to 5-fold stimulation of pE activity relative to plasmid-directed synthesis of α containing the K271E substitution (Fig. 2A).
Figure 2.
Identification of αCTD residues important for CII-dependent activation of pE in vivo. (A) Strain WAM105, containing plasmids pTJSpE, pJMH1 and pMO23, was transformed with one of a set of plasmids encoding the RNAP α subunit in which each residue of αCTD (positions 255–329) was changed individually to alanine. Cultures were grown in LB medium containing appropriate antibiotics at 37°C until OD ∼0.2, whereupon IPTG was added to a final concentration of 0.1 mM. After 75 min induction, the β-galactosidase activities were determined. The activities are presented relative to the activity of the strain harbouring pLAW2, encoding wild-type (WT) α (100% = 18 293 Miller units), and are averages of at least three independent experiments. The green bar shows the activity for the strain harbouring pLAW2phs (specifying α K271E). Blue bars and pink bars indicate positions within αCTD at which alanine substitution causes a decrease in activity of ≥40% compared with wild-type α, with the pink bars corresponding to residues within the DNA-binding determinant (7,8). Grey bars indicate positions where alanine occurs naturally (i.e. same as WT α). (B) Structure of αCTD (37), showing amino acid residues that are important for CII-dependent activation of pE. Residues comprising part of the DNA-binding determinant are shown in magenta; other residues are shown in blue (residues 281 and 303 are buried within the αCTD structure and so are not visible in this representation). Lys271 is also highlighted (green).
The results show that alanine substitutions at residues 265, 268, 281, 295, 296, 303 and 320 in αCTD most strongly impaired CII-dependent activation of pE [due to the higher sensitivity of our system in comparison with previous alanine scan experiments with αCTD, which required dominant-negative effects of plasmid-encoded α (11), we regard substitutions that give rise to ≤60% of the activity afforded by plasmid-encoded wild-type α as exerting strong inhibitory effects on pE] (Fig. 2A). The location of these residues on the αCTD structure is shown in Figure 2B. Residues 265, 268, 295 and 296 fall within the 265 DNA-binding determinant (7,8), suggesting that DNA binding of αCTD is important for CII-dependent activation of pE. Interestingly, in contrast to the rpoA341-encoded K271E substitution, substitution of K271 by alanine exerted only a small inhibitory effect on activation of pE by CII. Concerning the effects of alanine substitutions at residues 281, 303 and 320, these could be due to either direct or indirect effects. Residues 281 and 303 are buried within αCTD and so the latter possibility appears more likely. Regarding residue 320, the side chain is surface exposed but is located some distance from the other important residues (Fig. 2B).
Determinants in αCTD important for CII-dependent activation of pE in vitro
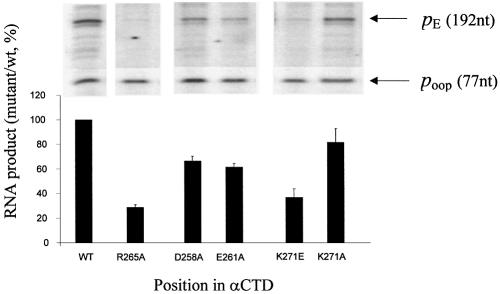
In the next set of experiments, we reconstituted RNAP containing wild-type or mutant α subunits and used run-off transcription assays to measure its activity at pE. Our primary aim was to quantify the effect of disrupting DNA binding by αCTD, and thus we compared wild-type RNAP with RNAP carrying α subunits harbouring the R265A substitution. Results presented in Figure 3 show that CII-dependent activation of pE is greatly reduced by the R265A substitution (while transcription from the control oop promoter is unaffected). Since our analysis with Fe·BABE suggested that one αCTD was positioned such that it could contact the RNAP σ subunit, we also reconstituted RNAP with α subunits carrying the D258A and E261A substitutions in the 261 determinant, known to be involved in αCTD–σ interactions. Results in Figure 3 show that these preparations of RNAP were also impaired for CII-dependent activation, although to a lesser degree than with the R265A substitution. Finally, we also measured CII-dependent activation at pE with RNAP containing α subunits with the K271E or K271A substitutions. Consistent with the in vivo analysis (Fig. 2A), RNAP reconstituted with K271Eα was severely impaired for CII-mediated activation of pE, whereas RNAP reconstituted with K271Aα supported efficient activation (Fig. 3).
Figure 3.
Identification of αCTD residues important for activation of pE by CII in vitro. Single-round in vitro transcription experiments were performed using linear template DNA containing pE and poop, CII, and RNAP reconstituted with hexahistidine-tagged α derivatives containing alanine substitutions at the positions indicated. The activities of purified RNAPs were normalized at the poop promoter. The efficiency of transcription from pE in the presence of each reconstituted mutant RNAP is shown below the corresponding lane from a typical transcription gel. Values (with standard deviation) are expressed as percentages of the transcript yield obtained with wild-type RNAP.
Effect of the rpoA341 mutation on CII-mediated transcription activation at pI and paQ
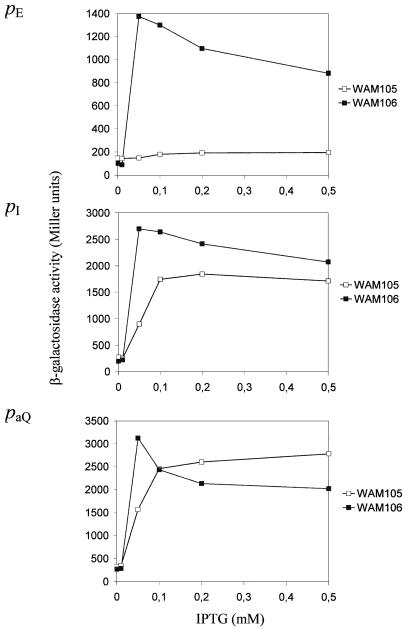
To determine whether the K271Eα substitution affects the other CII-dependent promoters, pI and paQ, reporter gene fusions to all three CII-dependent promoters were constructed in single copy using the same genetic system, and promoter activity was measured in the presence and absence of CII. For this experiment, levels of induced CII production were varied by using different concentrations of IPTG. We found that the maximum induced activities of the three lysogenic promoters in the rpoA+ background occurred following induction with 0.05 mM IPTG (Fig. 4). With this level of CII synthesis, the degree of activation afforded by CII at each promoter was similar, i.e. 12.5- to 14-fold. Consistent with previous observations, in the rpoA341 mutant, the activity of the pE promoter was approximately equal to that observed in the wild-type strain in the absence of CII synthesis, i.e. CII activation of pE was negligible (Fig. 4A) (16). The pI promoter was ∼30% as active in the rpoA341 mutant in comparison with the wild-type strain when CII synthesis was induced by 0.05 mM IPTG. However, this difference became less pronounced with higher levels of CII induction (Fig. 4B). The efficiency of CII-mediated stimulation of paQ was least affected by the rpoA341 mutation (Fig. 4C). Only when CII synthesis was induced with 0.05 mM IPTG was there a significant difference between paQ activity in the wild-type and mutant strains, suggesting that it may be slightly less sensitive to CII in the mutant background. These results suggest differences in the role of αCTD at each of these promoters.
Figure 4.
CII-dependent and CII-independent activities of the phage λ pE (A), pI (B) and paQ (C) promoters in rpoA+ (WAM106) and rpoA341 (WAM105) hosts. Each host strain (rpoA+ and rpoA341), carrying a single copy pE–lacZ, pI–lacZ or paQ–lacZ fusion together with plasmids pMO23 and pJMH1, was grown in LB medium containing chloramphenicol (34 µg/ml) and kanamycin (50 µg/ml) at 37°C to OD ∼0.2. IPTG was added to the indicated final concentrations and the activities of β-galactosidase (in Miller units) were measured 60 min later.
DISCUSSION
Previous biochemical and genetic studies have suggested a role for αCTD in CII-dependent activation of the λ pE promoter (14,15). Using a DNA cleavage reagent that was tagged to two different locations in αCTD, we deduced the location and orientation of αCTD in RNAP–promoter open complexes at pE. These experiments showed that one αCTD binds near position –41 and the other αCTD binds further upstream. The αCTD near position –41 must be bound on the opposite face of the DNA helix to CII (Fig. 5A), in accord with previous investigators who showed that CII binds to the opposite face of the DNA helix to σ (38). The results of the Fe·BABE analysis are also consistent with previous DMS protection studies (38) showing that protection occurs at positions –40 and –41 at pE in the presence of CII and RNAP. Our results show that the αCTD near position –41 is bound with its 261 determinant pointing downstream such that it could interact with region 4 of the RNAP σ subunit bound to the –35 element (Fig. 5A). Our observation that substitutions in the 261 determinant slightly impair CII-dependent activation of pE is consistent with such an interaction, which may be required for full activation of pE by CII. In this regard, it is noteworthy that, at certain activator-independent promoters such as rrnB P1, where αCTD binds at the same location, functional interactions occur between two residues within the 261 determinant, D259 and E261, and the side chain of R603 in region 4.2 of σ (12). Also, at the class I CRP-dependent lac promoter, where αCTD binds to the –42/–43 region, the 261 determinant also interacts with σ and this interaction is required for activation by CRP (10,11,13).
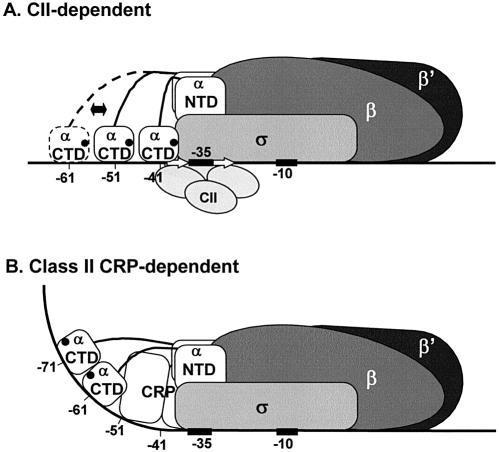
Figure 5.
Model for activation of the λ pE promoter by CII. (A) Location and orientation of αCTD at pE as determined in this work. The approximate location of the 261 determinant in αCTD is shown as a filled circle. (B) For comparison, a model for class II CRP-dependent transcription activation is also shown (5).
The location of αCTD in CII-dependent open complexes at pE is very similar to that found at factor-independent promoters such as rrnB P1 and lacUV5 (39–41). This contrasts sharply with the situation at most class II activator-dependent promoters, where the activator is positioned on the same side of the DNA helix as σ, and αCTD is displaced to a site upstream of the bound activator (Fig. 5B). An exception is found at the Bordetella pertussis fha promoter, where three dimers of the activator, BvgA, occupy a region extending from positions –35 to –100 on the same face of the DNA helix as σ. At this promoter, αCTD binds to the same segment of DNA as BvgA, but to a different face of the DNA helix (42). One common pattern which is emerging from studies of class II or class II-like promoters is that, at this class of promoter, αCTD appears to bind to the nearest ‘vacant’ segment of upstream DNA to the promoter, with a preference for binding to the same face of the DNA as RNAP (43).
The alanine scanning analysis identified four residues in the αCTD DNA-binding determinant where alanine substitution causes at least a 40% decrease in activation in our assays, suggesting that DNA binding by αCTD is important for CII-dependent activation of pE. This conclusion was supported by run-off transcription assays in vitro. However, the mechanism of activation of the pE promoter by CII still remains unclear. In particular, it is not apparent whether there is any direct contact between CII and αCTD at this promoter. The alanine scanning analysis showed that substitutions of residues at positions 281, 303 and 320 caused large decreases in activation, but, for reasons discussed above, we think it is unlikely that L281 and I303 contact CII. We are unable to conclude whether N320 is involved in contacts with CII. However, due to its location on αCTD, this residue would not be expected to contact an activator bound to the opposite face of the DNA. Interestingly, although the K271E substitution severely inhibits CII- mediated activation of pE, alanine substitution at position 271 had very little effect. This suggests that the effect of the K271E substitution might be to create an interaction (or ‘clash’) which impairs CII function. In the last part of our study, we showed that each of the three CII-dependent λ promoters is affected differently by the K271E substitution. This suggests that, although CII binds to identical sequences at identical locations at these promoters, the mechanisms of transcription activation, particularly with respect to the role of αCTD, are distinct. This is consistent with previous observations that the initial binding and isomerization steps in the transcription initiation pathway are differentially affected by CII at each of these promoters (2,44). Thus, the KB step is stimulated 103- to 104-fold at pI and paQ, whereas at pE, which is distinguished by having an ‘extended’ –10 sequence, KB is increased by only 15-fold (44–46).
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Wenmao Meng, Tamara Belyaeva and Lars Westblade for their help. This work was supported by the Polish State Committee for Scientific Research (project no. 3 P04A 049 24) and by WT project grants 050794 (to M.S.T. and S.J.W.B.) and 059718 (to S.J.W.B). G.W. also acknowledges financial support from the Foundation for Polish Science (subsidy 14/2000). B.K. acknowledges support from EMBO (scholarship ASTF 9531) and FEBS for visits to the UK.
REFERENCES
- 1.Echols H. (1986) Bacteriophage λ development: temporal switches and the choice of lysis or lysogeny. Trends Genet., 2, 26–30. [Google Scholar]
- 2.Ho Y.-S., Wulff,D. and Rosenberg,M. (1986) Protein–nucleic acid interactions involved in transcription activation by the phage lambda regulatory protein cII. In Booth,I.R. and Higgins,C.F. (eds), Regulation of Gene Expression—25 Years On. Cambridge University Press, Cambridge, pp. 79–103. [Google Scholar]
- 3.Ishihama A. (1992) Role of the RNA polymerase α subunit in transcription activation. Mol. Microbiol., 6, 3283–3288. [DOI] [PubMed] [Google Scholar]
- 4.Dove S.L., Darst,S.A. and Hochschild,A. (2003) Region 4 of σ as a target for transcription regulation. Mol. Microbiol., 48, 863–874. [DOI] [PubMed] [Google Scholar]
- 5.Busby S. and Ebright,R.H. (1999) Transcription activation by catabolite activator protein (CAP). J. Mol. Biol., 293, 199–213. [DOI] [PubMed] [Google Scholar]
- 6.Gourse R.L., Ross,W. and Gaal,T. (2000) UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol., 37, 687–695. [DOI] [PubMed] [Google Scholar]
- 7.Gaal T., Ross,W., Blatter,E.E., Tang,H., Jia,X., Krishnan,V.V., Assa-Munt,N., Ebright,R.H. and Gourse,R.L. (1996) DNA-binding determinants of the α subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev., 10, 16–26. [DOI] [PubMed] [Google Scholar]
- 8.Murakami K., Fujita,N. and Ishihama,A. (1996) Transcription factor recognition surface on the RNA polymerase α subunit is involved in contact with the DNA enhancer element. EMBO J., 15, 4358–4367. [PMC free article] [PubMed] [Google Scholar]
- 9.Ross W., Ernst,A. and Gourse,R.L. (2001) Fine structure of E.coli RNA polymerase–promoter interactions: α subunit binding to the UP element minor groove. Genes Dev., 15, 491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benoff B., Yang,H., Lawson,C.L., Parkinson,G., Liu,J., Blatter,E., Ebright,Y.W., Berman,H.M. and Ebright,R.H. (2002) Structural basis of transcription activation: the CAP–αCTD–DNA complex. Science, 297, 1562–1566. [DOI] [PubMed] [Google Scholar]
- 11.Savery N.J., Lloyd,G.S., Busby,S.J.W., Thomas,M.S., Ebright,R.H. and Gourse,R.L. (2002) Determinants of the C-terminal domain of the Escherichia coli RNA polymerase α subunit important for transcription at class I cyclic AMP receptor protein-dependent promoters. J. Bacteriol., 184, 2273–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross W., Schneider,D.A., Paul,B.J., Mertens,A. and Gourse,R.L. (2003) An inter-subunit contact stimulating transcription initiation by E.coli RNA polymerase: interaction of the α C-terminal domain and σ region 4. Genes Dev., 17, 1293–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H., Tang,H. and Ebright,R.H. (2003) Functional linkage of upstream-promoter and core-promoter regions: functional interaction between RNA polymerase α subunit C-terminal domain and σ70 in UP-element- and activator-dependent transcription. Mol. Cell, 11, 1621–1633. [DOI] [PubMed] [Google Scholar]
- 14.Gussin G.N., Olson,C., Igarashi,K. and Ishihama,A. (1992) Activation defects caused by mutations in Escherichia coli rpoA are promoter specific. J. Bacteriol., 174, 5156–5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Węgrzyn G., Glass,R.E. and Thomas,M.S. (1992) Involvement of the α subunit of E.coli RNA polymerase in transcriptional activation by the bacteriophage λ regulatory proteins CI and CII. Gene, 122, 1–7. [DOI] [PubMed] [Google Scholar]
- 16.Obuchowski M., Giladi,H., Koby,S., Szalewska-Pałasz,A., Węgrzyn,A., Oppenheim,A.B., Thomas,M.S. and Węgrzyn,G. (1997) Impaired lysogenisation of the Escherichia coli rpoA341 mutant by bacteriophage λ is due to the inability of CII to act as a transcriptional activator. Mol. Gen. Genet., 254, 304–311. [DOI] [PubMed] [Google Scholar]
- 17.Thomas M.S. and Glass,R.E. (1991) Nucleotide sequence of an E.coli rpoA mutation which impairs transcription of positively regulated systems. Mol. Microbiol., 5, 2719–2725. [DOI] [PubMed] [Google Scholar]
- 18.Giladi H., Goldenberg,D., Koby,S. and Oppenheim,A.B. (1995) Enhanced activity of the bacteriophage λ pL promoter at low temperature. Proc. Natl Acad. Sci. USA, 92, 2184–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latała B., Obuchowski,M. and Węgrzyn,G. (2001) Bacteriophage λ cIII gene product has an additional function apart from inhibition of cII degradation. Virus Genes, 22, 127–132. [DOI] [PubMed] [Google Scholar]
- 20.Giladi H., Koby,S., Gottesman,M.E. and Oppenheim,A.B. (1992) Supercoiling, integration host factor and a dual promoter system participate in the control of the bacteriophage λ pL promoter. J. Mol. Biol., 224, 937–948. [DOI] [PubMed] [Google Scholar]
- 21.Schmidhauser T.J., Filutowicz,M. and Helinski,D.R. (1983) Replication of derivatives of the broad host range plasmid RK2 in two distantly related bacteria. Plasmid, 9, 325–330. [DOI] [PubMed] [Google Scholar]
- 22.Tang H., Severinov,K., Goldfarb,A., Fenyo,D., Chait,B. and Ebright,R.H. (1994) Location, structure and function of the target of a transcription activator protein. Genes Dev., 8, 3058–3067. [DOI] [PubMed] [Google Scholar]
- 23.Blatter E.E., Ross,W., Tang,H., Gourse,R.L. and Ebright,R.H. (1994) Domain organization of RNA polymerase alpha subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell, 78, 889–896. [DOI] [PubMed] [Google Scholar]
- 24.Wood L.F., Tszine,N.Y. and Christie,G.E. (1997) Activation of P2 late transcription by P2 Ogr protein requires a discrete contact site on the C-terminus of the α subunit of Escherichia coli RNA polymerase. J. Mol. Biol., 274, 1–7. [DOI] [PubMed] [Google Scholar]
- 25.Kainz M. and Gourse,R.L. (1998) The C-terminal domain of the alpha subunit of Escherichia coli RNA polymerase is required for efficient Rho-dependent transcription termination. J. Mol. Biol., 284, 1379–1390. [DOI] [PubMed] [Google Scholar]
- 26.Zou C., Fujita,N., Igarashi,K. and Ishihama,A. (1992) Mapping the cAMP receptor protein contact site on the α subunit of Escherichia coli RNA polymerase. Mol. Microbiol., 6, 2599–2605. [DOI] [PubMed] [Google Scholar]
- 27.Miller J. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 28.Shotland Y., Shifrin,A., Ziv,T., Teff,D., Koby,S., Kobiler,O. and Oppenheim,A.B. (2000) Proteolysis of bacteriophage lambda CII by Escherichia coli FtsH (HflB). J. Bacteriol., 182, 3111–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang H., Severinov,K., Goldfarb,A. and Ebright,R.H. (1995) Rapid RNA polymerase genetics: one-day, no-column preparation of reconstituted recombinant Escherichia coli RNA polymerase. Proc. Natl Acad. Sci. USA, 92, 4902–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savery N.J., Lloyd,G.S., Kainz,M., Gaal,T., Ross,W., Ebright,R.H., Gourse,R.L. and Busby,S.J. (1998) Transcription activation at class II CRP-dependent promoters: identification of determinants in the C-terminal domain of the RNA polymerase α subunit. EMBO J., 17, 3439–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee D.J., Busby,S.J.W. and Lloyd,G.S. (2004) Exploitation of a chemical nuclease to investigate the location and orientation of the Escherichia coli RNA polymerase alpha subunit C-terminal domains at simple promoters that are activated by CRP. J. Biol. Chem., 278, 52944–52962. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 33.Kolb A., Kotlarz,D., Kusano,S. and Ishihama,A. (1995) Selectivity of the Escherichia coli RNA polymerase E σ38 for overlapping promoters and ability to support CRP activation. Nucleic Acids Res., 23, 819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murakami K., Kimura,M., Owens,J.T., Meares,C.F. and Ishihama,A. (1997) The two α subunits of Escherichia coli RNA polymerase are asymmetrically arranged and contact different halves of the DNA upstream element. Proc. Natl Acad. Sci. USA, 94, 1709–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakami K., Owens,J.T., Belyaeva,T.A., Meares,C.F., Busby,S.J.W. and Ishihama,A. (1997) Positioning of two alpha carboxy-terminal domains of RNA polymerase at promoters by two transcription factors. Proc. Natl Acad. Sci. USA, 94, 11274–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimatake H. and Rosenberg,M. (1981) Purified λ regulatory protein cII positively activates promoters for lysogenic development. Nature, 292, 128–132. [DOI] [PubMed] [Google Scholar]
- 37.Jeon Y.H., Negishi,T., Shirakawa,M., Yamazaki,T., Fujita,N., Ishihama,A. and Kyogoku,Y. (1995) Solution structure of the activator contact domain of the RNA polymerase α subunit. Science, 270, 1495–1497. [DOI] [PubMed] [Google Scholar]
- 38.Ho Y-S., Wulff,D.L. and Rosenberg,M. (1983) Bacteriophage λ protein cII binds promoters on the opposite face of the DNA helix from RNA polymerase. Nature, 304, 703–708. [DOI] [PubMed] [Google Scholar]
- 39.Estrem S.T., Gaal,T., Ross,W. and Gourse,R.L. (1998) Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl Acad. Sci. USA, 95, 9761–9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Estrem S.T., Ross,W., Gaal,T., Chen,Z.W.S., Niu,W., Ebright,R.H. and Gourse,R.L. (1999) Bacterial promoter architecture: subsite structure of UP elements and interactions with the C-terminal domain of the RNA polymerase alpha subunit. Genes Dev., 13, 2134–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naryshkin N., Revyakin,A., Kim,Y., Mekler,V. and Ebright,R.H. (2000) Structural organization of the RNA polymerase–promoter open complex. Cell, 101, 601–611. [DOI] [PubMed] [Google Scholar]
- 42.Boucher P.E., Maris,A.E., Yang,M-S. and Stibitz,S. (2003) The response regulator BvgA and RNA polymerase α subunit C-terminal domain bind simultaneously to different faces of the same segment of promoter DNA. Mol. Cell, 11, 163–173. [DOI] [PubMed] [Google Scholar]
- 43.Grainger D.C, Belyaeva,T.A., Lee,D.J., Hyde,E.I. and Busby,S.J.W. (2004) Transcription activation at the Escherichia coli melAB promoter: interactions of MelR with the C-terminal domain of the RNA polymerase α subunit. Mol. Microbiol., in press. [DOI] [PubMed] [Google Scholar]
- 44.McClure W. and Hoopes,B.C. (1987) Regulation of transcription initiation in Escherichia coli. In Reznikoff,W.S., Burgess,R.R., Dahlberg,J.E., Gross,C.A., Record,M.T.,Jr and Wickens,M.P. (eds), RNA Polymerase and the Regulation of Transcription. Elsevier Science Publishing Co., Inc., New York, NY, pp. 85–93. [Google Scholar]
- 45.Shih M.-C. and Gussin,G.N. (1984) Role of cII protein in stimulating transcription initiation at the λ PRE promoter: enhanced formation and stabilization of open complexes. J. Mol. Biol., 172, 489–506. [DOI] [PubMed] [Google Scholar]
- 46.Keilty S. and Rosenberg,M. (1987) Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J. Biol. Chem., 262, 6389–6395. [PubMed] [Google Scholar]